1. Introduction
Soil salinization is one of the important factors affecting crop yield worldwide. In addition to natural land salinization, irrigation methods and climate change can lead to land salinization [
1]. At present, more than 20% of cultivated land and 33% of irrigated farmland are affected by land salinization [
2]. The accumulation of sodium and chloride ions in the soil will reduce water availability and affect the original ion balance in plants, ultimately resulting in osmotic stress and ionic toxicity to plants. These factors collectively lead to delayed plant growth, hindered developmental processes, and adaptation of metabolic activities [
3]. Alfalfa (
Medicago sativa L.), also known as the king of forage, is a perennial leguminous forage with strong stress resistance, high quality and economic value, widely planted worldwide. [
4]. However, severe salt stress can lead to a decline in the growth of alfalfa roots, stems, and leaves, as well as nutritional value [
5]. Research on the molecular regulation mechanism of salt tolerance in alfalfa can help accelerate the cultivation of new varieties of salt-tolerant alfalfa, effectively solve the constraints of soil salinization on alfalfa planting areas, thereby improving the development scale and level of the forage industry, and playing a crucial role in promoting the development of circular ecological agriculture with forage and animal husbandry as the core. [
6]. Current studies have clarified the mechanism of salt-alkali tolerance of alfalfa from many aspects, such as seed germination, plant growth and development, physiological and biochemical reactions, and molecular biology [
7]. Most of the research literature focuses on key processes, such as osmotic regulation, ion balance and repair, reactive oxygen species (ROS) production, and its effects on downstream molecular targets [
2,
8,
9]. In addition, studies have shown that salt stress can trigger changes in gene expression, affect mRNA stability, regulate the translation process, and ultimately lead to changes in protein abundance [
10].
Plant hormones such as gibberellin, abscisic acid, ethylene, auxin, and cytokinin play indispensable roles in different stages of plant growth, organ development, internal homeostasis, and plant response to external environmental changes. [
11,
12]. Melatonin is a common indole heterocyclic compound found in plants and animals [
13]. Previous studies have demonstrated that melatonin plays an important role in regulating plant growth and responding to biotic and abiotic stresses, and is involved in regulating many physiological processes, including seed germination, flowering and fruiting, and mineral element absorption. [
14]. Furthermore, exogenous application of melatonin can enhance plant stress resistance and help plants maintain health under abiotic stress conditions. In particular, as an antioxidant, melatonin can activate the expression of antioxidant oxidase genes and improve enzyme activity, thereby improving the plant's tolerance to stress [
15]. We hypothesized that melatonin combats salt stress by mediating the expression of genes associated with plant hormones and antioxidant capacity and conducted transcriptomic analyses aimed at revealing key genes and pathways of melatonin-mediated salt tolerance in alfalfa.
2. Materials and Methods
2.1. Experimental Materials and Design
Cultivar Zhongmu No. 3 alfalfa used in this research was obtained by the Chinese Academy of Agricultural Sciences. The seeds were surface-sterilized in 75% ethanol for 10 minutes, and then rinsed three times with deionized water. The seeds were germinated in a petri dish (ΦA=90mm) containing 4 mL of 0, 10μM MT with 200 mM NaCl or deionized water. There were 6 replicates per treatment and 30 seeds per replicate. Then, the petri dish placed in a constant temperature incubator at 25℃ with relative humidity under a 16h light/ 8h dark cycle. After 7 days, the germination rate, fresh weight and root length were measured, and the germinated seedlings were frozen in liquid nitrogen and stored at -80 ℃. The seedlings treated with the 0 (also known as CK), 200 mM NaCl (also known as ZMN), 200 mM NaCl+10 μM MT (also known as ZMNMT) were used for subsequent physiological index determination and transcriptomic analysis.
2.2. Measurement of the Germination Rate, Root Length, and Fresh Weight
The germination rate was counted every day for seven consecutive days. The vertical distance of 10 seedlings per biological replicate from the cotyledon node to the top bud was measured with a calibration ruler and recorded as bud length. Fresh weights were determined for 10 seedlings per biological replicate.
2.3. Measurement of Physiological and Biochemical Indicators
GSH and O2- were respectively determined by referring to the Solarbio kit (BC1175, Beijing, China, BC1295, Beijing, China) SOD was determined according to Abbkine kit (KTB1030, Wuhan, China), POD was determined by reference to Abbkine kit (KTB1150, Wuhan, China), and MDA was determined by reference to Abbkine kit (KTB1050, Wuhan, China).
2.4. Transcriptomic Analysis
Total RNA was isolated from alfalfa root samples and cut into short fragments using the MJZol total RNA extraction kit (Shanghai Majorbio Biomedicine Technology Co., LTD., China), then cDNA was synthesized using the short fragment as a template using random hexamer primers and reverse transcriptase. Then, the Kit Biowest Agarose (Biowest, Spain) and RNA Purification Kit (Shanghai Majorbio Biomedicine Technology Co., LTD.) were used to purify the library fragments, and bridge PCR was performed on the cBot to generate clusters. Finally, high-throughput sequencing was performed on the Illumina NovaSeq 6000 platform (Illumina, USA). After the sequencing was completed, FASTp was used to filter the quality of the original readings, removing sequencing connector sequences, low-quality reads, sequences with high N (N indicates uncertain base information) rates, and sequences with concise length. Then, mapped data (reads) for subsequent transcript assembly and expression calculation were obtained by comparing them with reference genomes [
16].
2.5. Differential Gene GO and KEGG Annotation Analysis
The Blast2GO program (
https://www.blast2go.com/)[
17,
18] was used for function annotation and classification of the differentially expressed genes. The KOBAS 3.0 online program (
http://kobas.cbi.pku.edu.cn/) was used to determine enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among salt stress-responsive genes (
http://www.genome.jp/kegg/) [
19,
20,
21]. The genes in the gene concentration were enriched and analyzed by software, and Fisher was used for an accurate test. The BH method was used to adjust the
P-value, and when the corrected P-value (
P-adjust) < 0.05, it was considered that this function was significantly enriched.
2.6. Weighted Gene Co-Correlation Network Analysis and Protein Interaction Network Analysis
The co-expression network was constructed using WGCNA (weighted gene Co-expression network analysis), and modules containing closely related genes were identified. After obtaining the commonly expressed gene module (module), the module is associated with the phenotypic information of concern to explore the correlation between the gene network and phenotype and the core genes in the network. (
https://cran.r-project.org/web/packages/WGCNA/index.html). Protein interaction network analysis applied the interaction relationship in the STRING protein interaction database to construct the protein interaction network of differential genes to show the relationship between differential genes. (
http://string-db.org/).
2.7. qRT-PCR Analysis
Twelve differential genes were selected for qRT-PCR analysis. Full-length cDNA was then reverse transcribed using the cDNA synthesis kit PrimeScript RT Reagent Kit with gDNA Eraser (Cat# RR047A, TaKaRa, Tokyo, Japan). The qRT-PCR analysis was performed using a one-step qRT-PCR kit (Cat# RR420A; TaKaRa) according to the manufacturer’s instructions, using three independent RNA preparations as biological replicates.
3. Result Analysis
3.1. Effects of Melatonin on Alfalfa Plant Growth under NaCl Stress
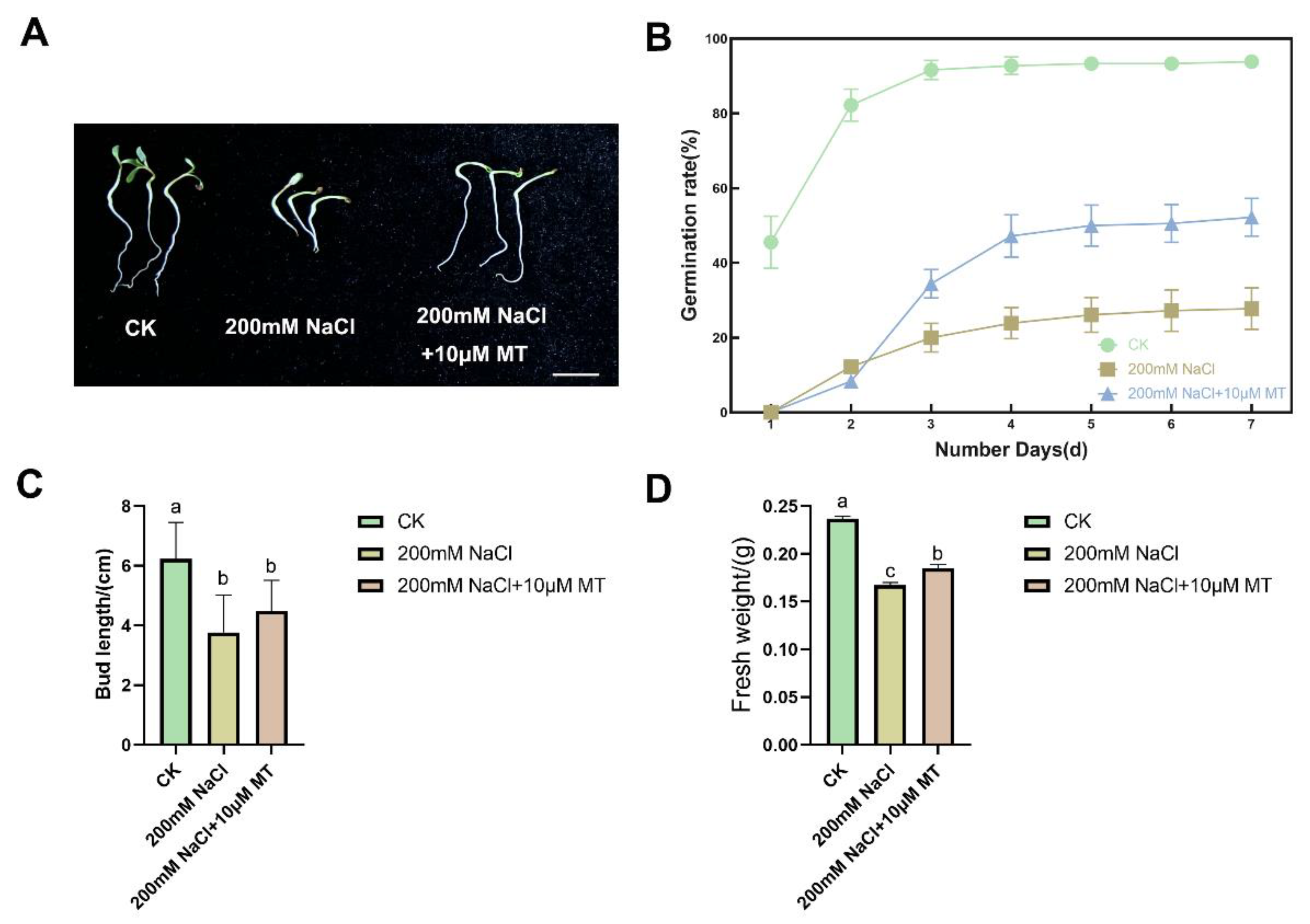
We found that under the same conditions, the germination rate of Zhongmu No. 3 seed under salt stress decreased significantly, and the germination rate of the salt stress group was only 27.8% on the 7th day, which had a strong inhibitory effect compared with 93.9% in CK group (
Figure 1A). Middle alone Zhongmu No. 3 seed germination rate in the melatonin group reached 34.4% on the 3rd day, which exceeded the germination rate in 7 days of the NaCl treatment group (
Figure 1B). NaCl treatment could significantly inhibit the growth of the plants; both the bud length of 3.77cm and fresh weight of 0.1673g were significantly lower than those of 6.23cm and 0.2370g in the CK group. Similarly, compared with NaCl alone, the bud length of 4.49cm and fresh weight of 0.1849g in the melatonin group were significantly increased (
Figure 1C-D). This indicates that melatonin treatment can alleviate the adverse effects of NaCl stress on plant growth.
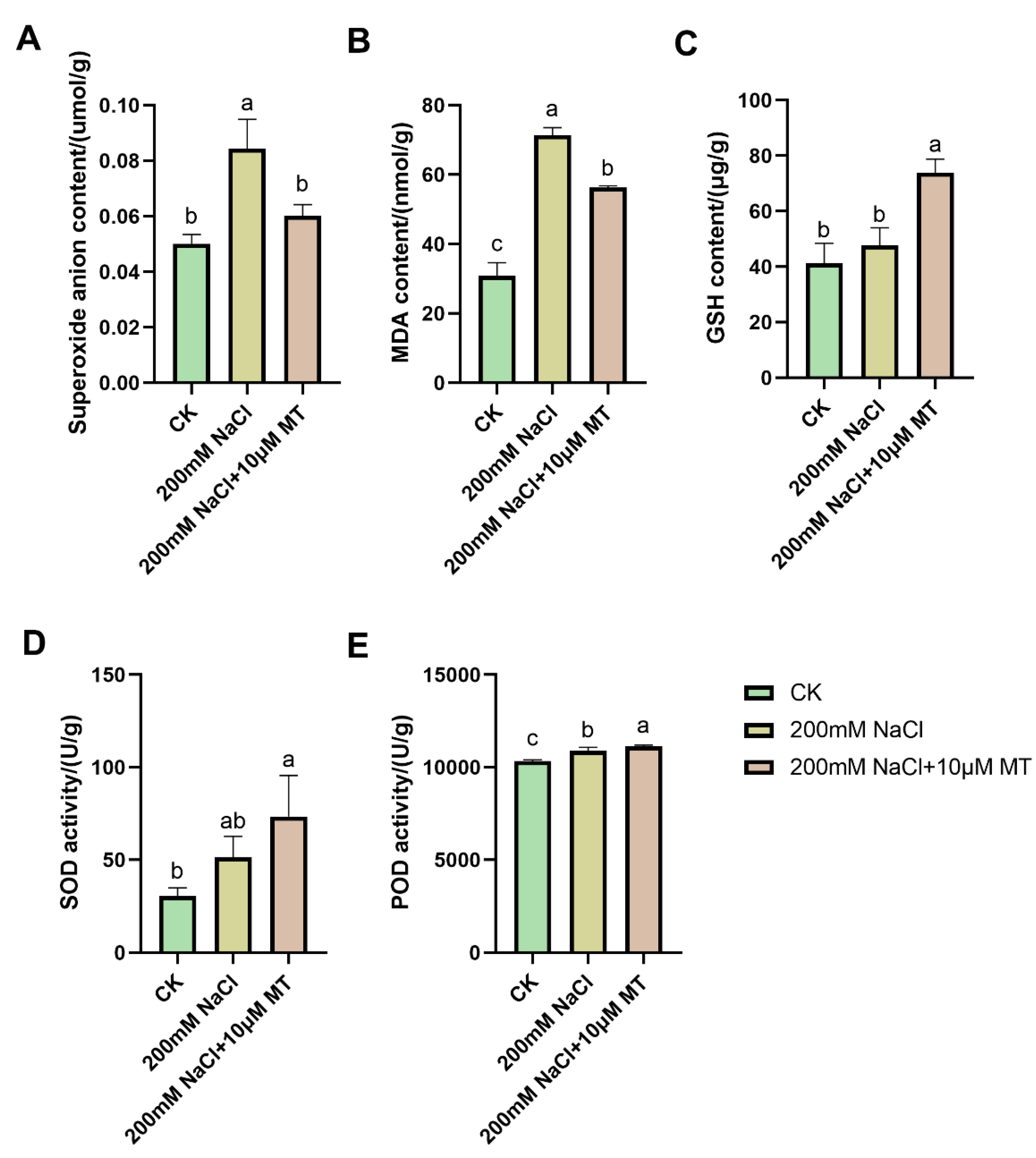
3.2. Changes in Oxidation System Indexes
To further reveal the mechanism by which melatonin enhances salt tolerance in alfalfa, we measured the index changes in oxidation systems under different treatments. Compared with the CK group, NaCl stress significantly increased the contents of O
2-, MDA and GSH and increased the activities of SOD and POD. Under combined melatonin and NaCl treatment, the accumulation of O
2- and MDA decreased by 28.7% and 21.1%(
Figure 2A-A), respectively, while the content of GSH, activity of SOD and POD increased by 54.7%, 43.1% and 2.5%(
Figure 2C-B), respectively.
3.3. Identification of Salt-Responsive Genes in Alfalfa Seedlings
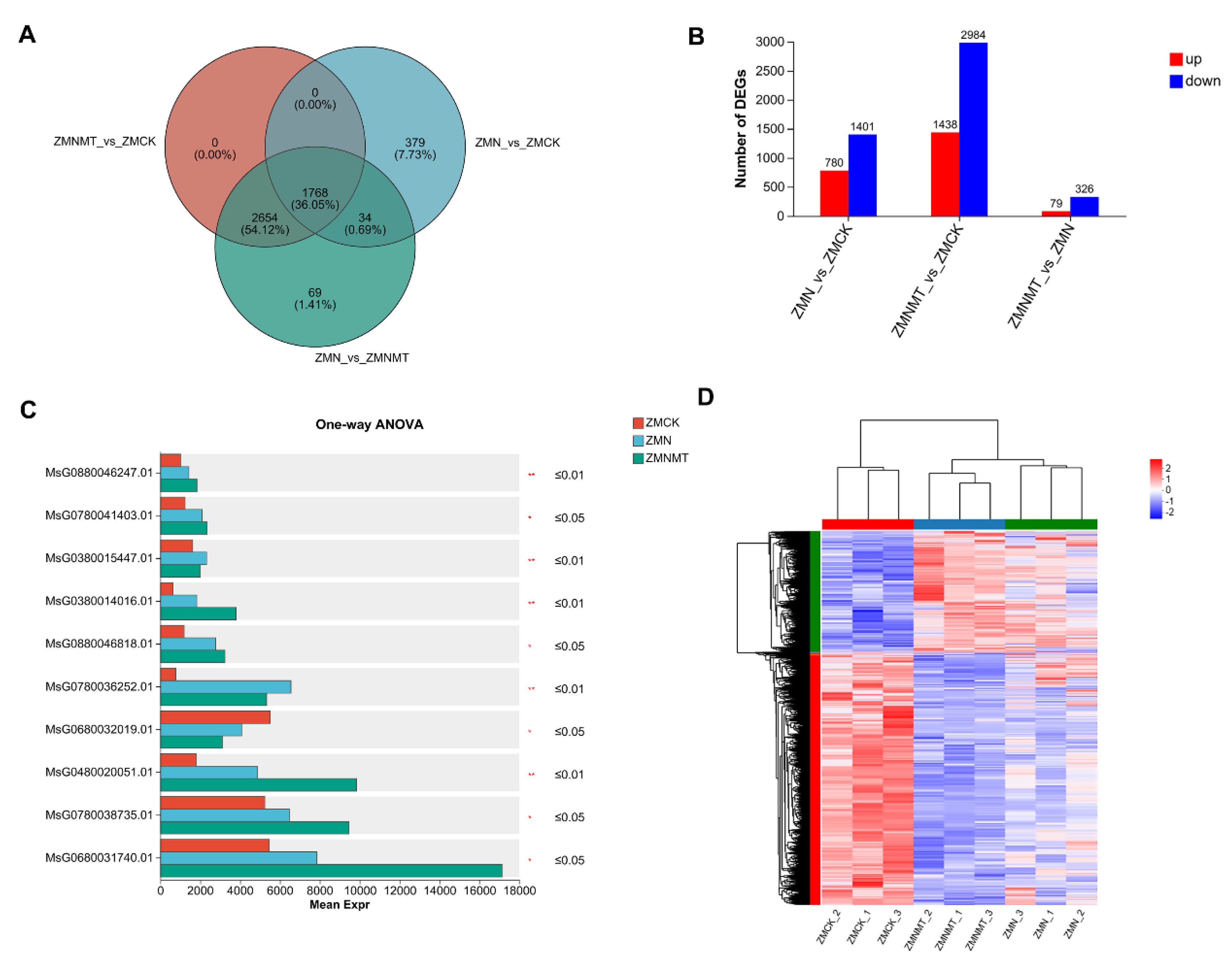
In addition, transcriptomic analysis was performed on alfalfa seedlings treated with ZMCK, ZMN and ZMNMT. A total of 4,904 genes were identified by comparison between the two different treatments, of which 1,768 genes were shared by the three treatments, accounting for 36.05% of the total genes, and 2,654 genes were shared by ZMNMT and ZMCK, ZMN and ZMNMT, accounting for 54.12% of the total genes. 34 genes were shared between ZMN and ZMNMT and between ZMN and ZMCK, accounting for 0.69% of the total genes; 69 genes were shared between ZMNMT and ZMCK, and between ZMN and ZMNMT, accounting for 1.41% of the total genes; 379 genes were unique to ZMCK and ZMN, accounting for 7.73% of the total genes (
Figure 3A;
Table S1). Comparisons of ZMN vs. ZMCK, ZMNMT vs. ZMCK, and ZMNMT vs. ZMN, consist of 780, 1,438 and 79 upregulated genes, and 1,401, 2,984 and 328 downregulated genes, respectively (
Figure 3B;
Table S1). Moreover, ten DEGs were detected based on the analysis of variance between different groups, which probably be related to melatonin mediated salt tolerance in alfalfa. Meanwhile, a heat map is drawn by clustering all DEGs to show the different regulatory modes for each treatment (
Figure 3D;
Table S1).
3.4. Functional Annotation of Differentially Expressed Genes
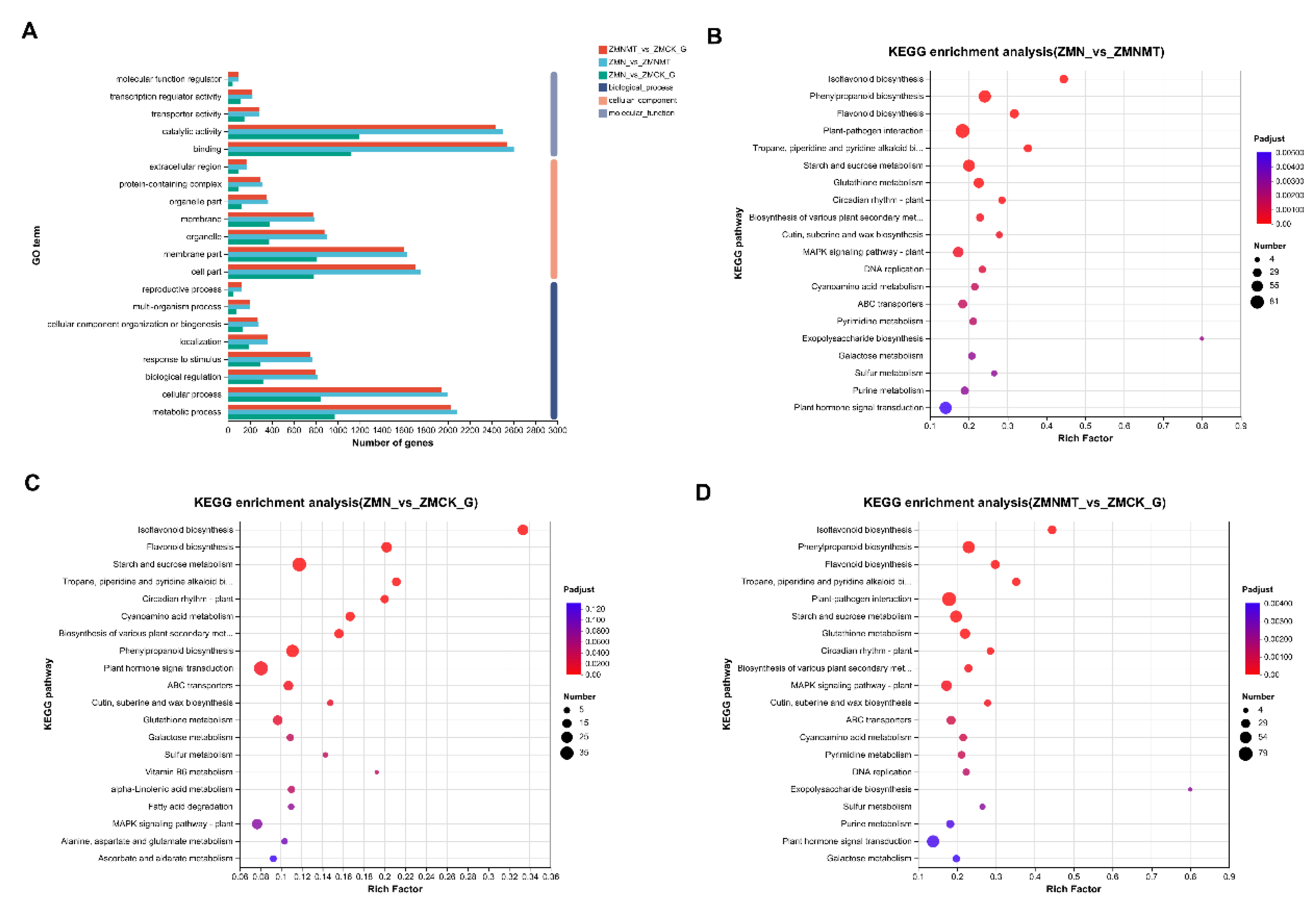
Functional annotation of differential proteins was carried out using the GO database. In the molecular function classification of GO, differentially expressed genes were assigned four high-order terms: molecular function regulator, transcriptional regulator activity, transport activity, and binding activity. The DEGs were divided into 7 components: extracellular region, protein complex, organelle part, membrane, organelle part, membrane part and cell part. Biological processes include reproductive, multi-organism, cellular component organization or biogenesis, localization, stimulus-response, biological regulation, cellular processes, and metabolic processes. In most GO terms, the number of annotated proteins of ZMN and ZMNMT is higher than that of ZMN and ZMCK and ZMNMT and ZMCK (
Figure 4A;
Table S2). KEGG pathway annotation results showed that ZMN and ZMNMT had 1,234 DEGs annotations in 124 pathways(
Figure 4B;
Table S3), ZMN and ZMCK had 591 differential gene annotations in 113 pathways(
Figure 4C;
Table S4), and ZMNMT and ZMCK had 1,194 differential gene annotations in 124 pathways(
Figure 4D;
Table S5). In the salt treatment group, there were 10 extremely significantly enriched KEGG pathways, and in the melatonin treatment group, there were 21 extremely significantly enriched KEGG pathways. It is worth noting that 10 extremely significantly enriched KEGG pathways in the salt treatment group were all reflected in the melatonin treatment group, including the synthesis pathways of flavonoids, isoflavones and various secondary metabolites. ABC transfer vehicle and plant signal transduction. MAPK signaling pathway, glutathione and galactose metabolism pathway were enriched in the melatonin treatment group, and these pathways may be closely related to stress response.
4. Discussion
Multiple genes control the response of alfalfa under stress. This study obtained over 10,000 differentially expressed genes (DEGs) through transcriptomic analysis of Zhongmu NO.3 seedlings under different treatments. By WGCNA, we investigated the correlation between DEGs and physiological characteristics related to salt stress. The analysis identified 12 gene co-expression modules. Module-trait association analysis showed that phenotypic characteristics such as germination rate, bud length and fresh weight were significantly positively correlated with gene expression levels in turquoise modules, with correlation coefficients ranging from 0.85 to 0.886. Meanwhile, germination rate, bud length and fresh weight were significantly negatively correlated with gene expression levels in blue modules, with correlation coefficients ranging from 0.783 to 0.836. This finding suggests that genes in the turquoise module may play a role in MT-mediated salt resistance. In addition, genes in the blue module were significantly positively correlated with MDA content and O
2- levels, suggesting they may accumulate in large quantities under salt stress. In addition, indicators of antioxidant capacity in the black modules, including SOD and POD activity, were also positively correlated with gene expression levels (
Figure 5A;
Table S6). The turquoise module contains 6,847 DEGs, the blue module contains 3,505 DEGs, and the black module has 290 DEGs. The top 30 genes based on connectivity were identified as key hub genes in these three modules, representing the main function of the corresponding module (
Figure 5B;
Table S6). Using Cytoscape software, we visualized these top 30 key hub genes and found that these key genes play important roles in flavonoid, plant hormone, soluble sugar biosynthesis, GSH metabolism, and ABA transporters (
Figure 5C-D;
Table S7).
4.1. Biosynthesis of Flavonoids and Isoflavones
Flavonoids are a class of secondary metabolites widely distributed in plants. According to their molecular structure, flavonoids can be divided into six categories: flavonols, anthocyanins, proanthocyanidins, isoflavones, chalones, flavonoids, flavanols, and aurones [
22]. Studies have shown that the antioxidant system in plants plays a vital role in eliminating the accumulation of excess ROS caused by stress [
23]. In
Arabidopsis thaliana, enhancing the expression of the CHS gene in Okra can increase the content of flavonoids and the expression of SOD and POD genes under salt stress, which helps to enhance the resistance of Arabidopsis to salt [
24]. In the comparison between salt treatment and melatonin treatment, we found that the flavonoid synthesis the gene expression abundance of DEGs related to flavonoid biosynthesis (Such as CHS, E5.5.1.6, CYP73A, CYP75B1, E2.1.1.104, and CYP75A) were increased, which indicating that melatonin can alleviate the stress effect of salt on alfalfa seedlings. In addition, upregulating the ANR gene in tobacco can promote the accumulation of flavan-3-ols (catechins and epicatechins), directly enhancing tobacco plants' antioxidant capacity [
25]. In the salt treatment group, the epicatechin synthesis gene ANR was significantly up-regulated, which played an important role in coping with salt stress. In the KEGG pathway, it is worth noting that CHS is related to isoliquiritin synthesis. Isoliquiritin is converted into liquiritigenin under the action of E5.5.1.6, which is related to the synthesis of naringin, and both liquiritigenin and naringin are involved in the biosynthesis of isoflavones. Therefore, we speculate that some essential genes in the flavonoid synthesis pathway are regulated. The content of flavonoids and isoflavones in alfalfa can be changed to enhance the plant's stress resistance. (
Figure 6A).
4.2. Glutathione Metabolic Pathway
When plants are exposed to salt stress, oxidative stress and cell damage are often caused [
26], and glutathione (GSH) is an essential non-enzymatic antioxidant in cell defense [
27]. Glutathione-s-transferase (GST) plays a vital role in the metabolic pathway of glutathione. GST can promote the binding of GSH to electrophilic substrates, at the same time, GST can catalyze the reaction of hydrogen peroxide (H
2O
2) and GSH, thereby reducing cell oxidative damage [
28,
29]. In this study, 18 and 41 differential genes were enriched in the glutathione pathway in the NaCl and MT treatment groups, among which 16 and 27 genes were related to GST, respectively. However, only one gene was upregulated, which was inconsistent with previous research reports. We speculated that this upregulated gene played a key role in combating salt stress. Subsequently, we used quantitative verification of this upregulated gene [
30,
31,
32]. Studies have shown that GSSG (oxidized glutathione) can be reconverted to GSH through the ascorbate-glutathione cycle to help maintain the reduced state within cells. This process helps protect cell membranes from oxidative damage [
33]. In the glutathione biosynthesis pathway, we found that DHAR, an essential gene involved in the AScorbate-glutathione (ASA-GSH) cycle, was significantly upregulated in the MT treatment group, which may accelerate the process of GSSG conversion to GSH to achieve the purpose of antioxidant. This provides a new basis for MT to enhance the salt tolerance of alfalfa. (
Figure 6B).
4.3. Plant Hormone Synthesis and Signal Transduction
Plants can accurately regulate the production, transport and decomposition of various hormones to combat adverse conditions such as salt stress [
34]. Salt stress can induce plant synthesis of ABA, and ABA has many functions, such as controlling the size of leaf stomata, limiting the growth of plant lateral roots, participating in seed germination, accelerating fruit ripening, and stimulating the production of intracellular protective proteins [
35,
36,
37]. In our experiment, in the salt-treated group, NCED, a key gene for ABA synthesis, was significantly upregulated, crtB, a key gene for catalytic ABA synthesis, was upregulated, and CYP707A, a key gene for the ABA degradation pathway, was down-regulated. Nguyen et al. [
38] found that 2C protein phosphatase (PP2Cs) constituted a key class of protein phosphatase in plants, and the regulatory role of PP2Cs was also observed in the process of ABA signal transduction, which revealed that members of the PP2C family played an important role in coping with adversity in plants. The expression of ABF and PP2C genes is increased in ABA signal transduction. We hypothesized that the up-regulation of ABF and PP2C genes accelerates the action time of ABA, thereby regulating seed dormancy and enhancing plant resistance to salt stress, the same as the result of bud length measured in our phenotype indicators. At the same time, the expressions of NCED and crtB were decreased in comparing melatonin and salt treatment, indicating that adding melatonin can help reduce the synthesis of ABA (
Figure 6C).
Under salt stress, the concentration of gibberellin (GA) in plants decreases, and this change is related to the decreased expression of GA20ox, a key gene for gibberellin synthesis [
39]. Salt stress inhibits plant growth by reducing GA levels, but when exogenous GA is applied, it can stimulate the synthesis of endogenous GA in plants and promote plant growth [
40]. In the signaling process of gibberellin, GA affects the elongation of cotyle cells by regulating the interaction between the DELLA protein and PIF3 and PIF4 genes [
41]. In this study, the expressions of KAO and GA20ox, key genes in GA synthesis, were significantly down-regulated under NaCl stress. However, in the gibberellin signaling pathway, the expression of the PIF4 gene was upregulated. This may be because alfalfa inhibits plant growth by reducing GA content to better adapt to a stressful environment. With alfalfa's adaptation to salt stress, GA promotes root elongation by regulating PIF4 expression. In the melatonin-treated group, the gene expression of synthetic DELLA protein was upregulated to enhance the effect of GA, which was consistent with the results of previous studies (
Figure 6D).
4.4. Biosynthesis of Physiological Regulatory Substances
Proline is a recognized osmoregulatory substance in plants. Some amino acids (such as phenylalanine, tryptophan, tyrosine and other aromatic amino acids) are precursors of natural products such as alkaloids, flavonoids, plant auxin and cell wall components and play an important role in plant growth and response to environmental stress [
42]. Guo et al. [
43] compared wheat's metabolic response and adaptation mechanism to salt-alkali stress. They found that salt stress would increase sugar content in wheat, and the metabolic process tended to respond to osmotic stress through gluconeogenesis. Under salt stress, various differential metabolites were detected in the roots and leaves of wheat seedlings, which were involved in the tricarboxylic acid (TCA) cycle, glycolysis, amino acids and sugars. By KEGG pathway analysis, we found that the expression of P5CS, the key enzyme encoding gene for proline synthesis, was significantly increased, and the expression of ADT and PAT, several key genes in the phenylalanine and tyrosine synthesis pathway, were increased. In addition, in the gluconeogenic pathway, the expression of the gene E5.1.3.15, which is responsible for synthesizing fructose-6-phosphate, was significantly increased. In addition, we noted that the expressions of genes AKR1A1 and ADH5, which are involved in the synthesis of fructose-6-phosphate, were upregulated in both glycolysis and gluconeogenesis in the MT group. This is consistent with the study of Guo et al. [
43].
4.5. ABC Transporter
The ABC transporter family is widely distributed in the biological world and is the largest and most functional transmembrane transporter currently known, which plays a crucial role in the transmembrane transport of substances between eukaryotes and prokaryotes [
44]. Some studies have pointed out that under salt stress, the expression level of ABC transporter in plant leaves increases, suggesting that ABC protein may play an important role in regulating plants' internal and external balance, helping plants make internal regulations to cope with salt stress [
45]. Most of the members of the ABCB subfamily identified are mainly involved in transporting and regulating hormone-like substances such as auxin [
46]. When multiple phosphatase inhibitors were used to inhibit the activity of the ABCG transporter, the use of inhibitors of the PDR transporter could significantly reduce the number of flavonoids and design secreted by soybean roots, and EST database analysis showed that PDR transporter coding genes were abundant in the roots. Therefore, it is speculated that PDR transporters may be key proteins in the root secretion of flavonoids [
47,
48]. In addition, the ABCC2 transporter is involved in ABA transport, which in turn is involved in flavonoid transport [
49]. In our study, the expression of ABCB1 was significantly upregulated, the expression of the PDR5 gene was down-regulated, and the expression of ABCC2 was upregulated under salt stress, suggesting that the synthesis and transport of flavonoids and plant hormones were important mechanisms for alfalfa to adapt to salt stress. After the addition of MT, the expression of the ABC transporter gene was upregulated, indicating that MT can alleviate salt stress by accelerating the transport of flavonoids and plant hormones.
4.6. MAPK Signal Path
Mitogen-activated protein kinase (MAPK) signaling pathway is a key signal transduction system in eukaryotic cells, involving three core protein kinase components: The cascade reaction of MAPK (also known as MPK), MAPK kinase (also known as MAPKK, MKK, or MEK), and MAPKK kinase (also known as MAPKKK, MKKK, or MEKK) progressively transfers signals from upstream sensors of cells to downstream effector molecules through continuous phosphorylation[
50,
51,
52]. MAPK can intervene in the salt stress response mechanism by sensing the external high salt signal and transmitting the signal to the interior of the cell, which helps to improve the salt stress tolerance of plants [
53]. MAPK signaling pathway plays an important role in plant response to salt stress, and its role is dependent on the ABA signaling pathway [
54]. The expression of differential genes involved in the PYL-PP2C-SnRK2 signaling pathway in alfalfa seedlings was significantly changed under salt stress, and most of the differential genes were upregulated. SnRK2 could activate MAPKKK17_18, and then MKK3, MPK1_2 and downstream stress adaptation genes were activated through a series of phosphorylation reactions. Moreover, the expression of the CALM gene, which synthesizes MAPK8, is down-regulated to maintain the balance between ROS production and clearance. In the melatonin treatment group, the expression of MAPK3 and WRKY22, which can lead to cell death and produce H
2O
2, was down-regulated, once again proving that melatonin can alleviate the stress effect of salt on alfalfa (
Figure 6E).
5. Conclusion
In conclusion, our physiochemical and transcriptomic analyses revealed the roles of MT in mediating the salt tolerance of alfalfa. MT enhanced salt tolerance by rebuilding plant growth, improving antioxidant capacity. Moreover, MT addition to salt stress triggered a large-scale transcriptomic remodeling. Especially many differentially expressed genes related to flavonoid biosynthesis, plant hormone signaling transduction, and MAPK signaling pathways, were altered by MT under salt stress. The potential hub genes positively associated with salt-responsive traits are identified through WGCNA, suggesting that these genes might participate in MT-regulated salt tolerance. Taken together, our transcriptomic study results elucidate the molecular mechanism of MT mediated salt tolerance in alfalfa and provide candidate genes for breeding salt-tolerant alfalfa.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, X.R., H.S. and X.Z.; methodology, X.R., H.S. and X.Z.; data curation, Z.L., W.Z., G.L. and Y.L.; Validation, Z.L., W.Z., G.L. and Y.L.; Software, Z.L.; writing—original draft preparation, Z.L.; Funding acquisition, H.S.; Investigation, H.S., X.Z. C.W., Y.S.; Writing – original draft, Z.L.; Writing – review & editing, H.S., X.Z. C.W., Y.S.
Funding
This work was supported by the earmarked fund for the National Science Foundation of China (No. 32001393), the China Postdoctoral Science Foundation (2022M721043), the Science and Technology Innovation 2030-Major Project (2022ZD04011), the Henan Science and Technology Research Projects (222103810006), the China Agriculture Research System (CARS-34). The funding body played no role in the design of the study, the collection, analysis and interpretation of the data or the writing of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data presented in this study are available upon reasonable request to the corresponding author.
Acknowledgments
We thank Shanghai Majorbio Biotechnology Co., Ltd. for their help with sequencing.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zelm, E.V.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annual Review of Plant Biology 2020, 71. [Google Scholar] [CrossRef]
- Ludwiczak, A.; Osiak, M.; Cárdenas-Pérez, S.; Lubińska-Mielińska, S.; Piernik, A. Osmotic Stress or Ionic Composition: Which Affects the Early Growth of Crop Species More? Agronomy 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Israelsen, K.R.; Waldron, R.B.L. Salinity Tolerance of Foxtail Barley (Hordeum jubatum) and Desirable Pasture Grasses. Weed Science. 2011, 59, 500–505. [Google Scholar] [CrossRef]
- Singer, S.D.; Hannoufa, A.; Acharya, S. Molecular improvement of alfalfa for enhanced productivity and adaptability in a changing environment. Plant Cell Environ. 2018, 41, 1955–1971. [Google Scholar] [CrossRef]
- Stritzler, M.; Elba, P.; Berini, C.; Gomez, C.; Ayub, N.; Soto, G. High quality forage production under salinity using a salt-tolerant AtNXH1-expressing transgenic alfalfa combined with a natural stress-resistant nitrogen-fixing bacterium. Biotechnol 2018, 276-277, 42–45. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Ma, X.; Cai, W.; Wang, Y.; Gao, X.; Fu, B.; Li, S. Exogenous Proline Improves Salt Tolerance of Alfalfa through Modulation of Antioxidant Capacity, Ion Homeostasis, and Proline Metabolism. Plants 2022, 11, 2994. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yin, J.; Wang, J.; Li, J. Integrative analysis of transcriptome and metabolome revealed the mechanisms by which flavonoids and phytohormones regulated the adaptation of alfalfa roots to NaCl stress. Front. Plant Sci. 2023, 14, 1117868–10. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Gómez, S.; Mesa-Marín, J.; Pérez-Romero, J.A.; López-Jurado, J.; García-López, J.V.; Mariscal, V.; Molina -Heredia, F.P.; Pajuelo, E.; Rodríguez-Llorente, I.D.; Flowers, T.J.; et al. Consortia of Plant-Growth-Promoting Rhizobacteria Isolated from Halophytes Improve Response of Eight Crops to Soil Salinization and Climate Change Conditions. Agronomy 2021, 11, 1609. [Google Scholar] [CrossRef]
- Gul, Z.; Tang, Z.-H.; Arif, M.; Ye, Z. An Insight into Abiotic Stress and Influx Tolerance Mechanisms in Plants to Cope in Saline Environments. Biology 2022, 11, 597. [Google Scholar] [CrossRef]
- Li, N.; Wang, Z.Y.; Wang, B.K.; Wang, J.; Xu, R.Q.; Yang, T.; Huang, S.Y.; Wang, H.; Yu, Q. Identification and Characterization of Long Non-coding RNA in Tomato Roots Under Salt Stress. Front. Plant Sci. 2022, 13, 834027. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biology. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. International Journal of Molecular Sciences. 2018, 19, 3206–10. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and its relationship to plant hormones. Annals of Botany. 2018, 121, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: plant growth regulator and/or biostimulator during stress? Trends Plant Sci. 2014, 19, 789–797. [Google Scholar] [CrossRef]
- Park, H.S.; Kazerooni, E.A.; Kang, S.M.; Al-Sadi, A.M.; Lee, I.J. Melatonin Enhances the Tolerance and Recovery Mechanisms in Brassica juncea (L.) Czern. Under Saline Conditions. Front. Plant Sci. 2021, 12, 593717. [Google Scholar] [CrossRef]
- Shen, C.; Du, H.; Chen, Z.; Wang, T. The Chromosome-Level Genome Sequence of the Autotetraploid Alfalfa and Resequencing of Core Germplasms Provide Genomic Resources for Alfalfa Research. Molecular Plant. 2020, 13, 12. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Ashburner, M.M.; Ball, C.; Blake, J.; Sherlock, G. Gene ontology: a tool for the unification of biology. Nat Genet. 2000, 25, 25–9. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Brunetti, C. Flavonoids as Antioxidants in Plants Under Abiotic Stresses. Springer New York, 2012. [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev 2011, 10, 397–412. [Google Scholar] [CrossRef]
- Mahajan, M.; Yadav, S.K. Overexpression of a tea flavanone 3-hydroxylase gene confers tolerance to salt stress and Alternaria solani in transgenic tobacco. Plant Mol.Biol 2014, 85, 551–573. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev.Plant Biol 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Chawla, S.; Jain, S.; Jain, V. Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativaL.). Plant Biochem. Biotechnol. 2013, 22, 27–34. [Google Scholar] [CrossRef]
- Nazar, R.; Iqbal, N.; Syeed, S.; Khan, N.A. Salicylic acid alleviates decreases in photosynthesis under salt stress by enhancing nitrogen and sulfur assimilation and antioxidant metabolism differentially in two mungbean cultivars. Plant Physiol. 2011, 168, 807–815. [Google Scholar] [CrossRef]
- Rausch, T.; Gromes, R.; Liedschulte, V.; Müller, I.; Bogs, J.; Galovic, V.; Wachter, A. Novel insight into the regulation of GSH biosynthesis in higher plants. Plant Biol. 2007, 9, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Stavridou, E.; Michailidis, M.; Gedeon, S.; Ioakeim, A.; Kostas, S.; Chronopoulou, E.; Labrou, N.E.; Edwards, R.; Day, A.; Nianiou-Obeidat, I.; et al. Tolerance of Transplastomic Tobacco Plants Overexpressing a Theta Class Glutathione Transferase to Abiotic and Oxidative Stresses. Front. Plant Sci. 2019, 9, 1861. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, L.; Liu, Y.; Shen, X.; Guo, Y.; Ma, X.; Zhang, X.; Li, X.; Cheng, T.; Wen, H.; et al. RNA-Seq-Based WGCNA and Association Analysis Reveal the Key Regulatory Module and Genes Responding to Salt Stress in Wheat Roots. Plants 2024, 13, 274. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Yu, X.; Wang, Y.; Fu, W.; Cao, R.; Yang, L.; Ye, X. Genome-wide identification and expression analysis of glutathione S-transferase gene family to reveal their role in cold stress response in cucumber. Front. Genet. 2022, 13, 1009883. [Google Scholar] [CrossRef]
- Madhu, K.A.; Tyagi, S.; Shumayla; Upadhyayet, S.K. Exploration of glutathione reductase for abiotic stress response in bread wheat (Triticum aestivum L.). Plant Cell Reports. 2022, 41, 639–654. [Google Scholar] [CrossRef] [PubMed]
- Sabagh, A.E.; Mbarki, S.; Hossain, A.; Iqbal, M.A.; Islam, M.S.; Raza, A.; Analía, L.; Reginato, M.; Rahman, M.A.; Mahboob, W.; et al. Potential role of plant growth regulators in administering crucial processes against abiotic stresses. Front. Agron 2021, 3, 648694. [Google Scholar] [CrossRef]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar]
- Mittler, R.; Blumwald, E. The roles of ROS and ABA in systemic acquired acclimation. Plant Cell. 2015, 27, 64–70. [Google Scholar] [CrossRef]
- Marusig, D.; Tombesi, S. Abscisic acid mediates drought and salt stress responses in Vitis vinifera: A review. International Journal of Molecular Sciences. 2020, 21, 8648. [Google Scholar] [CrossRef]
- Chuong, N.N.; Hoang, X.L.T.; Nghia, D.H.T.; Daiet, T.N.T.; Thao, N.P. Protein Phosphatase Type 2C Functions in Phytohormone-Dependent Pathways and in Plant Responses to Abiotic Stresses. Current Protein and Peptide Science. 2021, 22. [Google Scholar] [CrossRef]
- Zhu, T.; Lin, J.; Zhang, M.; Li, L.; Chen, M. Phytohormone involved in salt tolerance regulation of elaeagnus angustifolia l. seedlings. For. Res. 2019, 24, 235–242. [Google Scholar] [CrossRef]
- Niharika, S.; Singh, A.; Khare, S.; Yadav, R.K. Mitigating strategies of gibberellins in various environmental cues and their crosstalk with other hormonal pathways in plants: a review. Plant Mol. Biol. Rep. 2020, 39, 34–49. [Google Scholar] [CrossRef]
- Lucas, M.D.; Salomé, P. PIFs get BRright: phytochrome interacting factors as integrators of light and hormonal signals. New Phytologist. 2014, 202, 4. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annual Review of Plant Biology. 2012, 63, 73. [Google Scholar] [CrossRef]
- Guo, R.; Yang, Z.; LI, F.; Yan, C.R.; Zhong, X.L.; Liu, Q.; Xia, X.; Li, H.R.; Zhao, L. Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol. 2015, 15, 170–2022. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Jin, J.Y.; Alejandro, S.; Martinoia, E.; Lee, Y. Overexpression of AtABCG36 improves drought and salt stress resistance inArabidopsis. Physiol.Plant. 2010, 139, 170–180. [Google Scholar] [CrossRef]
- Terasaka, K.; Blakeslee, J.J.; Titapiwatanakun, B.; Peer, W.A.; Bandyopadhyay, A.; Makam, S.N.; Lee, O.R.; Richards. E.L.; Murphy,A.S. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell. 2005, 17, 2922–2939. [Google Scholar] [CrossRef]
- Sugiyama, A.; Shitan, N.; Yazaki, K. Signaling from soybean roots to Rhizobium: An ATP-binding cassette-type transporter mediates genistein secretion. Plant Signaling&Behavior. 2008, 3, 38–40. [Google Scholar] [CrossRef]
- Sugiyama, A.; Shitan, N.; Yazaki, K. Signaling from soybean roots to rhizobium: An ATP-binding cassette-type transporter mediates genistein secretion. Plant Signaling&Behavior. 2008, 3, 38. [Google Scholar] [CrossRef]
- Behrens, C.E.; Smith, K.E.; Iancu, C.V.; Choe, J.Y.; Dean, J.V. Transport of anthocyanins and other flavonoids by the arabidopsis ATP-binding cassette transporter AtABCC2. Sci. Rep. 2016, 9, 437. [Google Scholar] [CrossRef]
- Manna, M.; Rengasamy, B.; Sinha, A.K. Revisiting the role of MAPK signalling pathway in plants and its manipulation for crop improvement. Plant Cell Environ. 2023, 46, 2277–2295. [Google Scholar] [CrossRef]
- Rohila, J.S.; Yang, Y. Rice mitogen-activated protein kinase gene family and its role in biotic and abiotic stress response. Integr. Plant Biol. 2007, 49, 751–759. [Google Scholar]
- Zhang, M.; Su, J.B.; Zhang, Y.; Xu, J.; Zhang, S.Q. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. Current opinion in plant biology. 2018, 45, 1–10. [Google Scholar] [CrossRef]
- Sadau, S.B.; Mehari, T.G.; Ahmad, A.; Tajo, S.M.; Ibrahim, S.; Iqbal, M.S.; Elasad, M.; Zhang, J.; Wei, H.; Yu, S. Genome wide identification and characterization of MAPK genes reveals their potential in enhancing drought and salt stress tolerance in Gossypium hirsutum. Cotton Res. 2022, 5, 23. [Google Scholar] [CrossRef]
- Bai, G.; Xie, H.; Yao, H.; Li,F. ; Chen, X.; Zhang, Y.; Xiao, B.; Yang, J.; Li, Y.; Yang, D.H. Genome-wide identification and characterization of ABA receptor PYL/RCAR gene family reveals evolution and roles in drought stress in Nicotiana tabacum. BMC Genomics, 2019, 20, 575. [Google Scholar] [CrossRef]
Figure 1.
Melatonin eliminates the inhibitory effect of salt stress on seed germination, bud length and fresh weight. (A) Alfalfa seedling phenotypes. Bar=2 cm; (B) Seed germination rate after 7-day treatment. Line chart represent average of six biological replicates with standard deviation, and each replicate has 30 seeds. (C) Bud length, columns with different lowercase letters indicate significant differences among treatments at p < 0.05 using Duncan’s test. Data represent average of three biological replicates with standard deviation, and each replicate has 10 plants. (D) Fresh weight of alfalfa seedlings. the column represents the mean of the three biological replicates. Different lowercase letters indicate significant differences between treatments by Duncan test. CK indicates control.
Figure 1.
Melatonin eliminates the inhibitory effect of salt stress on seed germination, bud length and fresh weight. (A) Alfalfa seedling phenotypes. Bar=2 cm; (B) Seed germination rate after 7-day treatment. Line chart represent average of six biological replicates with standard deviation, and each replicate has 30 seeds. (C) Bud length, columns with different lowercase letters indicate significant differences among treatments at p < 0.05 using Duncan’s test. Data represent average of three biological replicates with standard deviation, and each replicate has 10 plants. (D) Fresh weight of alfalfa seedlings. the column represents the mean of the three biological replicates. Different lowercase letters indicate significant differences between treatments by Duncan test. CK indicates control.
Figure 2.
Exogenous melatonin can alleviate the oxidative stress caused by NaCl treatment. O2- content (A), MDA content (B), GSH content (C), SOD activity (D), POD activity (E). The columns represent the average of three biological replicates, each with ten individual plants. Using the Duncan test, different lowercase letters indicated significant differences between treatments. *p < 0.05, ** p < 0.01.
Figure 2.
Exogenous melatonin can alleviate the oxidative stress caused by NaCl treatment. O2- content (A), MDA content (B), GSH content (C), SOD activity (D), POD activity (E). The columns represent the average of three biological replicates, each with ten individual plants. Using the Duncan test, different lowercase letters indicated significant differences between treatments. *p < 0.05, ** p < 0.01.
Figure 3.
Comprehensive visualization of control differentially expressed genes (DEGs)CK, NaCl, and MT combined with NaCl (MT+NaCl). (A) The Venn diagram shows the unique and shared DEGs. (B) The number of differentially expressed genes and the number of up-regulated and down-regulated genes. (C) Analysis of variance for multiple groups. * 0.01 < p ≤ 0.05,** 0.001 < p ≤ 0.01. (D) Hierarchical clustering analysis based on the log2 RPFK expression trends of DEGs under salt alone or with melatonin treatment.
Figure 3.
Comprehensive visualization of control differentially expressed genes (DEGs)CK, NaCl, and MT combined with NaCl (MT+NaCl). (A) The Venn diagram shows the unique and shared DEGs. (B) The number of differentially expressed genes and the number of up-regulated and down-regulated genes. (C) Analysis of variance for multiple groups. * 0.01 < p ≤ 0.05,** 0.001 < p ≤ 0.01. (D) Hierarchical clustering analysis based on the log2 RPFK expression trends of DEGs under salt alone or with melatonin treatment.
Figure 4.
Functional annotation and KEGG enrichment analysis of differentially expressed genes. (A) According to the GO classification of DEGs, DEGs is divided into metabolic classes according to biological processes, cell components, and molecular functions, and the three colors from top to bottom represent ZMNMT vs CK, ZMN vs CK, and ZMNMT vs ZMN, respectively. The functional classification of KEGG is that the spot size represents the number of genes, and the color represents the q-value. (B-D) KEGG functional classification of DEGs in ZMNMT vs CK(B), ZMN vs CK(C), and ZMNMT vs ZMN(D).
Figure 4.
Functional annotation and KEGG enrichment analysis of differentially expressed genes. (A) According to the GO classification of DEGs, DEGs is divided into metabolic classes according to biological processes, cell components, and molecular functions, and the three colors from top to bottom represent ZMNMT vs CK, ZMN vs CK, and ZMNMT vs ZMN, respectively. The functional classification of KEGG is that the spot size represents the number of genes, and the color represents the q-value. (B-D) KEGG functional classification of DEGs in ZMNMT vs CK(B), ZMN vs CK(C), and ZMNMT vs ZMN(D).
Figure 5.
Weighted gene co-expression network analysis (WGCNA) revealed the correlation between salt-stress-related physiological indices regulated by melatonin and differentially expressed genes. (A) The heat map shows the correlation between the modules and the physiological parameters. (B) Module classification tree using WGCNA hierarchical clustering. (C) Co-expression network of DEGs in MEturquoise. (D) Co-expression network of DEGs in MEblue.
Figure 5.
Weighted gene co-expression network analysis (WGCNA) revealed the correlation between salt-stress-related physiological indices regulated by melatonin and differentially expressed genes. (A) The heat map shows the correlation between the modules and the physiological parameters. (B) Module classification tree using WGCNA hierarchical clustering. (C) Co-expression network of DEGs in MEturquoise. (D) Co-expression network of DEGs in MEblue.
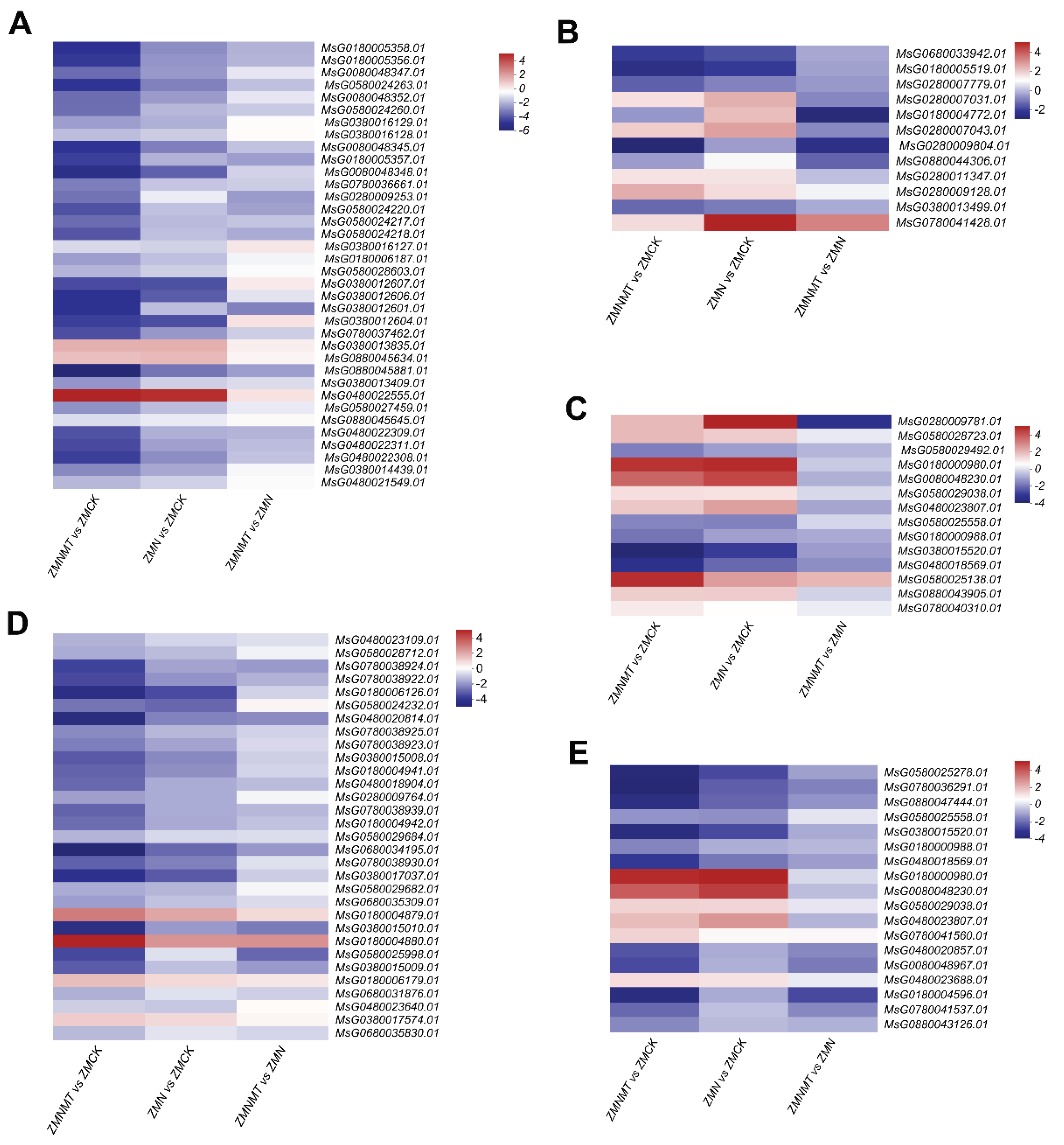
Figure 6.
According to KEGG and related literature, the log2FC of essential genes for the synthesis of important metabolites under three different treatments was plotted for cluster analysis. Italics represent gene ids of labeled enzymes or transcription factors, including essential genes involved in flavonoid biosynthesis (A), gibberellin (B), abscisic acid synthesis and signal transduction (C), GSH biosynthesis (D), and MAPK signaling pathway (E).
Figure 6.
According to KEGG and related literature, the log2FC of essential genes for the synthesis of important metabolites under three different treatments was plotted for cluster analysis. Italics represent gene ids of labeled enzymes or transcription factors, including essential genes involved in flavonoid biosynthesis (A), gibberellin (B), abscisic acid synthesis and signal transduction (C), GSH biosynthesis (D), and MAPK signaling pathway (E).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).