1. Introduction
Rheumatoid arthritis (RA) is a systemic condition affecting the joints and their surrounding tissues. RA presents a significant medical concern, and, to date, constitutes a major therapeutic challenge. Approximately, 1% of the global population suffer from RA, making it the most prevalent chronic autoimmune disorder [
1]. Typically, the age of initial symptoms is estimated between 35 to 60. It is characterized by severe joint pain, stiffness, and swelling, and if not treated effectively, it can lead to disability, chronic pain, and distress. Proper treatment for RA alleviates pain, enhances functionality, and improves the overall quality of life. However, a substantial number of RA patients do not achieve remission [
2]. The exact cause of RA remains unknown, with the primary risk factors encompassing genetics, environmental factors, lifestyle, and hormonal influences. Recent scientific reports indicate that Endoplasmic Reticulum (ER) stress constitutes a significant etiological factor in various human diseases, including conditions associated with the development of inflammation and oxidative stress [
3]. Specifically examining the PERK (protein kinase RNA-like ER kinase) cellular signaling pathway, there is promising potential for understanding the pathological mechanisms related to rheumatoid arthritis. Specifically examining the PERK cellular signaling pathway, there is promising potential for understanding the pathological mechanisms related to the mentioned health condition.
It is known that rheumatoid arthritis is a chronic inflammatory disease characterized by abnormal proliferation of synoviocytes, leukocyte infiltration, and angiogenesis. Despite new methods of diagnostics and treatment as well as extensive biological and immunosuppressive treatment, which significantly slow down the course of the disease, the problem of diagnosis and treatment of RA patients is still current and affects a large group of patients. The etiology of RA is not fully understood. It has been suggested that its development is influenced by autoimmune, environmental, and genetic factors.
The endoplasmic reticulum (ER) is the site of biosynthesis for all secreted and membrane proteins. The accumulation of unfolded proteins leads to a condition of ER stress. Eukaryotic cells respond to the accumulation of unfolded proteins in the endoplasmic reticulum by activating the unfolded protein response (UPR) [
4]. One of the transducers of the mammalian UPR is PERK kinase, which upon ER stress causes the global attenuation of protein synthesis mediated by the phosphorylation of eIF2α. However, protein synthesis largely recovers while stress ensues, indicating an adpatation process. A group of genes, termed the ER-adaptosome, was induced transcriptionally and escaped translation repression under chronic ER stress conditions [
5]. Eukaryotic cells with developed ER, express different ER-translated mRNAs than normal cells [
6]. It is suggested that these ER-related features may impair adaptation to chronic stress, inferring the risk of rheumatoid arthritis (RA). Thus, we are going to applied Real-Time qPCR expression of mRNA (the genes of the PERK-UPR) to determine global changes in mRNA translation specific for chronic ER stress conditions in rheumatoid arthritis patients.
3. Discussion
Rheumatoid arthritis (RA) is a chronic autoimmune inflammatory disease of the joints of unknown etiopathogenesis, manifested by an inflammatory process mainly in the small metacarpophalangeal and metatarsophalangeal joints, as well as in the wrists, leading to deformation and permanent damage to the joints, as well as to simultaneous multiorgan changes. Despite new methods of diagnostics and treatment as well as extensive biological and immunosuppressive treatment, which significantly slow down the course of the disease, the problem of diagnosis and treatment of RA patients is still current and affects a large group of patients [
7].
RA is the most common inflammatory joint disease, affecting approximately 0.3-1.5% of the population. The incidence is 0.1-0.5% in adults. The peak incidence occurs in the 4th and 5th decade of life. Women get sick 3 times more often than men, and the age of onset for women is usually over 50. Burdensome family history increases the risk of developing the disease many times over. The attempt to classify RA is made using the ACR / EULAR 2010 criteria. The quality of life in RA patients is assessed using the AIMS-2 scale [
8].
Despite numerous studies, the etiology of RA is not fully understood. It has been suggested that its development is influenced by genetic, autoimmune, and environmental factors. The heredity of rheumatoid arthritis is estimated to be 66%. There is an increased risk of developing rheumatoid arthritis in first-degree relatives. The presence of the disease in a parent increases the risk of its development in a child 2-5 times [
7]. Genome sequencing revealed the association of many genes with an increased risk of disease development [
8].
Rheumatoid arthritis, due to the course of the disease and its scope, as well as treatment costs, is a social disease that requires constant introduction of new therapies tailored to the patient’s needs. Due to the large group of disabled and invalids, there is a need to assess the accuracy of diagnostics and personalized treatment with the use of new targeted molecular drugs. To better understand the etiopathogenesis of the disease and act on the initiating factor of the avalanche of the progressive RA process, it is necessary to continue research at the molecular level so that the applied therapy protects the patient against the advanced effects of the disease [1-3abc].
Most secretory and transmembrane proteins fold and mature in the endoplasmic reticulum (ER). The flux of proteins entering the ER is dynamic and regulated. In demanding states, the protein load in the ER is increased and must be met by the organelle folding capacity. Adaptation to the load requires quality control mechanisms which monitor the levels of unfolded proteins and prevent their accumulation for risks of aggregation. Once these quality control mechanisms are compromised, or when cells undergo an insult that changes the physiology of the secretory pathway, such as viral infection or in response to hypoxia [
9,
10], the balance between folded and unfolded proteins in the ER is tipped, resulting in the accumulation of misfolded proteins, a state referred to as ER stress.
Eukaryotic cells respond to ER stress by activating a signaling pathway coined the Unfolded Protein Response (UPR). The UPR is a collection of signaling pathways which can resolve ER stress by integrating mRNA translation control with the regulation of gene transcription. If ER stress persists despite the activation of these feedback, the UPR will initiate apoptosis [
4]. In mammalian cells the UPR is comprised of three major branches: inositol requiring enzyme 1 (IRE1); double-stranded RNA-activated protein kinase (PKR)- like ER kinase (PERK) and activating transcription factor 6 (ATF6), each termed after the ER-transmembrane sensors which gauge the levels of misfolded proteins in the ER lumen and consequently activate their respective downstream signaling cascades. Briefly, upon ER stress, the ATF6 transmembrane sensor travels from the ER to the Golgi, where it is cleaved in a manner that liberates the N-terminus domain of ATF6 (ATF6(N)). ATF6(N) translocates to the nucleus and functions as a transcription factor. PERK is activated by oligomerization, and once activated, phosphorylates the translation initiation factor eIF2α. This reduces translation initiation, leading to a global decrease in protein synthesis. Paradoxically, eIF2α phosphorylation increases synthesis of select transcripts some of which contain short overlapping open reading frames in their 5′UTR, such as ATF4, a transcription factor that coordinates transcription of genes that determine cell fate following ER stress [
5]. The third UPR sensor, IRE1, is kinase and endonuclease. Once activated it splices the mRNA of the transcription factor XBP1, excising a 26-nucleotide intron. This non-canonical splicing causes a shift in the reading frame, yielding the spliced form of XBP1 (XBP1s) [
11,
12]. XBP1s is a highly potent transcription factor that promotes the level of a large variety of ER chaperones and induces expansion of the ER [
13].
In addition to its roles in development and cell function, the UPR modulates prominent diseases, such as diabetes, liver steatosis, inflammatory bowel disease, cancers and more [
14,
15,
16,
17]. A role for the ER stress in rheumatoid arthritis was initially suggested [
18,
19]. However, the involvement of the UPR in RA patients has turned out to be much more general with important contributions to disease initiation, progression, and response to therapy [
20,
21].
The cellular program controlled by PERK in mouse embryonic fibroblasts (MEFs), represent in normal cells, is widespread and includes many downstream genes regulated by multiple mechanisms [
22]. However, PERK affects its targets in a cell type and physiological context-dependent manner [
23]. For instance, PERK has been found to be essential to the progression of BRAF-mutated melanoma, less of non- BRAF mutated tumors [
24]. PERK regulates cellular redox by directly phosphorylating and activating NRF2 [
25,
26]. PERK interphases with the circadian oscillations by induction of miRNA that represses major circadian genes in a manner that affects Burkitt lymphoma progression [
27]. Hence, it is not surprising that multiple pharmaceutical companies developed high affinity inhibitors of PERK. Glaxo Smith Kline developed GSK2606414 (GSK414); Amgen developed AMG PERK 44; Eli Lilly developed Ly4. In our laboratory we developed specific inhibitors for PERK treatment in neurodegenerative disorders including glaucoma (termed PERKi) [
28,
29,
30,
31]. The intricacies of the transcription and translation program controlled by PERK invites research in different tissue types and in combinations with other drugs to assess its role as an efficacious therapeutic target.
Translation repression by PERK in response to chronic ER stress is reversed by adaptation. The phosphorylation of eIF2α by PERK and by additional kinases attenuates translation initiation by the sequestration of the multi- subunit GEF eIF2B. This results in reduction of the ternary complex of translation initiation and leads to a global repression in protein synthesis [
32]. Since cells cannot survive under prolonged translation repression, homeostatic mechanisms are engaged to gradually restore protein synthesis during chronic ER stress. One such mechanism, described by Hatzoglou et al, is a conversion from the classical and efficient CAP-dependent mRNA translation, driven by eIF4E, to a less favorable mechanism that relies on recruitment of the ribosome by eIF3 [
33]. This adaptation process is dependent on the constant repression of eIF2B activity [
34]. Analysis of the stress specific transcriptome and translatome in MEFs subjected to a prolonged ER stress identified a set of 567 genes that were induced at the level of transcription and were translated under the chronic conditions [
6]. Remarkably, 35 of these genes encode proteins that function in the ER in protein folding, glycosylation, trafficking and degradation. Genes were indentified under ER stress conditions using thapsigargin (Tg:16h vs. Tg:1h). Because the ER protein processing pathway includes in total approximately 141 genes (according to KEGG, PATHWAY: ko04141 in
www.genome.jp), it is of note that a relatively large subset of genes specifically in this category was upregulated during adaptation to ER chronic stress. When PERKi was applied post-establishment of the transcriptional and translational reprogramming, this led to a 50% larger shift in fold changes for ER-translated mRNAs, which was determined by the comparison of polysome-associated to total mRNAs. Consistently, these mRNAs were regulated via both, changes in translation efficiency (61 genes) and mRNA abundance (105 genes). We use the term ER- adaptosome to describe the group of genes congruently induced during chronic ER stress.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that the group of genes congruently up-regulated during chronic ER stress is enriched in those that encode proteins involved in ER functions, including genes listed in reference to PERK-dependent UPR [
6]. ER-adaptosome genes (including 35 genes of the ER protein processing pathway) are known targets of UPR-induced transcription factors, including ATF6, which exhibits protective functions during chronic ER stress and is negatively regulated by PERKi [
35]. One of these factors, the ER transmembrane glycoprotein wolframin, is a regulator of ER calcium levels, which plays a crucial role in ER homeostasis [
36]. Because ATF4 induction requires PERK and is necessary for maximal induction of ATF6 [
35], it is suggested that sustained PERK activity during chronic ER stress maintains ER proteostasis in concert with congruently up-regulated genes. This, in combination with the effects on the ER-localized translation, suggests that chronic ER stress is tailored to maintain ER function by coordinating protein load and processing capacity of the ER. Transcripts that are predominantly translated at the ER [
37] showed congruent decreases in polysome-association and cytosolic mRNA levels during chronic ER stress and in large part reversed by PERKi. Thus, ER-associated mRNA translation is modulated during chronic ER stress in a PERK-dependent manner.
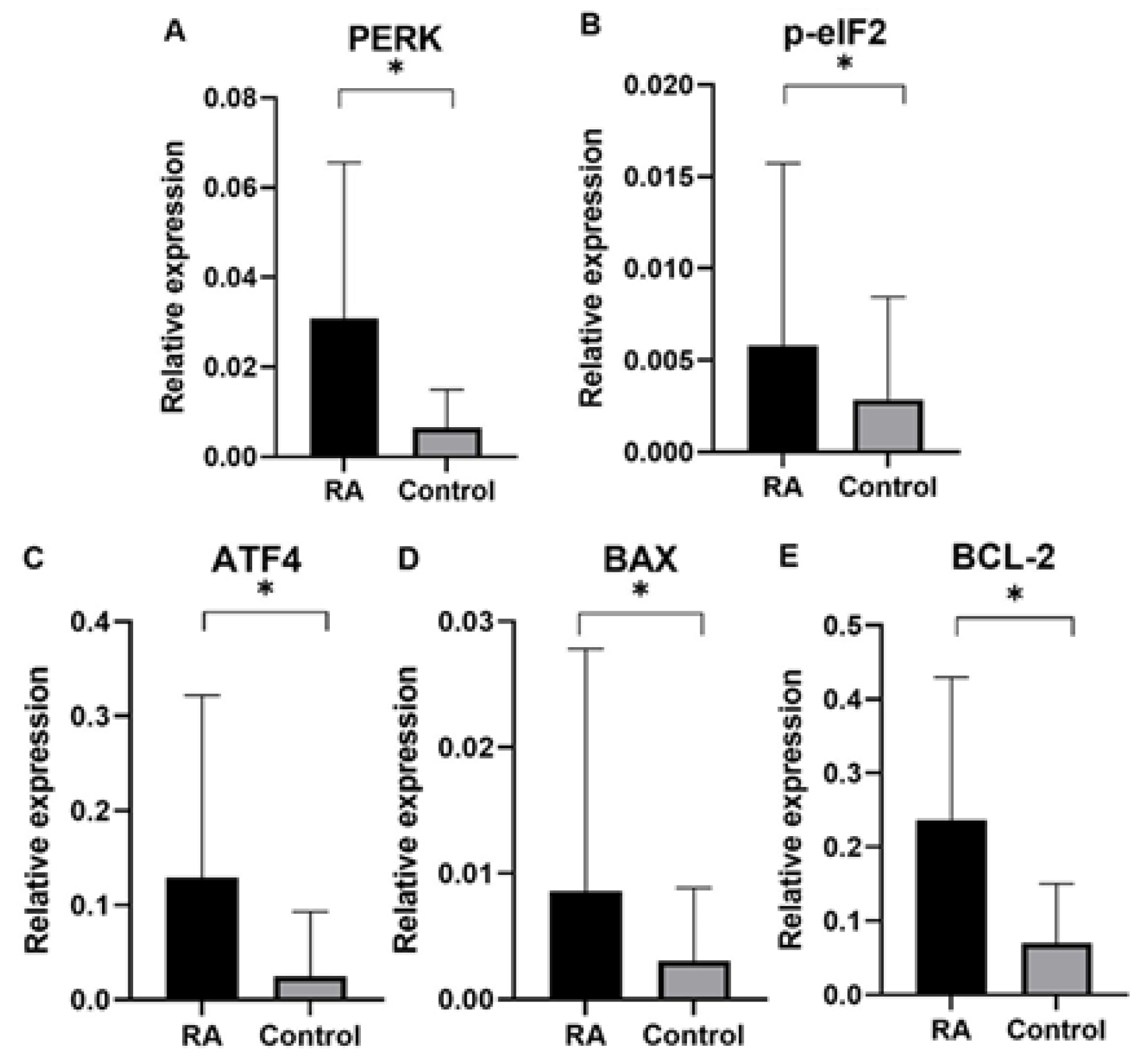
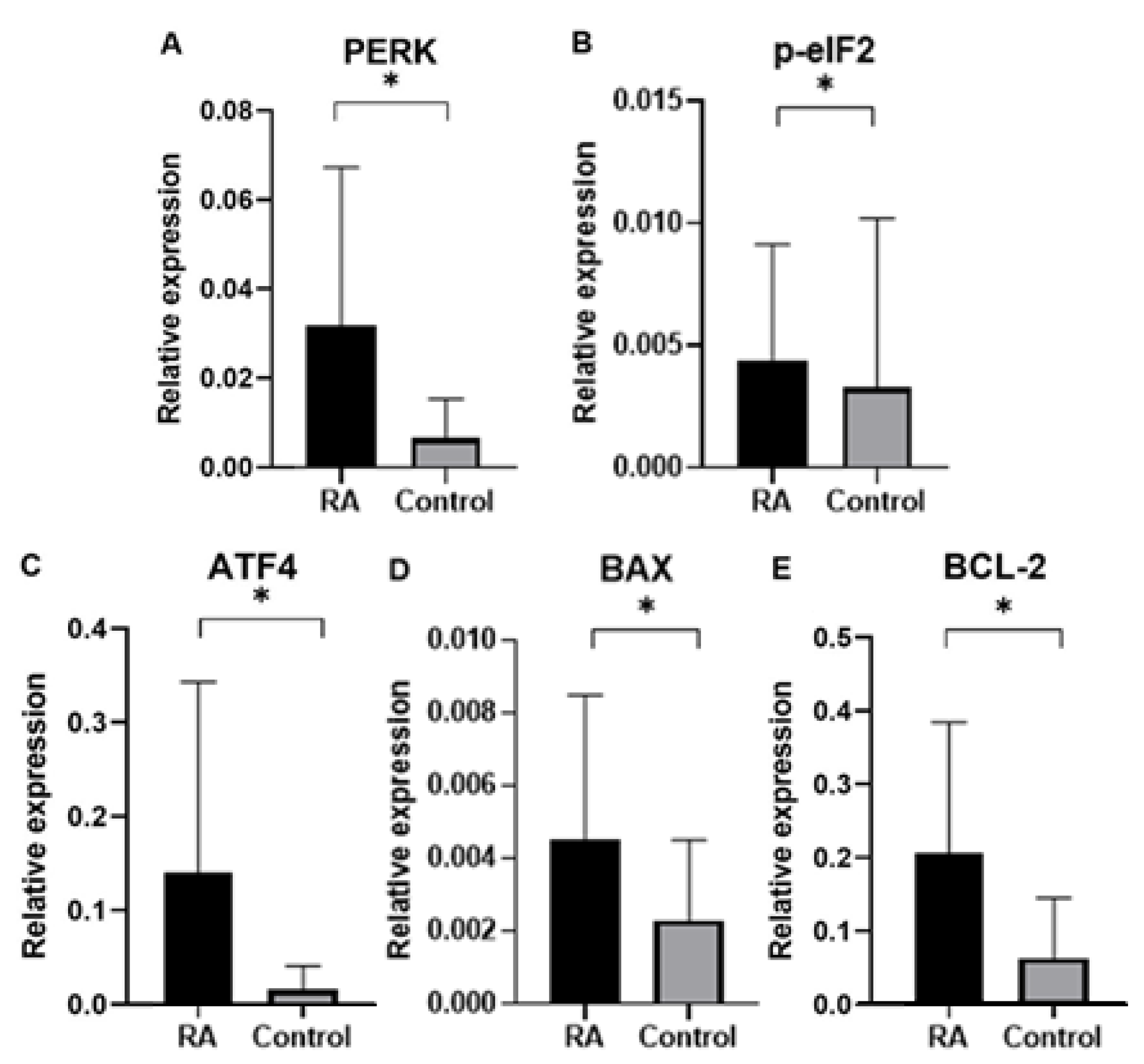
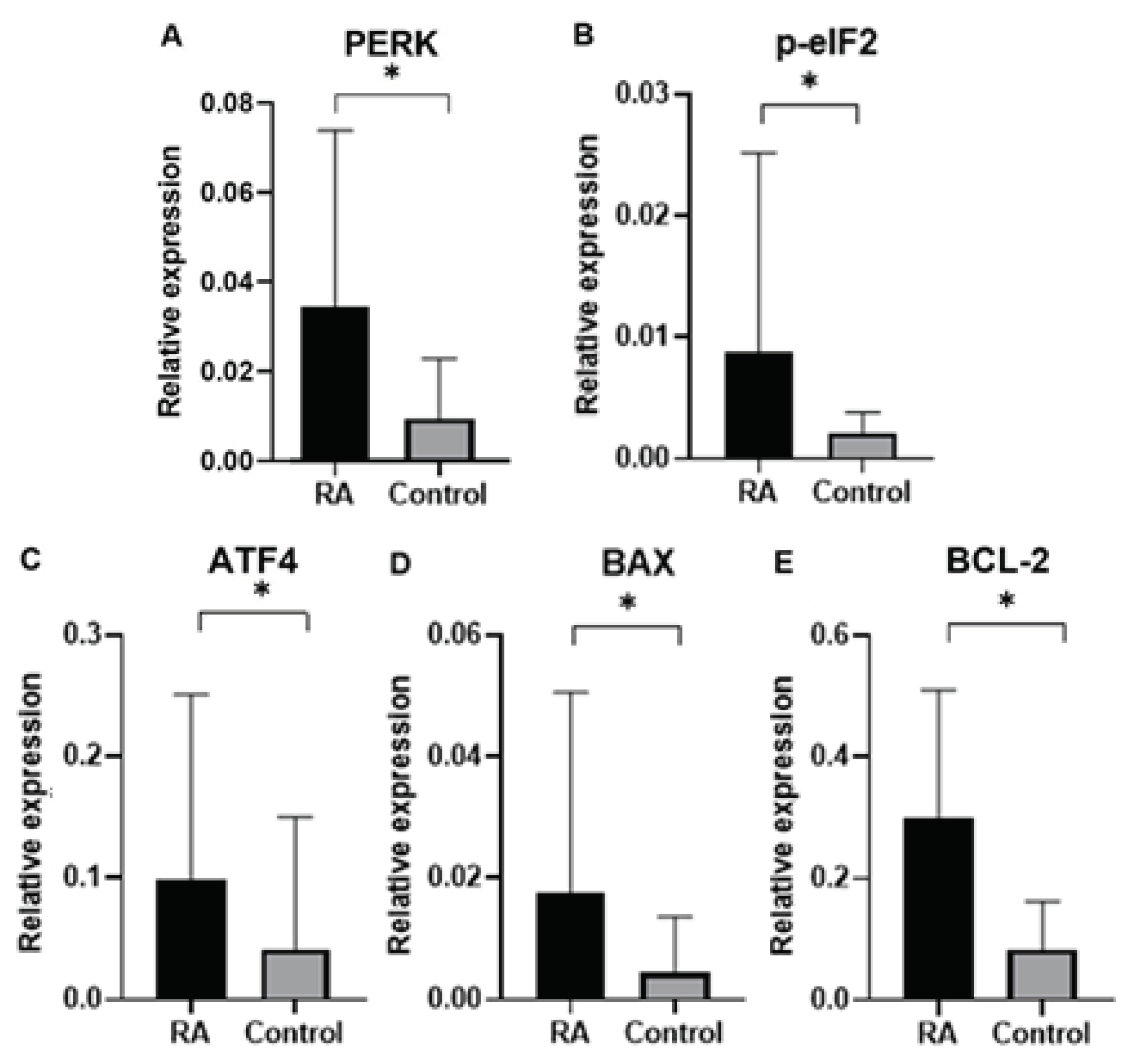
Finally, in our study we analyzed gene expression of ER proteins processing pathway in blood samples of rheumatoid arthritis (RA) patients in comparison to control group subjects. Gene expression analysis was performed for genes of
PERK, BCL-2, p-eIF2, ATF4, BAX as well as endogenous control gene
GAPDH. An expression of ER stress genes of
PERK, BCL-2, p-eIF2, ATF4, BAX were found to be significantly higher in RA patients than in the control group. Interestingly, ER stress genes of
PERK, BCL-2, p-eIF2, ATF4, BAX in patients divided by gender was found higher according to controls in both male and female group, which suggested that UPR-unfolded proteins response is a global process involved in RA pathogenesis. In agreement with the PERK-mediated maintenance of the adaptive state during chronic stress conditions, inclusion of PERKi resulted in the formation of large membrane distensions, associated with perturbation of the ER-adaptosome. These intracellular distensions gradually ballooned, and the foamy cells eventually died in a manner that coincided with the rupture of the vacuoles [
27]. However, in agreement with the idea that the intracellular ballooning is a sign of ER dysfunction and not a programmed cell death, was finding that the vacuoles were reversed back to normal ER structures and perfectly functioning cells if the stress was removed shortly after the appearance of ER distension. It is named ER-dysfunction mediated cell death mechanism, Ballooning Endoplasmic Reticulum cell Death (BERD). Thus, how the adaptation to chronic ER stress is modulated in rheumatoid arthritis cells and whether a failure to adapt can be used for BERD-mediated treatment of patients with rheumatoid arthritis should be the subject of future studies, to determine the unfolded protein response pathway as a modulator of rheumatoid arthritis initiation, progression, and therapy.
4. Materials and Methods
4.1. Patients and Study Specimens
The material was collected from a total 56 rheumatoid arthritis patients, including 39 women and 17 men. The average age among women was 73 years, while among men it was 63 years. Study participants were patients with newly diagnosed rheumatoid arthritis according to ACR/EULAR 2010 guidelines. Positive family history of inflammatory joint diseases was present in 21 women and 10 men. The mean erythrocyte sedimentation rate (ESR) before treatment initiation in the female group was 47 mm/h, whereas in the male group it was 64 mm/h. The mean C-reactive protein (CRP) level before treatment initiation in the female group was 31 mg/l, while among men it was 38 mg/l. The Disease Activity Score (DAS28) was also analyzed, which is commonly used in clinical practice to assess disease activity and joint damage. The score is calculated using a calculator that considers: 1) The number of swollen joints; 2) The number of tender joints (28 joints are considered: wrists, metacarpophalangeal, proximal interphalangeal, elbows, shoulders, and knees); 3) ESR or CRP; 4) The patient’s overall assessment of disease activity on a visual analogue scale (VAS, 0–100).
The possible values on this scale range from 0 to 9.4. This scale allows us to assess disease activity as follows: <2.6 indicates remission; ≤3.2 indicates low activity; 3.2 and ≤5.1 indicate moderate activity; 5.1 is interpreted as high activity. This scale also facilitates an objective assessment of treatment response, where: a good response is defined by a change in activity of ≥1.2 and low activity; a moderate response is defined by a change of >0.6 and <1.2 and low or moderate activity, or in the case of a change of ≥1.2 and high or moderate activity; no response is observed when the change is <0.6 or <1.2 and the activity is high.
The mean DAS28 score before treatment initiation in the female group was 5.76, while in the male group it was 6.23. All patients were treated according to current ACR/EULAR recommendations. Methotrexate was the first-choice drug among disease-modifying antirheumatic drugs (DMARDs) at an initial dose of 10 mg/week. Subsequently, the dose was gradually increased by 5 mg every 2 or 4 weeks to a maximum dose of 25-30 mg/week or to a dose tolerated by the patient. In cases where methotrexate was contraindicated or early intolerance occurred (within 6 weeks of starting treatment), leflunomide or sulfasalazine was used as the first choice DMARD (5 women and 1 man). Short-term oral glucocorticoid (methylprednisolone) therapy was administered to 27 women and 16 men during the initiation or modification of DMARD therapy at various doses, but they were tapered as quickly as the clinical condition of the patients allowed. The mean DAS28 score after 3 months of treatment initiation was 2.69 among women and 2.58 among men, indicating achievement of remission. After treatment initiation, a significant decrease in inflammatory parameters was also observed. In the female group, the mean ESR was 19 mm/h, while in the male group it was 12 mm/h. The mean CRP level among women was 6 mg/l, while among men it was 4 mg/l.
Table 1.
Clinical characteristic of Rheumatoid Arthritis patients.
Table 1.
Clinical characteristic of Rheumatoid Arthritis patients.
| Category |
Mean value |
|
| Number of patients (n) |
56 |
|
| Number of patients treated with methotrexate (MTX) (n) |
50 |
|
| Positive family history (n) |
31 |
|
| Number of patients bridging with GCS (n) |
43 |
|
| Mean ESR before treatment initiation (mm/h) |
55,5 |
|
| Mean ESR after treatment initiation (mm/h) |
15,5 |
|
| Mean CRP before treatment initiation (mg/l) |
34,5 |
|
| Mean CRP after treatment initiation (mg/l) |
5 |
|
| Mean DAS-28 before treatment |
5,99 |
|
| Mean DAS-28 after treatment |
2,63 |
|
Overall, the research encompassed a group of 86 individuals. The study group comprised 56 patients with diagnosed Rheumatoid Arthritis selected from patients of the the Vadimed Medical Center in Krakow, Poland. Concurrently, the control group included 30 volunteers selected from healthy subjects admitted to the Department for other reasons not associated with chronic inflammatory, cancer nor neurodegenerative disorders. The control group was matched to the study group regarding sex and age. Blood samples were collected from both groups for gene expression assessment. The study received approval from the Institutional Bioethics Committee (protocol no. 7/KBL/OIL/2022.). All participants provided written informed consent to participate in the study. Prior to commencing the experiments, all participants underwent comprehensive medical examinations.
4.2. RNA Isolation
Total RNA was extracted from whole blood in sterile environment using a commercially available RiboPure™-Blood Kit (Invitrogen™, ThermoFisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s protocol. The concentration and quality of the extracted RNA were evaluated by spectrophotometric measurement of samples at 260 and 280 nm using a Multiskan SkyHigh Microplate Spectrophotometer (Thermo Scientific™, ThermoFisher Scientific, Waltham, MA, USA).
4.3. Generation of Single-Stranded cDNA
Obtained RNA was used for the quantitative conversion of 100 ng of total RNA into cDNA in single 10 μl reaction using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™, ThermoFisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions. To perform reverse transcription the thermal cycler VeritiPro (Applied Biosystems™, ThermoFisher Scientific, Waltham, MA, USA) was used using the conditions suggested by the manufacturer.
4.4. Quantitative Real-Time PCR (qPCR)
The expression of genes associated with selected PERK signaling pathway in the study and control groups was evaluated by TaqMan technique. For qPCR reactions, 10ng of generated cDNA was used for analysis on the CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Quantification of mRNA expression was performed using the TaqMan™ gene expression assays including TaqMan Universal PCR Master Mix (Applied Biosystems™, ThermoFisher Scientific, Waltham, MA, USA) with the predeveloped TaqMan assays for EIF2A (Assay ID HS00230684_m1), BCL-2 (Assay ID HS00708019_s1), PERK (Assay ID HS00984003_n1), p-eIF2 (Assay ID Hs00909569_g1), BAX (Assay ID Hs00180269_m1) and GAPDH (Assay ID HS02786624_g1). GAPDH expression was used as an endogenous standard. The targeted transcripts were run in triplicate. Real-Time PCR conditions were as universal cycling conditions in accordance with standard protocol. The quantities of selected genes in relation to the housekeeping gene were assessed using the comparative Ct method by Livak.
4.5. Statistical Analysis of Gene Expression
Statistical analyses were performed using Statistica 13.1 software (StatSoft, Tulsa, OK, USA). To evaluate the significance of expression differences between the study and control groups, a non-parametric test, U Mann-Whitney was used. The distribution of variables was examined using the Shapiro–Wilk test. Statistical significance was considered for p-values less than 0.05.