1. Introduction
Hepatitis B virus (HBV) remains a global health concern even in the era of potent vaccines and antiretroviral therapy (ART) that can greatly reduce morbidity and mortality. HBV global prevalence is 3.8% and HBV infection causes 820,000 deaths annually [
1]. Of 296 million people living with chronic hepatitis B (CHB) globally, 82 million are in Africa [
1]. There is a call to eliminate HBV by the year 2030 with specific targets to reduce HBV incidence by 95% and mortality by 65% [
1]. The use of nucleos(t)ide analogues (NA) can greatly contribute to these targets, but the development of resistance-associated mutations (RAMs) remains one of the challenges that hinder HBV elimination. Prolonged antiviral use without adequate monitoring may lead to the selection of variants with RAMs that reduce ART susceptibility [
2].
The World Health Organization recommends treatment of CHB patients who have a viral load of greater or equal to 20,000IU/mL[
3]. NAs with a high barrier to drug resistance are highly recommended for treatment. These include entecavir for children aged 2-11 years and tenofovir (TFV) for individuals aged 12 years or older [
3]. In individuals with HBV/HIV coinfection, TFV + lamivudine (3TC) or emtricitabine (FTC) is recommended. Although other drugs such as 3TC and telbivudine are active against HBV, they have a low barrier to resistance and therefore are not recommended for HBV treatment [
4,
5]. TFV is considered the most effective drug against HBV, however, there is emerging evidence of possible amino acid substitutions that may reduce susceptibility to TFV, such as rtS78T, rtA194T and rtN236T [
6,
7,
8,
9].
Botswana has a HIV prevalence of 20.8% in the general population [
10]. Hepatitis B surface (HBsAg) prevalence ranges from 1.1% to 10.6% while occult hepatitis B infection (OBI) prevalence ranges from 6.6% to 33% with predominant subgenotypes being A1, D3 and E [
11,
12,
13,
14,
15]. Occult hepatitis B is defined as the presence of replication-competent HBV DNA in the blood and/or liver of individuals who test negative for HBsAg [
16]. The country has a robust HIV treatment program that has seen the country surpass the United Nations Programme on HIV/AIDS (UNAIDS) 95-95-95 targets at 95-98-98 [
10]. In 2016, Botswana adopted the HIV Treat-all program with the first line regimen being Truvada and dolutegravir [
17]. The prior regimen was TFV/FTC/EFV as the first line, while the earliest first-line regimen had 3TC as the only HBV active drug. The widespread use of ART in the country has improved patient health outcomes among people living with HIV (PLWH), however, HBV viral load and RAMs are not monitored in this setting. We therefore aimed to determine the prevalence of HBV RAMs in PLWH coinfected with HBV in Botswana.
2. Materials and Methods
2.1. Study Population
Plasma samples from PLWH recruited in the Botswana Combination Prevention Project (BCPP) (2013 – 2018) were used in this study. Details of BCPP are described elsewhere [
18]. In brief, the BCPP study was a cluster-randomized trial conducted in 15 paired communities matched by size, pre-existing health services, population, age structure, and geographic location. The study enrolled 12610 participants, 3596 of whom were PLWH [
18]. Participants signed written informed consent. Our study was approved by the Human Research Development Committee at the Botswana Ministry of Health (HPDME 13/18/1) with a waiver of consent.
2.2. Nanopore Sequencing
Participant plasma samples previously screened for HBsAg and OBI [
13] were used. Briefly, HBsAg screening was performed using
(Murex Version 2, Diasorin, Dartford, United Kingdom) while OBI screening was conducted using the
COBAS AmpliPrep/COBAS TaqMan HBV Test version 2.0 (Roche Diagnostics, Mannheim, Germany) following the manufacturer’s instructions [
13]. The QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) was used to extract DNA from 200μL of plasma with a final elution volume of 30μL. DNA concentration and quality were determined using the Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
Library preparation was adopted from an already established protocol [
19] and modified as previously described [
20]. In brief, a two-step polymerase chain reaction (PCR) was performed to amplify the whole HBV genome. Master mixes were prepared for two HBV primer pools which covered the whole HBV genome. We used Q5
® Hot Start High Fidelity 2× master mix (New England Biolabs, Ipswich, MA, USA) for PCR. Library preparation was carried out as per the Oxford Nanopore PCR tiling with rapid barcoding and midnight expansion protocol (version MRT_9127_v110_revH_14Jul2021) [
21], replacing SARS-CoV-2 primers with HBV primers. The library was quantified using the Qubit fluorometer and loaded into version R9.4.1 flow cells (Oxford Nanopore Technologies, Oxford, UK). HBV sequences were generated using GridION (Oxford Nanopore Technologies, Oxford, UK).
2.3. Sequencing Analysis
Raw FASTQ files were exported and subsequently processed using Guppy, employing dual-indexed reads for base calling and demultiplexing. FASTQ were uploaded into Genome Detective (version 2.64, last accessed 19 August 2023) for reference assembly. Generated consensus HBV sequences were viewed, aligned and trimmed using AliView alignment viewer [
22]. Geno2pheno (
https://hbv.geno2pheno.org) (last accessed 11 December 2023) and the Stanford HBVseq (
https://hivdb.stanford.edu/HBV/HBVseq/development/HBVseq.html) (last accessed 11 December 2023) online databases were used to determine HBV genotypes and RAMs. Furthermore, genotypes were confirmed using phylogenetic analysis. We constructed a maximum-likelihood tree (ML-tree) using the best-fitting model of nucleotide substitution [TVM+F+I+G4] using IQ-TREE with 1000 bootstrap replicates [
23,
24]. For mutational analysis, 98 HBV sequences had evaluable reverse transcriptase (RT) region coverage. We trimmed the RT position to only have amino acid positions rt1-250 and these amino acid RT sequences were analyzed for HBV RAMs using the Sandford HBVseq tool (
https://hivdb.stanford.edu/HBV/HBVseq/development/HBVseq.html).
2.4. Statistical Analysis
Participants’ sociodemographic and clinical characteristics were summarized in proportions and medians with interquartile ranges (IQR). Categorical data was analyzed using Fishers’ exact test or Chi-squared test where appropriate while continuous variables were compared using the Wilcoxon rank-sum test. All statistical analyses were conducted using Stata version 18.0 (StataCorp LLC, College Station, Texas, USA) and p-values less than 0.05 were deemed statistically significant.
3. Results
3.1. Participant Description
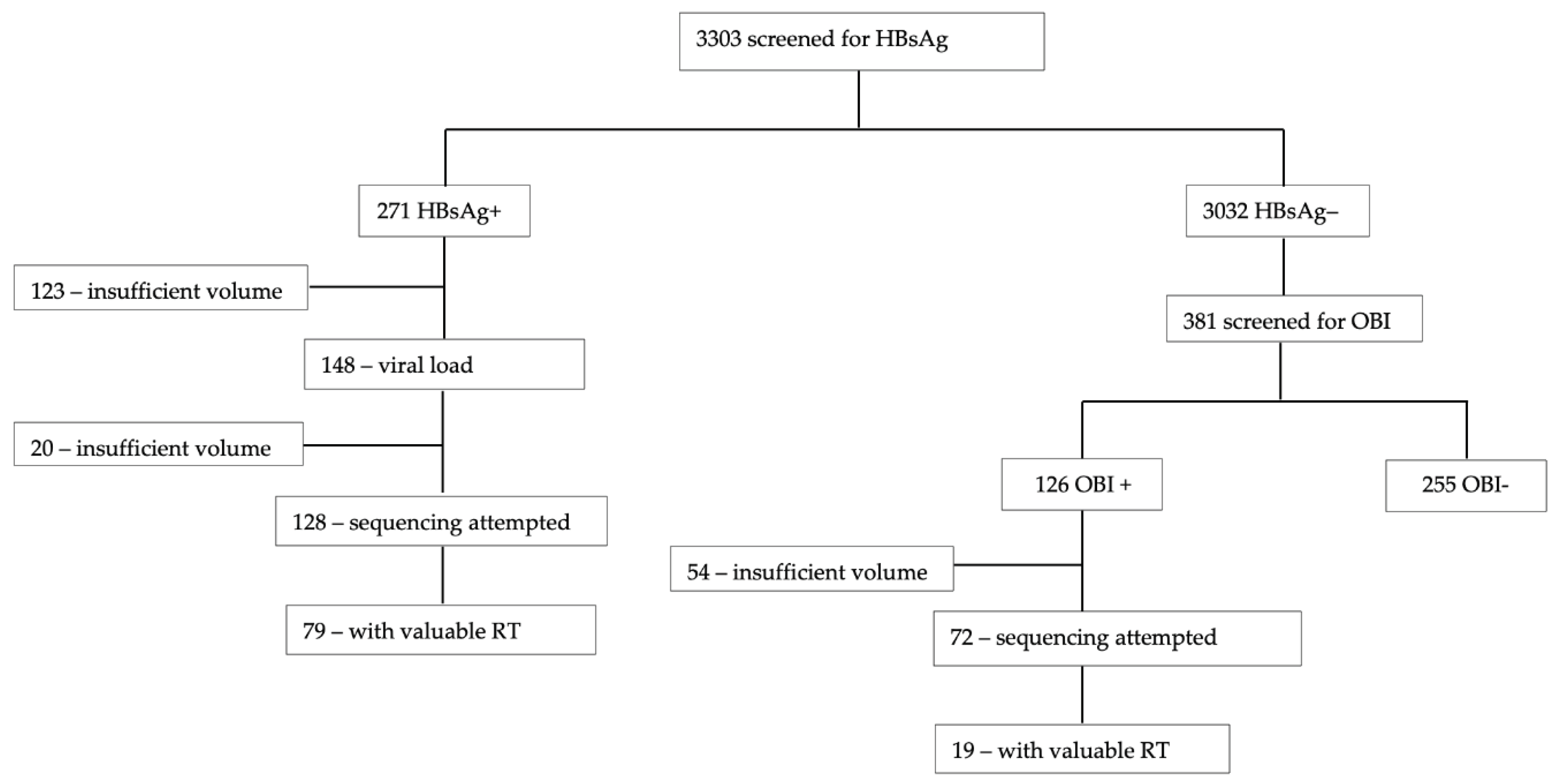
Participant plasma samples with positive HBsAg and OBI were used. From 271 samples with positive HBsAg, we attempted to sequence 128 and 79/128 (61.7%) had a valuable RT region. Out of the 72 samples with OBI, only 19/72 (26.4%) had a valuable RT region, hence a much lower sequencing success rate for samples with OBI. Therefore, we had a total of 98 RT sequences for downstream analysis (
Figure 1).
Most participants had suppressed HIV viral load (VL) (85/97, 87.6%), were on ART (93/98, 94.9%) and mostly on tenofovir disoproxil fumarate (TDF)-containing regimen (40/67, 59.7%). There was no statistically significant difference between participants with and without RAMs in all variables except for HBV viral load categories,
Table 1. HBV sequences with RAMs were also generated from participants with low HBV viral loads and some whose viral load results indicated “target not detected”,
Table 1.
3.2. Resistance Associated Mutations
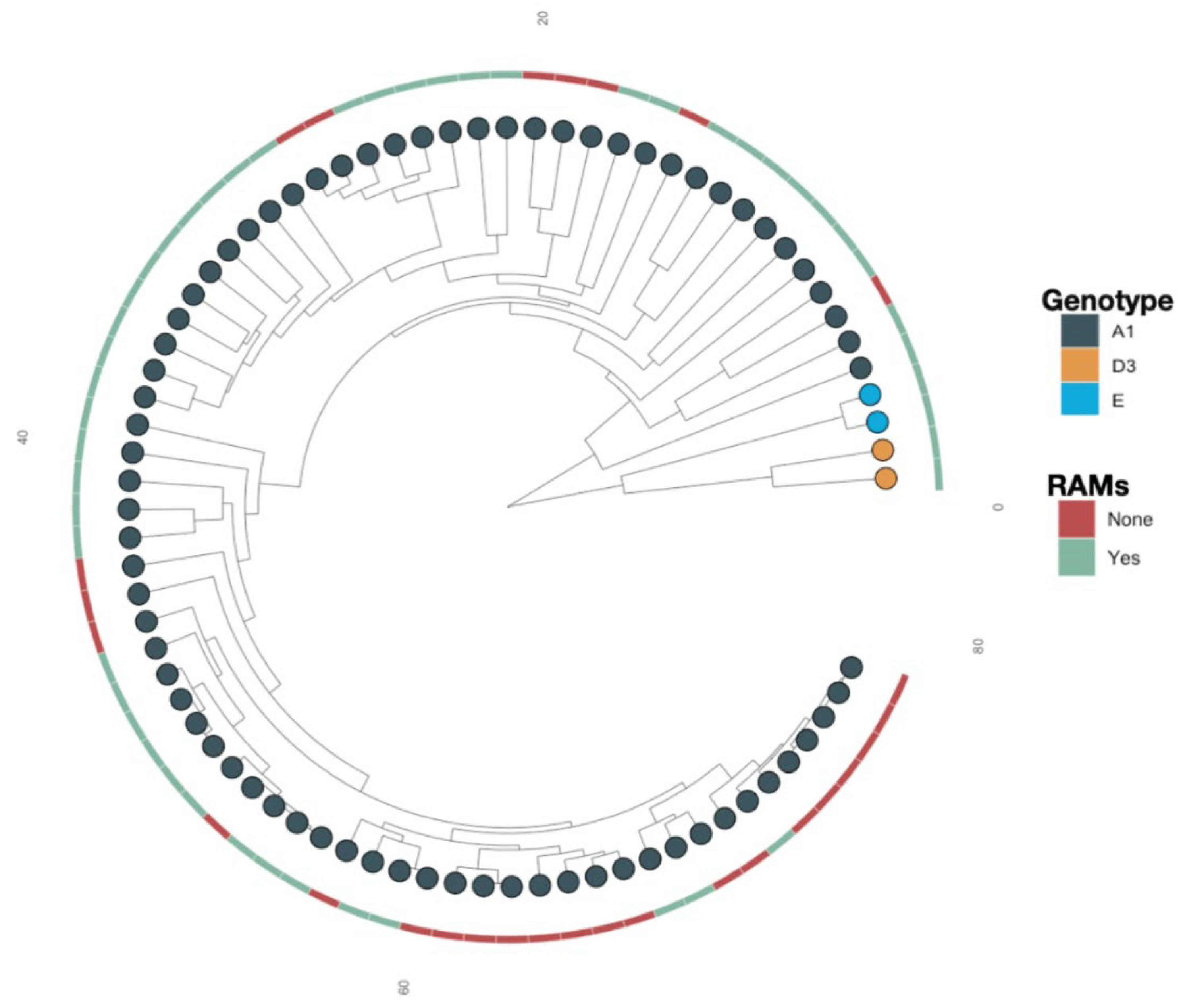
Overall, subgenotypes A1 (93.9%), D3 (3.1%) and E (3.1%) were identified among the 98 participants. A similar distribution was observed among participants with RAMs (n=61); A1 (90.2%), D3 (4.9%), E (4.9%). Phylogenetic analysis based on maximum-likelihood with 1000 bootstrap replicates was used to construct a tree.
Some of the sequences
with RAMs were clustering closely together (supported by posterior probability >0.90), suggesting a possible transmission, Figure 2.
In this study, we identified RAMs which confer resistance to 3TC in 61/98 (62.2%) participants. There were no RAMs identified which confer resistance to tenofovir in our study, therefore, among participants on TDF-containing regimen, none had HBV TDF RAMs. However, there was one participant who had an amino acid substitution ((
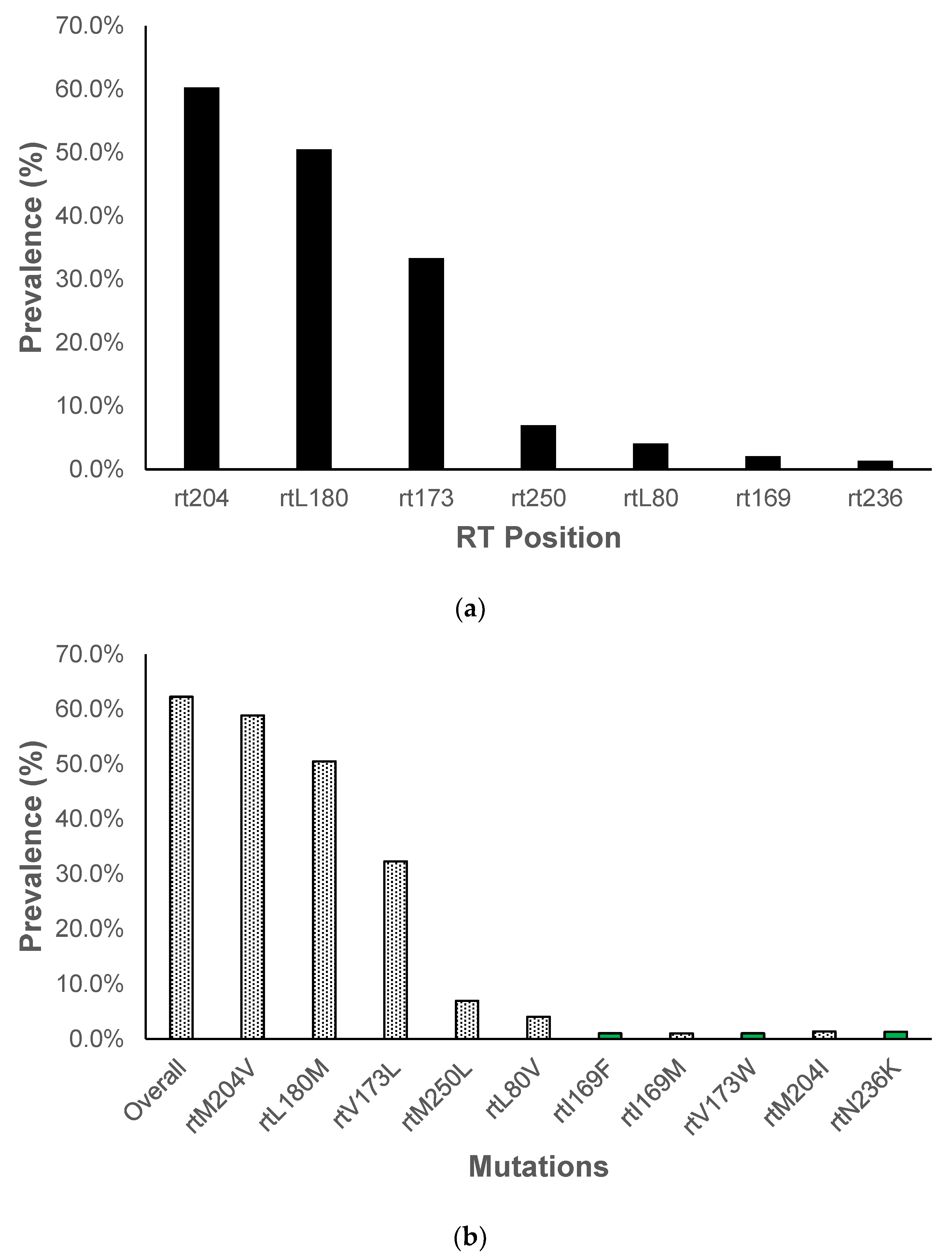
N236K) at a position that has been reported to confer resistance against tenofovir, rt236. Amino acid substitutions were identified at 7 RT positions, most being at position rt204 (60.3%), rt180 (50.5%), rt173 (33.3%) and rt250 (6.9%),
Figure 3A and B. We observed already characterized amino acid substitutions, rtM204V, rtL180M and rtV173L being the most prevalent. Other amino acid substitutions were observed in positions known for resistance associated mutations, however, they had not been well characterized,
Figure 3B.
Uncharacterized mutations mostly appeared in one participant,
Table 2. Some participants [17/61 (27.9%) and 10/61 (16.4%)] had the triple mutations (rtV173L, rtL180M and rtM204V) and the double mutations (rtL180M and M204V). Of participants with the triple mutations, 5/12 (41.7%) were on the 3TC regimen and among those with the double mutations, 5/8 (62.5%) were on a regimen with 3TC-only backbone,
Table 2.
4. Discussion
Botswana has a robust and successful HIV treatment program resulting in the country achieving UNAIDS 95-95-95 goals [
10]. However, HBV screening before ART initiation is not a robust and there is also no monitoring of HBV response to ART once people with HIV/HBV are initiated on ART for HIV. This widespread and prolonged use of ART for HIV without HBV screening and monitoring has potentially led to the selection of drug resistant HBV variants. In this study we report the prevalence of HBV RAMs in participants with HBV/HIV in Botswana. We generated HBV sequences from samples with varying HBV viral loads, even those deemed to have undetectable HBV viral load by the assay used in the study. This is not common as HBV sequencing is generally done for high viral load samples limiting the identification of the full spectrum of mutations in a population [
6].
HBV sequences generated in this study were mostly subgenotype A1 (93.9%) which is considerably different from other previous studies in the country. In blood donors, subgenotype A1 prevalence was 36.1% [
11], 45.5% in pregnant women [
25] and 48% in ART-naïve PLWH [
26]. In our current study, our sample size is larger, and sequences are generated from diverse communities across Botswana as opposed to previous studies that focused mainly on the capital city Gaborone and surrounding areas.
We detected RAMs in participants with varying HBV viral load, some with a TND result from the assay that we used which has also been reported elsewhere [
27,
28].
Therefore, HBV drug resistance is not to only be associated with treatment failure, as it also occurs at low viraemia. Most participants in this study were on TDF-containing or 3TC-containing regimens, which reflects the first line ART regimens at the time the BCPP study was conducted as well as historic first line ART regimen. Most participants had RAMs associated with 3TC therapy. Botswana adopted the Treat-All strategy in 2016 with DTG-based ART as the first line regimen. However, earlier first-line regimens included 3TC, and many participants in our study had been on 3TC-containing regimens, especially zidovudine/3TC plus EFV or NVP. With the ART duration of our study participants, it is not surprising that they would harbor HBV variants with mutations associated with 3TC resistance.
3TC is a low genetic barrier drug for HBV [
29] , hence as expected, we identified a high prevalence of 3TC associated resistance in this study. M204V (58.9%), L180M (50.5%) and V173L (32.3%) were the predominant mutations and they are known to confer resistance to 3TC and enhance viral replication [
30]. A similar pattern has been observed in Gabon [
31] and also similarly to our study, the predominant RAM was M204V/I in a systematic review of HBV RAMS in Africa [
9] and globally [
32]. The M204V mutation as shown in our results, occurred either alone or in combination with other mutations, most commonly with L180M and V173L which are described as compensatory mutations to M204V as they enhance viral replication [
31,
33].
Some of the study participants were on TDF-containing regimen and we report no RAM conferring resistance to TDF. HBV resistance against TFV is still controversial. A recent review suggests that resistance to TFV may require more than one resistance mutation that confers resistance to other NAs being L180M, A181V/T, M204I/V and N236T [
6]. A study in the United States showed that participants who had been exposed to 3TC took a longer time to achieve HBV viral suppression while on TFV compared to 3TC-naïve participants [
34]. Therefore, it is likely that 3TC-associated mutations may eventually lead to decreased susceptibility to TFV. Some of the study participants with 3TC-associated mutations were on a TDF-containing regimen, this could be because they started on 3TC-based ART before switching to a TDF-based regimen. It could also be due to the transmission of variants with 3TC resistance mutations [
35]. We report an uncharacterized polymorphism
N236K in a position known to confer resistance to TFV.
5. Conclusions
In a population of ART-experienced individuals with concomitant HBV/HIV, the prevalence of HBV RAMs was high, particularly those known to confer resistance to 3TC. The high prevalence of 3TC RAMs in this population discourages the use of ART regimens with 3TC as the only HBV-active drug in people living with HIV/HBV. The presence of HBV RAMs hinders HBV elimination efforts hence the need to monitor HBV drug resistance mutations in Botswana and globally. TDF-associated resistance mutations were not observed while most participants were on TDF hence supporting the effectiveness of TDF in treating HBV.
Author Contributions
Conceptualization, B.B.P., M.A., M.M., I.G.; methodology, B.B.P., G.M. T.R., B.P., validation, B.B.P., W.T.C.; formal analysis, B.B.P.,; investigation, B.B.P., resources, M.A., J.M., S.M., S.G.; data curation, B.B.P.,; writing—original draft preparation, B.B.P.; writing—review and editing, B.B.P, M.A., I.G., M.M., W.T.C., T.R., B.P., G.M., R.M., J.M., S.L., R.S., S.M., S.G.; visualization, B.B.P.; supervision, M.A., M.M., I.G., S.G.; project administration, B.B.P.; funding acquisition, M.A., R.M., S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Wellcome Trust (grant number 218770/Z/19/Z). W. T. C., S. M., and S. G. are partly supported through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE 2.0) from the Bill & Melinda Gates Foundation (INV-033558). S. G. and B.B.P are supported by the Fogarty International Center at the US National Institutes of Health (D43 TW009610). B. B. P. and S. G. were supported by the National Institutes of Health (NIH) Common Fund, award number U41HG006941 (H3ABioNet). H3ABioNet is an initiative of the Human Health and Heredity in Africa Consortium (H3Africa) program of the African Academy of Science. B. B. P., R. M., and S. M. are also supported by Trials of Excellence in Southern Africa (TESAIII), which is part of the EDCTP2 program supported by the European Union (grant number CSA2020NoE-3104 TESAIII). S. L., R. S. and S.M. received support from the NIH (award numbers K24 AI131928, K24 AI131924 and K43 TW012350 respectively).
Institutional Review Board Statement
The study was approved by the Ministry of Health Research and Development Committee (HDPME 13/18/1, 25 June 2021).
Informed Consent Statement
Written informed consent was obtained from all subjects involved in the BCPP study.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author. The data are not publicly available as the sequences are currently being analyzed for other objectives of the bigger project.
Acknowledgments
The authors thank the Botswana Prevention Combination Project study participants, Dikgosi and other community leaders, the clinic staff, District Health Management Teams, and Community Health Facilities at study sites; the Ya Tsie Study Team at the Botswana Harvard AIDS Institute Partnership, the Harvard T. H. Chan School of Public Health, the Centers for Disease Control and Prevention (CDC) Botswana, CDC Atlanta, and the Botswana Ministry of Health. The authors also thank those who served on the Ya Tsie Community Advisory Board, Laboratory Staff, and Management of Botswana Harvard HIV Reference Laboratory.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- WHO. Interim Guidance for Country Validation of Viral Hepatitis Elimination; World Health Organization: Geneva, Switzerland, 2021.
- Pal, A.; Sarkar, N.; Saha, D.; Guha, S.K.; Saha, B.; Chakrabarti, S.; Chakravarty, R. High incidence of lamivudine-resistance-associated vaccine-escape HBV mutants among HIV-coinfected patients on prolonged antiretroviral therapy. Antivir Ther 2015, 20, 545-554. [CrossRef]
- WHO. Guidelines for the Prevention, Care and Treatment of Persons with Chronic Hepatitis B Infection. 2015, 29.
- Woo, H.Y.; Park, J.Y.; Bae, S.H.; Kim, C.W.; Jang, J.Y.; Tak, W.Y.; Kim, D.J.; Kim, I.H.; Heo, J.; Ahn, S.H. Entecavir+tenofovir vs. lamivudine/telbivudine+adefovir in chronic hepatitis B patients with prior suboptimal response. Clin Mol Hepatol 2020, 26, 352-363. [CrossRef]
- Msomi, N.; Parboosing, R.; Wilkinson, E.; Giandhari, J.; Govender, K.; Chimukangara, B.; Mlisana, K.P. Persistent Hepatitis B Viraemia with Polymerase Mutations among HIV/HBV Co-Infected Patients on HBV-Active ART in KwaZulu-Natal, South Africa. Viruses 2022, 14. [CrossRef]
- Mokaya, J.; McNaughton, A.L.; Bester, P.A.; Goedhals, D.; Barnes, E.; Marsden, B.D.; Matthews, P.C. Hepatitis B virus resistance to tenofovir: fact or fiction? A systematic literature review and structural analysis of drug resistance mechanisms. Wellcome Open Res 2020, 5, 151. [CrossRef]
- Amini-Bavil-Olyaee, S.; Herbers, U.; Sheldon, J.; Luedde, T.; Trautwein, C.; Tacke, F. The rtA194T polymerase mutation impacts viral replication and susceptibility to tenofovir in hepatitis B e antigen-positive and hepatitis B e antigen-negative hepatitis B virus strains. Hepatology 2009, 49, 1158-1165. [CrossRef]
- Shirvani-Dastgerdi, E.; Winer, B.Y.; Celia-Terrassa, T.; Kang, Y.; Tabernero, D.; Yagmur, E.; Rodriguez-Frias, F.; Gregori, J.; Luedde, T.; Trautwein, C.; et al. Selection of the highly replicative and partially multidrug resistant rtS78T HBV polymerase mutation during TDF-ETV combination therapy. J Hepatol 2017, 67, 246-254. [CrossRef]
- Mokaya, J.; McNaughton, A.L.; Hadley, M.J.; Beloukas, A.; Geretti, A.M.; Goedhals, D.; Matthews, P.C. A systematic review of hepatitis B virus (HBV) drug and vaccine escape mutations in Africa: A call for urgent action. PLoS Negl Trop Dis 2018, 12, e0006629. [CrossRef]
- Mine, M.; Stafford, K.; Laws, R.L.; Marima, M.; Lekone, P.; Ramaabya, D.; Makhaola, K.; Mapondera, P.; Wray-Gordon, F.; Akbakwuru, C.; et al. Botswana achieved the Joint United Nations Programme on HIV/AIDS (UNAIDS) 95-95-95 targets: results from the Fifth Botswana HIV/AIDS Impact Survey (BAIS V), 2021. In Proceedings of the The 24th International AIDS Conference, Montreal, Canada & Virtual, 29th July - 2nd August 2022; p. 231.
- Choga, W.T.; Anderson, M.; Zumbika, E.; Moyo, S.; Mbangiwa, T.; Phinius, B.B.; Melamu, P.; Kayembe, M.K.; Kasvosve, I.; Sebunya, T.K.; et al. Molecular characterization of hepatitis B virus in blood donors in Botswana. Virus Genes 2018. [CrossRef]
- Wester, C.W.; Bussmann, H.; Moyo, S.; Avalos, A.; Gaolathe, T.; Ndwapi, N.; Essex, M.; MacGregor, R.R.; Marlink, R.G. Serological evidence of HIV-associated infection among HIV-1-infected adults in Botswana. Clin Infect Dis 2006, 43, 1612-1615. [CrossRef]
- Phinius, B.B.; Anderson, M.; Gobe, I.; Mokomane, M.; Choga, W.T.; Mutenga, S.R.; Mpebe, G.; Pretorius-Holme, M.; Musonda, R.; Gaolathe, T.; et al. High Prevalence of Hepatitis B Virus Infection Among People With HIV in Rural and Periurban Communities in Botswana. Open Forum Infect Dis 2023, 10, ofac707. [CrossRef]
- Baruti, K.; Lentz, K.; Anderson, M.; Ajibola, G.; Phinius, B.B.; Choga, W.T.; Mbangiwa, T.; Powis, K.M.; Sebunya, T.; Blackard, J.T.; et al. Hepatitis B virus prevalence and vaccine antibody titers in children HIV exposed but uninfected in Botswana. PLoS One 2020, 15, e0237252. [CrossRef]
- Anderson, M.; Gaseitsiwe, S.; Moyo, S.; Thami, K.P.; Mohammed, T.; Setlhare, D.; Sebunya, T.K.; Powell, E.A.; Makhema, J.; Blackard, J.T.; et al. Slow CD4+ T cell Recovery in Human Immunodeficiency Virus/Hepatitis B Virus-Coinfected Patients Initiating Truvada-Based Combination Antiretroviral Therapy in Botswana. Open Forum Infect Dis 2016, 3. [CrossRef]
- Raimondo, G.; Locarnini, S.; Pollicino, T.; Levrero, M.; Zoulim, F.; Lok, A.S.; Taormina Workshop on Occult, H.B.V.I.F.M. Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J Hepatol 2019, 71, 397-408. [CrossRef]
- Ministy-of-Health. Handbook of the Botswana 2016 Integrated HIV Clincial Care Guidelines. 2016.
- Makhema, J.; Wirth, K.E.; Pretorius Holme, M.; Gaolathe, T.; Mmalane, M.; Kadima, E.; Chakalisa, U.; Bennett, K.; Leidner, J.; Manyake, K.; et al. Universal Testing, Expanded Treatment, and Incidence of HIV Infection in Botswana. N Engl J Med 2019, 381, 230-242. [CrossRef]
- Choga, W. Complete Hepatitis B Virus Sequencing using an ONT-Based Next-Generation Sequencing Protocol v1; 2023.
- Phinius, B.B.; Anderson, M.; Mokomane, M.; Gobe, I.; Choga, W.T.; Ratsoma, T.; Phakedi, B.; Mpebe, G.; Ditshwanelo, D.; Musonda, R.; et al. Atypical Hepatitis B Virus Serology Profile-Hepatitis B Surface Antigen-Positive/Hepatitis B Core Antibody-Negative-In Hepatitis B Virus/HIV Coinfected Individuals in Botswana. Viruses 2023, 15. [CrossRef]
- ONT. PCR Tiling of Sars Cov 2 Virus With Rapid Barcoding SQK rbk110 PCTR - 9125 - v110 - Revh - 24mar2021 Minion. Available online: https://community.nanoporetech.com/docs/prepare/library_prep_protocols/pcr-tiling-of-sars-cov-2-virus-with-rapid-barcoding-and-midnight/v/mrt_9127_v110_revw_14jul2021 (accessed on 28 March 2023).
- Larsson, A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276-3278. [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015, 32, 268-274. [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 2017, 14, 587-589. [CrossRef]
- Mbangiwa, T.; Kasvosve, I.; Anderson, M.; Thami, P.K.; Choga, W.T.; Needleman, A.; Phinius, B.B.; Moyo, S.; Leteane, M.; Leidner, J.; et al. Chronic and Occult Hepatitis B Virus Infection in Pregnant Women in Botswana. Genes (Basel) 2018, 9. [CrossRef]
- Anderson, M.; Choga, W.T.; Moyo, S.; Bell, T.G.; Mbangiwa, T.; Phinius, B.B.; Bhebhe, L.; Sebunya, T.K.; Lockman, S.; Marlink, R.; et al. Molecular Characterization of Near Full-Length Genomes of Hepatitis B Virus Isolated from Predominantly HIV Infected Individuals in Botswana. Genes (Basel) 2018, 9. [CrossRef]
- Svicher, V.; Alteri, C.; Gori, C.; Salpini, R.; Marcuccilli, F.; Bertoli, A.; Longo, R.; Bernassola, M.; Gallinaro, V.; Romano, S.; et al. Lamivudine-resistance mutations can be selected even at very low levels of hepatitis B viraemia. Dig Liver Dis 2010, 42, 902-907. [CrossRef]
- Wang, S.; Li, H.; Kou, Z.; Ren, F.; Jin, Y.; Yang, L.; Dong, X.; Yang, M.; Zhao, J.; Liu, H.; et al. Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clinical Microbiology and Infection 2021, 27, 443-450. [CrossRef]
- Ghany, M.G.; Doo, E.C. Antiviral resistance and hepatitis B therapy. Hepatology 2009, 49, S174-184. [CrossRef]
- Shaw, T.; Bartholomeusz, A.; Locarnini, S. HBV drug resistance: mechanisms, detection and interpretation. J Hepatol 2006, 44, 593-606. [CrossRef]
- Bivigou-Mboumba, B.; Amougou-Atsama, M.; Zoa-Assoumou, S.; M’Boyis Kamdem, H.; Nzengui-Nzengui, G.F.; Ndojyi-Mbiguino, A.; Njouom, R.; Francois-Souquiere, S. Hepatitis B infection among HIV infected individuals in Gabon: Occult hepatitis B enhances HBV DNA prevalence. PLoS One 2018, 13, e0190592. [CrossRef]
- Mokaya, J.; Vasylyeva, T.I.; Barnes, E.; Ansari, M.A.; Pybus, O.G.; Matthews, P.C. Global prevalence and phylogeny of hepatitis B virus (HBV) drug and vaccine resistance mutations. J Viral Hepat 2021, 28, 1110-1120. [CrossRef]
- Delaney, W.E.t.; Yang, H.; Westland, C.E.; Das, K.; Arnold, E.; Gibbs, C.S.; Miller, M.D.; Xiong, S. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J Virol 2003, 77, 11833-11841. [CrossRef]
- Kim, H.N.; Rodriguez, C.V.; Van Rompaey, S.; Eron, J.J.; Thio, C.L.; Crane, H.M.; Overton, E.T.; Saag, M.S.; Martin, J.; Geng, E.; et al. Factors associated with delayed hepatitis B viral suppression on tenofovir among patients coinfected with HBV-HIV in the CNICS cohort. J Acquir Immune Defic Syndr 2014, 66, 96-101. [CrossRef]
- Mantovani, N.; Cicero, M.; Santana, L.C.; Silveira, C.; do Carmo, E.P.; Abrão, P.R.; Diaz, R.S.; Caseiro, M.M.; Komninakis, S.V. Detection of lamivudine-resistant variants and mutations related to reduced antigenicity of HBsAg in individuals from the cities of Santos and São Paulo, Brazil. Virol J 2013, 10, 320. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).