Submitted:
13 March 2024
Posted:
14 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
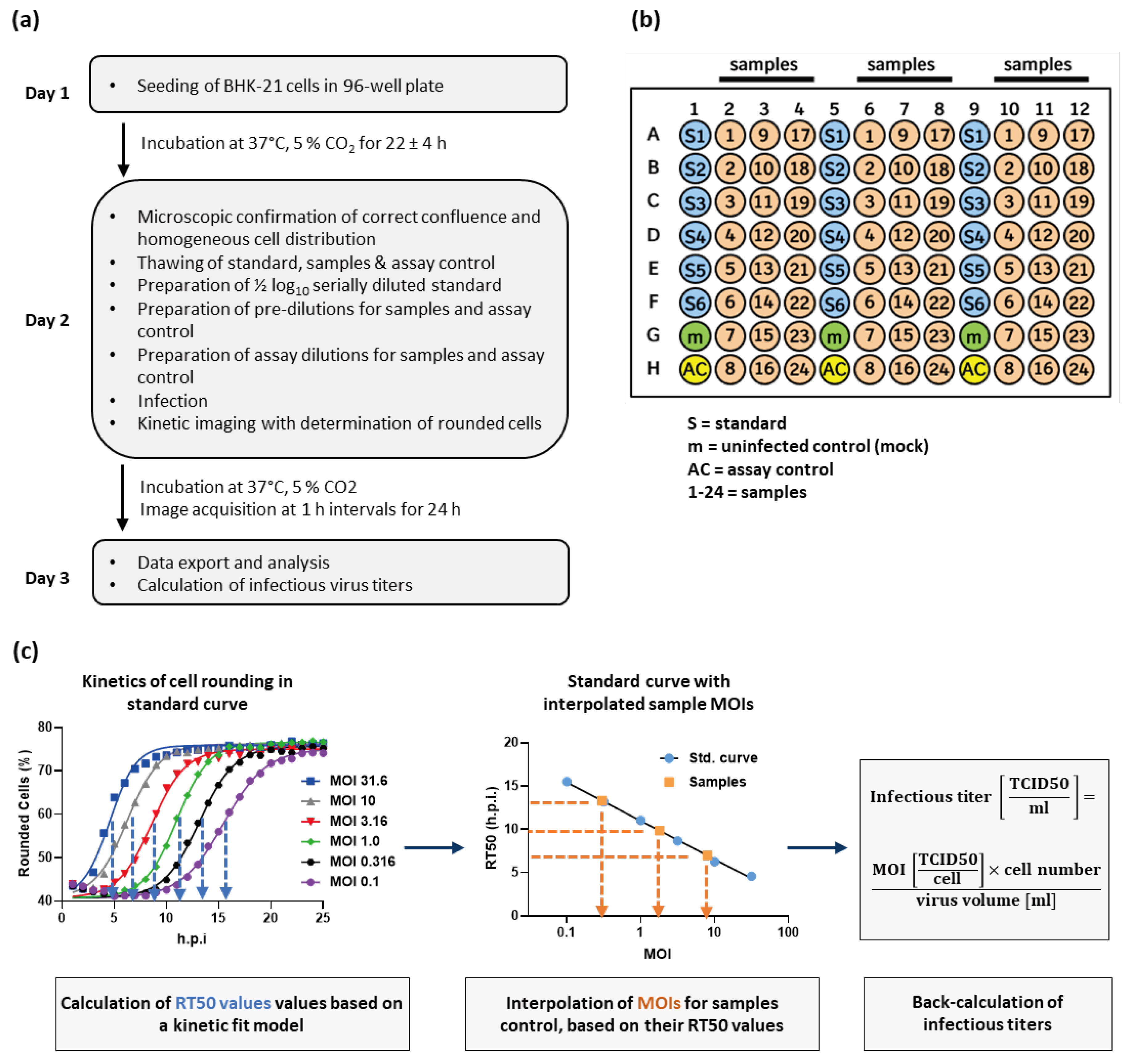
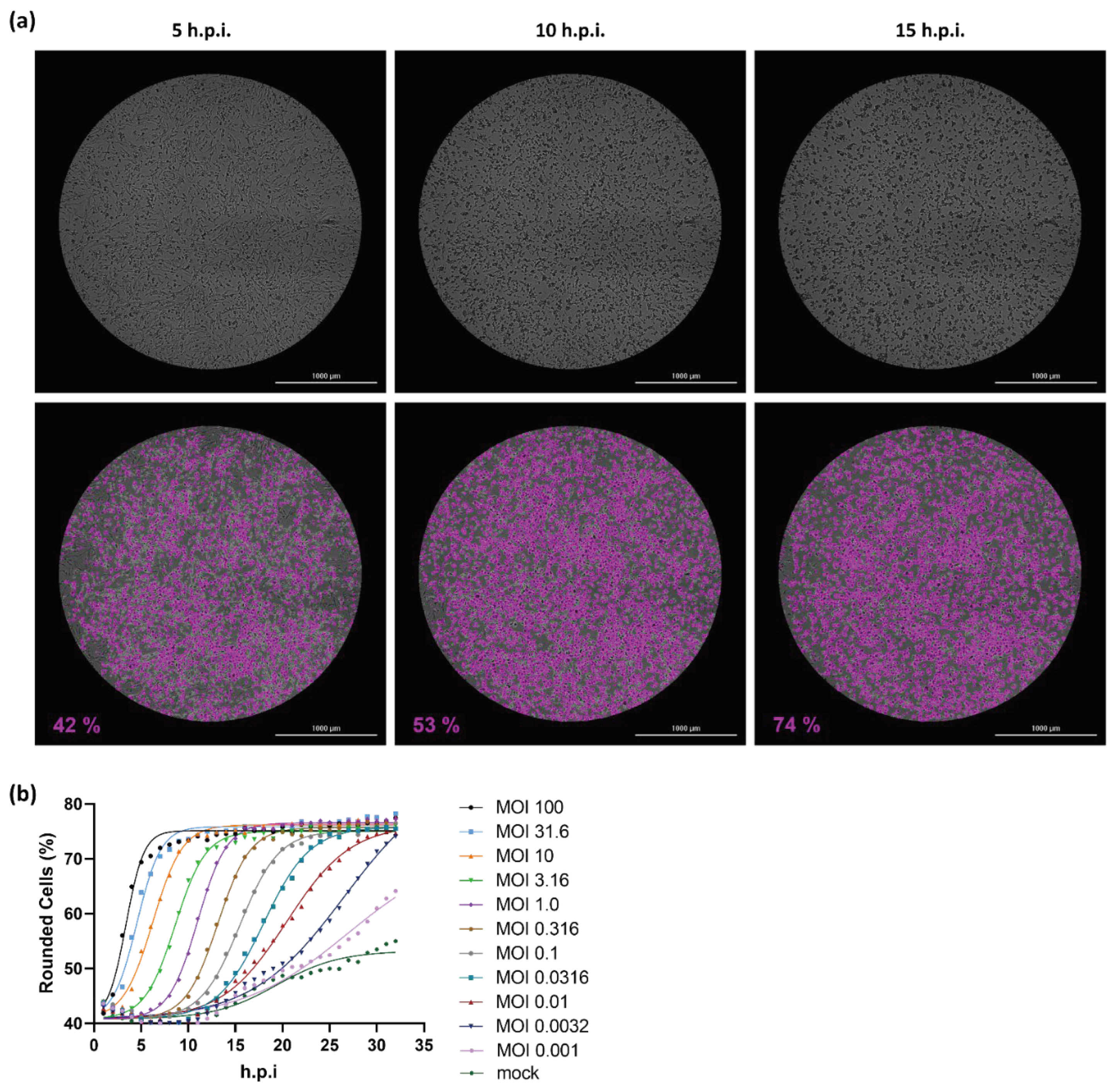
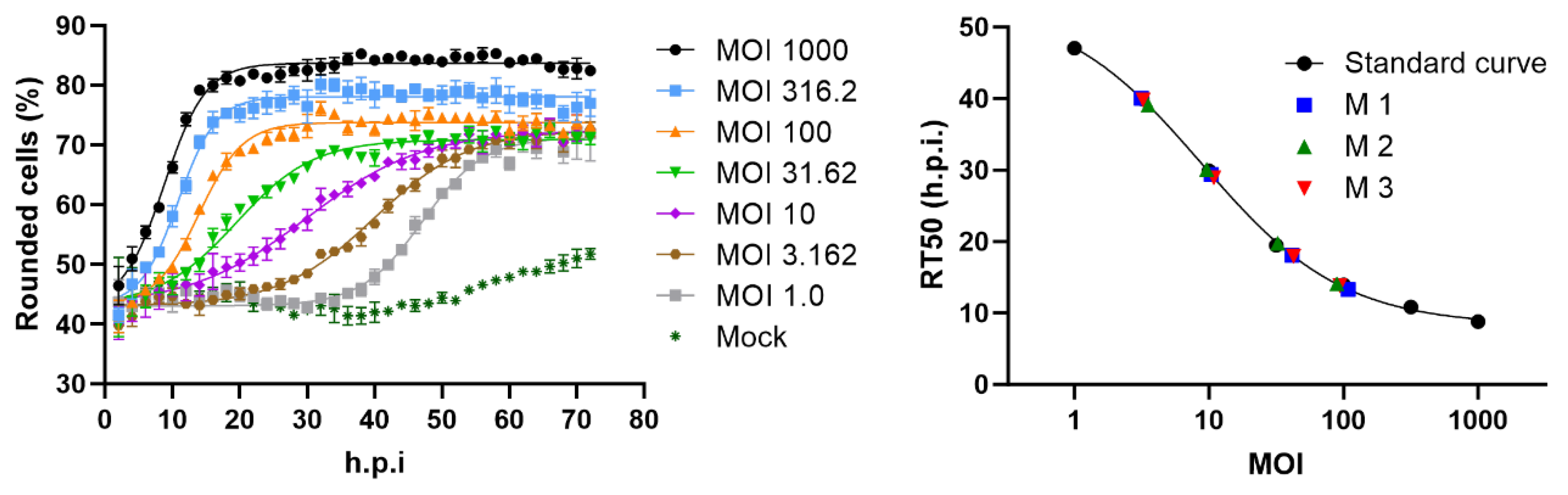
2.1. Cell rounding Occurs Early after Infection and Correlates with the Applied Virus Dose
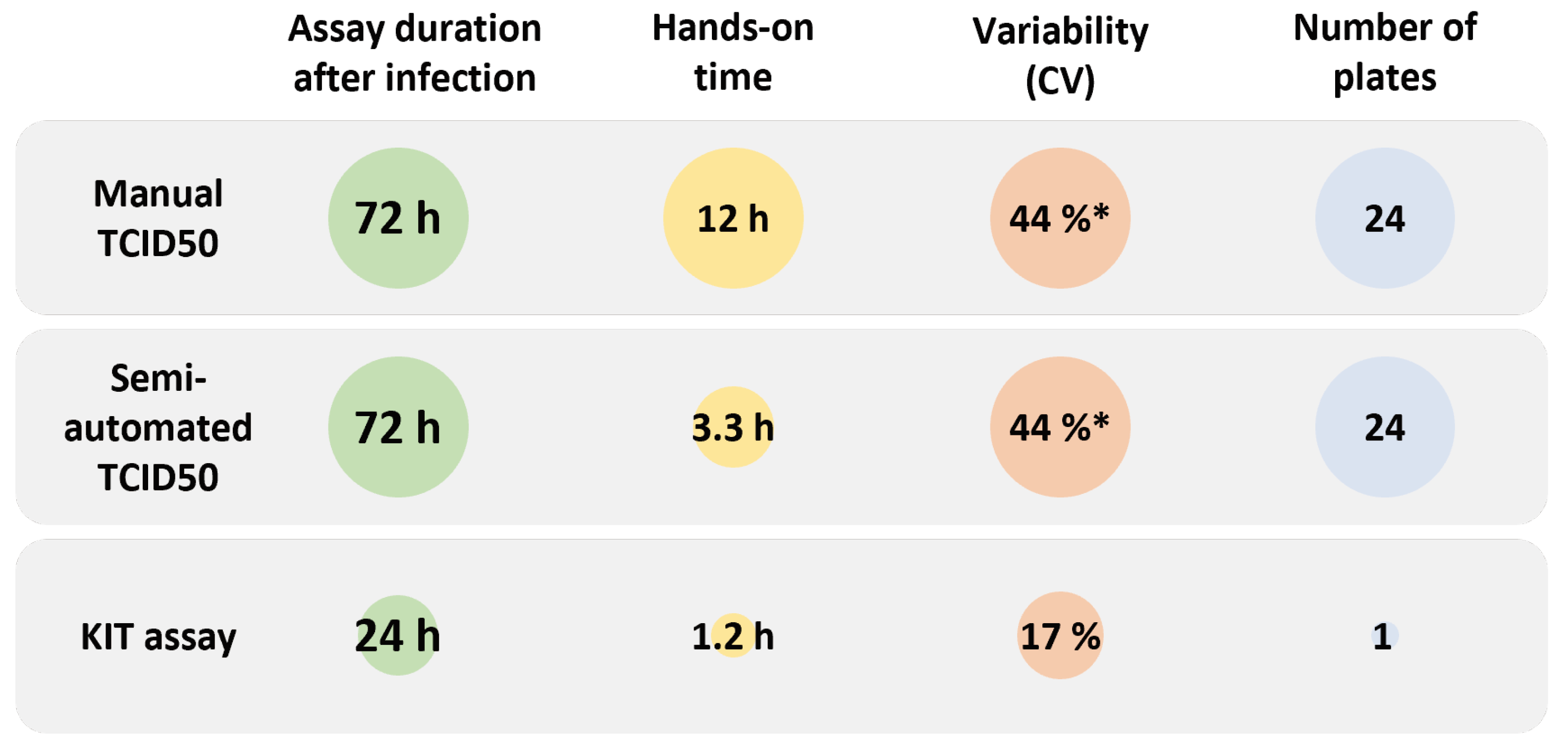
2.2. Titer Determination with the Kinetic Infectious Virus Titer (KIT) Assay
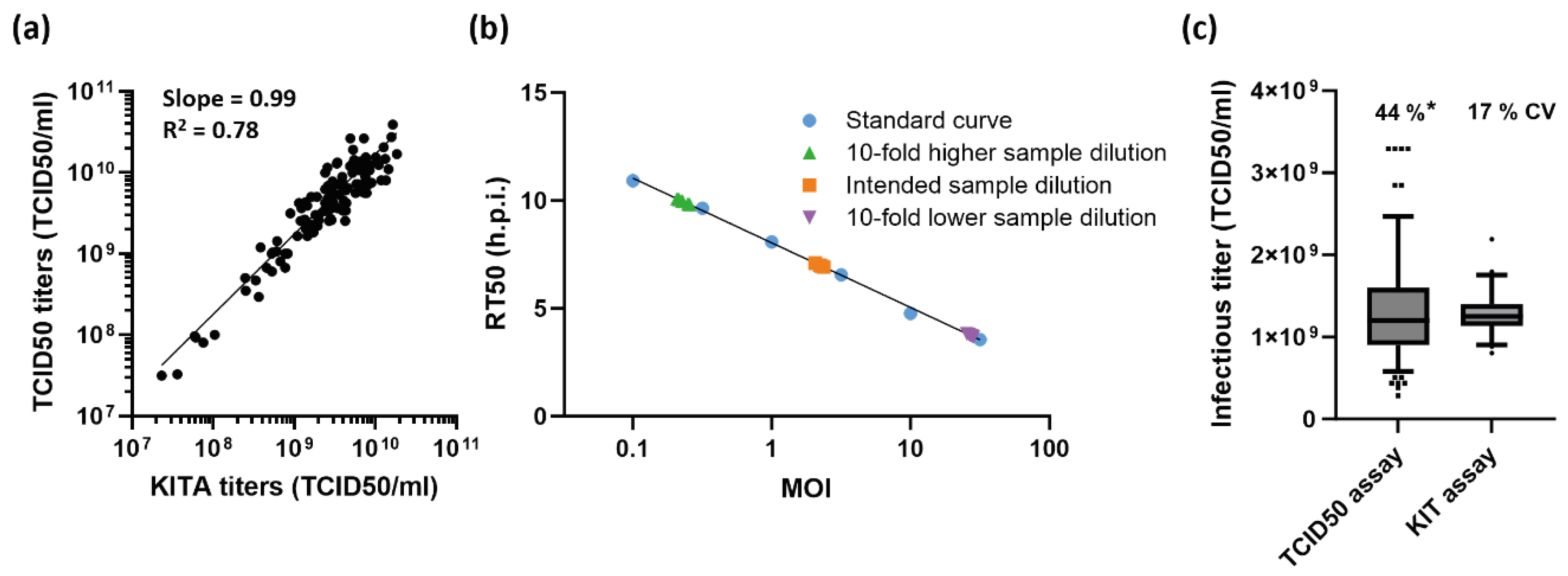
2.3. Performance Characteristics of the KIT Assay
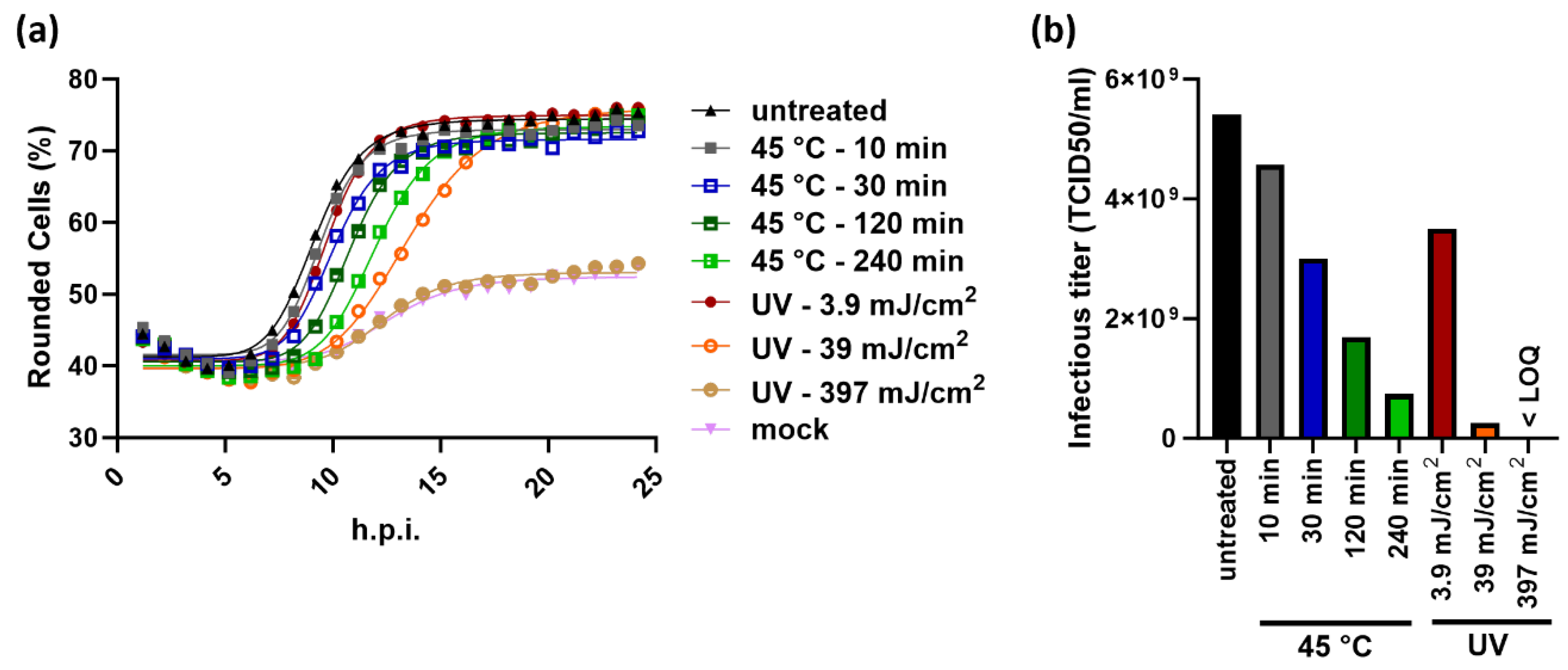
2.4. The Principle of the KIT Assay Can be Applied to Different Virus-Cell-Combinations
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Kinetic Infectious Virus Titer (KIT) Assay
4.3. TCID50 Assay
4.4. qRT-PCR
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, Z.; Anselmo, A.C.; Mitragotri, S. Viral Vector-based Gene Therapies in the Clinic. Bioeng Transl Medicine 2021, 7, e10258. [CrossRef]
- Fraikin, J.-L.; Teesalu, T.; McKenney, C.M.; Ruoslahti, E.; Cleland, A.N. A High-Throughput Label-Free Nanoparticle Analyser. Nat. Nanotechnol. 2011, 6, 308–313. [CrossRef]
- Kärber, G. Beitrag Zur Kollektiven Behandlung Pharmakologischer Reihenversuche. Naunyn-Schmiedebergs Arch. für Exp. Pathol. Pharmakol. 1931, 162, 480–483. [CrossRef]
- SPEARMAN, C. THE METHOD OF ‘RIGHT AND WRONG CASES’ (‘CONSTANT STIMULI’) WITHOUT GAUSS’S FORMULAE. Br. J. Psychol., 19041920 1908, 2, 227–242. [CrossRef]
- REED, L.J.; MUENCH, H. A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS12. Am. J. Epidemiology 1938, 27, 493–497. [CrossRef]
- Zapata-Cardona, M.I.; Flórez-Álvarez, L.; Gómez-Gallego, D.M.; Moncada-Díaz, M.J.; Hernandez, J.C.; Díaz, F.; Rugeles, M.T.; Aguilar-Jiménez, W.; Zapata, W. Comparison among Plaque Assay, Tissue Culture Infectious Dose (TCID50) and Real-Time RT-PCR for SARS-CoV-2 Variants Quantification. Iran. J. Microbiol. 2022, 14, 291–299. [CrossRef]
- Shurtleff, A.C.; Bloomfield, H.A.; Mort, S.; Orr, S.A.; Audet, B.; Whitaker, T.; Richards, M.J.; Bavari, S. Validation of the Filovirus Plaque Assay for Use in Preclinical Studies. Viruses 2016, 8, 113. [CrossRef]
- Hochdorfer, D.; Businger, R.; Hotter, D.; Seifried, C.; Solzin, J. Automated, Label-Free TCID50 Assay to Determine the Infectious Titer of Virus-Based Therapeutics. J Virol Methods 2022, 299, 114318. [CrossRef]
- Masci, A.L.; Menesale, E.B.; Chen, W.-C.; Co, C.; Lu, X.; Bergelson, S. Integration of Fluorescence Detection and Image-Based Automated Counting Increases Speed, Sensitivity, and Robustness of Plaque Assays. Mol Ther - Methods Clin Dev 2019, 14, 270–274. [CrossRef]
- Hebert, C.G.; DiNardo, N.; Evans, Z.L.; Hart, S.J.; Hachmann, A.-B. Rapid Quantification of Vesicular Stomatitis Virus in Vero Cells Using Laser Force Cytology. Vaccine 2018, 36, 6061–6069. [CrossRef]
- Hebert, C.G.; Rodrigues, K.L.; DiNardo, N.; Hachmann, A.-B. Viral Infectivity Quantification and Neutralization Assays Using Laser Force Cytology. Methods Mol. Biol. (Clifton, NJ) 2020, 2183, 575–585. [CrossRef]
- Dodkins, R.; Delaney, J.R.; Overton, T.; Scholle, F.; Frias-De-Diego, A.; Crisci, E.; Huq, N.; Jordan, I.; Kimata, J.T.; Findley, T.; et al. A Rapid, High-Throughput, Viral Infectivity Assay Using Automated Brightfield Microscopy with Machine Learning. SLAS Technol. 2023, 28, 324–333. [CrossRef]
- Lebourgeois, S.; Fraisse, A.; Hennechart-Collette, C.; Guillier, L.; Perelle, S.; Martin-Latil, S. Development of a Real-Time Cell Analysis (RTCA) Method as a Fast and Accurate Method for Detecting Infectious Particles of the Adapted Strain of Hepatitis A Virus. Front. Cell. Infect. Microbiol. 2018, 8, 335. [CrossRef]
- Melzer, M.; Lopez-Martinez, A.; Altomonte, J. Oncolytic Vesicular Stomatitis Virus as a Viro-Immunotherapy: Defeating Cancer with a “Hammer” and “Anvil.” Biomedicines 2017, 5, 8. [CrossRef]
- Porosnicu, M.; Quinson, A.-M.; Crossley, K.; Luecke, S.; Lauer, U.M. Phase I Study of VSV-GP (BI 1831169) as Monotherapy or Combined with Ezabenlimab in Advanced and Refractory Solid Tumors. Futur. Oncol. 2022, 18, 2627–2638. [CrossRef]
- Muik, A.; Stubbert, L.J.; Jahedi, R.Z.; Geiβ, Y.; Kimpel, J.; Dold, C.; Tober, R.; Volk, A.; Klein, S.; Dietrich, U.; et al. Re-Engineering Vesicular Stomatitis Virus to Abrogate Neurotoxicity, Circumvent Humoral Immunity, and Enhance Oncolytic Potency. Cancer Res. 2014, 74, 3567–3578. [CrossRef]
- Muik, A.; Stubbert, L.J.; Jahedi, R.Z.; Geiβ, Y.; Kimpel, J.; Dold, C.; Tober, R.; Volk, A.; Klein, S.; Dietrich, U.; et al. Re-Engineering Vesicular Stomatitis Virus to Abrogate Neurotoxicity, Circumvent Humoral Immunity, and Enhance Oncolytic Potency. Cancer Res 2014, 74, 3567–3578. [CrossRef]
- Ogino, M.; Fedorov, Y.; Adams, D.J.; Okada, K.; Ito, N.; Sugiyama, M.; Ogino, T. Vesiculopolins, a New Class of Anti-Vesiculoviral Compounds, Inhibit Transcription Initiation of Vesiculoviruses. Viruses 2019, 11, 856. [CrossRef]
- Jayakar, H.R.; Whitt, M.A. Identification of Two Additional Translation Products from the Matrix (M) Gene That Contribute to Vesicular Stomatitis Virus Cytopathology. J. Virol. 2002, 76, 8011–8018. [CrossRef]
- Kobbe, C. von; Deursen, J.M.A. van; Rodrigues, J.P.; Sitterlin, D.; Bachi, A.; Wu, X.; Wilm, M.; Carmo-Fonseca, M.; Izaurralde, E. Vesicular Stomatitis Virus Matrix Protein Inhibits Host Cell Gene Expression by Targeting the Nucleoporin Nup98. Mol. Cell 2000, 6, 1243–1252. [CrossRef]
- Petersen, J.M.; Her, L.-S.; Varvel, V.; Lund, E.; Dahlberg, J.E. The Matrix Protein of Vesicular Stomatitis Virus Inhibits Nucleocytoplasmic Transport When It Is in the Nucleus and Associated with Nuclear Pore Complexes. Mol. Cell. Biol. 2000, 20, 8590–8601. [CrossRef]
- Quan, B.; Seo, H.-S.; Blobel, G.; Ren, Y. Vesiculoviral Matrix (M) Protein Occupies Nucleic Acid Binding Site at Nucleoporin Pair (Rae1•Nup98). Proc. Natl. Acad. Sci. 2014, 111, 9127–9132. [CrossRef]
- Kunzelmann, M.; Wittmann, A.; Nold, V.; Presser, B.; Schreiber, J.; Gehrig, T.; Sadlers, S.; Scholz, R.; Solzin, J.; Berger, A.; et al. Functional Design of Experiment for Potency Assay Optimization and In-Silico Simulation. J. Pharm. Biomed. Anal. 2023, 234, 115584. [CrossRef]
- Solzin, J.; Eppler, K.; Knapp, B.; Buchner, H.; Bluhmki, E. Optimising Cell-Based Bioassays via Integrated Design of Experiments (IxDoE) - A Practical Guide. SLAS Discov. 2023, 28, 29–38. [CrossRef]
- Anam, S.; Rahman, S.U.; Ali, S.; Saeed, M.; Goyal, S.M. Comparative Growth Kinetic Study of Newcastle Disease Virus, Infectious Bursal Disease Virus and Avian Influenza Virus in Chicken Embryo Fibroblast and DF-1 Cell Lines. Pol. J. Vet. Sci. 2021, 24, 287-292-287–292. [CrossRef]
- Hollý, J.; Fogelová, M.; Jakubcová, L.; Tomčíková, K.; Vozárová, M.; Varečková, E.; Kostolanský, F. Comparison of Infectious Influenza A Virus Quantification Methods Employing Immuno-Staining. J. Virol. Methods 2017, 247, 107–113. [CrossRef]
- Taylor, M.P.; Koyuncu, O.O.; Enquist, L.W. Subversion of the Actin Cytoskeleton during Viral Infection. Nat. Rev. Microbiol. 2011, 9, 427–439. [CrossRef]
- Werner, J.; Kronberg, R.M.; Stachura, P.; Ostermann, P.N.; Müller, L.; Schaal, H.; Bhatia, S.; Kather, J.N.; Borkhardt, A.; Pandyra, A.A.; et al. Deep Transfer Learning Approach for Automatic Recognition of Drug Toxicity and Inhibition of SARS-CoV-2. Viruses 2021, 13, 610. [CrossRef]
- Wang, T.-E.; Chao, T.-L.; Tsai, H.-T.; Lin, P.-H.; Tsai, Y.-L.; Chang, S.-Y. Differentiation of Cytopathic Effects (CPE) Induced by Influenza Virus Infection Using Deep Convolutional Neural Networks (CNN). PLoS Comput. Biol. 2020, 16, e1007883. [CrossRef]
- Piszczatoski, C.R.; Gums, J.G. Ervebo (Ebola Zaire Vaccine, Live/RVSVΔG-ZEBOV-GP): The First Licensed Vaccine for the Prevention of Ebola Virus Disease. J Pharm Technology 2020, 36, 243–250. [CrossRef]
- Saphire, E.O. A Vaccine against Ebola Virus. Cell 2020, 181, 6. [CrossRef]
- Yahalom-Ronen, Y.; Tamir, H.; Melamed, S.; Politi, B.; Shifman, O.; Achdout, H.; Vitner, E.B.; Israeli, O.; Milrot, E.; Stein, D.; et al. A Single Dose of Recombinant VSV-∆G-Spike Vaccine Provides Protection against SARS-CoV-2 Challenge. Nat. Commun. 2020, 11, 6402. [CrossRef]
- Das, K.; Belnoue, E.; Rossi, M.; Hofer, T.; Danklmaier, S.; Nolden, T.; Schreiber, L.-M.; Angerer, K.; Kimpel, J.; Hoegler, S.; et al. A Modular Self-Adjuvanting Cancer Vaccine Combined with an Oncolytic Vaccine Induces Potent Antitumor Immunity. Nat. Commun. 2021, 12, 5195. [CrossRef]
- Merchan, J.R.; Patel, M.; Cripe, T.P.; Old, M.O.; Strauss, J.F.; Thomassen, A.; Diaz, R.M.; Peng, K.W.; Russell, S.J.; Russell, L.; et al. Relationship of Infusion Duration to Safety, Efficacy, and Pharmacodynamics (PD): Second Part of a Phase I-II Study Using VSV-IFNβ-NIS (VV1) Oncolytic Virus in Patients with Refractory Solid Tumors. J. Clin. Oncol. 2020, 38, 3090–3090. https://doi.org/10.1200/jco.2020.38.15_suppl.3090. [CrossRef]

| Sample ID | Expected titer (TCID50/ml | Rounded titer (TCID50/ml) | Target MOI | Required pure virus per well (µl) | Number of 10-1 sample pre-dilutions | Volume pre-diluted virus (µl)* | Volume medium (µl)* |
|---|---|---|---|---|---|---|---|
| Sample 1 | 2.72E+08 | 1E+08 | 1.78 | 0.32 | 2 | 192 | 408 |
| Sample 2 | 8.16E+08 | 1E+09 | 1.78 | 0.032 | 3 | 192 | 408 |
| Sample 3 | 5.09E+09 | 1E+10 | 1.78 | 0.0032 | 4 | 192 | 408 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





