1. Introduction
Microfluidics is the scientific technology that manipulates minute fluid volumes, typically between 10
-9 and 10
-18 liters [
1]. This is made possible by utilizing small channels; these channels’ dimensions range from ten to a couple of hundred micrometers. Microchannels of this size have a large relative surface area and, consequentially, high mass transfer. Other inherent benefits of microfluidics include faster mixing rates, increased control of volumes, and other properties such as low reagent use and prompt response [
2,
3]. Microfluidics has been impacting biological research for some time, having been integrated into processes for sample preparation and different cell culture applications [
4].
Organ-on-a-chip technology utilizes microfluidics to simulate
in vivo research while maintaining the controlled parameters
in vitro research provides. Standard cell biology research relies on
in vitro research, that is, research performed in a test tube, culture dish, or elsewhere outside the living organism. OOC technology aims to overcome these challenges of
in vivo and
in vitro research [
2].
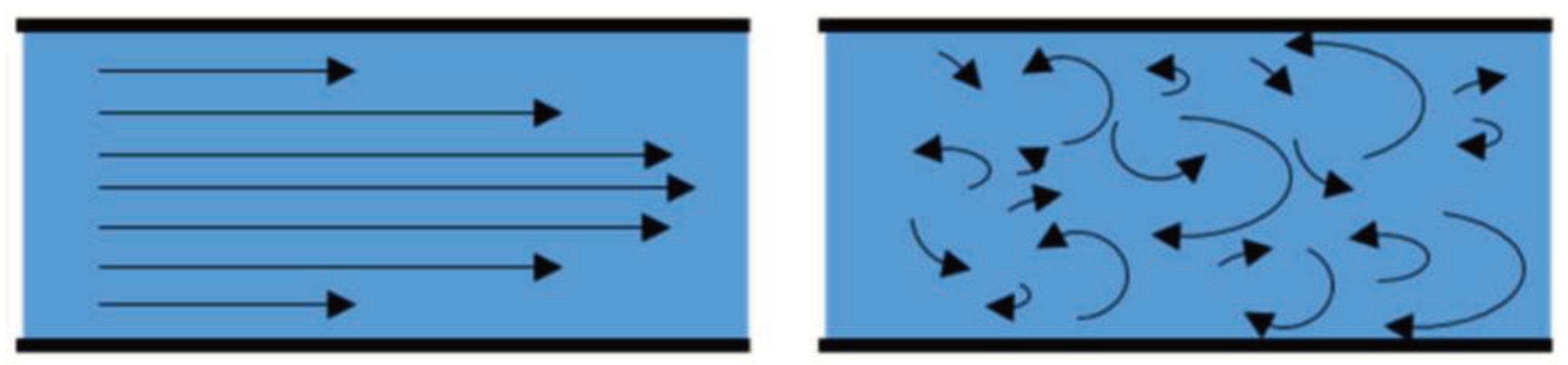
OOC technology utilizes low sample volumes, ranging anywhere from microliters to femtoliters. Such small volumes of liquid drastically reduce its Reynolds Number. Reynolds Number is a dimensionless quality used to predict fluid flow patterns under various flow situations. A low Reynolds Number indicates laminar flow, whereas a high Reynolds Number indicates turbulent flow, as shown in
Figure 1. A liquid under turbulent flow conditions follows no defined flow pattern; flow lines are seen intersecting and even moving counter to the overall direction of the flow in eddy currents. Contrarily, a fluid under laminar conditions has a streamlined flow pattern, allowing it to move more efficiently through the microchannels. It has been widely observed and proven that fluids flowing in the microchannel are often laminar, while turbulent flow is found only in macroscale processes [
5]. In cell culture, the Reynolds number is important as it can affect the mass transfer and shear stress that cells experience [
6]. Cells lacking a cell wall and not being evolutionarily adapted to life exposed to a free-flowing liquid phase are more sensitive to hydrodynamic forces in their environment than fungi or bacteria.
Microfluidics has significant advantages over working on a macro scale, including increased efficiency, lower cost, and increased control of volume and other experimental parameters [
7]. OOC technology utilizes microfluidics to provide a state-of-the-art platform for biological research that combines the advantages of
in vivo and
in vitro research [
8].
2. Methods
Microfluidics was first implemented in gas chromatography systems and printing machines to perform fast, precise analysis on small volumes [
9]. Later, microelectronic lithography was adapted to construct devices from biocompatible materials for medical purposes. Since then, increasing interaction between microelectromechanical systems (MEMS) and microfluidics has led to novel fluidic device fabrication method that uses photo-sensible polymers to produce molds using UV light, which is then used to cast a pattern onto a biocompatible polymer [
10]. The growing utilization of microfluidics in technology led to the development of many other fabrication processes using novel materials. Replica molding, injection molding, and microprinting are a few techniques used to create microfluidic platforms [
11,
12,
13]. Polydimethylsiloxane remains the standard material used in fabrication due to its transparency, flexibility, permeability, and biocompatibility.
Recently, three-dimensional (3D) bioprinting technology has revolutionized the printing of microfluidic devices. It can integrate channels and connections in organ-on-a-chip models [
14]. 3D printing technology reduces the fabrication process of microfluidic devices to a shorter time, making it less cumbersome. It also has considerable advantages over traditional fabrication processes. These include the ability to incorporate multiple biomaterials, including living cells, controlled porosity of tissue scaffolding, and record accuracy. This technique can be divided into nozzle-based and optical-based methods [
14]. Nozzle-based methods involve the use of a nozzle that applies the ink onto a substrate, typically with adequate pressure to ensure ink transfer, while optical-based 3D printing can be either direct printing or transferring the ink onto a receiving substrate through ejection or by utilizing light-material interactions to crosslink the ink [
14]. 3D printing technology utilizes various starting materials, including photocurable resin, photopolymers, thermoplastic polymers, and hydrogels [
15]. PDMS remains the preferred choice in microfluidics, but other potential materials are continuously developing for various biological research applications. Some of these materials are tabulated below in
Table 1.
To choose the best material according to the organ-on-a-chip application, one should consider surface properties such as surface roughness and wettability, which affect cell adhesion and migration, and mechanical properties such as stiffness, which influence the later stages of cell growth [
45]. Analyses such as tensile strength, compressive stress, and wettability are commonly conducted to assess the suitability of polymer properties for the intended application [
45]. For the fabrication of porous-compartment-separation membranes, synthetic polymers like polydimethylsiloxane (PDMS), polycarbonate (PC), polyethylene terephthalate (PET), aliphatic polyesters, polyurethanes, among others, have been utilized due to their ability to adjust porosity, surface roughness, and mechanical properties easily. In contrast, natural polymers such as collagen, gelatin, or polysaccharides have gained recent attention for their enhanced biocompatibility and replication of native tissues due to facilitation in the creation of channel interconnections, enabling the perfusion of oxygen and nutrients to mimic the natural behaviors of cell differentiation, spreading, and adhesion [
45].
2.1. The History of Organ-on-a-Chip
Huh et al. [
46] published the first "lung-on-a-chip" article in 2010, introducing the idea of "organ-on-a-chip" as microfluidic in vitro cell culture systems that mimic the physiology of the basic functional units of an organ. A public-private partnership assigned Ingber's lab to develop ten human organs-on-chips two years later. The business has created liver and intestine models in addition to a lung chip, according to Hamilton, and is currently working on the next generation, which will include brain, kidney, and skin chips [
47]. Organ-on-a-chip technology is growing in popularity for biological research due to the high expectations for reproducing essential elements of the human physiological environment and the ability to replace animal models. Organs-on-chip technology could speed up the time it takes for therapeutic compounds to enter clinical trials by reducing the reliance on animal testing, which is now the primary method used to evaluate the kinetics, efficacy, and safety of medication candidates. Also, studying pharmacokinetics in particular persons, i.e., personalized, is possible by cultivating human cells on chips [
48].
2.2. Surface Treatment of OOC Devices
To ensure biocompatibility or improve cell adherence, microfluidic device surfaces that are in contact with cells must be treated. Treatment with pluronic acid is often used in 3D spheroid or organoid cultures to passivate the chip surface and avoid unwanted spheroid or organoid breakup through possible cell attachment. This is critical because the loss of 3D tissue architecture may result in a loss of physiological organ function. On the other hand, protein and extracellular matrix (ECM) coatings may be employed to increase cell adhesion to the chip substrate. These coatings are required to produce connected, confluent monolayers of cells, which mimic the intestinal epithelium in the gut or the endothelium in the blood-brain barrier (BBB) OOC [
49,
50,
51]. Tissue-specific and disease-specific matrices may be utilized to produce higher-fidelity patho physiological models, such as generating crypt-like formations in gut-on-a-chip systems [
52]. They may be basic biomatrices formed from fibrin or collagen or more complicated biomatrices like matrigel. The liquid form of these biomatrices may be combined with cells before being fed into the OOC device and allowed to polymerize into a gel. Biomatrices may act as a helpful framework for cells to remodel into a 3D structure since certain organs, like the liver and skin, need a 3D microenvironment to achieve physiological function. Additionally, biomatrices could be necessary for the upkeep or differentiation of certain cell types. For instance, it has been shown that matrigel encourages neural stem cells to maintain and differentiate their neurons, while myogenic differentiation was increased when myoblasts were cultured on a suspended overhanging fibrin-based gel [
53,
54].
Another method, micropatterning, is a powerful tool to control the spatial organization of cells and tissues to mimic the in vivo microenvironment closely. A recent study by Fang et al. demonstrated the potential of micropatterning technology to study tubulogenesis in epithelial organs and the differentiation of mesenchymal stem cells [
55]. Moreover, Nguyen et al. quantified the electrical interaction between cardiomyocytes and other cells in micropatterned cell pairs [
56]. These two studies showed the diverse application of micropatterning in OOC systems.
3. Supporting Life inside the Device
3.1. Cell Culture Media Selection
It is also possible to use culture media designed for traditional cultures for a single-OOC platform involving just one kind of cell. When developing single or multi-OOC platforms containing numerous cell types, each with a different nutritional demand, a new level of complexity for media selection is introduced. Each unique cell population must retain its viability and functional phenotype in the optimum co-culture medium. This condition must be met before assessing the appropriateness of the media for supporting downstream analysis without unwanted interference. Many research groups have employed combinations of the original culture media used for each distinct cell type to optimize a suitable co-culture medium for OOC applications, with mostly positive results. These include multicellular OOC models simulating adipose tissue and multi-organ OOC models simulating liver-kidney and liver-adipose-skin-lung interactions. However, optimizing a good co-culture media becomes increasingly difficult when more diverse cell types are co-cultured. Through the usage of permeable membranes, cells can be divided into distinct compartments without the need for a common co-culture medium. This allows for the delivery of cell-specific media to each compartment while allowing paracrine interactions between the compartments, as demonstrated by OOC devices that mimic the skin [
57] and liver [
58]. Since sera are a significant source of variance and may affect tests, a serum-free medium should be considered when practical. Starting with the medium that is most often used for non-OOC work with the same cells, such as minimum essential medium (MEM)-based medium for epithelial cell cultures or endothelial cell growth medium (ECGM) for endothelial cells, media should be improved in research with the OOC [
59].
3.2. Cell Microenvironment Control
The notion that cells may be manipulated by their immediate surroundings is well-established in cell biology. Over-the-chamber (OoC) systems have been widely used to manipulate various environmental factors (including shear stress, autocrine/paracrine soluble factors, cell-cell, and cell-ECM interactions) at physiologically relevant lengths and timescales to elucidate their effects on cell phenotypes and functions. This concept is best exemplified in the context of stem cell and tumor microenvironments, as seen by the large number of reviews covering these areas [
60]. OoC systems provide a more precise method to study the effects of paracrine and autocrine signaling than conventional techniques, such as conditioned medium or changing cell density, because soluble biochemical factors can be controlled in OoC systems by manipulating fluid mass transport properties at microscale resolution. A switch may be made between diffusion-dominated transport, in which secreted factors stay close to cells for receptor binding, and convection-dominated transport, in which secreted factors are carried away from cells [
61]. OoC devices are useful for stimulating cells in various physical environments. The shear stresses produced by the body's fluids, such as blood and interstitial fluids, are often mimicked with OoC systems since fluid flow is an intrinsic part of many of these systems. One may create multiplexed channels with variable geometries to simultaneously apply shear stress across a range of magnitudes to investigate the impact of shear stress on cell proliferation, differentiation, and function. In contrast, fluid shear promotes microvascular development and function in various organotypic tissues. Mechanical tensile or compressive pressures may be applied to cells using OoC devices to simulate the stretching of the airway during breathing, the movements of the digestive system during peristalsis, or the contractility of cardiac tissues. To accomplish this, OoC devices often feature thin, flexible PDMS membranes stretched cyclically by vacuum or pressure pneumatic actuators, allowing cells to be grown in a fluidic environment. OoC devices have included in situ microfabricated microelectrode array as part of the device or electrodes placed into the OoC system to better mimic the electrophysiological activities of neuronal and cardiac tissues via electrical stimulation [
62,
63].
3D culture, multi-cell type co-culture, and tri-culture systems aided researchers in creating various in vitro complex models to mimic in vivo environments and provide valuable information. In the context of cell culture, adding a third dimension requires knowledge about the design of the scaffolds to support the cell organization or the utilization of bioreactors to control the nutrient/waste exchange [
64]. 3D cell culture is widely used in drug screening and tissue engineering [
64].
Huh et al. studied the importance of 3D cell culture, which enhances cell differentiation and tissue organization compared to 2D models [
65]. Other groups underscored the significance of multi-cell-type co-culture to produce multi-organoid-on-chip-a-chip to design human models with better physiological mimicry [
66,
67]. The study by Wagner et al. showcased the interaction of natural and drug-induced liver-testis model derived from primary adult testicular cells and liver spheroids consisting of cultured HepaRG cells and hepatic stellate cells to demonstrate the implications of multi-cell-type co-culture [
68].
3.3. Post-Experiment Analysis
Intermittent simultaneous measurements of different cell parameters are ideal for profiling OOC homeostasis and response to externally applied chemical or physical stressors. Some standard techniques include multiplexed (bead-based) protein-binding/DNA-binding assays or offline high-performance liquid chromatography/mass spectrometry (HPLC/MS), which enable simultaneous measurement of various chemicals, many being responsive biomarkers [
69]. Both offline methods may be carried out using microliter sample quantities, consistent with the typical flow rates used in one-pass perfusion, or by periodically removing recirculating medium circuits. For some biomarkers (such as albumin, lactate dehydrogenase, alanine aminotransferase, aspartate aminotransferase, and urea for liver systems), different commercial assay kits can be used as an alternative; however, any potential interference from medium components (particularly serum) must be carefully considered. The number of measurements that can be done with these kits in extremely low-volume systems is somewhat restricted, although modern microarray methods enable the use of very low volumes (<5 µL) and measurement parallelization (cell health assays) [
70]. Supernatant measurements, either online or in-person, may provide measurements of nutrients and metabolites with little to no time lag. Tiny sensors that rely on optical detection using dyes are often utilized [
71]. A thorough study of in situ dissolved oxygen measurements in OOC was conducted by Rivera et al. [
72]. A crucial component of metabolism is dissolved oxygen, particularly in the action of the liver's CYP450 enzyme [
73]. Microprobes or pH-sensitive patches may be used to monitor pH and dissolved oxygen. Microscopy or optical fibers are required for optical detection and automated solutions [
74]. Biosensors play a crucial role in organ-on-a-chip systems by enabling real-time monitoring of various biological parameters, which is essential for understanding cellular responses and evaluating the performance of these platforms [
75]. Electrochemical enzyme-based biosensors may be used to test specific analytes, including lactate and glucose. These online, almost continuous observations are preferred since periodic measurements could exclude significant response variations. Analyte concentrations may be spatially resolved if sensors or patches are incorporated as arrays at close range or in direct contact with tissue structures [
76,
77].
Visual examination is the most popular way to test the functioning of cells and tissues in OOC devices since microscopy and high-content imaging are two of the most widely used analytical techniques in cell biology. Most OOC devices can be built to fit inside the imaging depth of common epifluorescence or confocal microscopes since they are made of optically transparent materials, including glass, PDMS, and thermoplastics. Thus, by using the organ-on-a-chip’s microfluidic capability to supply the required chemicals, it is possible to label the cells and tissues on-chip using fluorescent dyes or antibodies to evaluate cell viability or the expression of certain biomarkers [
78]. In recent years, imaging sample preparation processes (cleaning, staining, etc.) and analysis have evolved and been modified to cope with complex, 3D multicellular structures. These techniques include confocal microscopy and light-sheet microscopy. High-content imaging can provide spatial and temporal information on the morphology of cells and tissues, which may be used to evaluate innovative treatments and aid in understanding disease pathogenesis more effectively. An optical window to the cells and a design specification that permits positioning and alignment of the OOC on a microscope stage are necessary for visualizing the OOC systems. The tissue model, preparation techniques (fixing, staining, and clearing), and microscopy must be perfectly matched to the methodology for image analysis and data processing to be successful [
79].
Other ways to examine cell functionality are measuring cells’ force and electrical activity. Cantilevers detect the force generated by cells, whereas microelectrode arrays (MEAs) assess electrical activity in cells and tissues. Compared to the patch clamp, the MEA system is well suited for use with OOCs. Using the heart model as an example, conduction velocity, beat frequency, and field potential length estimations can be made using a 2D array of cells, making this approach a good approximation of the QT interval, the period from ventricular depolarization to ventricular repolarization, which can be measured by electrocardiography as well. Several systems, including the cardiovascular, neuromuscular, skeletal-muscle contraction, and nervous systems, may benefit from MEAs [
80]. In particular, micro tunnels have been used to characterize the propagation of action potentials from presynaptic to postsynaptic chambers [
81]. Several organ activities would not be possible without mechanical forces, such as the formation of contractile force by cells or tissues like the lung and heart. Cells are etched onto silicon cantilevers, and their deflection may be monitored optically or electronically in OOC devices, allowing for non-destructive, near-real-time measurements. A TEER-MEA (Transepithelial Electrical Resistance) cardiac chip has been shown to be capable of multifunctional measurement due to the integration of MEAs and electrodes for TEER measurements during chip manufacture [
82]. Integrity and permeability of any barrier tissue may be evaluated by transepithelial electrical resistance (TEER) (such as the gastrointestinal tract, the kidney, and the BBB). TEER levels are linked to the function of several native barrier tissues in the body. The BBB is the most impermeable barrier, with values of 1,500 - 8,000 cm
2, in contrast to the proximal tube in the kidney, which has a value of around 70 cm
2. Values shown in models of organs and tissues should match biologically (in vitro). Tissue models may sometimes provide far higher results than physiological values, while results might be significantly lower in other circumstances. In these cases, the completeness of the model is crucial, particularly when the physiological system is composed of several cell types, but the model only employs a subset of them [
83,
84].
4. Standardization
In the context of OOC platforms, standardization involves consistent protocols and benchmarks to ensure the reproducibility and reliability of OOC experiments. There are several standardized test methods and requirements, particularly for sterilization, the safety of pumping systems, and the characterization of materials. There are many standards on components commonly used during the design and prototyping phases (such as needles, connections, and syringes) and on compatibility (such as microplate geometry and pitch-spacing) that are already available from the medical device, plastics, and in vitro diagnostic industries [
85].
4.1. Radiation-Based Health Risk Assessment and Limitation
Individual radiobiological effects are essential to research as they cause DNA damage, cell killing, and point mutations, and they depend on age, gender, and genetic background. By knowing this factor, researchers can optimize radiotherapy or radiation diagnostics doses to lessen malignant neoplasm risk. OOCs enable the culture of primary cells originating from persons to perform studies of personal damage response. OOCs could be utilized to develop personal chemotherapies and understand how low-dose radiation generates bio-effects. Technically, judging radiation damage on the chip and correlating cellular responses to real organs is crucial. Biomarkers, genetic analyses, and physiological responses can be used to analyze radiation damage [
86].
OOCs are currently not at the stage where they can study long-term consequences, such as disease progression. Typical organ-on-a-chip cell culture durations are weeks, which is insufficient to examine low-dose radiation-induced chronic illnesses. Chronic disorders generated by low-dose radiation, like heart disease and neoplasms, should not be studied on organ-on-a-chip systems. Low-dose radiation may cause cognitive damage, which cannot be studied in organ-on-a-chip systems. Radiation may be used to evaluate neural activity. Organ-on-a-chip technology has demonstrated successful utilization across different research fields. We firmly believe that its capabilities, including emulating human organ functions, integrating with microfluidics, and compatibility with diverse endpoint assays, can offer exceptional and groundbreaking tools for radiobiology investigations. This, in turn, lays the foundation for advancing precision radiobiology studies in the future [
86,
87,
88].
5. Organ-on-a-Chip
OOC technology can simulate various organs, including the kidneys, lungs, heart, and liver, which are discussed below. All OOCs utilize microfluidics, biocompatible materials, and sensing components. These sensing components can include an automated imaging system, an embedded output sensing component, or various other microsensors [
2].
5.1. Kidney-on-a-Chip
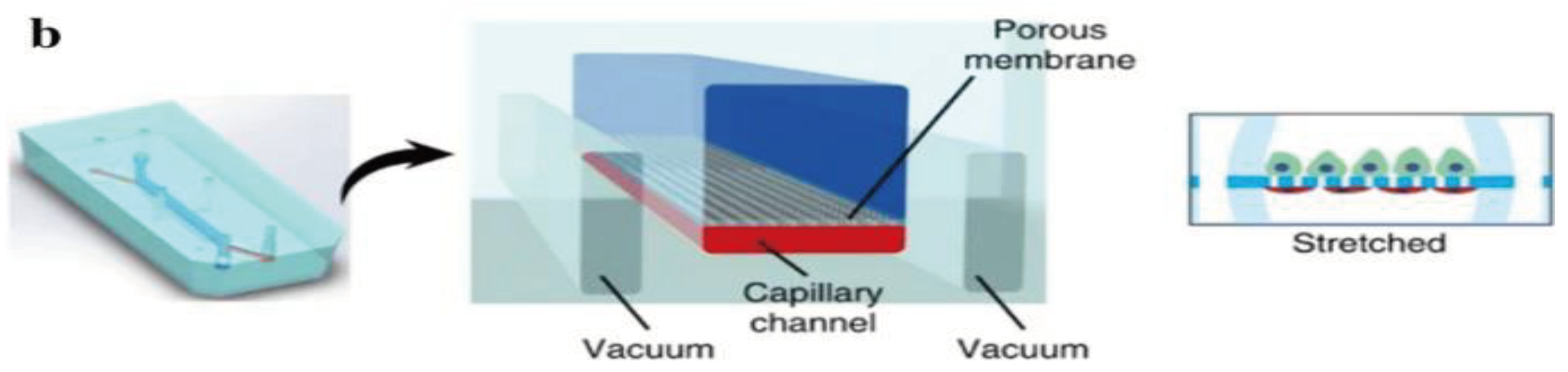
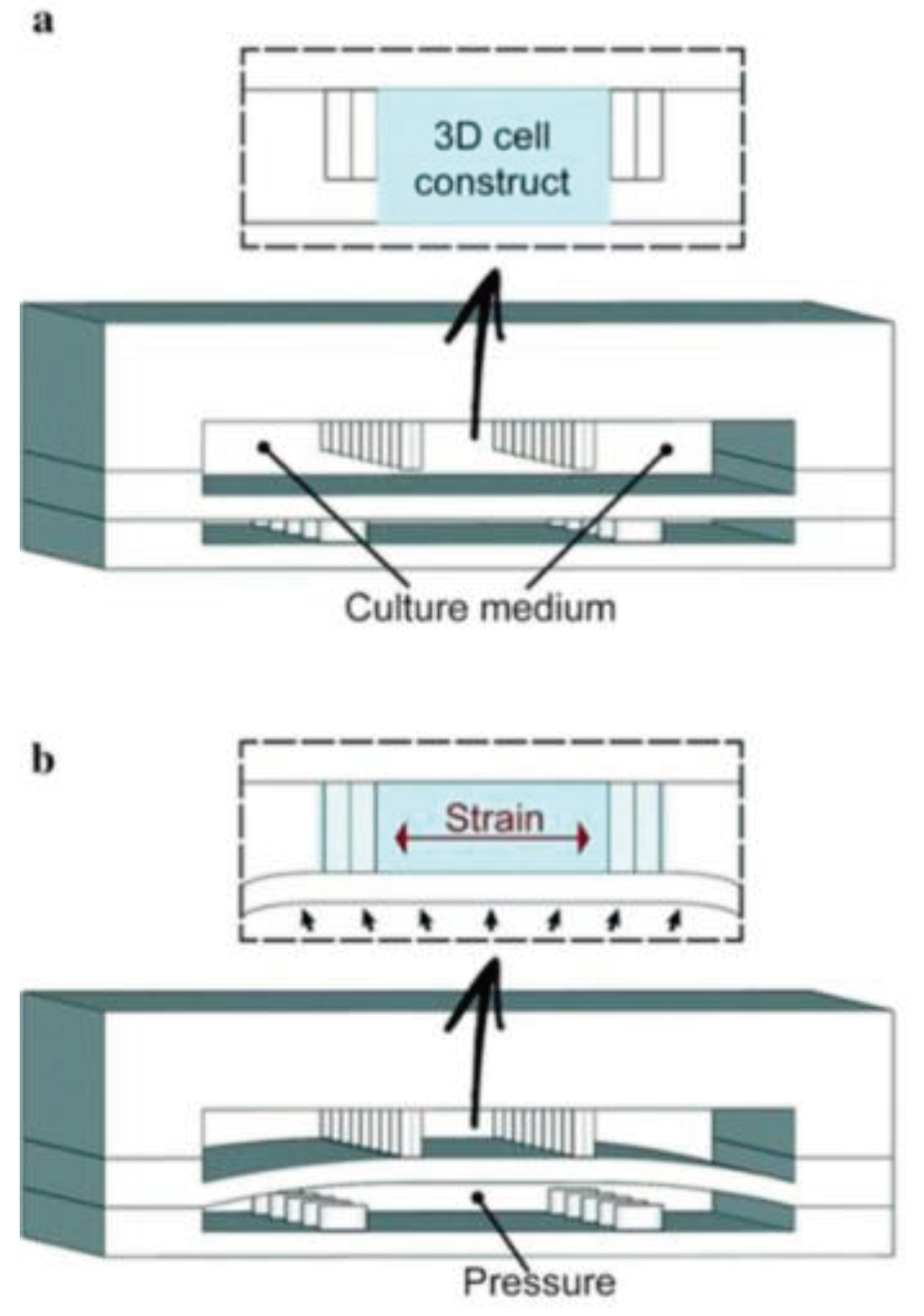
The kidneys remove waste and extra water from the blood as urine and help keep the chemical balance of sodium, potassium, and calcium in the body. They also produce hormones that help control blood pressure and stimulate the bone marrow to make red blood cells. Jang et al.'s [
89] group was the first to create a multilayered microfluidic system that simulated renal filtration using mouse kidney cells. The same device was later used to culture human kidney cells; this was among the first toxicity studies of primary kidney epithelial cells [
90]. Musah et al. [
91] created a kidney-on-a-chip that mimicked the structure and function of the capillary wall, as seen in
Figure 2. Mechanical pressure to the cell layer via vacuum chambers on either side can be produced. Many other studies revolve around disease modeling, drug screening, and toxicology assessment. Kim et al. tested the nephrotoxicity of gentamycin in two different pharmacokinetic profiles using kidney-on-a-chip. They found that continuous infusion has less nephrotoxicity compared to bolus injection. They also suggested using microfluidic cell culture models for further pharmacokinetic and toxicity studies [
92].
5.2. Lung-on-a-Chip
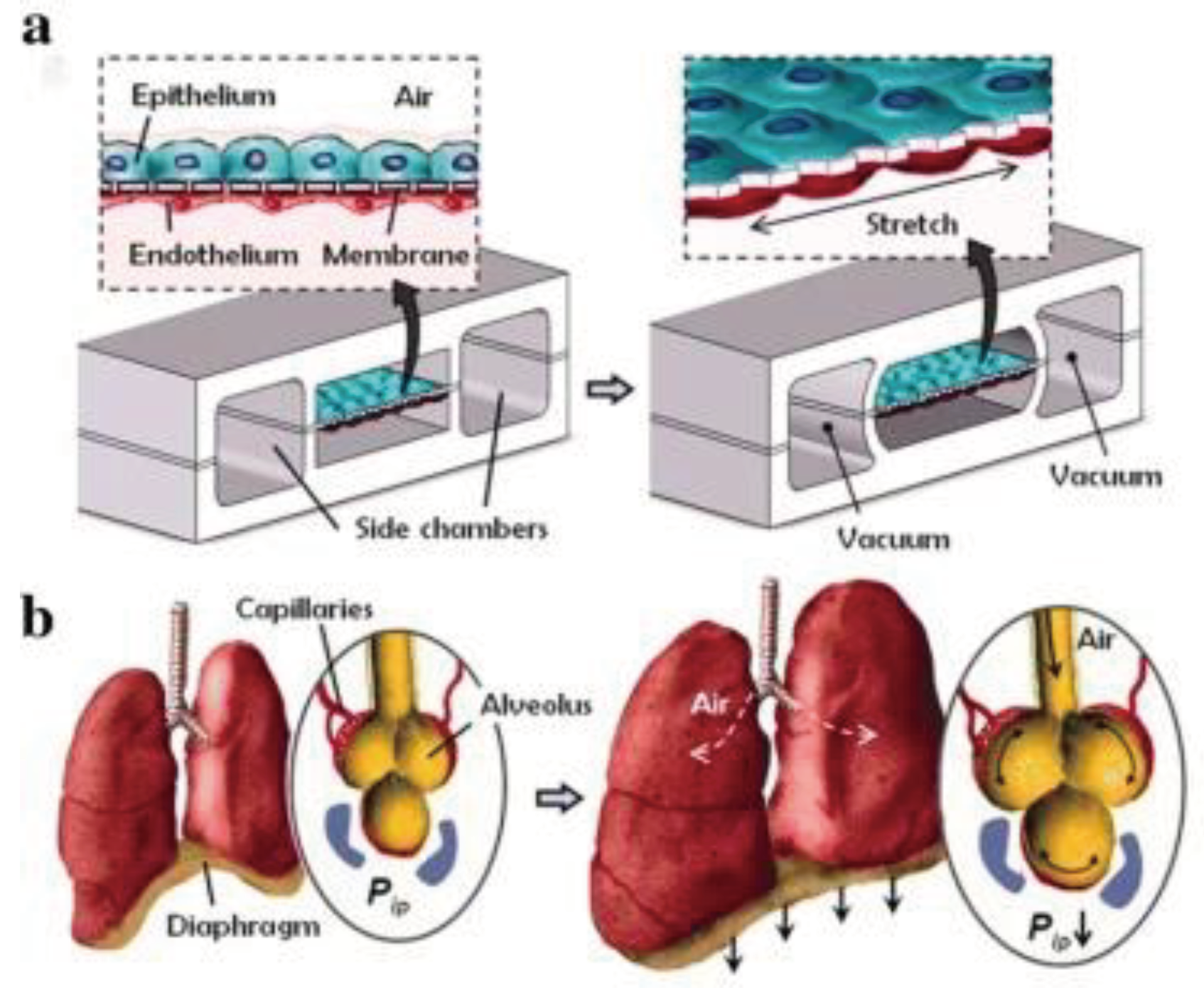
Gas exchange in the human lungs is regulated by alveoli, tiny sacs that allow for the blood exchange of oxygen and carbon dioxide during breathing. This process has been challenging to reproduce
in vitro. However, OOC technology has allowed researchers to create accurate lung microenvironments that sustain consistent gaseous exchange. Current studies focus on regulating mechanical airway pressure and the blood-brain barrier [
93]. Huh et al. [
87] created a lung microenvironment separated into regions, as shown in
Figure 3. The upper region contained alveolar cells, and the lower region had pulmonary cells; this simulated the alveolar-capillary barrier. The system was operated in a vacuum to simulate the expansion and contraction of alveoli during respiration. Inflammatory stimuli were also introduced to the system as neutrophils through the fluidic channels.
Blume et al. [
94] designed a chip that simulated pulmonary interstitial flow by utilizing a permeable filter and a single tissue culture chamber. Multiple chambers could be combined for improved integration. Pressure could be applied to the alveoli and attached capillaries, providing a shear flow profile and realistically simulating a lung microenvironment. Lung-on-a-chips are also useful as respiratory assistance devices. Another study on lung-assist devices allowed additional gas exchange in the placenta for unborn infants in the case of respiratory failure [
95]. Dabaghi and others [
96] utilized double-sided gas delivery on their microfluidic blood oxygenators to improve gas exchange. Oxygen uptake on the double-sided device increased by 343% compared to single-sided devices.
5.3. Heart-on-a-Chip
Microfluidics has enabled novel research on cardiac tissue. The myocardium is the heart's muscle tissue; the anatomical unit of the myocardium is cardiomyocytes (CMs). Grosberg et al. [
97] used a PDMS chip to implant rat CMs to form muscle membranes on a membrane. When the CMs contracted, the membrane curled, and the degree of the curl was measured to analyze cell contraction capabilities. A platform was designed by Marsane et al. [
98] that mimicked the physiological and mechanical microenvironment of the heart using CMs, as seen on the right in
Figure 4. The lower compartment is pressurized to deform the separating membrane and to compress the 3D structure, simulating the heart pumping. Other studies used CMs to directly assess the effects of various drugs related to the rate of the heart pumping [
99]. There have been multiple advances in drug development because of drug trials in cardiotoxicity performed on heart-on-a-chips.
5.4. Artery-on-a-Chip
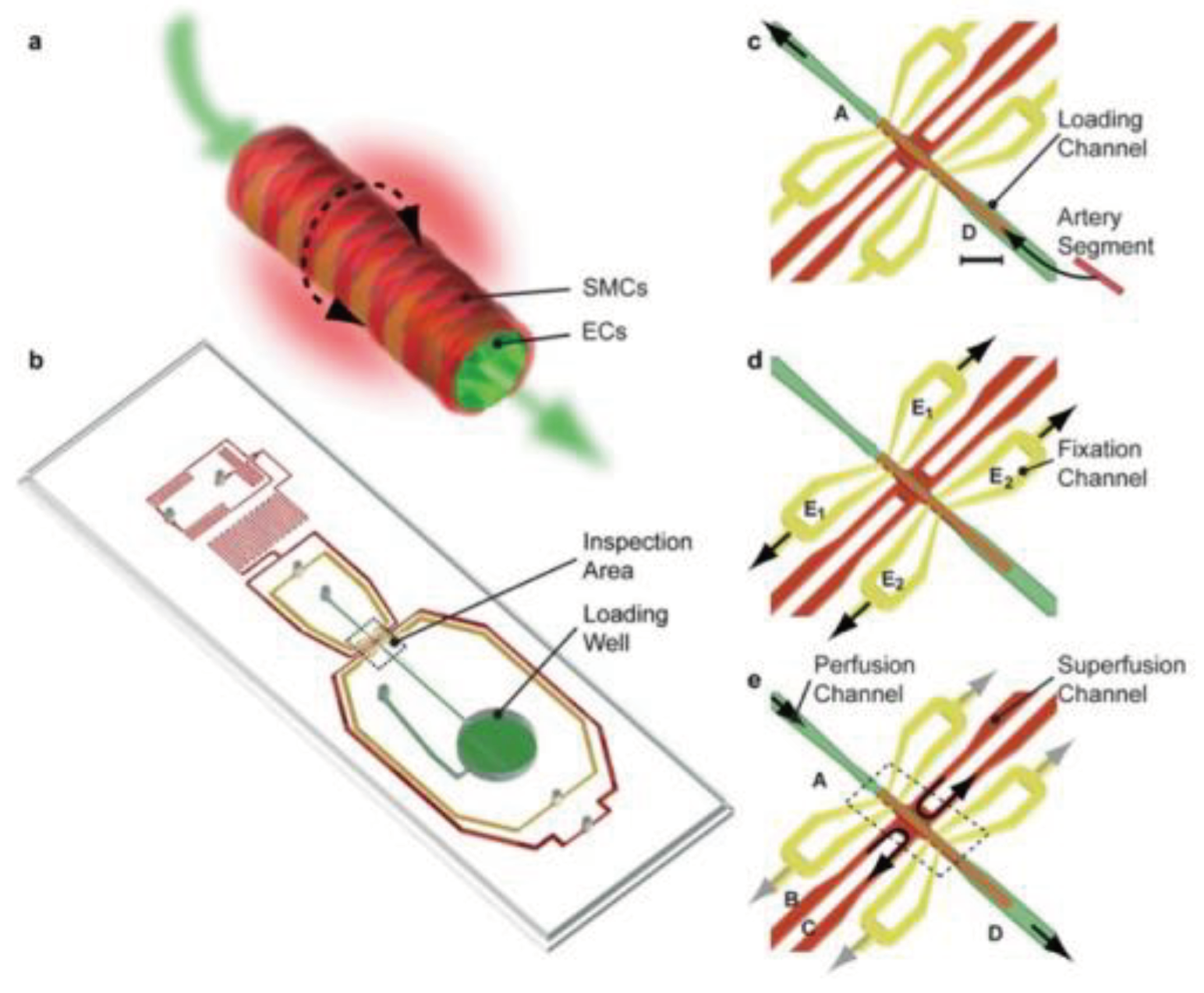
High blood pressure causes heart disease, stroke, and other conditions. Tiny artery pathologies cause and sustain cardiovascular disease. Analyzing tissue slices, single and co-cultured cell populations and bioengineered tissues with microfluidics can reveal underlying mechanisms. Better treatment procedures require scalable ways to study intact cardiovascular tissues in health and sickness [
100]. Most existing technologies perfuse small arteries with glass micropipettes, requiring skilled expertise and often not scalable. Günther et al. [
101] created a scalable organ-based microfluidic platform to load, position, fix, perfuse, and superfuse a frail resistance artery segment. Resistance arteries regulate organ blood flow and redistribution. Endothelial cells (ECs) and smooth muscle cells (SMCs) make up the terminal of the arterial vascular tree. ECs can produce vasoconstricting and vasodilating chemicals to control vascular tone. The platform featured a loading space, a microchannel network, and an inspection area. A thermoelectric heater and thermo resistor maintained the artery inspection region at 37 °C. Yellow microchannels were used for arterial fixation and perfusion, and red microchannels were used for superfusion, as shown in
Figure 5. A segment of the resistance artery was immersed in the loading well to begin the artery loading process. Fixation and superfusion inlet and outlet lines remained closed. The loading well remained open, and microchannel A was connected to a syringe pump dispensing a constant flow of buffer, transporting the artery segment to its dedicated position in the inspection area. Yellow microchannels apply sub-atmospheric pressure to the artery segment. The loading well mimics arterial blood supply to a capillary. The last pair of red microchannels maintain organ model physiology and metabolism with superfusion flow rates and a stable sustaining medium. This gadget can study artery structure and function in a well-defined spatiotemporal environment. According to the platform, the vasoconstrictor response did not propagate to the contralateral side [
102]; adding phenylephrine to one or two super fusion channels spatially confined the no-stimulated side. This scalable experimental platform could identify, validate, create, and test cardiovascular drugs. Another study created an artery-on-a-chip to explore two cardiovascular diseases: carotid artery disease (CAD) and abdominal aortic aneurysms (AAA). Both diseases share common characteristics of vessel wall remodeling and changes in hemodynamic shear stress. The chip consists of a membrane layer and two glass slides that could be resealed, forming the upper and lower chambers of the Artery-on-a-chip device. Flow sensors were strategically placed between the reservoirs and the chip to ensure consistent flow rates. The cells used in this study were primary human aortic endothelial cells (EC) and human aortic smooth muscle cells (SMC).
5.5. Liver-on-a-Chip
The liver is the leading site of drug/toxin metabolism. It comprises anatomic units called hepatic lobules; these hepatocytes work together to filter the blood from the digestive tract by detoxifying chemicals and metabolizing drugs [
103]. The first liver-based microfluidic system was designed by Kane et al. [
104], consisting of rat liver cells and mouse fibroblasts cultured to mimic an airway interface. Rat hepatocytes were cultured in the chip and were shown to synthesize and metabolize albumin stably. A chip established by Lee et al. [
105] imitated the interstitial space between liver endothelial and hepatocytes, facilitating new opportunities to carry out substance exchange studies. Using radical electric field gradients and electrophoresis, Ho et al. [
106] restored the hexagonal shape of the hepatic lobule that allows cell arrangement onto a circular PDMS chip. Chong et al. [
107] were able to scrutinize the metabolism process via assays that keep track of skin reactions to drugs. This study can impact drug screening research as it can identify compounds that produce systemic skin reactions. Several studies involving disease/injury research, such as Kang et al. [
108], analyzed the replication of Hepatitis B, and Zhou et al. [
109] modeled alcohol injury using a liver OOC. It is crucial to obtain further knowledge regarding Hepatitis B since more than 240 million people worldwide are infected with HBV, which may cause liver cancer. Noh et al. HBV infection was induced by transfecting HBV-genome cDNA onto a microfluidic platform and infecting the cells with an HBV genome expressed from a recombinant adenovirus. For HepG2 cells, transfection had a high infection efficiency; for primary rat hepatocytes (PRH), adenovirus infection had a higher infection rate [
110].
Table 2 demonstrates materials, sensors, and types of cells commonly used in liver-on-a-chip systems:
5.6. Intestine-on-a-Chip
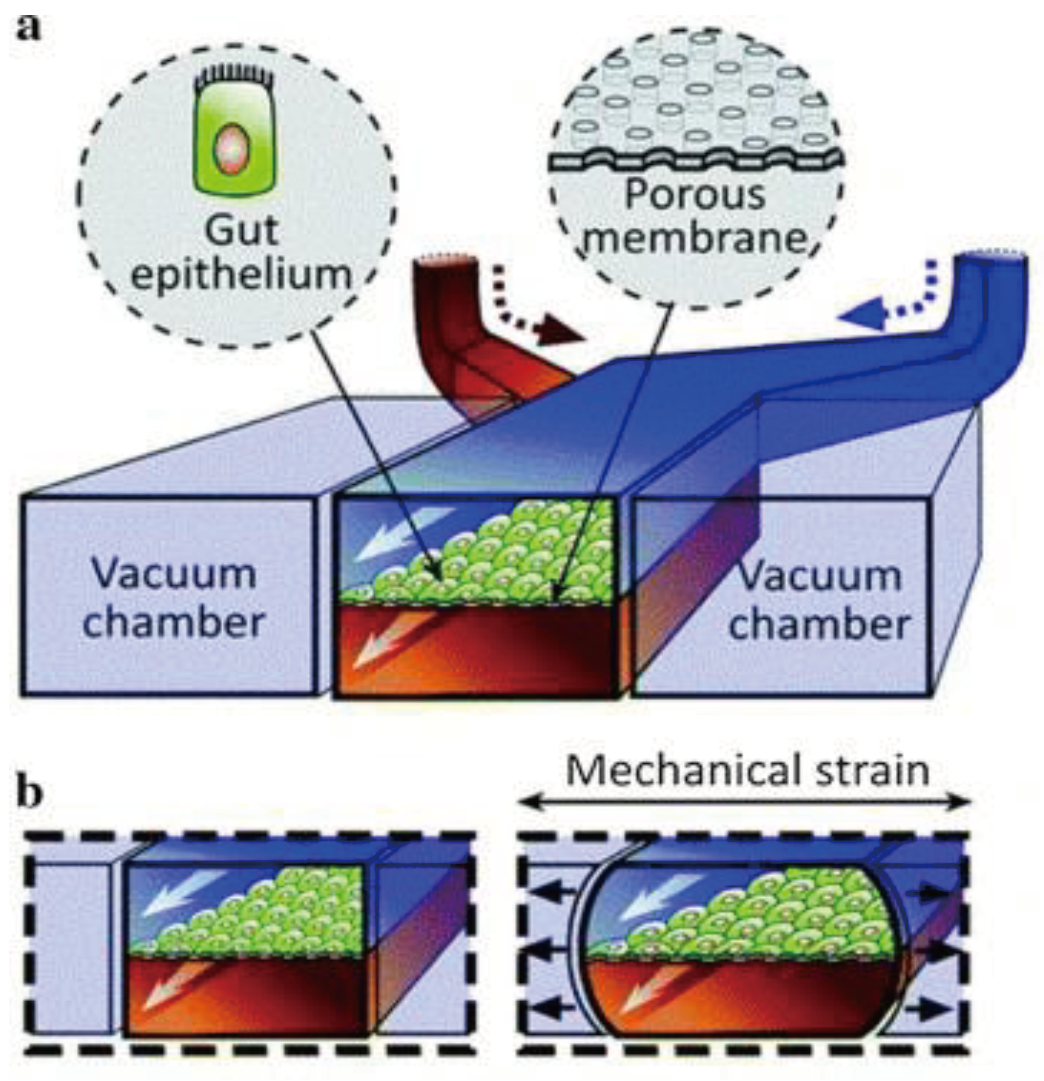
Similar to the lungs and their alveoli, the human small intestine is lined with small appendages called villi that are key to the adsorption of nutrients. Current studies involving intestine-on-a-chip include the adsorption of oral drugs into the bloodstream and a general analysis of microbial flora found in the gut. Kim et al. [
115] created an intestine-on-a-chip by constructing a chamber bilayer separated by a porous membrane lined with gut epithelium cells, as seen in
Figure 6. Drugs or inflammatory cells were introduced to the bottom chamber to analyze adsorption and cell reaction. Two vacuum chambers on either side of the cell bilayer simulated the cyclic strain that a human small intestine involuntarily does. This study demonstrated the potential of intestine-on-a-chip for personalized medicinal studies. Kasendral et al. [
116] developed a model of the human duodenum, the first part of the human small intestine, on a microchip to enhance the knowledge of the microbiome environment within the gut and intestinal morphology. To study the human gut microbiome, Firouzinezhad et al. co-cultured aerobic and anaerobic microbiota in the intestine-on-a-chip, which enables them to control and assess oxygen gradient. This platform may be a future discovery tool for microbiome-related therapeutic studies [
117].
5.7. Brain-on-a-Chip
The brain is a complex organ that includes a variety of functional units and cell types, and each is responsible for a different task. The aim is to model such an organ in a microchip to reduce the cost and time used to test certain drugs that correlate to central nervous system (CNS) diseases. Modeling a brain onto a chip that incorporates all aspects of brain functionalities is nearly impossible due to the different cell culturing needed in the same system. Therefore, the brain-on-a-chip needs to be designed with a unique platform for modeling only one specific compartment of the brain [
118]. According to Dr. Maoz, there is no “one-brain-on-a-chip fits all” model, and each application requires a different platform, which may require different considerations in terms of practicality. Researchers interested in developing and using such a platform should first consider the problem they are trying to solve, define the most critical parameters that might affect the result, and identify which platforms will allow them to incorporate the parameters of interest [
119].
The blood-brain barrier is a huge challenge in drug development for neurological diseases, and animal models do not provide enough information about mechanistic studies on BBB function and its interaction with the therapeutic. Researchers mimicked BBB on a chip to study the interaction between BBB and a nanoparticle at the cellular and molecular level [
120].
Research conducted by Tong et al. used a microfluidic approach to study the regeneration of the axons after axotomy, which is essential for patients who suffer from neural injuries. Using this approach, they experimented to mimic the
in vivo microenvironment in the chip and expose the axons to the inhibitory molecule after axotomy to observe the axon’s reaction [
121]. Another process in the CNS is myelination, which forms myelin sheath around the axons to enable neurons to transmit the signals faster and more efficiently. A study applied a microfluidic device to examine different stages of myelination using embryonic stem cells derived from the neurons [
122].
Despite advances in disease modeling and the development of effective treatments, the exact pathological mechanisms of initiation and progression of Alzheimer’s disease remain unclear. Studies have revealed that Alzheimer's disease is caused by an abnormal accumulation of amyloid-β (Aβ) and the hyperphosphorylated microtubule-associated protein
Tau. Microfluidic devices have been used recently to gain new insights into the trafficking and accumulation of pathological Aβ and
Tau in human cells [
123].
Table 3.
Various Brain-on-a-chip materials, sensors, and cell types.
Table 3.
Various Brain-on-a-chip materials, sensors, and cell types.
| Material |
Sensor |
Cell type |
Reference |
| PDMS |
Electrical |
hCMEC/D3 human brain endothelial cell line |
[124] |
| PMMA |
Electrical |
bEnd.3 murine brain endothelial cell line |
[124] |
| polyethylene terephthalate and PDMS |
Electrical |
Human iPSC-derived endothelial cells, human astrocytes |
[125] |
5.8. Nerve-on-a-Chip
Experimental models of neurodegenerative illnesses have not been successful in predicting outcomes since a large number of clinical trials are unsuccessful, and less than 7% of neurological medications are commercially available [
126]. Because of the complexity of the nervous system and the interaction between neurons and glia in the brain, micro-physiological systems used to assess possible human responses to novel treatment options lag behind those used to assess reactions in other organs. There are inadequate human-relevant
in vitro models, particularly for peripheral nerves [
127]. Animal research remains the fundamental basis since they are the only ones to validate the histopathology and electrophysiology of a complex biological system, which are the most often used clinically significant measures for evaluating peripheral neurotoxicity [
128]. Utilizing
in vitro neural cells, nerve-on-a-chip models have recently been created to simplify the complexity observed
in vivo. Microfabrication is used to create
in vitro extracellular recording interfaces to track action potentials, ensuring reproducibility and enabling statistically significant sample volumes. Planar microelectrode arrays or microchannel electrodes integrate axonal transmission with high signal-to-noise ratio (SNR) recordings to form nerve-on-a-chip models. High-density complementary metal-oxide-semiconductor microelectrode arrays with a built-in microfluidic channel could assist in the detection of complicated impulses along individual axons at the microscopic level [
129]. A recent study by Sharma et al. [
130] designed a three-dimensional functional human peripheral nerve
in vitro using the nerve-on-a-chip platform. They effectively created human neurons and primary human Schwann cell spheroids, which showed long-term culture and robust neurite outgrowth in the dual-hydrogel system. This was the first biomaterial micro-physiological system of the myelinated human peripheral nerve that could be utilized to evaluate electrophysiology, which was previously only achievable with
in vivo experiments.
5.9. Pancreas-on-a-Chip
Treatment with insulin has been recognized as a gold-standard therapy for Type 1 diabetic patients. However, this method cannot precisely mimic the effects of normal beta cells and possible hypoglycemic attacks [
131]. Thus, pancreas beta cell transplantation and islet replacement therapy were proposed as effective treatment techniques for insulin-dependent diabetes. Existing approaches for beta cell transplantation include separating and purifying the islets from the rest of the pancreatic tissue, frequently resulting in ischemia damage and an inflammatory signature response [
132], and evaluating the quantity and quality of islet cells before transplantation is vital. Pancreas-on-a-chip (POC) is a technology that enables researchers to evaluate islet potency and quality using a standardized platform. This platform has been demonstrated to be used as a tool for islet clustering to investigate the islet function in a standardized situation [
133]. Several POC systems were used in a microfluidic environment to evaluate the pancreas islet cell function in a specific pancreas dysfunction. In a study by Shik et al. in 2019, POC was utilized to evaluate islet cell function in cystic fibrosis [
134].
5.10. Placenta-on-a-Chip
The placenta is essential for a pregnancy to be successful. The primary role is the interchange of endogenous and external molecules, which permits appropriate oxygen and nutrition delivery, excretion of fetal metabolic waste, and defense against potentially dangerous agents. A multilayered structure commonly referred to as the "placental barrier, " made up of trophoblasts, connective tissue, basal lamina, and the fetal endothelium, allows substances to be transferred between the intervillous space and fetal capillaries [
134]. Several factors, including umbilical blood flow, metabolism, thickness, etc., can influence placental transfer. OOC technology can help researchers to evaluate placental transport using microfluidics and microfabrication. Placenta-on-a-chip enables researchers to build a physiological placental barrier
in vitro using human trophoblasts, umbilical vein endothelial cells, and extracellular matrix membranes. This technology might solve the primary drawbacks of existing placenta model systems, such as 3-dimensional organoids from human blastocysts, and provide research platforms for reproductive biology and medicine [
135].
5.11. Skin-on-a-Chip
In vitro and
ex vivo dermatological, pharmacological, and cosmetic investigations use artificial membranes from rebuilt skins or excised skin samples. Skin permeability, irritation, corrosion, toxicity, and different disease models can be evaluated through reconstructed skin tissue. Also, different formulations and pharmacologic features of different drugs could be tested in these environments. In the literature, many skin-on-a-chip platforms have been proposed, including epidermal and full-thickness skin models as well as diffusion surface materials (membranes—STRAT-M®, chitosan, parallel artificial membrane permeability assay (PAMPA), skins of both animal and human origin) [
136]. Based on how the skin is generated on the chip, Dr. Risueno and his colleagues classified the devices. In designing microfluidic chips for modeling the skin, two major approaches have been developed: the first is the direct introduction of a skin fragment from a biopsy or a human skin equivalent (HSE) directly into the chip (transferred skin-on-a-chip), while the second is the generation of tissue on the chip in situ (in situ skin-on-a-chip) [
137]. The transferred skin-on-a-chip model uses two primary sources of tissue fragments: a skin biopsy obtained from a donor or an in vitro-generated HSE. Both laboratory-made and commercially available HSEs have been used for skin microfluidic chips. Among HSEs, the ones with a dermal compartment are most commonly used for transferred skin-on-a-chip. As a result, more realistic models can be created of the various layers of skin present in these well-formed tissue fragments. The
in-situ skin-on-a-chip model focuses on skin generation on the chip, and it can also be divided into two groups. Jones et al. (2022) developed a unique skin-on-a-chip model incorporating human-derived primary and immortalized cells in a full-thickness skin analog. The model was contained in a microfluidic device with a previously built microvasculature. This genuine microvasculature could help evaluate the efficacy and safety of medications administered systemically to patients [
138]. It was shown by Kim et al. 2022 that human skin analogs might be utilized to simulate the main pathogenic aspects of atopic dermatitis, overcoming the shortcomings of prior studies that have relied primarily on mice models and have been unable to extrapolate their effects to people [
139].
5.12. Stomach-on-a-Chip
Existing experimental laboratory approaches and animal models have substantial limitations for mimicking human gastrointestinal disorders since they might not fully depict multicellular human primary tissues. Consequently, new medicines must be validated using model systems that mirror human
in vivo development, physiology, and disease processes. The production of three-dimensional (3D) human gastric organoids (hGOs) by guided development of human pluripotent stem cells was an important point toward this objective. Normal stomach activities and illnesses occur in the luminal epithelium; however, reaching the epithelium on the interior of organoids is difficult [
140]. Lee et al. [
141] described a novel microfluidic imaging device with the peristaltic luminal flow that houses hGOs
in vitro. This model enables stable, long-term 3D development of hGOs with luminal delivery capability through a peristaltic pump. Organoids were cannulated, and fluorescent dextran-containing media was supplied through the lumen with a peristaltic pump. This method also enabled the researchers to rhythmically add stretch and contraction to the organoid, simulating gastrointestinal motion. This technology can transport nutrients or pharmacological substances into the gastric lumen
in vitro to study human physiology, disease modeling, and drug screening. Li et al. presented an
in vitro microfluidic tool to study mucin secretion against acidic digestive juice and the protection of epithelial cells in the stomach. Their result demonstrated that the mucin secretion layer inhibits acid diffusion. This system may be beneficial to study mucus-drug interactions [
142].
5.13. Muscle-on-a-Chip
Skeletal muscle is mainly responsible for generating contractile forces and ensuring the functional performance of critical functions, including mastication and eye movements [
143]. Tissue-engineered skeletal muscle offers a potentially valuable option for physiologically relevant testing and study of the musculoskeletal system. For example, these tissues could be used
in vitro to study how the human musculoskeletal system responds to injury, disease, and treatment [
144]. The construction of a three-dimensional (3D) skeletal muscle tissue that mimics the morphological and structural intricacies of muscle inside a microfluidic device was described in a study by Agrawal et al. [
145]. Using a 3D photo-patterning technique, scientists spatially confined a cell-filled gelatin network around two bio-inert hydrogel pillars, which support the linear elastic alignment of the cells and act as attachment points for the encapsulated cells and muscle tissues as they develop and expand. The dose-dependent impact of cardiotoxin on the designed muscle tissue architecture was investigated, as was the subsequent effect of the cardiotoxin on the passive tension [
146]. Studies that need to analyze the structure and function of skeletal muscle, such as preclinical drug discovery and development, may find this tool intriguing because of its simplicity while maintaining its high level of effectiveness.
5.14. Testis-on-a-Chip
The testis-on-a-chip investigations bring up new avenues for the analysis and implementation of in-vitro testicular function. They may also facilitate incorporating microfluidic technology into future treatment techniques to maintain prepubescent male fertility and maturation arrest in adults. Finally, they may offer an environment close to the human body for evaluating drugs and toxins [
147]. For the advancement of
in vitro spermatogenesis, Abu Madighem et al. [
148] brought up new possibilities for a microfluidic 3D culture system, a testis-on-a-chip platform. From the seminiferous tubules of sexually immature mice, testicular cells were enzymatically extracted, seeded in a methylcellulose gel, and grown on a microfluidic chip. The cells continued to divide and differentiate as they grew into spheroids. Over 95% of the cells were still viable after seven weeks of cultivation. A structure that includes premeiotic, meiotic, and post-meiotic germ cells, representing each step of spermatogenesis up to and including meiosis II, was shown on the platform. The Sertoli and peritubular cells that serve as support were also present in the spheroid formation. The experiment also investigated how sensitive the system was to the culture medium's addition of retinoic acid and testosterone. A traditional 3D methylcellulose cell growth system in a well plate is contrasted with the testis-on-a-chip as a standard. Fluorescence-activated cell sorting analysis reveals more haploid cells on the chip than in the plates. Immunofluorescence labeling after seven weeks of culture reveals that the chip has more differentiated cells than the well plate.
5.15. Bone-on-a-Chip
All three types of bone cells (including osteoblasts, osteocytes, and osteoclasts) differentiation, activation, or apoptosis frequently depend on intracellular communication between them, which occurs via ligand-receptor interactions, molecules and ions moving via the extracellular matrix (ECM) or gap junctions [
149,
150] Riquelme et al. created a microfluidic technology to research bone cell-cell contact at physiologically significant distances. This platform examined how osteoclasts and osteocytes interact to cause pathogenic bone illnesses such as osteoporosis and metastasis. Two or three channels are separated by high resistance posts made up the microfluidic platform. Additionally, bone-on-a-chip is used for tissue engineering, vascularization, and bone regeneration. The bone-on-a-chip platform's relative newness compared to other organ-on-chip systems makes investigations on bone cancer metastasis rare. The role of bone-on-a-chip is still being investigated.
5.16. Bladder-on-a-Chip
The bladder-chip model contains critical bladder physiology elements essential to detect early uropathogenic Escherichia coli (UPEC) infection on a pressure-driven flow control platform. This platform is suitable for live-cell imaging and antibiotic delivery. A layer of bladder cells on the chip created by Sharma et al. sits at the bottom of a channel filled with diluted human urine. Infected cells were scanned to see how UPEC migrated, interacted with bladder cells, and aggregated [
151]. In addition to infection detection, a bladder-on-a-chip system has been used for noninvasive diagnostic purposes. Wang et al. developed a protein microarray chip to capture bladder cancer cells and biomarkers from urine samples [
152]. Similarly, Lin et al. were able to have a quantitative measurement of biomarker concentration using a bead-based enzyme-linked immunosorbent assay [
153]. Moreover, other research groups developed a microfluidic system to recognize bladder cancer cells at high processing speeds to study bladder cancer pathophysiology [
154].
5.17. Bone Marrow-on-a-Chip
Bone marrow contains hematopoietic stem cells that can become red blood cells, white blood cells, or platelets. It also contains stem cells that can become cartilage, fat, or bone cells. Bone marrow-on-a-chip sustains these types of stem cells and preserves spatial structure within an
in vitro developed 3D bone marrow niche. These characteristics of the bone marrow-on-a-chip are probably essential to its capacity to sustain the intricate functionalities of the entire organ that are incomparably difficult for traditional stroma-supported cultures to imitate. Notably, microfluidics also makes it possible to analyze responses under flow, which is crucial for controlling bone marrow physiology [
155,
156]. The bone-marrow-on-a-chip offers an intriguing alternative to animal models because it allows for the
in vitro manipulation of specific hematopoietic cell populations (either genetically or through the use of drugs) as well as the insertion of different cell types (such as tumor cells) before examining how the intact marrow responds to relevant clinical challenges, such as radiation or medications. Finally, the bone-marrow-on-a-chip is a potent technique to speed up research and development in various biomedical domains, including hematology, oncology, drug discovery, and tissue engineering [
157].
5.18. Uterus-on-a-Chip
The female reproductive system is developed at puberty and consists of the vagina, cervix, uterus, fallopian tubes, and ovaries. Each releases hormones that communicate with other parts and regulate the female body, including the organized menstrual cycles. During pregnancy, the uterus is where the eggs are fertilized after passing from the ovaries across the fallopian tubes. Being able to design a part or all of the female reproductive system on a chip can lead to a better understanding of physiology. It could encourage new methods of approaching medical cases related to the female reproductive system.
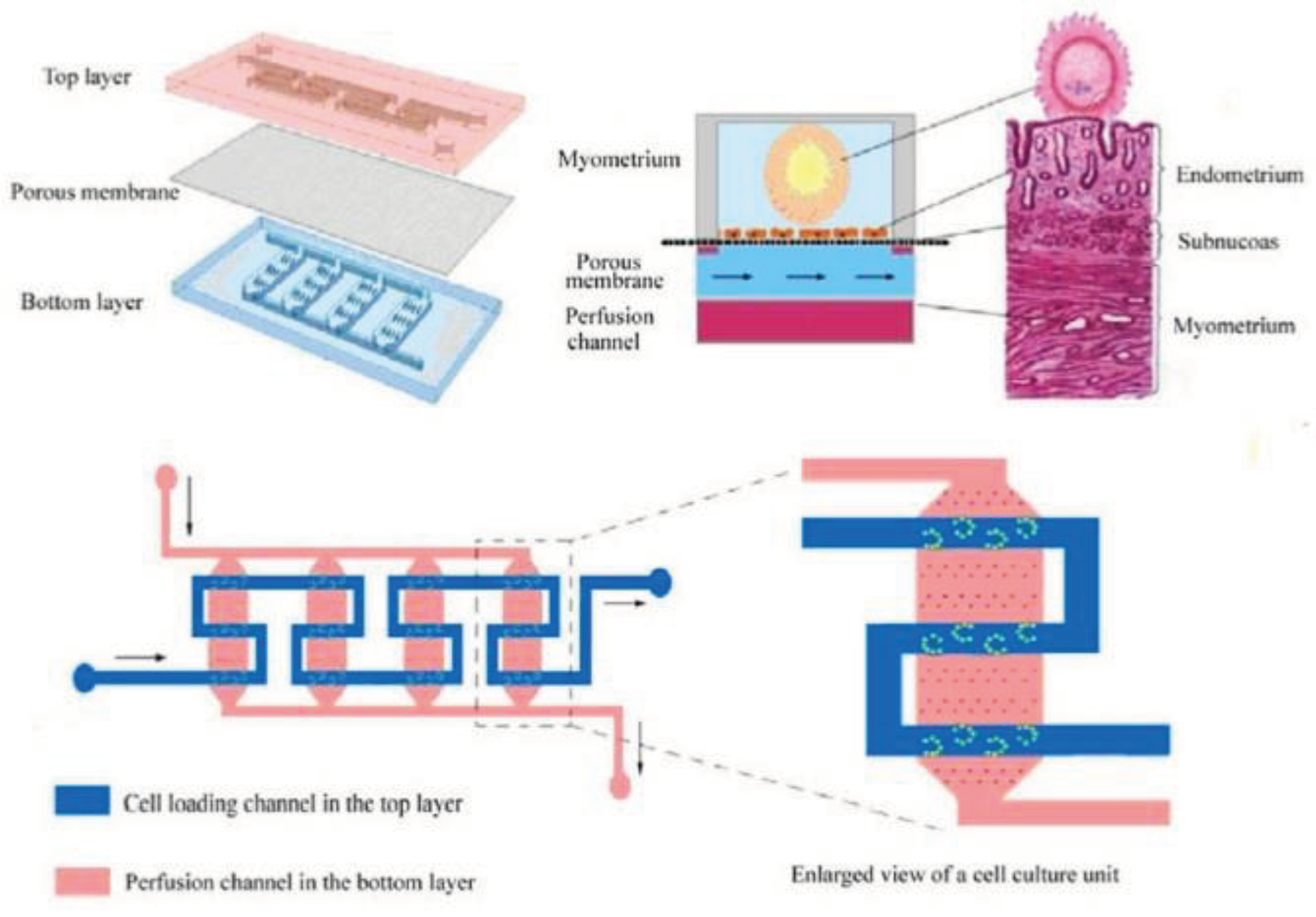
In vitro fertilization-embryo transplantation (IVF-ET) is a process of fertilizing the egg out of the female body and is one of the widespread treatments for infertility. However, IVF-ET has some limitations, such as a low feralization rate. To tackle these limitations, a microfluidic uterus is developed to perform processes in an embryo-like environment, including ovulation, insemination, and embryo development. As shown in
Figure 7, the microchip consisted of three layers; two PDMS layers were separated by a porous layer used to support endometrial cell culture. The top layer had a zigzag-shaped channel and several interlaced microsievers on the channel bed to capture oocytes. In contrast, the bottom layer was designed with four parallel channels with a micropillar array on each bed to support the porous membrane [
158].
5.19. Other Applications of OOC Chips
5.19.1. Cervix-on-a-Chip
A solid alternative to the
in vitro model for studying the physiology of the cervix in real-time using high-resolution imaging for analysis of biological reactions in the cervix, as well as medication discovery, can be modeled by uterine cervix-on-a-chip. This well-established uterine cervix-on-a-chip is transparent, efficient, and simple to use. It is anticipated to have significant implications for the customized management of human papillomavirus infection lesions and cervical cancer, as well as a possible role in tissue engineering and other clinical treatments [
159]. A different study showed that cervix-on-a-chip is effective for obstetrics and gynecology research. It can be used to study the dynamics of cervical epithelial cells in the benign and malignant cervix and the remodeling of the cervical cavity during pregnancy and parturition [
160].
5.19.2. Hair-on-a-Chip
Ataç et al. cultured ex vivo prepuce as a skin organ culture to evaluate the native state of the tissue. They compared their result in two different media culture conditions. In the first condition, they cultured the cells in Transwells (static culture), while in the second, they used a dynamically perfused microchannel system (multi-organ-on-chip). Results from their study showed a distinct difference between the parallel static and multi-organ-on-chip (MOC) cultures and that dynamic perfusion prevents tissue disintegration. MOC hair follicle cultures revealed many hair-fiber extensions from the epidermis while triple the culture period compared to the Philpott assay (using single, truncated hair follicles to study hair follicle biology
in vitro) for
ex vivo hair elongation and drug detection. These findings demonstrate that hair-on-a-chip may evaluate hair follicle biology [
161].
5.19.3. Lymph Nodes-on-a-Chip
Lymph nodes-on-chip are appealing platforms for examining immune system-related illnesses like cancer and inflammatory diseases. Biomimetic lymph node-on-chip systems can load healthy and diseased cells or tissues, and their behavior can be easily observed. Treatment regimens that use immunosuppressive medications to target the immune system to alter for treating certain illness conditions greatly benefit from lymph nodes-on-chip technology. Immunotherapy has long been used to treat allergies, lessen the chance of rejection of transplanted organs, and minimize autoimmune disorders [
162]. Additionally, it is now a typical kind of treatment for many tumors. Numerous solid and hematologic cancers have responded well to cancer immunotherapy, and it is well-known that lymph node draining is helpful [
163]. Additionally, it has been demonstrated that immunotherapies have distinctive toxicity profiles linked to particular modes of action [
164]. Lymph nodes-on-chip platforms that are physiologically appropriate can be used to explore these phenomena extensively. Lymph nodes on a chip are helpful in studying immune-related disorders like cancer and inflammation [
165]. Biomimetic lymph node-on-chip devices can easily monitor normal and sick cells or tissues [
165].
5.19.4. Vagina-on-a-Chip
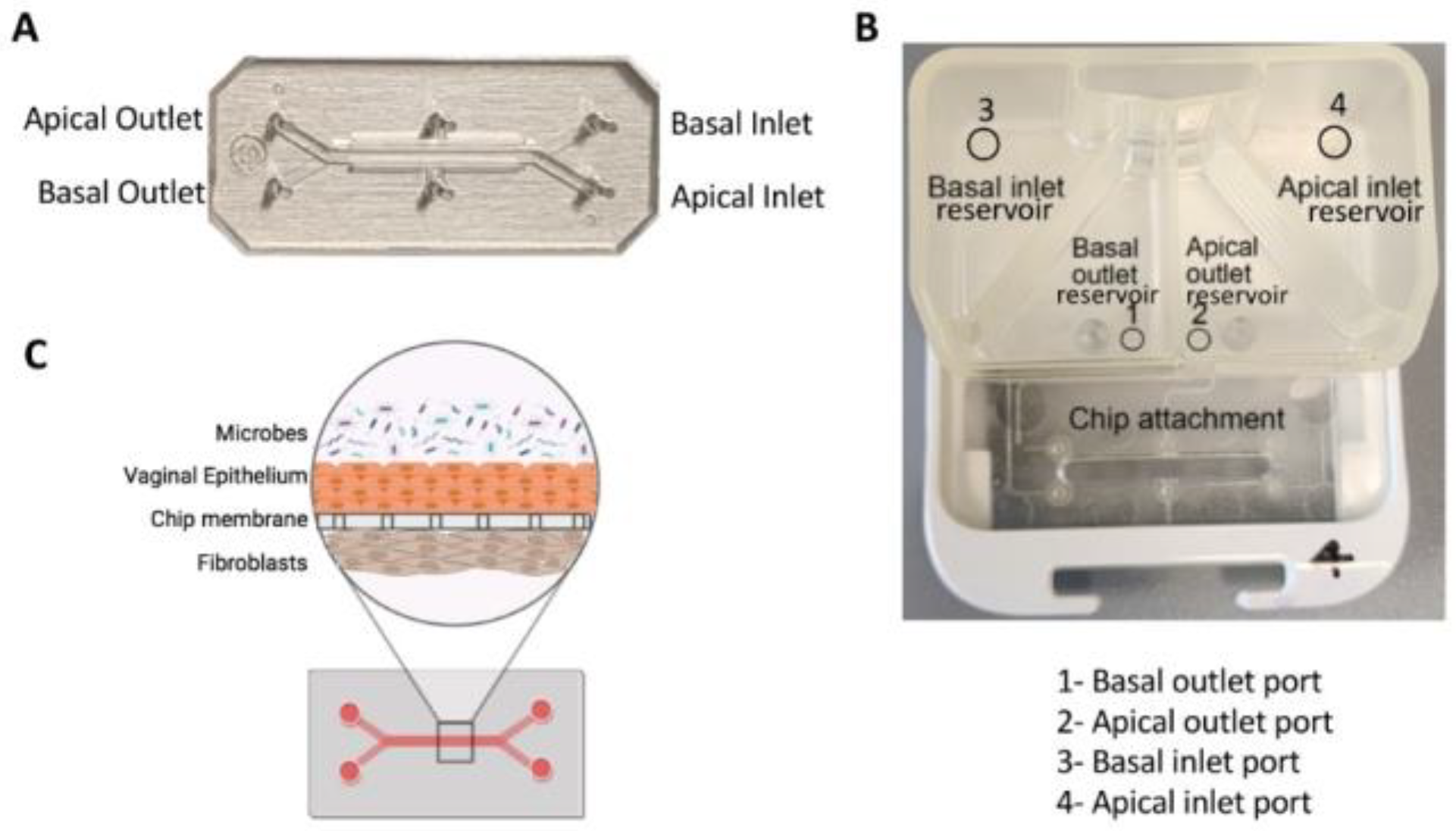
Mahajan et al. developed a Vagina-on-a-chip model to improve understanding of the interaction between the host and microbiome in the vagina. They co-cultured primary human vaginal epithelial cells and uterine fibroblasts in the microfluidic device to mimic the vaginal epithelial-stromal interface. Co-culture with
Lactobacillus crispatus containing consortia maintained epithelial cell viability and physiologically low pH, D-lactic and L-lactic acid accumulation, and downregulation of inflammatory cytokines. However, co-culture with Gardnerella vaginalis-containing consortia revealed injury in epithelial cells, increased pH, and upregulation of pro-inflammatory cytokines [
166]. A recent study utilized human vagina chips to co-culture human vaginal microbial consortia with primary human vaginal epithelium to investigate the physiological responses of the human vagina to healthy and dysbiotic microbiomes [
167]. The chip had parallel hollow microchannels hosting living cells under dynamic fluid flow [
167]. These chips facilitate the recreation of organ-level tissue-tissue interfaces by culturing diverse cell types, such as epithelium and stromal fibroblasts or epithelium and vascular endothelium, on opposite sides of a porous membrane within the channels.
Figure 8 demonstrates the design of the chip [
167].
5.20. Multi-Organs-on-a-Chip
The human body is complicated, and single organ chips cannot fully and accurately reflect a human organ without recognizing that organ’s interactions with other organs and systems within the body. The “multi-organ-on-a-chip,” also known as the “human-on-a-chip,” is an ongoing research project that would simultaneously construct multiple organs on a single chip, as seen in
Figure 9 [
168]. Multi-organ integration would theoretically be achieved by different organs and tissues connected by bionic blood channels [
169]. This would permit the analysis of whole-body system interactions. Ongoing studies include the design of two, three, four, and even ten-organ-chips. Midwoud et al. [
170] successfully combined the liver and intestines on a microfluidic chip to examine the regulation of bile acid synthesis. Inevitably, the more organs added to a chip, the higher the complexity and the greater the challenge of controlling the system's parameters. Overcoming these challenges would revolutionize the drug discovery research field.
In terms of drug safety, there are various challenges to overcome. Two-dimensional cultures and animal studies are not truly representative of the human body. The liver metabolizes an administered drug, and the resulting compound may cause unpredictable toxicity to organs not intended to be affected, such as the heart. Several liver and heart models have been widely used to assess the toxicity of new or recalled drugs. A multi-Organs-on-a-chip platform provides an innovative approach to studying drug-related effects, resulting in predictions of liver metabolism on off-target organs, ultimately improving drug safety testing during preclinical stages of the development process [
171].
Figure 9.
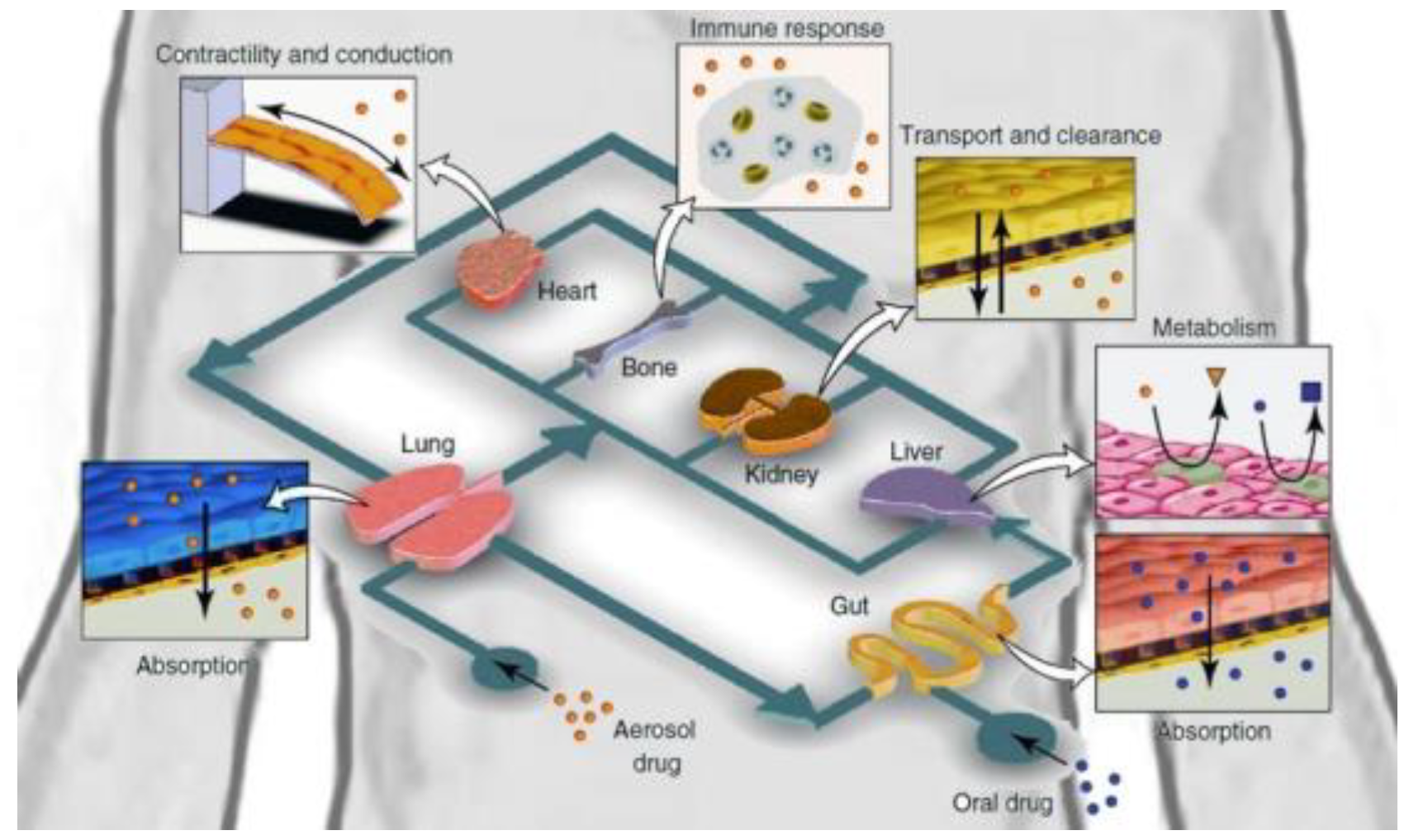
Diagram of Human-on-a-chip. This image has been taken from ELVESYS Group website (elveflow.com).
Figure 9.
Diagram of Human-on-a-chip. This image has been taken from ELVESYS Group website (elveflow.com).
6. Disease Models on Organ-on-a-Chip
In recent years, organ-on-a-chip technology has emerged as a promising platform for modeling and studying diseases in a physiologically relevant and accurate manner. Traditional methods to study disease states have relied on animal models and 2D cell culture, which often fail to provide the complexity of the human body. However, OOC technology enabled researchers to recreate the microenvironment of multiple organs in order to develop more realistic disease models. The table presented below provides a summary of disease models on the organ-on-a-chip platform.
Table 4.
Disease models on the organ-on-a-chip.
Table 4.
Disease models on the organ-on-a-chip.
| Organ modeled on chip |
Disease |
Reference |
| Lung |
Lung cancer, Asthma, SARS-CoV-2 |
[172,173] |
| Liver |
Hepatitis, Liver fibrosis |
[109,110] |
| Heart |
Myocardial infraction, Arrhythmias |
[174] |
| Brain |
Alzheimer’s disease |
[123] |
| Intestine |
Inflammatory bowel disease, Colorectal cancer, GI bacterial infection, GI viral infection |
[175] |
| BBB |
Stroke, Multiple sclerosis |
[176] |
7. Commercialization
During the translation and commercialization of organ-on-a-chip, the industrial perspective is prioritized, including technical standardization and dependability, simplicity of use, cost-effectiveness, and compliance with legal requirements [
177,
178,
179]. Therefore, it is necessary to conduct additional analytical validations of these chips to evaluate their diagnostic and therapeutic efficacy and repeatability and to satisfy the practical requirements of pharmacological and medical use and FDA regulations. During each stage of organ-on-a-chip development, early and close collaboration between academic institutions, industrial R&D divisions, and healthcare organizations will create a positive feedback loop to confirm the efficacy of these platforms and maximize their utility in the healthcare industry. For instance, Emulate Inc. has collaborated with AstraZeneca, Johnson & Johnson, Merck, Takeda, and FDA to analyze the efficacy and safety of drug candidates for human use in an industrial setting to validate the effectiveness of their various products.
Despite its vast potential, a significant challenge to the commercialization of organ-on-a-chip technology is insufficient venture capital funding. Only a few OOC startups, including Emulate Inc. (
$142.3M), InSphero (
$35.2M), and Mimetas (
$34.2M), have managed to secure substantial investments, according to public sources like Crunchbase. To overcome this challenge, national healthcare and research systems can play a vital role by fostering collaborations with academic institutions and companies, offering support for intellectual property protection, and providing funding avenues [
180].
8. Conclusions and Recommendations
This paper covered the kidney, lung, heart, liver, intestine, brain, and uterus-on-a-chip, but microfluidic technology has been used to reconstruct other organs, including blood vessels, skin [
181], skeletal muscle [
182], and the central nervous system [
183]. The development of these organs-on-a-chip has led to significant scientific advances because it allows researchers to study biological processes in a controlled yet accurate microenvironment.
This technology has developed rapidly and shows no signs of stopping, but the “human-on-a-chip” theory is far from a reality. A universal cell culture medium suitable for all organs would be necessary, and the practicality of collecting samples on a “human-on-a-chip” is shaky at best. This is due to the disruption during operation and resulting changes to the system. As the number of organs on the chip increases, so does the system's complexity. All generated data would carry artefactual and non-translatable risks. At this moment, this is currently unsolvable.
Finding a more suitable material to replace the most commonly used material, PDMS, would also be ideal. While it has its advantages, PDMS is thicker and denser than most
in vivo tissue, preventing microenvironments on the chip from being as accurate as possible. Manufacturing these chips and experimental procedures on them are relatively expensive. This is not conducive to the widespread use of organs-on-a-chip, so the components of which these chips can be made must be of lower cost and easily disposable. Expensive components should be reusable. There are many ongoing projects to improve OOC platforms. An article entitled "Pump-less, recirculating organ-on-a-chip (rOoC) platform" introduces a novel approach by eliminating the need for pumps [
184]. The authors of this study have developed a passive recirculation mechanism for their organ-on-a-chip device [
184]. Typical organ-on-a-chip devices require pumps to circulate fluids; however, a special porous membrane facilitates passive recirculation in this model. With pump-less systems, cell viability is improved, complexity is reduced, and reproducibility is enhanced. This platform brings a more reliable and cost-effective approach to studying human organs in vitro.
OOC technology has shown great promise and potential, as well as produced a plethora of biological research advancements due to its ability to provide the best of in vivo and in vitro research. Even so, OOCs have the ability to improve the cost and accuracy of microenvironments, but solving these issues in the future foretells a revolutionary turn in biological systems research.
Data Availability Statement
Data sharing not applicable to this article as no data sets were generated or analyzed in this review article.
Conflict of Interest
The authors declare no conflict of interest.
References
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: recent breakthroughs and future prospects. Biomed. Eng. Online 2020, 19, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Figeys, D.; Pinto, D. Lab-on-a-Chip: A Revolution in Biological and Medical Sciences. Anal. Chem. 2000, 72, 330 A–335 A. [Google Scholar] [CrossRef] [PubMed]
- Tehranirokh, M.; Kouzani, A.Z.; Francis, P.S.; Kanwar, J.R. Microfluidic devices for cell cultivation and proliferation. Biomicrofluidics 2013, 7, 051502. [Google Scholar] [CrossRef] [PubMed]
- Sosa-Hernández, J.E.; Villalba-Rodríguez, A.M.; Romero-Castillo, K.D.; Aguilar-Aguila-Isaías, M.A.; García-Reyes, I.E.; Hernández-Antonio, A.; Ahmed, I.; Sharma, A.; Parra-Saldívar, R.; Iqbal, H. Organs-on-a-Chip Module: A Review from the Development and Applications Perspective. Micromachines 2018, 9, 536. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Hernández, J.; Nicasio-Torres, M.d.P.; Sarmiento-López, L.G.; Rodríguez-Monroy, M. Production of anti-inflammatory compounds in Sphaeralcea angustifolia cell suspension cultivated in stirred tank bioreactor. Eng. Life Sci. 2019, 19, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Giduthuri, A.T. Microfluidic-Chip Technology for Disease Diagnostic Applications via Dielectrophoresis. In Nanosensors for Futuristic Smart and Intelligent Healthcare Systems, 1st edition; 2022; p. 318. [Google Scholar]
- Bravard, J.P.; Petit, F. Geomorphology of Streams and Rivers. In Encyclopedia of Inland Waters; Elsevier: The Netherlands, 2009; pp. 387–395. [Google Scholar]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Verpoorte, E.; de Rooij, N.F. Microfluidics meets MEMS. Proc. IEEE 2003, 91, 930–953. [Google Scholar] [CrossRef]
- Sia, S.K.; Whitesides, G.M. Microfluidic devices fabricated in Poly(dimethylsiloxane) for biological studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef]
- Whitesides, G.M.; Ostuni, E.; Takayama, S.; Jiang, X.; Ingber, D.E. Soft Lithography in Biology and Biochemistry. Annu. Rev. Biomed. Eng. 2001, 3, 335–373. [Google Scholar] [CrossRef]
- Adekanmbi, E.O.; Srivastava, S.K. Applications of Electrokinetics and Dielectrophoresis on Designing Chip-Based Disease Diagnostic Platforms. In Bio-Inspired Technology [Working Title]; IntechOpen, 2019. [Google Scholar] [CrossRef]
- Miri, A.K.; Mostafavi, E.; Khorsandi, D.; Hu, S.-K.; Malpica, M.; Khademhosseini, A. Bioprinters for organs-on-chips. Biofabrication 2019, 11, 042002. [Google Scholar] [CrossRef] [PubMed]
- Au, A.K.; Huynh, W.; Horowitz, L.F.; Folch, A. 3D-Printed Microfluidics. Angew. Chem. Int. Ed. 2016, 55, 3862–3881. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Wu, L.-Q.; Ghodssi, R.; Rubloff, G.W.; Payne, G.F.; Bentley, W.E. Signal-Directed Sequential Assembly of Biomolecules on Patterned Surfaces. Langmuir 2005, 21, 2104–2107. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Wu, L.-Q.; Bentley, W.E.; Ghodssi, R.; Rubloff, G.W.; Culver, J.N.; Payne, G.F. Biofabrication with Chitosan. Biomacromolecules 2005, 6, 2881–2894. [Google Scholar] [CrossRef] [PubMed]
- Osório, L.A.; Silva, E.; Mackay, R.E. A Review of biomaterials and scaffold fabrication for organ-on-a-chip (OOAC) systems. Bioengineering 2021, 8, 113. [Google Scholar] [CrossRef] [PubMed]
- Bettinger, C.J.; Cyr, K.M.; Matsumoto, A.; Langer, R.; Borenstein, J.T.; Kaplan, D.L. Silk Fibroin Microfluidic Devices. Adv. Mater. 2007, 19, 2847–2850. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rigueiro, J.; Viney, C.; Llorca, J.; Elices, M. Mechanical properties of single-brin silkworm silk. J Appl Polym Sci 2000, 75, 1270–1277. [Google Scholar] [CrossRef]
- Zhao, H.-P.; Feng, X.-Q.; Shi, H.-J. Variability in mechanical properties of Bombyx mori silk. Mater. Sci. Eng. C 2007, 27, 675–683. [Google Scholar] [CrossRef]
- Ling, Y.; Rubin, J.; Deng, Y.; Huang, C.; Demirci, U.; Karp, J.M.; Khademhosseini, A. A cell-laden microfluidic hydrogel. Lab Chip 2007, 7, 756–762. [Google Scholar] [CrossRef]
- Masuda, K.; Sah, R.L.; Hejna, M.J.; Thonar, E.J.-M.A. A novel two-step method for the formation of tissue-engineered cartilage by mature bovine chondrocytes: The alginate-recovered-chondrocyte (ARC) method. J. Orthop. Res. 2003, 21, 139–148. [Google Scholar] [CrossRef]
- Rahfoth, B.; Weisser, J.; Sternkopf, F.; Aigner, T.; von der Mark, K.; Bräuer, R. Transplantation of allograft chondrocytes embedded in agarose gel into cartilage defects of rabbits. Osteoarthr. Cartil. 1998, 6, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Shim, K.Y.; Kim, B.; Sung, J.H. Hydrogel-based three-dimensional cell culture for organ-on-a-chip applications. Biotechnol Prog 2017, 33, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Grover, W.H.; von Muhlen, M.G.; Manalis, S.R. Teflon films for chemically-inert microfluidic valves and pumps. Lab Chip 2008, 8, 913–918. [Google Scholar] [CrossRef]
- Sugioka, K.; Hanada, Y.; Midorikawa, K. 3D MICROSTRUCTURING OF GLASS BY FEMTOSECOND LASER DIRECT WRITING AND APPLICATION TO BIOPHOTONIC MICROCHIPS. Prog. Electromagn. Res. Lett. 2008, 1, 181–188. [Google Scholar] [CrossRef]
- Hanada, Y.; Sugioka, K.; Shihira-Ishikawa, I.; Kawano, H.; Miyawaki, A.; Midorikawa, K. 3D microfluidic chips with integrated functional microelements fabricated by a femtosecond laser for studying the gliding mechanism of cyanobacteria. Lab Chip 2011, 11, 2109–2115. [Google Scholar] [CrossRef]
- Quan, H.; Zhang, T.; Xu, H.; Luo, S.; Nie, J.; Zhu, X. Photo-curing 3D printing technique and its challenges. Bioact. Mater. 2020, 5, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Jiang, H. Autonomous microfluidics with stimuli-responsive hydrogels. Soft Matter 2007, 3, 1223–1230. [Google Scholar] [CrossRef]
- Rogers, C.I.; Qaderi, K.; Woolley, A.T.; Nordin, G.P. 3D printed microfluidic devices with integrated valves. Biomicrofluidics 2015, 9, 016501. [Google Scholar] [CrossRef]
- Pokharna, P.P.; Ghantasala, M.K.; Rozhkova, E.A. 3D printed polylactic acid and acrylonitrile butadiene styrene fluidic structures for biological applications: Tailoring bio-material interface via surface modification. Mater. Today Commun. 2021, 27, 102348. [Google Scholar] [CrossRef]
- Rogers, C.I.; Oxborrow, J.B.; Anderson, R.R.; Tsai, L.-F.; Nordin, G.P.; Woolley, A.T. Microfluidic valves made from polymerized polyethylene glycol diacrylate. Sensors Actuators B Chem. 2014, 191, 438–444. [Google Scholar] [CrossRef]
- Plegue, T.J.; Kovach, K.M.; Thompson, A.J.; Potkay, J.A. Stability of Polyethylene Glycol and Zwitterionic Surface Modifications in PDMS Microfluidic Flow Chambers. Langmuir 2018, 34, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Shakeri, A.; Jarad, N.A.; Khan, S.; Didar, T.F. Bio-functionalization of microfluidic platforms made of thermoplastic materials: A review. Anal. Chim. Acta 2022, 1209, 339283. [Google Scholar] [CrossRef] [PubMed]
- Cherpinski, A.; Torres-Giner, S.; Vartiainen, J.; Peresin, M.S.; Lahtinen, P.; Lagaron, J.M. Improving the water resistance of nanocellulose-based films with polyhydroxyalkanoates processed by the electrospinning coating technique. Cellulose 2018, 25, 1291–1307. [Google Scholar] [CrossRef]
- Cherpinski, A.; Torres-Giner, S.; Vartiainen, J.; Peresin, M.S.; Lahtinen, P.; Lagaron, J.M. Improving the water resistance of nanocellulose-based films with polyhydroxyalkanoates processed by the electrospinning coating technique. Cellulose 2018, 25, 1291–1307. [Google Scholar] [CrossRef]
- Chen, M.B.; Srigunapalan, S.; Wheeler, A.R.; Simmons, C.A. A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell–cell interactions. Lab Chip 2013, 13, 2591–2598. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.; Kim, S.H.; Lee, D.; Kim, B.; Kim, T.H.; Jung, Y.; Choi, N.; Sung, J.H. Fabrication of micrometer-scale porous gelatin scaffolds for 3D cell culture. J. Ind. Eng. Chem. 2017, 50, 183–189. [Google Scholar] [CrossRef]
- Rajabi, N.; Rezaei, A.; Kharaziha, M.; Bakhsheshi-Rad, H.R.; Luo, H.; RamaKrishna, S.; Berto, F. Recent Advances on Bioprinted Gelatin Methacrylate-Based Hydrogels for Tissue Repair. Tissue Eng. Part A 2021, 27, 679–702. [Google Scholar] [CrossRef] [PubMed]
- Zamboni, F.; Keays, M.; Hayes, S.; Albadarin, A.B.; Walker, G.M.; Kiely, P.A.; Collins, M.N. Enhanced cell viability in hyaluronic acid coated poly(lactic-co-glycolic acid) porous scaffolds within microfluidic channels. Int. J. Pharm. 2017, 532, 595–602. [Google Scholar] [CrossRef]
- Roy, E.; Geissler, M.; Galas, J.-C.; Veres, T. Prototyping of microfluidic systems using a commercial thermoplastic elastomer. Microfluid. Nanofluidics 2011, 11, 235–244. [Google Scholar] [CrossRef]
- Borysiak, M.D.; Yuferova, E.; Posner, J.D. Simple, Low-Cost Styrene-Ethylene/Butylene-Styrene Microdevices for Electrokinetic Applications. Anal. Chem. 2013, 85, 11700–11704. [Google Scholar] [CrossRef]
- Domansky, K.; Sliz, J.D.; Wen, N.; Hinojosa, C.; Thompson, G.; Fraser, J.P.; Hamkins-Indik, T.; Hamilton, G.A.; Levner, D.; Ingber, D.E. SEBS elastomers for fabrication of microfluidic devices with reduced drug absorption by injection molding and extrusion. Microfluid. Nanofluidics 2017, 21, 107. [Google Scholar] [CrossRef]
- Galateanu, B.; Hudita, A.; Biru, E.I.; Iovu, H.; Zaharia, C.; Simsensohn, E.; Costache, M.; Petca, R.-C.; Jinga, V. Applications of Polymers for Organ-on-Chip Technology in Urology. Polymers 2022, 14, 1668. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Hargrove-Grimes, P.; Low, L.A.; Tagle, D.A. Microphysiological systems: What it takes for community adoption. Exp. Biol. Med. 2021, 246, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Syama, S.; Mohanan, P.V. Microfluidic based human-on-a-chip: A revolutionary technology in scientific research. Trends Food Sci. Technol. 2021, 110, 711–728. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Maoz, B.M.; Herland, A.; A FitzGerald, E.; Grevesse, T.; Vidoudez, C.; Pacheco, A.R.; Sheehy, S.P.; Park, T.-E.; Dauth, S.; Mannix, R.; et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat. Biotechnol. 2018, 36, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Esch, M.B.; Sung, J.H.; Yang, J.; Yu, C.; Yu, J.; March, J.C.; Shuler, M.L. On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic ‘body-on-a-chip’ devices. Biomed. Microdevices 2012, 14, 895–906. [Google Scholar] [CrossRef]
- Afshar Bakooshli, M. A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction. Elife 2019, 8, e44530. [Google Scholar] [CrossRef]
- Uemura, M.; Refaat, M.M.; Shinoyama, M.; Hayashi, H.; Hashimoto, N.; Takahashi, J. Matrigel supports survival and neuronal differentiation of grafted embryonic stem cell-derived neural precursor cells. J. Neurosci. Res. 2009, 88, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Chen, Y.; Lu, H.; Jin, D. Advances in Spheroids and Organoids on a Chip. Adv. Funct. Mater. 2023, 33. [Google Scholar] [CrossRef]
- Nguyen, H.; Badie, N.; McSpadden, L.; Pedrotty, D.; Bursac, N. Quantifying Electrical Interactions Between Cardiomyocytes and Other Cells in Micropatterned Cell Pairs. Cardiac Tissue Engineering: Methods and Protocols 2014, 249–262. [Google Scholar] [CrossRef]
- Wufuer, M.; et al. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci Rep 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Otieno, M.A.; Ronxhi, J.; Lim, H.-K.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019, 11, eaax5516. [Google Scholar] [CrossRef] [PubMed]
- Even, M.S.; Sandusky, C.B.; Barnard, N.D. Serum-free hybridoma culture: ethical, scientific and safety considerations. Trends Biotechnol 2006, 24, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Sei, Y.J.; Jeon, N.L.; Kim, Y. Tumor microenvironment on a chip: the progress and future perspective. Bioengineering 2017, 4, 64. [Google Scholar] [CrossRef] [PubMed]
- Przybyla, L.; Voldman, J. Probing embryonic stem cell autocrine and paracrine signaling using microfluidics. Annu. Rev. Anal. Chem. 2012, 5, 293–315. [Google Scholar] [CrossRef]
- Visone, R.; Talò, G.; Occhetta, P.; Cruz-Moreira, D.; Lopa, S.; Pappalardo, O.A.; Redaelli, A.; Moretti, M.; Rasponi, M. A microscale biomimetic platform for generation and electro-mechanical stimulation of 3D cardiac microtissues. APL Bioeng. 2018, 2, 046102. [Google Scholar] [CrossRef]
- Duc, P.; Vignes, M.; Hugon, G.; Sebban, A.; Carnac, G.; Malyshev, E.; Charlot, B.; Rage, F. Human neuromuscular junction on micro-structured microfluidic devices implemented with a custom micro electrode array (MEA). Lab Chip 2021, 21, 4223–4236. [Google Scholar] [CrossRef]
- Haycock, J.W. 3D Cell Culture: A Review of Current Approaches and Techniques. 2011; pp. 1–15. [Google Scholar]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Hong, S.; Rhee, W.J. Microfluidic three-dimensional cell culture of stem cells for high-throughput analysis. World J. Stem Cells 2019, 11, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Xiao, Y.; Liu, H.; Fan, Y.; Dao, M. Patient-Specific Organoid and Organ-on-a-Chip: 3D Cell-Culture Meets 3D Printing and Numerical Simulation. Adv. Biol. 2021, 5. [Google Scholar] [CrossRef] [PubMed]
- Wagner, I.; Materne, E.-M.; Brincker, S.; Süßbier, U.; Frädrich, C.; Busek, M.; Sonntag, F.; Sakharov, D.A.; Trushkin, E.V.; Tonevitsky, A.G.; et al. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip 2013, 13, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Rissin, D.M.; Kan, C.W.; Song, L.; Rivnak, A.J.; Fishburn, M.W.; Shao, Q.; Piech, T.; Ferrell, E.P.; Meyer, R.E.; Campbell, T.G.; et al. Multiplexed single molecule immunoassays. Lab Chip 2013, 13, 2902–2911. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, J.J.; Mu, L.; Xue, Q.; Wu, Y.; Wu, P.-H.; Li, J.; Vortmeyer, A.O.; Miller-Jensen, K.; Wirtz, D.; et al. High-Throughput Secretomic Analysis of Single Cells to Assess Functional Cellular Heterogeneity. Anal. Chem. 2013, 85, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Grist, S.M.; Chrostowski, L.; Cheung, K.C. Optical oxygen sensors for applications in microfluidic cell culture. Sensors 2010, 10, 9286–9316. [Google Scholar] [CrossRef] [PubMed]
- Rivera, K.R.; Yokus, M.A.; Erb, P.D.; Pozdin, V.A.; Daniele, M. Measuring and regulating oxygen levels in microphysiological systems: Design, material, and sensor considerations. Analyst 2019, 144, 3190–3215. [Google Scholar] [CrossRef] [PubMed]
- Scheidecker, B.; Shinohara, M.; Sugimoto, M.; Danoy, M.; Nishikawa, M.; Sakai, Y. Induction of in vitro metabolic zonation in primary hepatocytes requires both near-physiological oxygen concentration and flux. Front. Bioeng. Biotechnol. 2020, 8, 524. [Google Scholar] [CrossRef]
- Sung, J.H.; Choi, J.; Kim, D.; Shuler, M.L. Fluorescence optical detection in situ for real-time monitoring of cytochrome P450 enzymatic activity of liver cells in multiple microfluidic devices. Biotechnol. Bioeng. 2009, 104, 516–525. [Google Scholar] [CrossRef]
- Ferrari, E.; Palma, C.; Vesentini, S.; Occhetta, P.; Rasponi, M. Integrating Biosensors in Organs-on-Chip Devices: A Perspective on Current Strategies to Monitor Microphysiological Systems. Biosensors 2020, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Wu, B.; Srivastava, S.K.; Ay, S.U. Comparative Study and Simulation of Capacitive Sensors in Microfluidic Channels for Sensitive Red Blood Cell Detection. Micromachines 2022, 13, 1654. [Google Scholar] [CrossRef] [PubMed]
- Misun, P.M.; Rothe, J.; Schmid, Y.R.F.; Hierlemann, A.; Frey, O. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsystems Nanoeng. 2016, 2, 16022. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Silva, P.N.; Syed, A.M.; Sindhwani, S.; Rocheleau, J.V.; Chan, W.C.W. Clarifying intact 3D tissues on a microfluidic chip for high-throughput structural analysis. Proc. Natl. Acad. Sci. 2016, 113, 14915–14920. [Google Scholar] [CrossRef] [PubMed]
- Wardwell-Swanson, J.; Suzuki, M.; Dowell, K.G.; Bieri, M.; Thoma, E.C.; Agarkova, I.; Chiovaro, F.; Strebel, S.; Buschmann, N.; Greve, F.; et al. A framework for optimizing high-content imaging of 3D models for drug discovery. SLAS Discov. Adv. Sci. Drug Discov. 2020, 25, 709–722. [Google Scholar] [CrossRef] [PubMed]
- van de Wijdeven, R.; et al. A novel lab-on-chip platform enabling axotomy and neuromodulation in a multi-nodal network. Biosens Bioelectron 2019, 140, 111329. [Google Scholar] [CrossRef] [PubMed]
- Moutaux, E.; Charlot, B.; Genoux, A.; Saudou, F.; Cazorla, M. An integrated microfluidic/microelectrode array for the study of activity-dependent intracellular dynamics in neuronal networks. Lab Chip 2018, 18, 3425–3435. [Google Scholar] [CrossRef] [PubMed]
- Maoz, B.M.; Herland, A.; Henry, O.Y.F.; Leineweber, W.D.; Yadid, M.; Doyle, J.; Mannix, R.; Kujala, V.J.; FitzGerald, E.A.; Parker, K.K.; et al. Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017, 17, 2294–2302. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.D.A.; Sayer, R.; Windass, A.S.; Haslam, I.S.; De Broe, M.E.; D'Haese, P.C.; Verhulst, A. Characterisation of human tubular cell monolayers as a model of proximal tubular xenobiotic handling. Toxicol. Appl. Pharmacol. 2008, 233, 428–438. [Google Scholar] [CrossRef]
- Wolff, A.; Antfolk, M.; Brodin, B.; Tenje, M. In vitro blood–brain barrier models—An overview of established models and new microfluidic approaches. J Pharm Sci 2015, 104, 2727–2746. [Google Scholar] [CrossRef]
- Piergiovanni, M.; Leite, S.B.; Corvi, R.; Whelan, M. Standardisation needs for organ on chip devices. Lab Chip 2021, 21, 2857–2868. [Google Scholar] [CrossRef] [PubMed]
- Maschmeyer, I.; et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015, 15, 2688–2699. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Syama, S.; Mohanan, P.V. Microfluidic based human-on-a-chip: A revolutionary technology in scientific research. Trends Food Sci. Technol. 2021, 110, 711–728. [Google Scholar] [CrossRef]
- Jang, K.-J.; Suh, K.-Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.-Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
- Musah, S.; Dimitrakakis, N.; Camacho, D.M.; Church, G.M.; Ingber, D.E. Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a Glomerulus Chip. Nat. Protoc. 2018, 13, 1662–1685. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; LesherPerez, S.C.; Kim, B.C.C.; Yamanishi, C.; Labuz, J.M.; Leung, B.; Takayama, S. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication 2016, 8, 015021. [Google Scholar] [CrossRef] [PubMed]
- Guenat, O.T.; Berthiaume, F. Incorporating mechanical strain in organs-on-a-chip: Lung and skin. Biomicrofluidics 2018, 12, 042207. [Google Scholar] [CrossRef]
- Blume, C.; Reale, R.; Held, M.; Millar, T.M.; Collins, J.E.; Davies, D.E.; Morgan, H.; Swindle, E.J. Temporal Monitoring of Differentiated Human Airway Epithelial Cells Using Microfluidics. PLoS ONE 2015, 10, e0139872. [Google Scholar] [CrossRef]
- Peng, J.; Rochow, N.; Dabaghi, M.; Bozanovic, R.; Jansen, J.; Predescu, D.; DeFrance, B.; Lee, S.-Y.; Fusch, G.; Selvaganapathy, P.R.; et al. Postnatal dilatation of umbilical cord vessels and its impact on wall integrity: Prerequisite for the artificial placenta. Int. J. Artif. Organs 2018, 41, 393–399. [Google Scholar] [CrossRef]
- Dabaghi, M.; Fusch, G.; Saraei, N.; Rochow, N.; Brash, J.L.; Fusch, C.; Selvaganapathy, P.R. An artificial placenta type microfluidic blood oxygenator with double-sided gas transfer microchannels and its integration as a neonatal lung assist device. Biomicrofluidics 2018, 12, 044101. [Google Scholar] [CrossRef]
- Grosberg, A.; Nesmith, A.P.; Goss, J.A.; Brigham, M.D.; McCain, M.L.; Parker, K.K. Muscle on a chip: in vitro contractility assays for smooth and striated muscle. J. Pharmacol. Toxicol. Methods 2012, 65, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016, 16, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Ribas, J.; Sadeghi, H.; Manbachi, A.; Leijten, J.; Brinegar, K.; Zhang, Y.S.; Ferreira, L.; Khademhosseini, A. Cardiovascular Organ-on-a-Chip Platforms for Drug Discovery and Development. Appl. in vitro Toxicol. 2016, 2, 82–96. [Google Scholar] [CrossRef]
- Choi, N.W.; Cabodi, M.; Held, B.; Gleghorn, J.P.; Bonassar, L.J.; Stroock, A.D. Microfluidic scaffolds for tissue engineering. Nat. Mater. 2007, 6, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Günther, A.; Yasotharan, S.; Vagaon, A.; Lochovsky, C.; Pinto, S.; Yang, J.; Lau, C.; Voigtlaender-Bolz, J.; Bolz, S.-S. A microfluidic platform for probing small artery structure and function. Lab Chip 2010, 10, 2341–2349. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Fu, F.; Cheng, Y.; Wang, C.; Zhao, Y.; Gu, Z. Organ-on-a-Chip Systems: microengineering to biomimic living systems. Small 2016, 12, 2253–2282. [Google Scholar] [CrossRef]
- Mccuskey, R.S. The Hepatic Microvascular System in Health and Its Response to Toxicants. The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology 2008, 291, 661–671. [Google Scholar] [CrossRef]
- Kane, B.J.; Zinner, M.J.; Yarmush, M.L.; Toner, M. Liver-Specific Functional Studies in a Microfluidic Array of Primary Mammalian Hepatocytes. Anal. Chem. 2006, 78, 4291–4298. [Google Scholar] [CrossRef]
- Lee, P.J.; Hung, P.J.; Lee, L.P. An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture. Biotechnol. Bioeng. 2007, 97, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-T.; Lin, R.-Z.; Chen, R.-J.; Chin, C.-K.; Gong, S.-E.; Chang, H.-Y.; Peng, H.-L.; Hsu, L.; Yew, T.-R.; Chang, S.-F.; et al. Liver-cell patterning Lab Chip: mimicking the morphology of liver lobule tissue. Lab Chip 2013, 13, 3578–3587. [Google Scholar] [CrossRef] [PubMed]
- Chong, L.H.; Li, H.; Wetzel, I.; Cho, H.; Toh, Y.-C. A liver-immune coculture array for predicting systemic drug-induced skin sensitization. Lab Chip 2018, 18, 3239–3250. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.B.; Sodunke, T.R.; Lamontagne, J.; Cirillo, J.; Rajiv, C.; Bouchard, M.J.; Noh, M. Liver sinusoid on a chip: Long-term layered co-culture of primary rat hepatocytes and endothelial cells in microfluidic platforms. Biotechnol. Bioeng. 2015, 112, 2571–2582. [Google Scholar] [CrossRef]
- Zhou, Q.; Patel, D.; Kwa, T.; Haque, A.; Matharu, Z.; Stybayeva, G.; Gao, Y.; Diehl, A.M.; Revzin, A. Liver injury-on-a-chip: microfluidic co-cultures with integrated biosensors for monitoring liver cell signaling during injury. Lab Chip 2015, 15, 4467–4478. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, D.; Lee, S.H.; Sung, J.H. Microtechnology-based in vitro models: Mimicking liver function and pathophysiology. APL Bioeng. 2021, 5, 041505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, N.; Xie, L.; Shu, F.; Shi, Q.; Shaheen, N. A New 3D Cultured Liver Chip and Real-Time Monitoring System Based on Microfluidic Technology. Micromachines 2020, 11, 1118. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered Liver-on-a-Chip Platform to Mimic Liver Functions and Its Biomedical Applications: A Review. Micromachines 2019, 10, 676. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, H.M.U.; Khalid, M.A.U.; Kim, K.H.; Lee, S.R.; Choi, K.H. Real-time physiological sensor-based liver-on-chip device for monitoring drug toxicity. J. Micromechanics Microengineering 2020, 30, 115013. [Google Scholar] [CrossRef]
- Liu, J.; Feng, C.; Zhang, M.; Song, F.; Liu, H. Design and Fabrication of a Liver-on-a-chip Reconstructing Tissue-tissue Interfaces. Front. Oncol. 2022, 12, 959299. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef] [PubMed]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Maoz, B.M. Brain-on-a-Chip: Characterizing the next generation of advanced in vitro platforms for modeling the central nervous system. APL Bioeng. 2021, 5, 030902. [Google Scholar] [CrossRef] [PubMed]
- Maoz, B.M. Brain-on-a-Chip: Characterizing the next generation of advanced in vitro platforms for modeling the central nervous system. APL Bioeng. 2021, 5, 030902. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.I.; Sei, Y.J.; Park, H.-J.; Kim, J.; Ryu, Y.; Choi, J.J.; Sung, H.-J.; MacDonald, T.J.; Levey, A.I.; Kim, Y. Microengineered human blood–brain barrier platform for understanding nanoparticle transport mechanisms. Nat. Commun. 2020, 11, 175. [Google Scholar] [CrossRef]
- Tong, Z.; Segura-Feliu, M.; Seira, O.; Homs-Corbera, A.; del Río, J.A.; Samitier, J. A microfluidic neuronal platform for neuron axotomy and controlled regenerative studies. RSC Adv. 2015, 5, 73457–73466. [Google Scholar] [CrossRef]
- Kerman, B.E.; Kim, H.J.; Padmanabhan, K.; Mei, A.; Georges, S.; Joens, M.S.; Fitzpatrick, J.A.J.; Jappelli, R.; Chandross, K.J.; August, P.; et al. In vitro myelin formation using embryonic stem cells. Development 2015, 142, 2213–2225. [Google Scholar] [CrossRef] [PubMed]
- Holloway, P.M.; Willaime-Morawek, S.; Siow, R.; Barber, M.; Owens, R.M.; Sharma, A.D.; Rowan, W.; Hill, E.; Zagnoni, M. Advances in microfluidic in vitro systems for neurological disease modeling. J. Neurosci. Res. 2021, 99, 1276–1307. [Google Scholar] [CrossRef]
- Kincses, A.; Vigh, J.P.; Petrovszki, D.; Valkai, S.; Kocsis, A.E.; Walter, F.R.; Lin, H.-Y.; Jan, J.-S.; Deli, M.A.; Dér, A. The Use of Sensors in Blood-Brain Barrier-on-a-Chip Devices: Current Practice and Future Directions. Biosensors 2023, 13, 357. [Google Scholar] [CrossRef]
- Motallebnejad, P.; Thomas, A.; Swisher, S.L.; Azarin, S.M. An isogenic hiPSC-derived BBB-on-a-chip. Biomicrofluidics 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Pangalos, M.N.; Schechter, L.E.; Hurko, O. Drug development for CNS disorders: strategies for balancing risk and reducing attrition. Nat. Rev. Drug Discov. 2007, 6, 521–532. [Google Scholar] [CrossRef]
- Muller, Q.; Beaudet, M.-J.; de Serres-Bérard, T.; Bellenfant, S.; Flacher, V.; Berthod, F. Development of an innervated tissue-engineered skin with human sensory neurons and Schwann cells differentiated from iPS cells. Acta Biomater. 2018, 82, 93–101. [Google Scholar] [CrossRef]
- Bespalov, A.; Steckler, T.; Altevogt, B.; Koustova, E.; Skolnick, P.; Deaver, D.; Millan, M.J.; Bastlund, J.F.; Doller, D.; Witkin, J.; et al. Failed trials for central nervous system disorders do not necessarily invalidate preclinical models and drug targets. Nat. Rev. Drug Discov. 2016, 15, 516. [Google Scholar] [CrossRef]
- Gribi, S.; du Bois de Dunilac, S.; Ghezzi, D.; Lacour, S.P. A microfabricated nerve-on-a-chip platform for rapid assessment of neural conduction in explanted peripheral nerve fibers. Nat. Commun. 2018, 9, 4403. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.D.; McCoy, L.; Jacobs, E.; Willey, H.; Behn, J.Q.; Nguyen, H.; Bolon, B.; Curley, J.L.; Moore, M.J. Engineering a 3D functional human peripheral nerve in vitro using the Nerve-on-a-Chip platform. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Basford, C.L.; Prentice, K.J.; Hardy, A.B.; Sarangi, F.; Micallef, S.J.; Li, X.; Guo, Q.; Elefanty, A.G.; Stanley, E.G.; Keller, G.; et al. The functional and molecular characterisation of human embryonic stem cell-derived insulin-positive cells compared with adult pancreatic beta cells. Diabetologia 2012, 55, 358–371. [Google Scholar] [CrossRef] [PubMed]
- Bruin, J.E.; Erener, S.; Vela, J.; Hu, X.; Johnson, J.D.; Kurata, H.T.; Lynn, F.C.; Piret, J.M.; Asadi, A.; Rezania, A.; et al. Characterization of polyhormonal insulin-producing cells derived in vitro from human embryonic stem cells. Stem Cell Res. 2014, 12, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Abadpour, S.; Aizenshtadt, A.; Olsen, P.A.; Shoji, K.; Wilson, S.R.; Krauss, S.; Scholz, H. Pancreas-on-a-chip technology for transplantation applications. Curr. Diabetes Rep. 2020, 20, 1–13. [Google Scholar] [CrossRef]
- Shik Mun, K.; Arora, K.; Huang, Y.; Yang, F.; Yarlagadda, S.; Ramananda, Y.; Abu-El-Haija, M.; Palermo, J.J.; Appakalai, B.N.; Nathan, J.D.; et al. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Lee, J.S.; Romero, R.; Han, Y.M.; Kim, H.C.; Kim, C.J.; Hong, J.-S.; Huh, D. Placenta-on-a-chip: a novel platform to study the biology of the human placenta. J. Matern. Neonatal Med. 2016, 29, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Ponmozhi, J.; Dhinakaran, S.; Varga-Medveczky, Z.; Fónagy, K.; Bors, L.A.; Iván, K.; Erdő, F. Development of skin-on-a-chip platforms for different utilizations: Factors to be considered. Micromachines 2021, 12, 294. [Google Scholar] [CrossRef] [PubMed]
- Józsa, L.; Nemes, D.; Pető, Á.; Kósa, D.; Révész, R.; Bácskay, I.; Haimhoffer, Á; Vasvári, G. Recent Options and Techniques to Assess Improved Bioavailability: in vitro and Ex Vivo Methods. Pharmaceutics 2023, 15, 1146. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.F.E.; di Cio, S.; Connelly, J.T.; Gautrot, J.E. Design of an Integrated Microvascularized Human Skin-on-a-Chip Tissue Equivalent Model. Front. Bioeng. Biotechnol. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, H.; Sung, G.Y. An Interleukin-4 and Interleukin-13 Induced Atopic Dermatitis Human Skin Equivalent Model by a Skin-On-A-Chip. Int. J. Mol. Sci. 2022, 23, 2116. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.-H.; Mayhew, C.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.; McCauley, H.A.; Broda, T.R.; Kofron, M.J.; Wells, J.M.; Hong, C.I. Human stomach-on-a-chip with luminal flow and peristaltic-like motility. Lab Chip 2018, 18, 3079–3085. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lieleg, O.; Jang, S.; Ribbeck, K.; Han, J. A microfluidic in vitro system for the quantitative study of the stomach mucus barrier function. Lab Chip 2012, 12, 4071–4079. [Google Scholar] [CrossRef] [PubMed]
- Ostrovidov, S.; Hosseini, V.; Ahadian, S.; Fujie, T.; Parthiban, S.P.; Ramalingam, M.; Bae, H.; Kaji, H.; Khademhosseini, A. Skeletal muscle tissue engineering: methods to form skeletal myotubes and their applications. Tissue Eng. Part B: Rev. 2014, 20, 403–436. [Google Scholar] [CrossRef]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell–ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef]
- Agrawal, G.; Aung, A.; Varghese, S. Skeletal muscle-on-a-chip: an in vitro model to evaluate tissue formation and injury. Lab Chip 2017, 17, 3447–3461. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.; Aung, A.; Varghese, S. Skeletal muscle-on-a-chip: an in vitro model to evaluate tissue formation and injury. Lab Chip 2017, 17, 3447–3461. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Venzac, B.; Burgers, T.; le Gac, S.; Schlatt, S. Microfluidics in male reproduction: is ex vivo culture of primate testis tissue a future strategy for ART or toxicology research? Mol. Hum. Reprod. 2020, 26, 179–192. [Google Scholar] [CrossRef] [PubMed]
- AbuMadighem, A.; Shuchat, S.; Lunenfeld, E.; Yossifon, G.; Huleihel, M. Testis on a chip—A microfluidic three-dimensional culture system for the development of spermatogenesis in-vitro. Biofabrication 2022, 14, 035004. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.A.; Cardenas, E.R.; Jiang, J.X. Osteocytes and bone metastasis. Front. Endocrinol. 2020, 11, 567844. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-H.V.; Middleton, K.; You, L.; Sun, Y. A review of microfluidic approaches for investigating cancer extravasation during metastasis. Microsystems Nanoeng. 2018, 4, 1–13. [Google Scholar] [CrossRef]
- Sharma, K.; et al. Dynamic persistence of UPEC intracellular bacterial communities in a human bladder-chip model of urinary tract infection. Elife 2021, 10, e66481. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, Y.; Song, Y. Microfluidics-Based Urine Biopsy for Cancer Diagnosis: Recent Advances and Future Trends. ChemMedChem 2022, 17. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Chen, Y.-J.; Lai, C.-S.; Chen, Y.-T.; Chen, C.-L.; Yu, J.-S.; Chang, Y.-S. A negative-pressure-driven microfluidic chip for the rapid detection of a bladder cancer biomarker in urine using bead-based enzyme-linked immunosorbent assay. Biomicrofluidics 2013, 7. [Google Scholar] [CrossRef]
- Lv, S.; Yu, J.; Zhao, Y.; Li, H.; Zheng, F.; Liu, N.; Li, D.; Sun, X. A Microfluidic Detection System for Bladder Cancer Tumor Cells. Micromachines 2019, 10, 871. [Google Scholar] [CrossRef]
- Wang, L.D.; Wagers, A.J. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat. Rev. Mol. Cell Biol. 2011, 12, 643–655. [Google Scholar] [CrossRef] [PubMed]
- di Maggio, N.; Piccinini, E.; Jaworski, M.; Trumpp, A.; Wendt, D.J.; Martin, I. Toward modeling the bone marrow niche using scaffold-based 3D culture systems. Biomaterials 2011, 32, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Csaszar, E.; Kirouac, D.C.; Yu, M.; Wang, W.; Qiao, W.; Cooke, M.P.; Boitano, A.E.; Ito, C.; Zandstra, P.W. Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell 2012, 10, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-X.; Liang, G.-T.; Yan, W.; Zhang, Q.; Wang, W.; Zhou, X.-M.; Liu, D.-Y. Artificial Uterus on a Microfluidic Chip. Chin. J. Anal. Chem. 2013, 41, 467–472. [Google Scholar] [CrossRef]
- Tantengco, O.A.G.; Richardson, L.; Radnaa, E.; Kammala, A.K.; Kechichian, T.; Ganguly, E.; Kim, S.; Han, A.; Medina, P.M.; Menon, R. Cervix-on-a-chip for investigating ascending Ureaplasma parvum infection in pregnancy. Am. J. Obstet. Gynecol. 2022, 226, S12–S13. [Google Scholar] [CrossRef]
- Tantengco, O.A.G.; Richardson, L.S.; Medina, P.M.B.; Han, A.; Menon, R. Organ-on-chip of the cervical epithelial layer: A platform to study normal and pathological cellular remodeling of the cervix. FASEB J. 2021, 35, e21463. [Google Scholar] [CrossRef] [PubMed]
- Ataç, B.; Wagner, I.; Horland, R.; Lauster, R.; Marx, U.; Tonevitsky, A.G.; Azar, R.P.; Lindner, G. Skin and hair on-a-chip: in vitro skin models versus ex vivo tissue maintenance with dynamic perfusion. Lab Chip 2013, 13, 3555–3561. [Google Scholar] [CrossRef]
- Boardman, D.A.; Levings, M.K. Cancer immunotherapies repurposed for use in autoimmunity. Nat. Biomed. Eng. 2019, 3, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Fransen, M.F.; et al. Tumor-draining lymph nodes are pivotal in PD-1/PD-L1 checkpoint therapy. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Kennedy, L.B.; Salama, A.K. A review of cancer immunotherapy toxicity. CA Cancer J Clin 2020, 70, 86–104. [Google Scholar] [CrossRef]
- Shanti, A.; Hallfors, N.; Petroianu, G.A.; Planelles, L.; Stefanini, C. Lymph Nodes-On-Chip: Promising Immune Platforms for Pharmacological and Toxicological Applications. Front. Pharmacol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Stone, L. A vagina on a chip to model microbiome–host interactions. Nat. Rev. Urol. 2023, 20, 64. [Google Scholar] [CrossRef] [PubMed]
- Gulati, A.; Jorgenson, A.; Junaid, A.; Ingber, D.E. Modeling Healthy and Dysbiotic Vaginal Microenvironments in a Human Vagina-on-a-Chip. Journal of Visualized Experiments 2024. [Google Scholar] [CrossRef] [PubMed]
- Marx, U.; Walles, H.; Hoffmann, S.; Lindner, G.; Horland, R.; Sonntag, F.; Klotzbach, U.; Sakharov, D.; Tonevitsky, A.; Lauster, R. ‘Human-on-a-chip’ Developments: A Translational Cutting-edge Alternative to Systemic Safety Assessment and Efficiency Evaluation of Substances in Laboratory Animals and Man? Altern. Lab. Anim. 2012, 40, 235–257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Montgomery, M.; Chamberlain, M.D.; Ogawa, S.; Korolj, A.; Pahnke, A.; Wells, L.A.; Massé, S.; Kim, J.; Reis, L.; et al. Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat. Mater. 2016, 15, 669–678. [Google Scholar] [CrossRef] [PubMed]
- van Midwoud, P.M.; Merema, M.T.; Verpoorte, E.; Groothuis, G.M.M. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip 2010, 10, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, E.; Rasponi, M. Liver–Heart on chip models for drug safety. APL Bioeng. 2021, 5, 031505. [Google Scholar] [CrossRef] [PubMed]
- Khalid, M.A.U.; Kim, Y.S.; Ali, M.; Lee, B.G.; Cho, Y.-J.; Choi, K.H. A lung cancer-on-chip platform with integrated biosensors for physiological monitoring and toxicity assessment. Biochem. Eng. J. 2020, 155, 107469. [Google Scholar] [CrossRef]
- Cho, S.; Lee, S.; Ahn, S.I. Design and engineering of organ-on-a-chip. Biomed. Eng. Lett. 2023, 13, 97–109. [Google Scholar] [CrossRef]
- Liu, H.; Bolonduro, O.A.; Hu, N.; Ju, J.; Rao, A.A.; Duffy, B.M.; Huang, Z.; Black, L.D.; Timko, B.P. Heart-on-a-Chip Model with Integrated Extra- and Intracellular Bioelectronics for Monitoring Cardiac Electrophysiology under Acute Hypoxia. Nano Lett. 2020, 20, 2585–2593. [Google Scholar] [CrossRef]
- Valiei, A.; Aminian-Dehkordi, J.; Mofrad, M.R.K. Gut-on-a-chip models for dissecting the gut microbiology and physiology. APL Bioeng. 2023, 7, 011502. [Google Scholar] [CrossRef] [PubMed]
- Phan, D.T.; Bender, R.H.F.; Andrejecsk, J.W.; Sobrino, A.; Hachey, S.J.; George, S.C.; Hughes, C.C. Blood–brain barrier-on-a-chip: Microphysiological systems that capture the complexity of the blood–central nervous system interface. Exp. Biol. Med. 2017, 242, 1669–1678. [Google Scholar] [CrossRef]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Livingston, C.A.; Fabre, K.M.; Tagle, D.A. Facilitating the commercialization and use of organ platforms generated by the microphysiological systems (Tissue Chip) program through public–private partnerships. Comput. Struct. Biotechnol. J. 2016, 14, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Radisic, M. Organ-on-a-chip devices advance to market. Lab Chip 2017, 17, 2395–2420. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Peng, Y.; Li, H.; Chen, W. Organ-on-a-chip: a new paradigm for drug development. Trends Pharmacol. Sci. 2021, 42, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Torras, N.; García-Díaz, M.; Fernández-Majada, V.; Martínez, E. Mimicking Epithelial Tissues in Three-Dimensional Cell Culture Models. Front. Bioeng. Biotechnol. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Khodabukus, A.; Madden, L.; Prabhu, N.K.; Koves, T.R.; Jackman, C.P.; Muoio, D.M.; Bursac, N. Electrical stimulation increases hypertrophy and metabolic flux in tissue-engineered human skeletal muscle. Biomaterials 2019, 198, 259–269. [Google Scholar] [CrossRef]
- Yildirimer, L.; Zhang, Q.; Kuang, S.; Cheung, C.-W.J.; Chu, K.A.; He, Y.; Yang, M.; Zhao, X. Engineering three-dimensional microenvironments towards in vitro disease models of the central nervous system. Biofabrication 2019, 11, 032003. [Google Scholar] [CrossRef]
- Busek, M.; Aizenshtadt, A.; Koch, T.; Frank, A.; Delon, L.; Martinez, M.A.; Golovin, A.; Dumas, C.; Stokowiec, J.; Gruenzner, S.; et al. Pump-less, recirculating organ-on-a-chip (rOoC) platform. Lab Chip 2023, 23, 591–608. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).