Submitted:
01 March 2024
Posted:
13 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Target Selection and Validation
2.2. Ligand Selection
2.3. Ligand Preparation
2.4. Protein Preparation
2.5. Site map Analysis
2.6. Receptor Grid Generation
2.7. Molecular Docking
2.7.1. Virtual Screening and Ligand Docking
2.7.2. Free Energy Calculation by MM-GBSA
2.7.3. ADMET Profiling of Novel Antiviral Compounds against LSDV
2.7.4. Molecular Dynamics (MD) Simulation
3. Results
3.1. LSDV DNA Polymerase Protein Has a Groove-Like Active Site
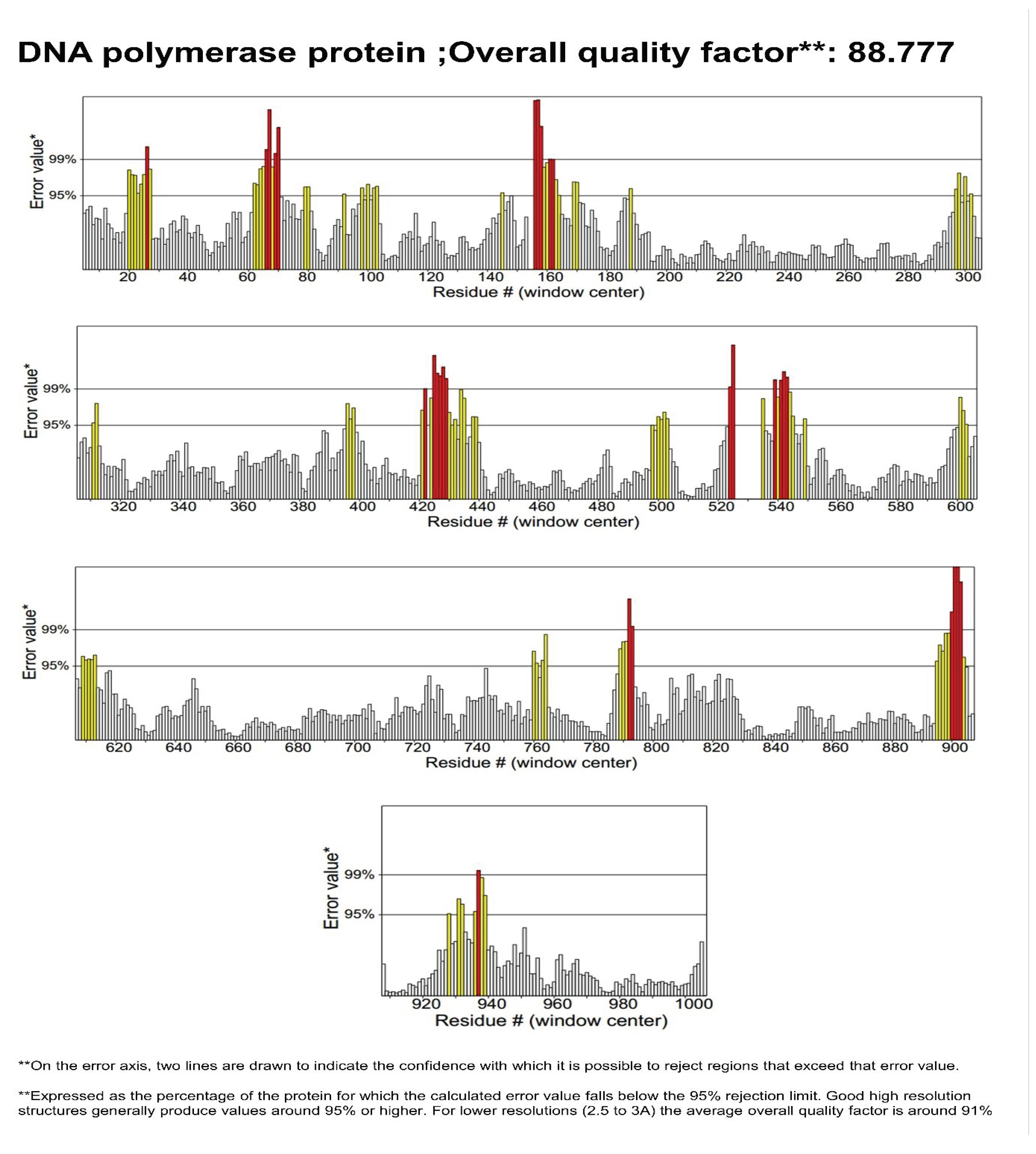
3.2. Structure Validation
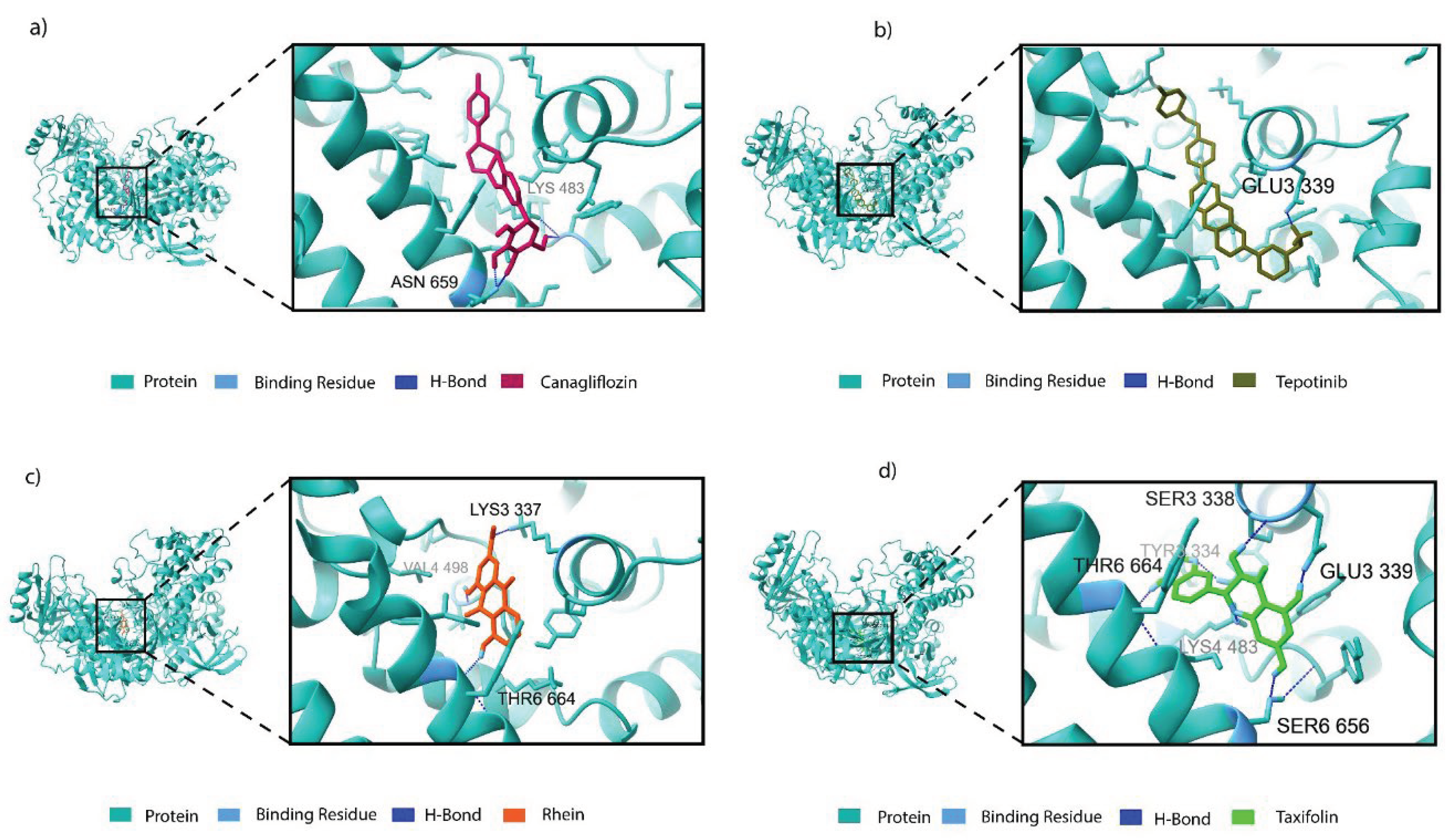
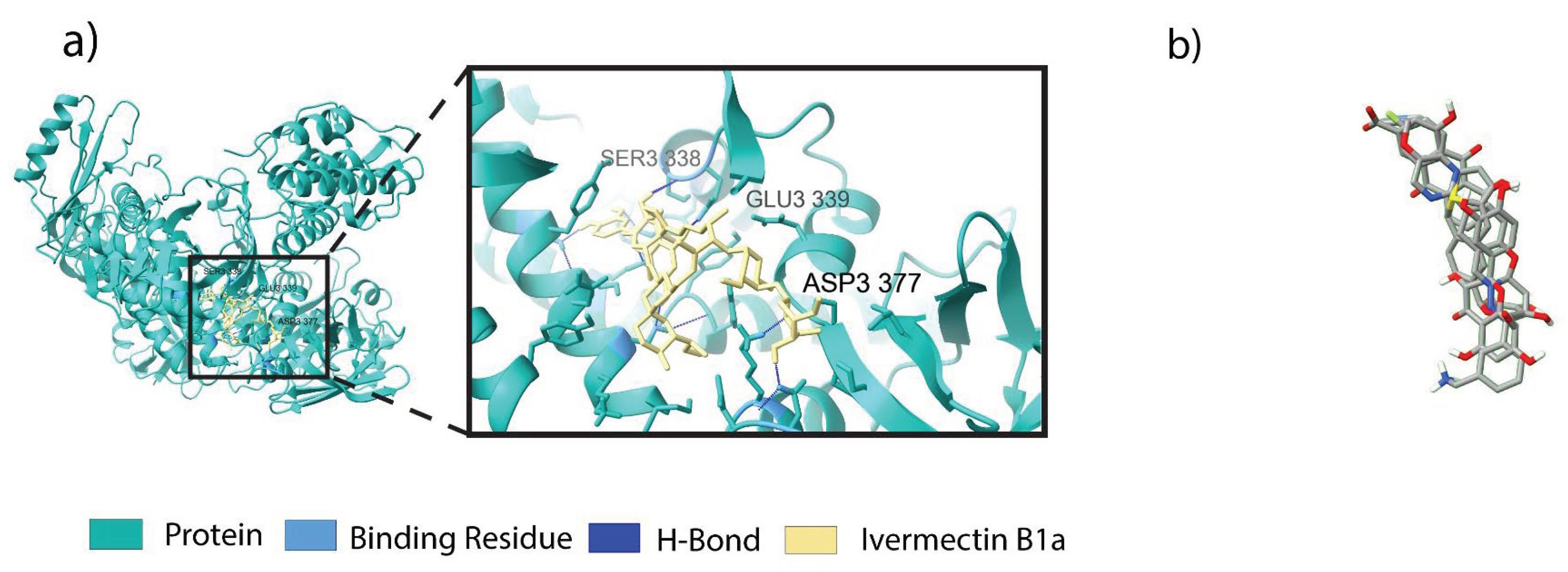
3.3. Binding Profile Analysis of Bonded Interactions
3.4. Binding Free Energy Calculations for Non-Bonded Interactions
3.5. ADMET Profiling of Novel Antiviral Compounds against LSDV
3.6. Molecular Dynamics (MD) Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding Statement
Data Availability
Acknowledgments
Conflicts of Interest
References
- Das, M.; Chowdhury, M.S.R.; Akter, S.; Mondal, A.K.; Uddin, M.J.; Rahman, M.M.; Rahman, M.M. An updated review on lumpy skin disease: perspective of Southeast Asian countries. J. adv. biotechnol. exp. ther 2021, 4(3), 322–333. [Google Scholar] [CrossRef]
- Molini, U.; Boshoff, E.; Niel, A.P.; Phillips, J.; Khaiseb, S.; Settypalli, T.B.; Dundon, W.G.; Cattoli, G.; Lamien, C.E. Detection of lumpy skin disease virus in an asymptomatic eland (Taurotragus oryx) in Namibia. The Journal of Wildlife Diseases 2021, 57(3), 708–711. [Google Scholar] [CrossRef]
- Ko, Y.S.; Oh, Y.; Lee, T.G.; Bae, D.Y.; Tark, D.; Cho, H.S. Serological and molecular prevalence of lumpy skin disease virus in Korean water deer, native and dairy cattle in Korea. Korean Journal of Veterinary Service 2022, 45(2), 133–137. [Google Scholar] [CrossRef]
- Dao, T.D.; Tran, L.H.; Nguyen, H.D.; Hoang, T.T.; Nguyen, G.H.; Tran KV, D.; Nguyen, H.X.; Van Dong, H.; Bui, A.N.; Bui, V.N. Characterization of Lumpy skin disease virus isolated from a giraffe in Vietnam. Transboundary and Emerging Diseases 2022, 69(5), e3268–e3272. [Google Scholar] [CrossRef] [PubMed]
- Buller, R.; Arif, B.; Black, D.; Dumbell, K.; Esposito, J.; Lefkowitz, E.; McFadden, G.; Moss, B.; Mercer, A.; Moyer, R. Family poxviridae. In Virus taxonomy: Classification and nomenclature of viruses. Eighth Report of the International Committee on Taxonomy of Viruses; 2005; pp. 117–133. [Google Scholar]
- Tulman, E.; Afonso, C.; Lu, Z.; Zsak, L.; Kutish, G.; Rock, D. Genome of lumpy skin disease virus. Journal of virology 2001, 75(15), 7122–7130. [Google Scholar] [CrossRef] [PubMed]
- Tuppurainen, E.S.; Venter, E.H.; Coetzer, J.A.; Bell-Sakyi, L. Lumpy skin disease: Attempted propagation in tick cell lines and presence of viral DNA in field ticks collected from naturally-infected cattle. Ticks and tick-borne diseases 2015, 6(2), 134–140. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Bernardo, B.; Haga, I.R.; Wijesiriwardana, N.; Basu, S.; Larner, W.; Diaz, A.V.; Langlands, Z.; Denison, E.; Stoner, J.; White, M. Quantifying and modeling the acquisition and retention of lumpy skin disease virus by hematophagus insects reveals clinically but not subclinically affected cattle are promoters of viral transmission and key targets for control of disease outbreaks. Journal of virology 2021, 95(9). [Google Scholar] [CrossRef] [PubMed]
- Sprygin, A.; Pestova, Y.; Wallace, D.; Tuppurainen, E.; Kononov, A. Transmission of lumpy skin disease virus: A short review. Virus research 2019, 269, 197637. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.; Patial, V.; Bali, D.; Angaria, S.; Sharma, M.; Chahota, R. A review: Lumpy skin disease and its emergence in India. Veterinary research communications 2020, 44, 111–118. [Google Scholar] [CrossRef]
- Liu, P.; Li, J.; Chen, R.; Cheng, Z.; Shi, Y. The first outbreak investigation of lumpy skin disease in China. China Animal Health Inspection 2020, 37(1), 1–5. [Google Scholar]
- Wang, Y.; Zhao, L.; Yang, J.; Shi, M.; Nie, F.; Liu, S.; Wang, Z.; Huang, D.; Wu, H.; Li, D. Analysis of vaccine-like lumpy skin disease virus from flies near the western border of China. Transboundary and Emerging Diseases 2022, 69(4), 1813–1823. [Google Scholar] [CrossRef]
- Farag, T.; El-Houssiny, A.; Abdel-Rahman, E.; Hegazi, A. A new approach to the treatment of lumpy skin disease infection in cattle by using propolis encapsulated within alg nps. Adv. Anim. Vet. Sci 2020, 8(12), 1346–1355. [Google Scholar] [CrossRef]
- Sprygin, A.; Artyuchova, E.; Babin, Y.; Prutnikov, P.; Kostrova, E.; Byadovskaya, O.; Kononov, A. Epidemiological characterization of lumpy skin disease outbreaks in Russia in 2016. Transboundary and Emerging Diseases 2018, 65(6), 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Ratyotha, K.; Prakobwong, S.; Piratae, S. Lumpy skin disease: A newly emerging disease in Southeast Asia. Veterinary World 2022, 15(12), 2764. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, L.; Ward, M.P. The Spread of Lumpy Skin Disease Virus across Southeast Asia: Insights from Surveillance. Transboundary and Emerging Diseases 2023. [Google Scholar] [CrossRef]
- Hasib FM, Y.; Islam, M.S.; Das, T.; Rana, E.A.; Uddin, M.H.; Bayzid, M.; Nath, C.; Hossain, M.A.; Masuduzzaman, M.; Das, S. Lumpy skin disease outbreak in cattle population of Chattogram, Bangladesh. Veterinary Medicine and Science 2021, 7(5), 1616–1624. [Google Scholar] [CrossRef] [PubMed]
- Gari, G.; Abie, G.; Gizaw, D.; Wubete, A.; Kidane, M.; Asgedom, H.; Bayissa, B.; Ayelet, G.; Oura, C.A.; Roger, F. Evaluation of the safety, immunogenicity and efficacy of three capripoxvirus vaccine strains against lumpy skin disease virus. Vaccine 2015, 33(28), 3256–3261. [Google Scholar] [CrossRef] [PubMed]
- Tuppurainen, E.S.; Pearson, C.R.; Bachanek-Bankowska, K.; Knowles, N.J.; Amareen, S.; Frost, L.; Henstock, M.R.; Lamien, C.E.; Diallo, A.; Mertens, P.P. Characterization of sheep pox virus vaccine for cattle against lumpy skin disease virus. Antiviral research 2014, 109, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Klement, E.; Broglia, A.; Antoniou, S.-E.; Tsiamadis, V.; Plevraki, E.; Petrović, T.; Polaček, V.; Debeljak, Z.; Miteva, A.; Alexandrov, T. Neethling vaccine proved highly effective in controlling lumpy skin disease epidemics in the Balkans. Preventive veterinary medicine 2020, 181, 104595. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, J.; Mohan, M.; Byrareddy, S.N. Drug repurposing approaches to combating viral infections. Journal of clinical medicine 2020, 9(11), 3777. [Google Scholar] [CrossRef]
- Mercorelli, B.; Palù, G.; Legian, A. Drug repurposing for viral infectious diseases: how far are we? Trends in microbiology 2018, 26(10), 865–876. [Google Scholar] [CrossRef]
- Punekar, M.; Kshirsagar, M.; Tellapragada, C.; Patil, K. Repurposing of antiviral drugs for COVID-19 and impact of repurposed drugs on the nervous system. Microbial Pathogenesis 2022, 168, 105608. [Google Scholar] [CrossRef]
- Toker, E.B.; Ates, O.; Yeşilbağ, K. Inhibition of bovine and ovine capripoxviruses (Lumpy skin disease virus and Sheeppox virus) by ivermectin occurs at different stages of propagation in vitro. Virus research 2022, 310, 198671. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Bhattu, M.; Tripathi, A.; Verma, M.; Acevedo, R.; Kumar, P.; …; Singh, J. Potential medicinal plants to combat viral infections: A way forward to environmental biotechnology. Environmental Research 2023, 115725. [Google Scholar] [CrossRef]
- Dastoli, S.; Nisticò, S.P.; Morrone, P.; Patruno, C.; Leo, A.; Citraro, R.; Gallelli, L.; Russo, E.; De Sarro, G.; Bennardo, L. Colchicine in managing skin conditions: A systematic review. Pharmaceutics 2022, 14(2), 294. [Google Scholar] [CrossRef]
- Malabadi, R.B.; Kolkar, P.; Chalannavar, K. Outbreak of lumpy skin viral disease of cattle and buffalo in india in 2022: Ethnoveterinary medicine approach. International Journal of Innovation Scientific Research and Review 2022, 4(11), 3562–3574. [Google Scholar]
- Yadav, J.V.; Lakshman, M.; Madhuri, D. A combination of conventional and alternative ethnoveterinary medicine for the treatment of lumpy skin disease in a she-buffalo: A case report. The Pharma Innovation 2021, SP-10(1), 83–84. [Google Scholar]
- Vijayasri, S.; Hopper, W. Towards the identification of novel phytochemical leads as macrodomain inhibitors of chikungunya virus using molecular docking approach. Journal of Applied Pharmaceutical Science 2017, 2017 7, 074–082. [Google Scholar]
- Jayant, V.; Ali, R. Potential Applications of Ivermectin (IVM) in Dermatology. In Chemistry and Biological Activities of Ivermectin; 2023; pp. 199–229. [Google Scholar]
- Badhy, S.C.; Chowdhury, M.G.A.; Settypalli TB, K.; Cattoli, G.; Lamien, C.E.; Fakir MA, U.; Akter, S.; Osmani, M.G.; Talukdar, F.; Begum, N. Molecular characterization of lumpy skin disease virus (LSDV) emerged in Bangladesh reveals unique genetic features compared to contemporary field strains. BMC veterinary research 2021, 17(1), 1–11. [Google Scholar] [CrossRef]
- Cramer, P. AlphaFold2 and the future of structural biology. Nature structural & molecular biology 2021, 28(9), 704–705. [Google Scholar]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of applied crystallography 1993, 26(2), 283–291. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein science 1993, 2(9), 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Banerjee, A.; Chakraborty, N.; Soren, K.; Chakraborty, P.; Bandopadhyay, R. Structural-functional analyses of textile dye degrading azoreductase, laccase and peroxidase: A comparative in silico study. Electronic Journal of Biotechnology 2020, 43, 48–54. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein science 2021, 30(1), 70–82. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L. InterPro: the integrative protein signature database. Nucleic acids research 2009, 37 (suppl_1), D211–D215. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: the protein families database. Nucleic acids research 2014, 42(D1), D222–D230. [Google Scholar] [CrossRef]
- Schrödinger. 2021. Schrödinger Release 2021-2: SiteMap. Schrödinger, LLC. New York, NY.
- Ali, M.C.; Munni, Y.A.; Das, R.; Akter, N.; Das, K.; Mitra, S.; Hannan, M.A.; Dash, R. In silico chemical profiling and identification of neuromodulators from Curcuma amada targeting Acetylcholinesterase. Network Modeling Analysis in Health Informatics and Bioinformatics 2021, 10, 1–16. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem 2023 update. Nucleic acids research 2023, 51(D1), D1373–D1380. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic acids research 2006, 34 (suppl_1), D668–D672. [Google Scholar] [CrossRef]
- O'Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. Journal of cheminformatics 2011, 3(1), 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger 2023 Schrödinger Release 2023-2: Maestro Schrödinger, L.L.C. New York, NY.
- Schrödinger. 2023. Schrödinger Release 2023-2: Protein Preparation Wizard. Epik, Schrödinger, LLC. New York, NY.
- Schrödinger 2023 Schrödinger Release 2023-3: SiteMap Schrödinger, L.L.C. New York, NY.
- Tahlan, S.; Kumar, S.; Ramasamy, K.; Lim, S.M.; Shah SA, A.; Mani, V.; Narasimhan, B. In-silico molecular design of heterocyclic benzimidazole scaffolds as prospective anticancer agents. BMC chemistry 2019, 13(1), 1–22. [Google Scholar] [CrossRef]
- David, T.I.; Adelakun, N.S.; Omotuyi, O.I.; Metibemu, D.S.; Ekun, O.E.; Inyang, O.K.; Adewumi, B.; Enejoh, O.A.; Owolabi, R.T.; Oribamise, E.I. Molecular docking analysis of phyto-constituents from Cannabis sativa with pfDHFR. Bioinformation 2018, 14(9), 574. [Google Scholar] [CrossRef]
- Singh, N.; Chaput, L.; Villoutreix, B.O. Virtual screening web servers: designing chemical probes and drug candidates in the cyberspace. Briefings in bioinformatics 2021, 22(2), 1790–1818. [Google Scholar] [CrossRef]
- Schrödinger 2019 Schrödinger Release 2019-3: Glide Schrödinger, L.L.C. New York, NY, USA.
- Li, J.; Abel, R.; Zhu, K.; Cao, Y.; Zhao, S.; Friesner, R.A. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins: Structure, Function, and Bioinformatics 2011, 79(10), 2794–2812. [Google Scholar] [CrossRef]
- Kadioglu, O.; Saeed, M.; Greten, H.J.; Efferth, T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Computers in biology and medicine 2021, 133, 104359. [Google Scholar] [CrossRef]
- Ghosh, S.; Nie, A.; An, J.; Huang, Z. Structure-based virtual screening of chemical libraries for drug discovery. Current opinion in chemical biology 2006, 10(3), 194–202. [Google Scholar] [CrossRef]
- Pattar, S.V.; Adhoni, S.A.; Kamanavalli, C.M.; Kumbar, S.S. In silico molecular docking studies and MM/GBSA analysis of coumarin-carbonodithioate hybrid derivatives divulge the anticancer potential against breast cancer. Beni-Suef University journal of basic and applied sciences 2020, 9(1), 1–10. [Google Scholar] [CrossRef]
- Sankar, M.; Ramachandran, B.; Pandi, B.; Mutharasappan, N.; Ramasamy, V.; Prabu, P.G.; Shanmugaraj, G.; Wang, Y.; Muniyandai, B.; Rathinasamy, S. In silico screening of natural phytocompounds towards identification of potential lead compounds to treat COVID-19. Frontiers in molecular biosciences 2021, 8, 637122. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-Y.; Wang, Q.; Zhang, J.; Wu, T.; Zhang, F. Trastuzumab-Peptide interactions: mechanism and application in structure-based ligand design. International Journal of Molecular Sciences 2013, 14(8), 16836–16850. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger 2021 QikProp In Schrödinger Release 2021-2 Schrödinger, L.L.C. New York, NY.
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular docking and structure-based drug design strategies. Molecules 2015, 20(7), 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, P.W.; Rose, A.S.; Tiemann, J.K. Bringing molecular dynamics simulation data into view. Trends in Biochemical Sciences 2019, 44(11), 902–913. [Google Scholar] [CrossRef]
- Rasheed, M.A.; Iqbal, M.N.; Saddick, S.; Ali, I.; Khan, F.S.; Kanwal, S.; Ahmed, D.; Ibrahim, M.; Afzal, U.; Awais, M. Identification of lead compounds against Scm (fms10) in Enterococcus faecium using computer aided drug designing. Life 2021, 11(2), 77. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. Journal of chemical theory and computation 2010, 6(5), 1509–1519. [Google Scholar] [CrossRef]
- Espinoza-Chávez, R.M.; Salerno, A.; Liuzzi, A.; Ilari, A.; Milelli, A.; Uliassi, E.; Bolognesi, M.L. Targeted Protein Degradation for Infectious Diseases: from Basic Biology to Drug Discovery. ACS bio & med Chem Au 2022, 3(1), 32–45. [Google Scholar]
- Kara, P.; Afonso, C.; Wallace, D.; Kutish, G.; Abolnik, C.; Lu, Z.; Vreede, F.; Taljaard, L.; Zsak, A.; Viljoen, G.J. Comparative sequence analysis of the South African vaccine strain and two virulent field isolates of lumpy skin disease virus. Archives of virology 2003, 148, 1335–1356. [Google Scholar] [CrossRef]
- Garcia-Diaz, M.; Bebenek, K. Multiple functions of DNA polymerases. Critical reviews in plant sciences 2007, 26(2), 105–122. [Google Scholar] [CrossRef]
- Berdis, A.J. DNA polymerases as therapeutic targets. Biochemistry 2008, 47(32), 8253–8260. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biology direct 2006, 1(1), 1–18. [Google Scholar] [CrossRef]
- Michel, M.; Visnes, T.; Homan, E.J.; Seashore-Ludlow, B.; Hedenström, M.; Wiita E e Vallin, K.; Paulin, C.B.; Zhang, J.; Wallner, O. Computational and experimental druggability assessment of human DNA glycosylases. ACS omega 2019, 4(7), 11642–11656. [Google Scholar] [CrossRef]
- Alzyoud, L.; Bryce, R.A.; Al Sorkhy, M.; Atatreh, N.; Ghattas, M.A. Structure-based assessment and druggability classification of protein–protein interaction sites. Scientific Reports 2022, 12(1), 7975. [Google Scholar] [CrossRef]
- Ali, M.C.; Nur, A.J.; Khatun, M.S.; Dash, R.; Rahman, M.M.; Karim, M.M. Identification of potential SARS-CoV-2 main protease inhibitors from Ficus Carica Latex: An in-silico approach. Journal of Advanced Biotechnology and Experimental Therapeutics 2020, 3(4), 57–67. [Google Scholar] [CrossRef]
- Whittle, L.; Chapman, R.; Williamson, A.-L. Lumpy Skin Disease—An Emerging Cattle Disease in Europe and Asia. Vaccines 2023, 11(3), 578. [Google Scholar] [CrossRef]
- Chouhan, C.S.; Parvin, M.S.; Ali, M.Y.; Sadekuzzaman, M.; Chowdhury MG, A.; Ehsan, M.A.; Islam, M.T. Epidemiology and economic impact of lumpy skin disease of cattle in Mymensingh and Gaibandha districts of Bangladesh. Transboundary and Emerging Diseases 2022, 69(6), 3405–3418. [Google Scholar] [CrossRef]
- Lu, G.; Xie, J.; Luo, J.; Shao, R.; Jia, K.; Li, S. Lumpy skin disease outbreaks in China, since 3 August 2019. Transboundary and Emerging Diseases 2021, 68(2), 216–219. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, S.B.; Mishra, N.; Kalaiyarasu, S.; Jhade, S.K.; Hemadri, D.; Sood, R.; Bal, G.C.; Nayak, M.K.; Pradhan, S.K.; Singh, V.P. Lumpy skin disease (LSD) outbreaks in cattle in Odisha state, India in August 2019: Epidemiological features and molecular studies. Transboundary and Emerging Diseases 2020, 67(6), 2408–2422. [Google Scholar] [CrossRef]
- Acharya, K.P.; Subedi, D. First outbreak of lumpy skin disease in Nepal. Preventive veterinary medicine 2020, 102(4), 274–283. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Chowdhury MS, R.; Akter, S.; Mondal, A.K.; Uddin, M.J.; Rahman, M.M.; Rahman, M.M. An updated review on lumpy skin disease: perspective of Southeast Asian countries. J. adv. biotechnol. exp. Ther 2021, 4(3), 322–333. [Google Scholar] [CrossRef]
- Wilhelm, L.; Ward, M.P. The Spread of Lumpy Skin Disease Virus across Southeast Asia: Insights from Surveillance. Transboundary and Emerging Diseases 2023. [CrossRef]
- Moudgil, G.; Chadha, J.; Khullar, L.; Chhibber, S.; Harjai, K. Lumpy skin disease: A comprehensive review on virus biology, pathogenesis, and sudden global emergence. 2023. [Google Scholar]
- Kayesh ME, H.; Hussan, M.T.; Hashem, M.A.; Eliyas, M.; Anower, A.M. Lumpy skin disease virus infection: An emerging threat to cattle health in Bangladesh. Hosts and Viruses 2020, 7(4), 97. [Google Scholar] [CrossRef]
- Parvin, R.; Chowdhury, E.H.; Islam, M.T.; Begum, J.A.; Nooruzzaman, M.; Globig, A.; Dietze, K.; Hoffmann, B.; Tuppurainen, E. Clinical Epidemiology, Pathology, and Molecular Investigation of Lumpy Skin Disease Outbreaks in Bangladesh during 2020–2021 Indicate the Re-Emergence of an Old African Strain. Viruses 2022, 14(11), 2529. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, R.E.; Haider, M.K. Hydrogen Bonds in Proteins: Role and Strength. In Encyclopedia of Life Science 2010; John Wiley & Sons, Ltd.
- Wade, R.C.; Goodford, P.J. The role of hydrogen-bonds in drug binding. Progress in clinical and biological research 1989, 289, 433–444. [Google Scholar] [PubMed]
- Bissantz, C.; Kuhn, B.; Stahl, M. A medicinal chemist’s guide to molecular interactions. Journal of medicinal chemistry 2010, 53(14), 5061–5084. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; Farley, A.R.; Wright, G.D.; Cox, J.R. Analysis of the π-π stacking interactions between the aminoglycoside antibiotic kinase APH (3′)-IIIa and its nucleotide ligands. Chemistry & biology 2002, 9(11), 1209–1217. [Google Scholar]
- Spassov, D.S.; Atanasova, M.; Doytchinova, I. A role of salt bridges in mediating drug potency: A lesson from the N-myristoyltransferase inhibitors. Frontiers in molecular biosciences 2023, 9, 1066029. [Google Scholar] [CrossRef]
- Dougherty, D.A. Cation-π interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 1996, 271(5246), 163–168. [Google Scholar] [CrossRef]
- Ma, J.C.; Dougherty, D.A. The cation− π interaction. Chemical reviews 1997, 97(5), 1303–1324. [Google Scholar] [CrossRef]
- Scrutton, N.S.; Raine, A.R. Cation-π bonding and amino-aromatic interactions in the biomolecular recognition of substituted ammonium ligands. Biochemical Journal 1996, 319(1), 1–8. [Google Scholar] [CrossRef]
- Wouters, J. Cation-π (Na+-Trp) interactions in the crystal structure of tetragonal lysozyme. Protein science 1998, 7(11), 2472–2475. [Google Scholar] [CrossRef]
- Gallivan, J.P.; Dougherty, D.A. Cation-π interactions in structural biology. Proceedings of the National Academy of Sciences 1999, 96(17), 9459–9464. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, N.; Dougherty, D.A. Cation–π interactions in ligand recognition and catalysis. Trends in pharmacological sciences 2002, 23(6), 281–287. [Google Scholar] [CrossRef]
- Wang, C.; Dai, S.; Gong, L.; Fu, K.; Ma, C.; Liu, Y.; Zhou, H.; Li, Y. A review of pharmacology, toxicity and pharmacokinetics of 2, 3, 5, 4′-tetrahydroxystilbene-2-O-β-D-glucoside. Frontiers in Pharmacology 2022, 12, 791214. [Google Scholar] [CrossRef]
- Zhang, S.-Q.; Chen, F. Factors Influencing ADME Properties of Drugs: Advances and Applications (Part I). Current Drug Metabolism 2023, 24(1), 3–4. [Google Scholar] [CrossRef]
- Glassman, P.M.; Muzykantov, V.R. Pharmacokinetic and pharmacodynamic properties of drug delivery systems. Journal of Pharmacology and Experimental Therapeutics 2019, 370(3), 570–580. [Google Scholar] [CrossRef]
- Ahmed, S.; Ali, M.C.; Ruma, R.A.; Mahmud, S.; Paul, G.K.; Saleh, M.A.; Alshahrani, M.M.; Obaidullah, A.J.; Biswas, S.K.; Rahman, M.M. Molecular docking and dynamics simulation of natural compounds from betel leaves (Piper betle L.) for investigating the potential inhibition of alpha-amylase and alpha-glucosidase of type 2 diabetes. Molecules 2022, 27(14), 4526. [Google Scholar] [CrossRef]
- Peele, K.A.; Durthi, C.P.; Srihansa, T.; Krupanidhi, S.; Ayyagari, V.S.; Babu, D.J.; Indira, M.; Reddy, A.R.; Venkateswarulu, T. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: A computational study. Informatics in Medicine Unlocked 2020, 19, 100345. [Google Scholar] [CrossRef]
- Munni, Y.A.; Ali, M.C.; Selsi, N.J.; Sultana, M.; Hossen, M.; Bipasha, T.H.; Rahman, M.; Uddin, M.N.; Hosen, S.Z.; Dash, R. Molecular simulation studies to reveal the binding mechanisms of shikonin derivatives inhibiting VEGFR-2 kinase. Computational Biology and Chemistry 2021, 90, 107414. [Google Scholar] [CrossRef]
- Dash, R.; Choi, H.J.; Moon, I.S. Mechanistic insights into the deleterious roles of Nasu-Hakola disease associated TREM2 variants. Scientific Reports 2020, 10(1), 3663. [Google Scholar] [CrossRef] [PubMed]
- Dash, R.; Ali, M.C.; Dash, N.; Azad MA, K.; Hosen, S.Z.; Hannan, M.A.; Moon, I.S. Structural and dynamic characterizations highlight the deleterious role of SULT1A1 R213H polymorphism in substrate binding. International Journal of Molecular Sciences 2019, 20(24), 6256. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk Gabr, N.; Hassan Mohammed, I. A comparative study of canagliflozin (INVOKANA) on type-I and type-II diabetes mellitus on adult male albino rat. Al-Azhar Medical Journal 2020, 49(1), 15–32. [Google Scholar] [CrossRef]
- Deeks, E.D.; Scheen, A.J. Canagliflozin: a review in type 2 diabetes. Drugs 2017, 77, 1577–1592. [Google Scholar] [CrossRef] [PubMed]
- Pratley, R.E.; Cersosimo, E. Use of canagliflozin in combination with and compared to incretin-based therapies in type 2 diabetes. Clinical Diabetes 2017, 35(3), 141–153. [Google Scholar] [CrossRef]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; …; Le, X. Tepotinib in non–small-cell lung cancer with MET exon 14 skipping mutations. New England Journal of Medicine 2020, 383(10), 931–943. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacological research 2022, 175, 106037. [Google Scholar] [CrossRef]
- Amir, M.; Javed, S. Elucidation of binding dynamics of tyrosine kinase inhibitor tepotinib, to human serum albumin, using spectroscopic and computational approach. International Journal of Biological Macromolecules 2023, 241, 124656. [Google Scholar] [CrossRef]
- Vijayasri, S.; Hopper, W. Towards the identification of novel phytochemical leads as macrodomain inhibitors of chikungunya virus using molecular docking approach. Journal of Applied Pharmaceutical Science 2017, 7, 074–082. [Google Scholar]
| Protein name | Binding site residues |
|---|---|
| DNA Polymerase (LSDV 039) | Lys 4, Glu 122, Gly 123, Cys 124, Arg 155, Phe156, Asn 157, Ile 158, Asn 159, Arg 160, Tyr 162, Phe 164, Ile 191, Asn 195, Leu 304, Phe 329, Thr 333, Tyr 334, Lys 337, Ser 338, Glu 339, Lys 340, Asn 352, Ala 353, Phe 354, Ser 355, Cys 356, Asn 374, Ile 379, Gly 380, Lys 381, Ile 382, Ser 383, Ser 384, Phe 385, Glu 387, Val 388, Asp 412, Tyr 473, Trp 475, Asn 476, Tyr 477, Tyr 478, Gly 479, Ile 480, Glu 481, Thr 482, Lys 483, Asp 485, Ala 486, Gly 487, Phe 489, Tyr 491, Val 498, Phe 499, Glu 500, Tyr 501, Arg 502, Ala 503, Leu 506, Tyr642, Tyr646, Leu 651, Ser 652, Thr 653, Lys 655, Ser 656, Ile 657, Tyr 658, Asn 659, Ser 660, Met 661, Glu 662, Tyr 663, Thr 664, Tyr 665, Ile 667, Ile 668, Ser 671 |
| Gene | Uniprot ID | Site Map Analysis | Errat value | Ramachandran plot | |||
|---|---|---|---|---|---|---|---|
| Most favored regions | Additional allowed regions | Generously allowed regions | Disallowed region | ||||
| LSDV039 | Q91MW8 | 1.048 | 88.777 | 91% | 8.0% | 0.6% | 0.3% |
| Compound type | Compound ID | Name | Glide Gscore (kcal/mol) | Glide emodel (kcal/mol) | MMGBSA dGbind (kcal/mol) |
|---|---|---|---|---|---|
| Positive control Antiviral drugs |
CID6321424 | Ivermectin B1a | −5.630 | −67.888 | −26.28 |
| DB08907 | Canagliflozin | −9.858 | −69.368 | −45.68 | |
| DB15133 | Tepotinib | −8.856 | −88.691 | −47.99 | |
| Phytocompounds | CID 10168 | Rhein | −8.965 | −43.078 | −44.72 |
| CID 439533 | Taxifolin | −7.195 | −51.868 | −44.48 |
| Compound type | Compound ID | Name | Residues in interaction | Bond distance (Å) | Bond type |
|---|---|---|---|---|---|
| Positive control Antiviral drugs |
CID6321424 (positive control) | Ivermectin B1a | Asp 337 Ser 338 Glu 339 |
1.83 2.16 1.55 |
H-bond H-bond H-bond |
| DB08907 | Canagliflozin | Lys 483 Asn 659 Asn 659 |
2.61 1.80 1.73 |
Salt bridge H-bond H-bond |
|
| DB15133 | Tepotinib | Glu 339 Glu 339 Tyr 477 Lys 483 Phe 499 Tyr 663 |
1.93 2.94 5.49 4.01 4.89 5.38 |
H-bond Salt bridge Pi-Pi Stacking Pi-cation Pi-Pi Stacking Pi-Pi Stacking |
|
| Phytocompounds | CID 10168 | Rhein | Lys 337 Val 498 Thr 664 |
2.71 2.12 2.09 |
Salt bridge H-bond H-bond |
| CID 439533 | Taxifolin | Ser 338 Glu 339 Lys 483 Lys 483 Ser 656 Thr 664 |
2.10 1.78 2.00 3.90 2.02 2.05 |
H-bond H-bond H-bond Pi-cation H-bond H-bond |
| compound type | Compound ID | Name | ∆Gbind(NS) | ∆Gcoulomb (NS) | ∆GLipo (NS) | ∆GvdW (NS) |
|---|---|---|---|---|---|---|
| Positive control Antiviral drugs |
CID6321424 | Ivermectin B1a | −39.70 | −15.52 | −17.01 | −63.90 |
| DB08907 | Canagliflozin | −51.44 | −46.81 | −25.21 | −42.95 | |
| DB15133 | Tepotinib | −54.06 | 50.09 | −23.28 | −58.20 | |
| Phytocompounds | CID 10168 | Rhein | −38.20 | −77.35 | −13.22 | −34.55 |
| CID 439533 | Taxifolin | −47.80 | −29.02 | −13.33 | −24.43 |
| Compound type | Name | #Star1 | Molecular Weight2 | SASA3 | FISA4 | QPlogPo/w5 | QPlogS6 | QPlogBB7 | QPlogHERG8 | QPlogKp9 | Percent Human OralAbsorption10 | Rule Of Five11 | Rule Of Tree12 |
| (Positive control) | Ivermectin B1a | 11 | 875.104 | 1232.663 | 137.092 | 6.398 | −8.094 | −2.277 | −5.978 | −2.113 | 73.189 | 3 | 2 |
| Antiviral drugs | Canagliflozin | 4 | 438.469 | 693.643 | 174.33 | 4.006 | −6.142 | −1.354 | −6.407 | −2.970 | 92.331 | 0 | 2 |
| Tepotinib | 1 | 494.595 | 881.973 | 94.452 | 4.588 | −6.747 | −0.547 | −8.583 | −3.171 | 100.00 | 0 | 2 | |
| Phytocompounds | Rhein | 0 | 284.225 | 478.121 | 271.668 | 0.979 | −2.658 | −1.968 | −2.686 | −5.511 | 47.411 | 0 | 1 |
| Taxifolin | 0 | 304.256 | 518.521 | 276.846 | 0.107 | −2.732 | −2.271 | −4.928 | −5.382 | 52.104 | 0 | 0 |
| System | RMSD (Å) | RMSF (Å) | Rg (Å) | SASA (Å) |
|---|---|---|---|---|
| Ivermectin B1a | 0.88 | 1.58 | 32.72 | 47186.72 |
| Canagliflozin | 0.88 | 1.69 | 32.97 | 47677.70 |
| Tepotinib | 1.44 | 1.26 | 32.99 | 47724.08 |
| Rhein | 0.33 | 1.62 | 33.18 | 48127.33 |
| Taxifolin | 0.51 | 1.63 | 32.89 | 47439.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





