Submitted:
08 March 2024
Posted:
12 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Non-Coding RNAs and Their Regulatory Role in Cancer
3. The Role of vtRNAs in Cancer Cells Proliferation and Tumorigenesis
4. vaultRNAs as Powerful Diagnostic and Cancer Therapy Monitoring Markers
5. Epigenetic Regulation of vtRNAs Expression in Cancer
6. Vault RNAs as a Regulator during the Autophagy
7. Vault RNAs Act as Apoptosis Modulators
8. vtRNAs and Drug Resistance
9. Vault RNAs in Aging
10. Vault RNAs and Immune Response
11. vtRNAs Role in Neural System Development and Pathology
12. Conclusions and Future Perspectives
List of abbreviations
| Ago2 | Argonaute 2 |
| AML | acute myeloid leukemia |
| AP1 | activator protein 1 |
| ARE | arecoline |
| ASD | autism spectrum disorder |
| ATF-2 | activating transcription factor 2 |
| ATG5 | autophagy related 5 protein |
| BC | breast cancer |
| CDC25A | cell division cycle 25 A |
| c-Jun | transcription factor Jun |
| CLEAR | coordinated lysosomal expression and regulation network |
| COX-2 | cyclooxygenase |
| CSCC | cervical squamous cel carcinoma |
| CYP3A4 | cytochrome P450 3A4 |
| DICER | endoribonuclease Dicer |
| DOX | doxorubicin |
| DROSHA | class 2 ribonuclease III enzyme |
| EBV | Epstein-Barr virus |
| eiF2α | eukaryotic initiation factor 2 |
| ERK1/ERK2 | extracellular signal-regulated kinases |
| ERKs | extracellular signal-regulated kinases |
| GAGE6 | G Antigen 6 |
| HCC | hepatocellular carcinoma cells |
| IBS | inflammatory bowel disease |
| IFN-β | interferon β |
| IRF3 | interferon regulatory factor 3 |
| JNK | c-Jun N-terminal kinases |
| LAMP1 | lysosomal associated membrane protein 1 |
| LC3II | autophagosome-associated protein LC3-II |
| LEF1 | lymphoid enhancer factor 1 |
| lncRNA | long non-coding RNA |
| MAPK | mitogen activated protein kinase |
| MDR | multidrug resistance |
| MDS | myelodysplastic syndrome |
| miRNA | micro RNA |
| MMP-9 | matrix metaloproteinase-9 |
| MVP | major vault protein |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NPC | nuclear pore complex |
| OSCC | oral squamous cel carcinoma |
| OXCT1-AS1 | OXCT1 antisense RNA 1 |
| p16INK4A | protein bound to cyclin dependent kinase 4 and 6 |
| p21Waf1/Cip1 | |
| p38 | p38 mitogen-activated protein kinases |
| p62 | nuclear pore glycoprotein p62 |
| PB1 | Phox and Bem1 domain |
| PI3K/Akt | phosphatidyl inositol 3-kinase/ v-akt murine thymoma viral oncogene homolog 1 |
| PKR | RNA-dependent protein kinase |
| pPKR | phosphorylated PKR |
| PSD95 | postsynaptic density protein 95 |
| PSF | polypyrimidine tract binding protein associated factor |
| RBD | RNA-binding domain |
| RNP | ribonucleoprotein |
| rRNA | ribosomal RNA |
| SA-β-gal | senescence-associated β-galactosidase |
| SQSTM1 | sequestosome 1 |
| TEP1 | telomerase associated protein 1 |
| TFEB | transcription factor EB |
| TME | tumor microenvironment |
| TNBC | triple-negative breast cancer |
| TSG | tumor suppressor gene |
| VEGF | vascular endothelial growth factor |
| vPARP | vault-associated poly (ADP-ribose) polymerase |
Author Contributions
Conflicts of Interest
References
- Zhou, S.; Van Bortle, K. The Pol III Transcriptome: Basic Features, Recurrent Patterns, and Emerging Roles in Cancer. WIREs RNA 2023, e1782. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Rome, L.H. Isolation and Characterization of a Novel Ribonucleoprotein Particle: Large Structures Contain a Single Species of Small RNA. J. Cell Biol. 1986, 103, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Abbondanza, C.; Rossi, V.; Roscigno, A.; Gallo, L.; Belsito, A.; Piluso, G.; Medici, N.; Nigro, V.; Molinari, A.M.; Moncharmont, B.; et al. Interaction of Vault Particles with Estrogen Receptor in the MCF-7 Breast Cancer Cell. J. Cell Biol. 1998, 141, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Heuser, J.E.; Chugani, D.C.; Rome, L.H. Vaults. III. Vault Ribonucleoprotein Particles Open into Flower-like Structures with Octagonal Symmetry. J. Cell Biol. 1991, 112, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.B.; Wadhwa, R.; Kumar, P.K.R. Expression of Noncoding Vault RNA in Human Malignant Cells and Its Importance in Mitoxantrone Resistance. Mol. Cancer Res. 2010, 8, 1536–1546. [Google Scholar] [CrossRef] [PubMed]
- Frascotti, G.; Galbiati, E.; Mazzucchelli, M.; Pozzi, M.; Salvioni, L.; Vertemara, J.; Tortora, P. The Vault Nanoparticle: A Gigantic Ribonucleoprotein Assembly Involved in Diverse Physiological and Pathological Phenomena and an Ideal Nanovector for Drug Delivery and Therapy. Cancers 2021, 13, 707. [Google Scholar] [CrossRef]
- Chugani, D.C.; Rome, L.H.; Kedersha, N.L. Evidence That Vault Ribonucleoprotein Particles Localize to the Nuclear Pore Complex. J. Cell Sci. 1993, 106, 23–29. [Google Scholar] [CrossRef]
- van Zon, A.; Mossink, M.H.; Schoester, M.; Scheffer, G.L.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A.C. Multiple Human Vault RNAs. J. Biol. Chem. 2001, 276, 37715–37721. [Google Scholar] [CrossRef]
- Van Zon, A.; Mossink, M.H.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A.C. The Vault Complex. Cell. Mol. Life Sci. CMLS 2003, 60, 1828–1837. [Google Scholar] [CrossRef]
- Hahne, J.C.; Lampis, A.; Valeri, N. Vault RNAs: Hidden Gems in RNA and Protein Regulation. Cell. Mol. Life Sci. 2021, 78, 1487–1499. [Google Scholar] [CrossRef]
- Kim, E.; Lee, S.; Mian, M.F.; Yun, S.U.; Song, M.; Yi, K.; Ryu, S.H.; Suh, P. Crosstalk between Src and Major Vault Protein in Epidermal Growth Factor-dependent Cell Signalling. FEBS J. 2006, 273, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.-H.; Ginn-Pease, M.E.; Eng, C. Phosphatase and Tensin Homologue Deleted on Chromosome 10 (PTEN) Has Nuclear Localization Signal–Like Sequences for Nuclear Import Mediated by Major Vault Protein. Cancer Res. 2005, 65, 4108–4116. [Google Scholar] [CrossRef]
- Slesina, M.; Inman, E.M.; Rome, L.H.; Volknandt, W. Nuclear Localization of the Major Vault Protein in U373 Cells. Cell Tissue Res. 2005, 321, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Pietras, P.; Leśniczak-Staszak, M.; Kasprzak, A.; Andrzejewska, M.; Jopek, K.; Sowiński, M.; Rucinski, M.; Lyons, S.M.; Ivanov, P.; Szaflarski, W. MVP Expression Facilitates Tumor Cell Proliferation and Migration Supporting the Metastasis of Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 12121. [Google Scholar] [CrossRef]
- Szaflarski, W.; Sujka-Kordowska, P.; Pula, B.; Jaszczyńska-Nowinka, K.; Andrzejewska, M.; Zawierucha, P.; Dziegiel, P.; Nowicki, M.; Ivanov, P.; Zabel, M. Expression Profiles of Vault Components MVP, TEP1 and vPARP and Their Correlation to Other Multidrug Resistance Proteins in Ovarian Cancer. Int. J. Oncol. 2013, 43, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Lara, P.C.; Pruschy, M.; Zimmermann, M.; Henríquez-Hernández, L.A. MVP and Vaults: A Role in the Radiation Response. Radiat. Oncol. 2011, 6, 148. [Google Scholar] [CrossRef]
- Lee, H.M.; Joh, J.W.; Seo, S.-R.; Kim, W.-T.; Kim, M.K.; Choi, H.S.; Kim, S.Y.; Jang, Y.-J.; Sinn, D.H.; Choi, G.S.; et al. Cell-Surface Major Vault Protein Promotes Cancer Progression through Harboring Mesenchymal and Intermediate Circulating Tumor Cells in Hepatocellular Carcinomas. Sci. Rep. 2017, 7, 13201. [Google Scholar] [CrossRef]
- Balan, S.; Lekshmi, S.; Radha, K.; Sathyan, S.; Vijai, J.; Banerjee, M.; Radhakrishnan, K. Major Vault Protein (MVP) Gene Polymorphisms and Drug Resistance in Mesial Temporal Lobe Epilepsy with Hippocampal Sclerosis. Gene 2013, 526, 449–453. [Google Scholar] [CrossRef]
- Mossink, M.H.; van Zon, A.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A. Vaults: A Ribonucleoprotein Particle Involved in Drug Resistance? Oncogene 2003, 22, 7458–7467. [Google Scholar] [CrossRef]
- Han, M.; Lv, Q.; Tang, X.-J.; Hu, Y.-L.; Xu, D.-H.; Li, F.-Z.; Liang, W.-Q.; Gao, J.-Q. Overcoming Drug Resistance of MCF-7/ADR Cells by Altering Intracellular Distribution of Doxorubicin via MVP Knockdown with a Novel siRNA Polyamidoamine-Hyaluronic Acid Complex. J. Controlled Release 2012, 163, 136–144. [Google Scholar] [CrossRef]
- Wojtowicz, K.; Januchowski, R.; Nowicki, M.; Zabel, M. vPARP Adjusts MVP Expression in Drug-Resistant Cell Lines in Conjunction with MDR Proteins. Anticancer Res. 2017, 37. [Google Scholar] [CrossRef]
- Jin, D.-H.; Kim, S.; Kim, D.-H.; Park, J. Two Genetic Variants in Telomerase-Associated Protein 1 Are Associated with Stomach Cancer Risk. J. Hum. Genet. 2016, 61, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Kickhoefer, V.A.; Liu, Y.; Kong, L.B.; Snow, B.E.; Stewart, P.L.; Harrington, L.; Rome, L.H. The Telomerase/Vault-Associated Protein Tep1 Is Required for Vault RNA Stability and Its Association with the Vault Particle. J. Cell Biol. 2001, 152, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.J.; An, H.J.; Oh, Y.S.; Choi, H.R.; Ha, M.K.; Park, S.C. On the Role of Major Vault Protein in the Resistance of Senescent Human Diploid Fibroblasts to Apoptosis. Cell Death Differ. 2008, 15, 1673–1680. [Google Scholar] [CrossRef]
- Dong, X.; Akuetteh, P.D.P.; Song, J.; Ni, C.; Jin, C.; Li, H.; Jiang, W.; Si, Y.; Zhang, X.; Zhang, Q.; et al. Major Vault Protein (MVP) Associated With BRAFV600E Mutation Is an Immune Microenvironment-Related Biomarker Promoting the Progression of Papillary Thyroid Cancer via MAPK/ERK and PI3K/AKT Pathways. Front. Cell Dev. Biol. 2022, 9, 688370. [Google Scholar] [CrossRef]
- Silva, P.; West, C.M.; Slevin, N.; Valentine, H.; Ryder, W.D.J.; Hampson, L.; Bibi, R.; Sloan, P.; Thakker, N.; Homer, J.; et al. Tumor Epression of Major Vault Protein Is an Adverse Prognostic Factor for Radiotherapy Outcome in Oropharyngeal Carcinoma. Int. J. Radiat. Oncol. 2007, 69, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Ramberg, H.; Richardsen, E.; De Souza, G.A.; Rakaee, M.; Stensland, M.E.; Braadland, P.R.; Nygård, S.; Ögren, O.; Guldvik, I.J.; Berge, V.; et al. Proteomic Analyses Identify Major Vault Protein as a Prognostic Biomarker for Fatal Prostate Cancer. Carcinogenesis 2021, 42, 685–693. [Google Scholar] [CrossRef]
- Gallo, S.; Kong, E.; Ferro, I.; Polacek, N. Small but Powerful: The Human Vault RNAs as Multifaceted Modulators of Pro-Survival Characteristics and Tumorigenesis. Cancers 2022, 14, 2787. [Google Scholar] [CrossRef]
- Nandy, C.; Mrázek, J.; Stoiber, H.; Grässer, F.A.; Hüttenhofer, A.; Polacek, N. Epstein–Barr Virus-Induced Expression of a Novel Human Vault RNA. J. Mol. Biol. 2009, 388, 776–784. [Google Scholar] [CrossRef]
- Mrazek, J.; Kreutmayer, S.B.; Grasser, F.A.; Polacek, N.; Huttenhofer, A. Subtractive Hybridization Identifies Novel Differentially Expressed ncRNA Species in EBV-Infected Human B Cells. Nucleic Acids Res. 2007, 35, e73–e73. [Google Scholar] [CrossRef]
- Bracher, L.; Ferro, I.; Pulido-Quetglas, C.; Ruepp, M.-D.; Johnson, R.; Polacek, N. Human vtRNA1-1 Levels Modulate Signaling Pathways and Regulate Apoptosis in Human Cancer Cells. Biomolecules 2020, 10, 614. [Google Scholar] [CrossRef] [PubMed]
- Stadler, P.F.; Chen, J.J.-L.; Hackermuller, J.; Hoffmann, S.; Horn, F.; Khaitovich, P.; Kretzschmar, A.K.; Mosig, A.; Prohaska, S.J.; Qi, X.; et al. Evolution of Vault RNAs. Mol. Biol. Evol. 2009, 26, 1975–1991. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.B.; Matsugami, A.; Katahira, M.; Kumar, P.K.R. Human Vault-Associated Non-Coding RNAs Bind to Mitoxantrone, a Chemotherapeutic Compound. Nucleic Acids Res. 2005, 33, 4874–4881. [Google Scholar] [CrossRef] [PubMed]
- Horos, R.; Büscher, M.; Kleinendorst, R.; Alleaume, A.-M.; Tarafder, A.K.; Schwarzl, T.; Dziuba, D.; Tischer, C.; Zielonka, E.M.; Adak, A.; et al. The Small Non-Coding Vault RNA1-1 Acts as a Riboregulator of Autophagy. Cell 2019, 176, 1054–1067e12. [Google Scholar] [CrossRef] [PubMed]
- Büscher, M.; Horos, R.; Huppertz, I.; Haubrich, K.; Dobrev, N.; Baudin, F.; Hennig, J.; Hentze, M.W. Vault RNA1–1 Riboregulates the Autophagic Function of P62 by Binding to Lysine 7 and Arginine 21, Both of Which Are Critical for P62 Oligomerization. RNA 2022, 28, 742–755. [Google Scholar] [CrossRef]
- Chen, J.; OuYang, H.; An, X.; Liu, S. Vault RNAs Partially Induces Drug Resistance of Human Tumor Cells MCF-7 by Binding to the RNA/DNA-Binding Protein PSF and Inducing Oncogene GAGE6. PLOS ONE 2018, 13, e0191325. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-Mediated Cytosine-5 Methylation of Vault Noncoding RNA Determines Its Processing into Regulatory Small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef]
- Persson, H.; Kvist, A.; Vallon-Christersson, J.; Medstrand, P.; Borg, Å.; Rovira, C. The Non-Coding RNA of the Multidrug Resistance-Linked Vault Particle Encodes Multiple Regulatory Small RNAs. Nat. Cell Biol. 2009, 11, 1268–1271. [Google Scholar] [CrossRef]
- Kickhoefer, V.A.; Searles, R.P.; Kedersha, N.L.; Garber, M.E.; Johnson, D.L.; Rome, L.H. Vault Ribonucleoprotein Particles from Rat and Bullfrog Contain a Related Small RNA That Is Transcribed by RNA Polymerase III. J. Biol. Chem. 1993, 268, 7868–7873. [Google Scholar] [CrossRef]
- Schramm, L.; Hernandez, N. Recruitment of RNA Polymerase III to Its Target Promoters. Genes Dev. 2002, 16, 2593–2620. [Google Scholar] [CrossRef]
- Bergeron, D.; Faucher-Giguère, L.; Emmerichs, A.-K.; Choquet, K.; Song, K.S.; Deschamps-Francoeur, G.; Fafard-Couture, É.; Rivera, A.; Couture, S.; Churchman, L.S.; et al. Intronic Small Nucleolar RNAs Regulate Host Gene Splicing through Base Pairing with Their Adjacent Intronic Sequences. Genome Biol. 2023, 24, 160. [Google Scholar] [CrossRef]
- Perreault, J.; Perreault, J.-P.; Boire, G. Ro-Associated Y RNAs in Metazoans: Evolution and Diversification. Mol. Biol. Evol. 2007, 24, 1678–1689. [Google Scholar] [CrossRef] [PubMed]
- Vabret, N.; Najburg, V.; Solovyov, A.; Gopal, R.; McClain, C.; Šulc, P.; Balan, S.; Rahou, Y.; Beauclair, G.; Chazal, M.; et al. Y RNAs Are Conserved Endogenous RIG-I Ligands across RNA Virus Infection and Are Targeted by HIV-1. iScience 2022, 25, 104599. [Google Scholar] [CrossRef] [PubMed]
- Driedonks, T.A.P.; Nolte-’t Hoen, E.N.M. Circulating Y-RNAs in Extracellular Vesicles and Ribonucleoprotein Complexes; Implications for the Immune System. Front. Immunol. 2019, 9, 3164. [Google Scholar] [CrossRef] [PubMed]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Boffelli, D.; Martin, D.I.K. Deep Sequencing of Serum Small RNAs Identifies Patterns of 5′ tRNA Half and YRNA Fragment Expression Associated with Breast Cancer. Biomark. Cancer 2014, 6, BICS20764. [Google Scholar] [CrossRef] [PubMed]
- Mosig, A.; Guofeng, M.; Stadler, B.M.R.; Stadler, P.F. Evolution of the Vertebrate Y RNA Cluster. Theory Biosci. 2007, 126, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Alagia, A.; Tereňová, J.; Ketley, R.F.; Di Fazio, A.; Chelysheva, I.; Gullerova, M. Small Vault RNA1-2 Modulates Expression of Cell Membrane Proteins through Nascent RNA Silencing. Life Sci. Alliance 2023, 6, e202302054. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding RNAs: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Kickhoefer, V.A.; Poderycki, M.J.; Chan, E.K.L.; Rome, L.H. The La RNA-Binding Protein Interacts with the Vault RNA and Is a Vault-Associated Protein. J. Biol. Chem. 2002, 277, 41282–41286. [Google Scholar] [CrossRef]
- Kickhoefer, V.A.; Rajavel, K.S.; Scheffer, G.L.; Dalton, W.S.; Scheper, R.J.; Rome, L.H. Vaults Are Up-Regulated in Multidrug-Resistant Cancer Cell Lines. J. Biol. Chem. 1998, 273, 8971–8974. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Ferro, I.; Gavini, J.; Gallo, S.; Bracher, L.; Landolfo, M.; Candinas, D.; Stroka, D.M.; Polacek, N. The Human Vault RNA Enhances Tumorigenesis and Chemoresistance through the Lysosome in Hepatocellular Carcinoma. Autophagy 2022, 18, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Amort, M.; Nachbauer, B.; Tuzlak, S.; Kieser, A.; Schepers, A.; Villunger, A.; Polacek, N. Expression of the Vault RNA Protects Cells from Undergoing Apoptosis. Nat. Commun. 2015, 6, 7030. [Google Scholar] [CrossRef] [PubMed]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef]
- Kato, H.; Hashimoto, Y.; Hatayama, Y.; Shimohiro, H.; Motokura, T. Serum Levels of Vault RNA Significantly Varied in Patients with Haematological Malignancies. Mol. Med. Rep. 2023, 28, 190. [Google Scholar] [CrossRef]
- Fort, R.S.; Garat, B.; Sotelo-Silveira, J.R.; Duhagon, M.A. vtRNA2-1/Nc886 Produces a Small RNA That Contributes to Its Tumor Suppression Action through the microRNA Pathway in Prostate Cancer. Non-Coding RNA 2020, 6, 7. [Google Scholar] [CrossRef]
- McDonald, A.C.; Vira, M.; Walter, V.; Shen, J.; Raman, J.D.; Sanda, M.G.; Patil, D.; Taioli, E. Circulating microRNAs in Plasma among Men with Low-grade and High-grade Prostate Cancer at Prostate Biopsy. The Prostate 2019, 79, 961–968. [Google Scholar] [CrossRef]
- Kunkeaw, N.; Lee, Y.-S.; Im, W.R.; Jang, J.J.; Song, M.-J.; Yang, B.; Park, J.-L.; Kim, S.-Y.; Ku, Y.; Kim, Y.; et al. Mechanism Mediated by a Noncoding RNA, Nc886, in the Cytotoxicity of a DNA-Reactive Compound. Proc. Natl. Acad. Sci. 2019, 116, 8289–8294. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-H.; Lee, H.-S.; Lee, J.-S.; Lee, Y.-S.; Park, J.-L.; Kim, S.-Y.; Hwang, J.-A.; Kunkeaw, N.; Jung, S.Y.; Kim, T.J.; et al. Nc886 Is Induced by TGF-β and Suppresses the microRNA Pathway in Ovarian Cancer. Nat. Commun. 2018, 9, 1166. [Google Scholar] [CrossRef]
- Lee, K.; Kunkeaw, N.; Jeon, S.H.; Lee, I.; Johnson, B.H.; Kang, G.-Y.; Bang, J.Y.; Park, H.S.; Leelayuwat, C.; Lee, Y.S. Precursor miR-886, a Novel Noncoding RNA Repressed in Cancer, Associates with PKR and Modulates Its Activity. RNA 2011, 17, 1076–1089. [Google Scholar] [CrossRef]
- Izquierdo, M.A.; Scheffer, G.L.; Schroeijers, A.B.; De Jong, M.C.; Scheper, R.J. Vault-Related Resistance to Anticancer Drugs Determined by the Expression of the Major Vault Protein LRP. Cytotechnology 1998, 27, 137–148. [Google Scholar] [CrossRef]
- Jeon, S.H.; Lee, K.; Lee, K.S.; Kunkeaw, N.; Johnson, B.H.; Holthauzen, L.M.F.; Gong, B.; Leelayuwat, C.; Lee, Y.S. Characterization of the Direct Physical Interaction of Nc886, a Cellular Non-Coding RNA, and PKR. FEBS Lett. 2012, 586, 3477–3484. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Liu, H. MicroRNA-886 Suppresses Osteosarcoma Cell Proliferation and Its Maturation Is Suppressed by Long Non-Coding RNA OXCT1-AS1. Bioengineered 2022, 13, 5769–5778. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. How Cancer Arises. Sci. Am. 1996, 275, 62–70. [Google Scholar] [CrossRef]
- Zhong, C.; Yu, Q.; Peng, Y.; Zhou, S.; Liu, Z.; Deng, Y.; Guo, L.; Zhao, S.; Chen, G. Novel LncRNA OXCT1-AS1 Indicates Poor Prognosis and Contributes to Tumorigenesis by Regulating miR-195/CDC25A Axis in Glioblastoma. J. Exp. Clin. Cancer Res. 2021, 40, 123. [Google Scholar] [CrossRef]
- Li, B.; Zhu, L.; Li, L.; Ma, R. lncRNA OXCT1-AS1 Promotes Metastasis in Non-Small-Cell Lung Cancer by Stabilizing LEF1, In Vitro and In Vivo. BioMed Res. Int. 2021, 2021, 1–15. [Google Scholar] [CrossRef]
- Kärkkäinen, E.; Heikkinen, S.; Tengström, M.; Kosma, V.-M.; Mannermaa, A.; Hartikainen, J.M. Expression Profiles of Small Non-Coding RNAs in Breast Cancer Tumors Characterize Clinicopathological Features and Show Prognostic and Predictive Potential. Sci. Rep. 2022, 12, 22614. [Google Scholar] [CrossRef]
- Bornstein, S.; Shapiro, I.; Mazumdar, A.; Zitzmann, K.; Nölting, S.; Luca, E.; Beuschlein, F.; Sharma, A.; Hantel, C. The Vault Complex Is Significantly Involved in Therapeutic Responsiveness of Endocrine Tumors and Linked to Autophagy under Chemotherapeutic Conditions. Cancers 2023, 15, 1783. [Google Scholar] [CrossRef] [PubMed]
- Treppendahl, M.B.; Qiu, X.; Søgaard, A.; Yang, X.; Nandrup-Bus, C.; Hother, C.; Andersen, M.K.; Kjeldsen, L.; Möllgaard, L.; Hellström-Lindberg, E.; et al. Allelic Methylation Levels of the Noncoding VTRNA2-1 Located on Chromosome 5q31. 1 Predict Outcome in AML. Blood 2012, 119, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.-C.; Lee, C.-W.; Lin, C.-H.; Wu, C.-H.; Lee, Y.-S.; Tsai, C.-L.; Tsai, C.-N. Differential Hypermethylation of the VTRNA2-1 Promoter in Hepatocellular Carcinoma as a Prognostic Factor: Tumor Marker Prognostic Study. Int. J. Surg. 2020, 79, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Dugué, P.-A.; Yu, C.; McKay, T.; Wong, E.M.; Joo, J.E.; Tsimiklis, H.; Hammet, F.; Mahmoodi, M.; Theys, D. ; kConFab; et al. VTRNA2-1: Genetic Variation, Heritable Methylation and Disease Association. Int. J. Mol. Sci. 2021, 22, 2535. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wang, M.; Zhou, Y.; Yang, J.; Wang, L.; Yang, R.; Wen, M.; Kong, L. Noncoding RNA 886 Alleviates Tumor Cellular Immunological Rejection in Host C57BL/C Mice. Cancer Med. 2020, 9, 5258–5271. [Google Scholar] [CrossRef]
- Fort, R.S.; Mathó, C.; Geraldo, M.V.; Ottati, M.C.; Yamashita, A.S.; Saito, K.C.; Leite, K.R.M.; Méndez, M.; Maedo, N.; Méndez, L.; et al. Nc886 Is Epigenetically Repressed in Prostate Cancer and Acts as a Tumor Suppressor through the Inhibition of Cell Growth. BMC Cancer 2018, 18, 127. [Google Scholar] [CrossRef] [PubMed]
- Kunkeaw, N.; Jeon, S.H.; Lee, K.; Johnson, B.H.; Tanasanvimon, S.; Javle, M.; Pairojkul, C.; Chamgramol, Y.; Wongfieng, W.; Gong, B.; et al. Cell Death/Proliferation Roles for Nc886, a Non-Coding RNA, in the Protein Kinase R Pathway in Cholangiocarcinoma. Oncogene 2013, 32, 3722–3731. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-S.; Shin, S.; Cho, E.; Im, W.K.; Jeon, S.H.; Kim, Y.; Park, D.; Fréchet, M.; Chajra, H.; Jung, E. Nc886, a Non-Coding RNA, Inhibits UVB-Induced MMP-9 and COX-2 Expression via the PKR Pathway in Human Keratinocytes. Biochem. Biophys. Res. Commun. 2019, 512, 647–652. [Google Scholar] [CrossRef]
- Lee, K.-S.; Park, J.-L.; Lee, K.; Richardson, L.E.; Johnson, B.H.; Lee, H.-S.; Lee, J.-S.; Kim, S.-B.; Kwon, O.-H.; Song, K.S.; et al. Nc886, a Non-Coding RNA of Anti-Proliferative Role, Is Suppressed by CpG DNA Methylation in Human Gastric Cancer. Oncotarget 2014, 5, 3944–3955. [Google Scholar] [CrossRef]
- Fort, R.S.; Duhagon, M.A. Pan-Cancer Chromatin Analysis of the Human vtRNA Genes Uncovers Their Association with Cancer Biology. F1000Research 2021, 10, 182. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, H.; Tang, L.; Lou, M.; Geng, Y. Silencing Nc886, a Non-Coding RNA, Induces Apoptosis of Human Endometrial Cancer Cells-1A In Vitro. Med. Sci. Monit. 2017, 23, 1317–1324. [Google Scholar] [CrossRef]
- Lei, J.; Xiao, J.-H.; Zhang, S.-H.; Liu, Z.-Q.; Huang, K.; Luo, Z.-P.; Xiao, X.-L.; Hong, Z.-D. Non-Coding RNA 886 Promotes Renal Cell Carcinoma Growth and Metastasis through the Janus Kinase 2/Signal Transducer and Activator of Transcription 3 Signaling Pathway. Mol. Med. Rep. 2017, 16, 4273–4278. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Yu, J.; Liu, X. Upregulation of miR-886 Indicates Poor Prognosis and Promotes Tumour Progression of Prostate Cancer. Andrologia 2022, 54. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Hao, Q.; Wang, Y.; Zhou, P.; Zou, B.; Zhang, Y. Regulation of P53 Expression and Apoptosis by Vault RNA2-1-5p in Cervical Cancer Cells. Oncotarget 2015, 6, 28371–28388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Han, S.; Wang, Y.; Liu, R.; Meng, F.; Su, Z.; Huo, F. Suppression of Mir-886-3P Mediated by Arecoline (Are) Contributes to the Progression of Oral Squamous Cell Carcinoma. J. Investig. Med. 2021, 69, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Helbo, A.; Treppendahl, M.; Aslan, D.; Dimopoulos, K.; Nandrup-Bus, C.; Holm, M.; Andersen, M.; Liang, G.; Kristensen, L.; Grønbæk, K. Hypermethylation of the VTRNA1-3 Promoter Is Associated with Poor Outcome in Lower Risk Myelodysplastic Syndrome Patients. Genes 2015, 6, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ye, X.; Zhao, T. The Physiological Roles of Autophagy in the Mammalian Life Cycle: Autophagy in the Mammal Life Cycle. Biol. Rev. 2019, 94, 503–516. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in Major Human Diseases. EMBO J. 2021, 40. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in Infection, Inflammation and Immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef]
- Büscher, M.; Horos, R.; Hentze, M.W. ‘High Vault-Age’: Non-Coding RNA Control of Autophagy. Open Biol. 2020, 10, 190307. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Marques, M.; Carvalho, A.; Cavadas, C.; Aveleira, C.A. PI3K/AKT/MTOR and ERK1/2-MAPK Signaling Pathways Are Involved in Autophagy Stimulation Induced by Caloric Restriction or Caloric Restriction Mimetics in Cortical Neurons. Aging 2021, 13, 7872–7882. [Google Scholar] [CrossRef]
- Cao, Z.; Liao, Q.; Su, M.; Huang, K.; Jin, J.; Cao, D. AKT and ERK Dual Inhibitors: The Way Forward? Cancer Lett. 2019, 459, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Moscat, J.; Diaz-Meco, M.T. P62 at the Crossroads of Autophagy, Apoptosis, and Cancer. Cell 2009, 137, 1001–1004. [Google Scholar] [CrossRef] [PubMed]
- Morana, O.; Wood, W.; Gregory, C.D. The Apoptosis Paradox in Cancer. Int. J. Mol. Sci. 2022, 23, 1328. [Google Scholar] [CrossRef] [PubMed]
- Kieser, A.; Sterz, K.R. The Latent Membrane Protein 1 (LMP1). In Epstein Barr Virus Volume 2; Münz, C., Ed.; Current Topics in Microbiology and Immunology; Springer International Publishing: Cham, 2015; ISBN 978-3-319-22833-4. [Google Scholar]
- Emran, T.B.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H. ; Siddiquee, Mohd. F.-R.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652. [Google Scholar] [CrossRef]
- Steiner, E.; Holzmann, K.; Elbling, L.; Micksche, M.; Berger, W. Cellular Functions of Vaults and Their Involvement in Multidrug Resistance. Curr. Drug Targets 2006, 7, 923–934. [Google Scholar] [CrossRef]
- Geisslinger, F.; Müller, M.; Vollmar, A.M.; Bartel, K. Targeting Lysosomes in Cancer as Promising Strategy to Overcome Chemoresistance—A Mini Review. Front. Oncol. 2020, 10, 1156. [Google Scholar] [CrossRef]
- Hong, H.; Wang, Q.; Li, J.; Liu, H.; Meng, X.; Zhang, H. Aging, Cancer and Immunity. J. Cancer 2019, 10, 3021–3027. [Google Scholar] [CrossRef]
- Li, S.; Min, J.-Y.; Krug, R.M.; Sen, G.C. Binding of the Influenza A Virus NS1 Protein to PKR Mediates the Inhibition of Its Activation by Either PACT or Double-Stranded RNA. Virology 2006, 349, 13–21. [Google Scholar] [CrossRef]
- Li, F.; Chen, Y.; Zhang, Z.; Ouyang, J.; Wang, Y.; Yan, R.; Huang, S.; Gao, G.F.; Guo, G.; Chen, J.-L. Robust Expression of Vault RNAs Induced by Influenza A Virus Plays a Critical Role in Suppression of PKR-Mediated Innate Immunity. Nucleic Acids Res. 2015, gkv1078. [Google Scholar] [CrossRef]
- Kim, Y.; Ji, H.; Cho, E.; Park, N.-H.; Hwang, K.; Park, W.; Lee, K.-S.; Park, D.; Jung, E. Nc886, a Non-Coding RNA, Is a New Biomarker and Epigenetic Mediator of Cellular Senescence in Fibroblasts. Int. J. Mol. Sci. 2021, 22, 13673. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-S.; Cho, E.; Weon, J.B.; Park, D.; Fréchet, M.; Chajra, H.; Jung, E. Inhibition of UVB-Induced Inflammation by Laminaria Japonica Extract via Regulation of Nc886-PKR Pathway. Nutrients 2020, 12, 1958. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Bao, X.; Lee, H.-H.; Jang, J.J.; Saruuldalai, E.; Park, G.; Im, W.R.; Park, J.-L.; Kim, S.-Y.; Shin, S.; et al. Nc886, a Novel Suppressor of the Type I Interferon Response Upon Pathogen Intrusion. Int. J. Mol. Sci. 2021, 22, 2003. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xiao, L.; Wen, S.J.; Yu, T.; Sharma, S.; Chung, H.K.; Warner, B.; Mallard, C.G.; Rao, J.N.; Gorospe, M.; et al. Small Noncoding Vault RNA2 -1 Disrupts Gut Epithelial Barrier Function via Interaction with HUR. EMBO Rep. 2023, 24, e54925. [Google Scholar] [CrossRef] [PubMed]
- Wakatsuki, S.; Araki, T. Novel Molecular Basis for Synapse Formation: Small Non-Coding Vault RNA Functions as a Riboregulator of MEK1 to Modulate Synaptogenesis. Front. Mol. Neurosci. 2021, 14, 748721. [Google Scholar] [CrossRef]
- Wakatsuki, S.; Takahashi, Y.; Shibata, M.; Adachi, N.; Numakawa, T.; Kunugi, H.; Araki, T. Small Noncoding Vault RNA Modulates Synapse Formation by Amplifying MAPK Signaling. J. Cell Biol. 2021, 220, e201911078. [Google Scholar] [CrossRef]
- Wakatsuki, S.; Ohno, M.; Araki, T. Human Vault RNA1-1, but Not Vault RNA2-1, Modulates Synaptogenesis. Commun. Integr. Biol. 2021, 14, 61–65. [Google Scholar] [CrossRef]
- Geoffray, M.-M.; Falissard, B.; Green, J.; Kerr, B.; Evans, D.G.; Huson, S.; Burkitt-Wright, E.; Garg, S. Autism Spectrum Disorder Symptom Profile Across the RASopathies. Front. Psychiatry 2021, 11, 585700. [Google Scholar] [CrossRef]
- Borrie, S.C.; Plasschaert, E.; Callaerts-Vegh, Z.; Yoshimura, A.; D’Hooge, R.; Elgersma, Y.; Kushner, S.A.; Legius, E.; Brems, H. MEK Inhibition Ameliorates Social Behavior Phenotypes in a Spred1 Knockout Mouse Model for RASopathy Disorders. Mol. Autism 2021, 12, 53. [Google Scholar] [CrossRef]
- Miñones-Moyano, E.; Friedländer, M.R.; Pallares, J.; Kagerbauer, B.; Porta, S.; Escaramís, G.; Ferrer, I.; Estivill, X.; Martí, E. Upregulation of a Small Vault RNA (svtRNA2-1a) Is an Early Event in Parkinson Disease and Induces Neuronal Dysfunction. RNA Biol. 2013, 10, 1093–1106. [Google Scholar] [CrossRef]
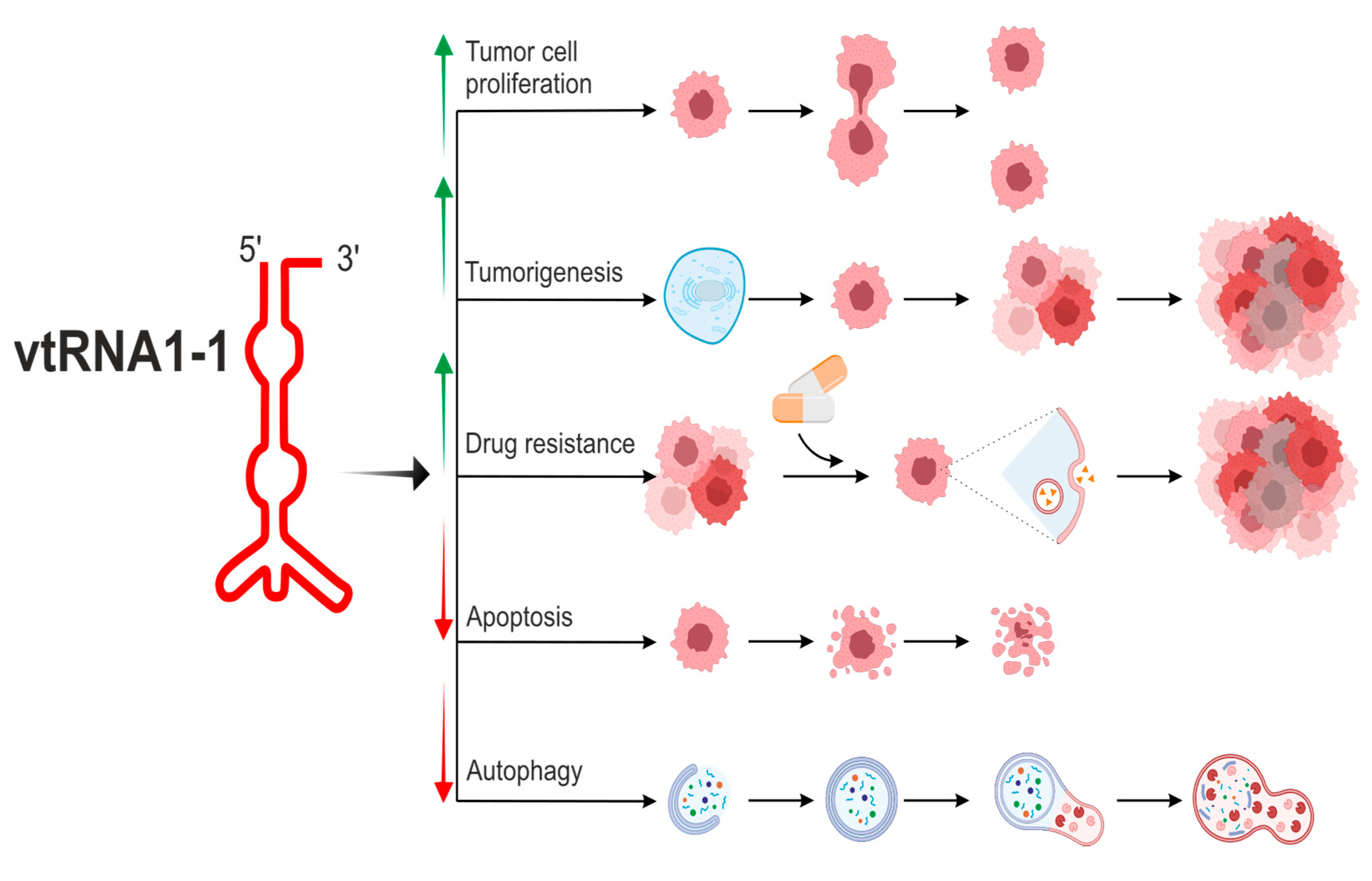

| vtRNA paralogue | structure | derivatives | role in cellular processes |
|---|---|---|---|
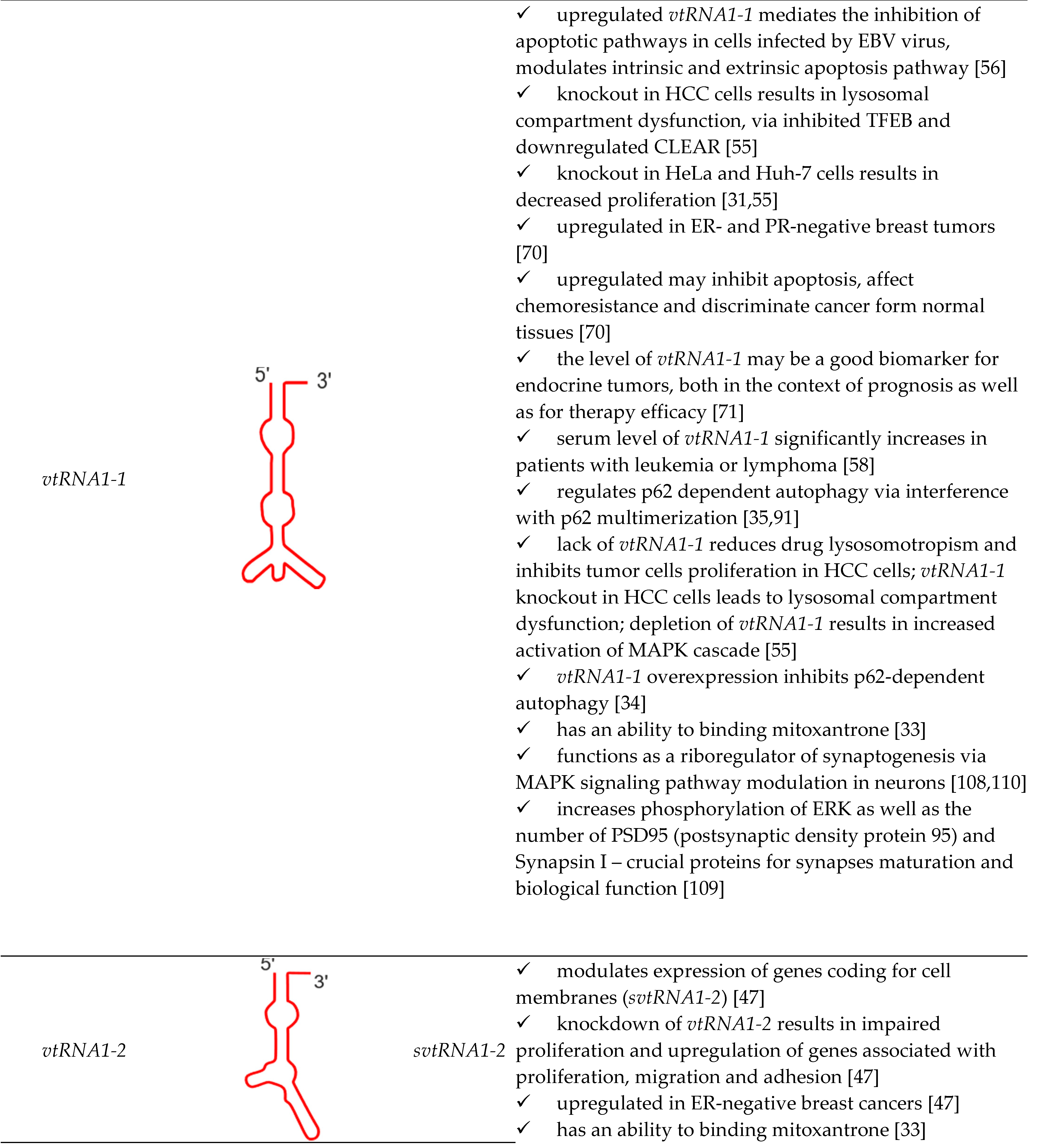 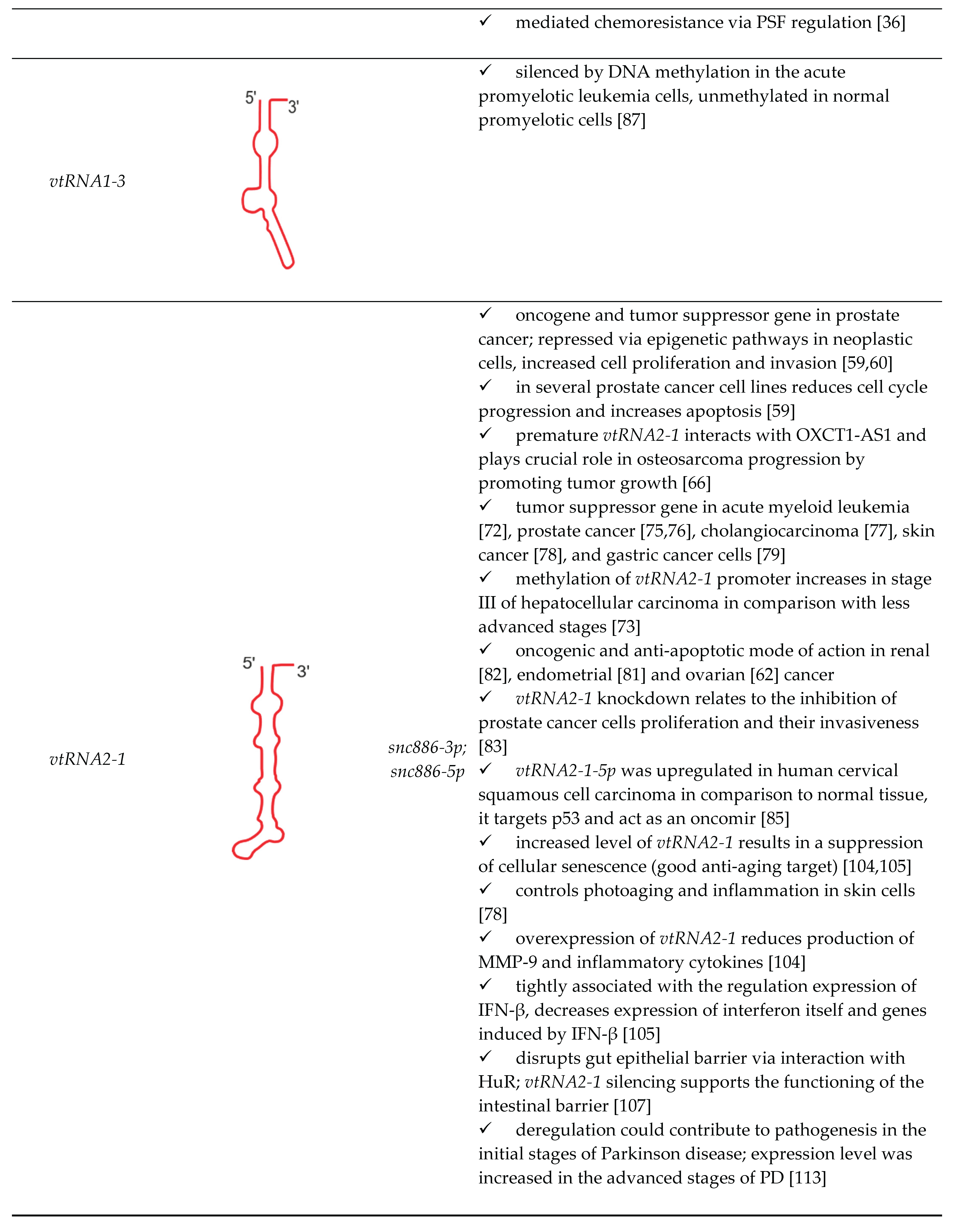
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





