2.5. Machine Learning Methods for Grain Yield Predictions
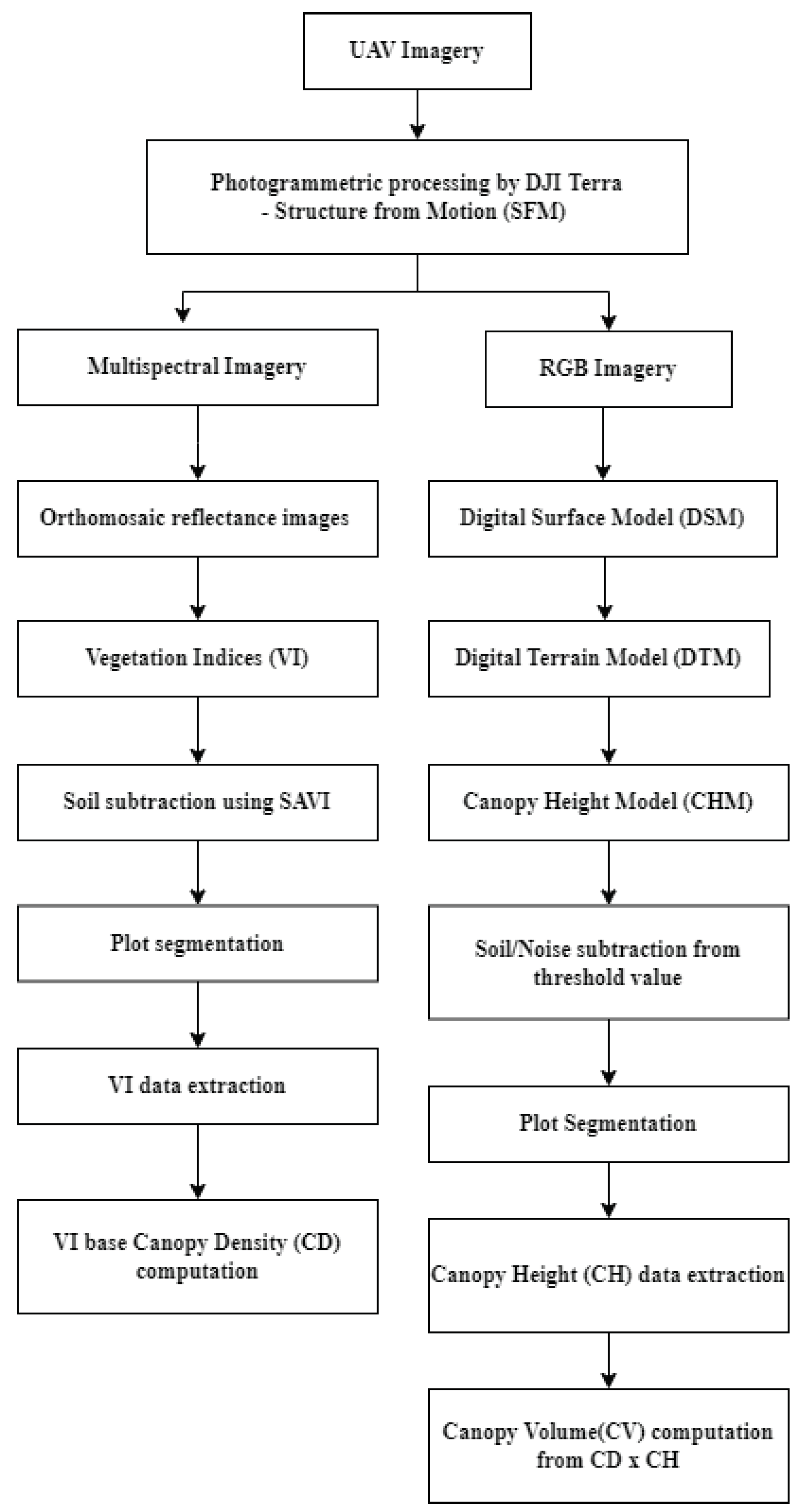
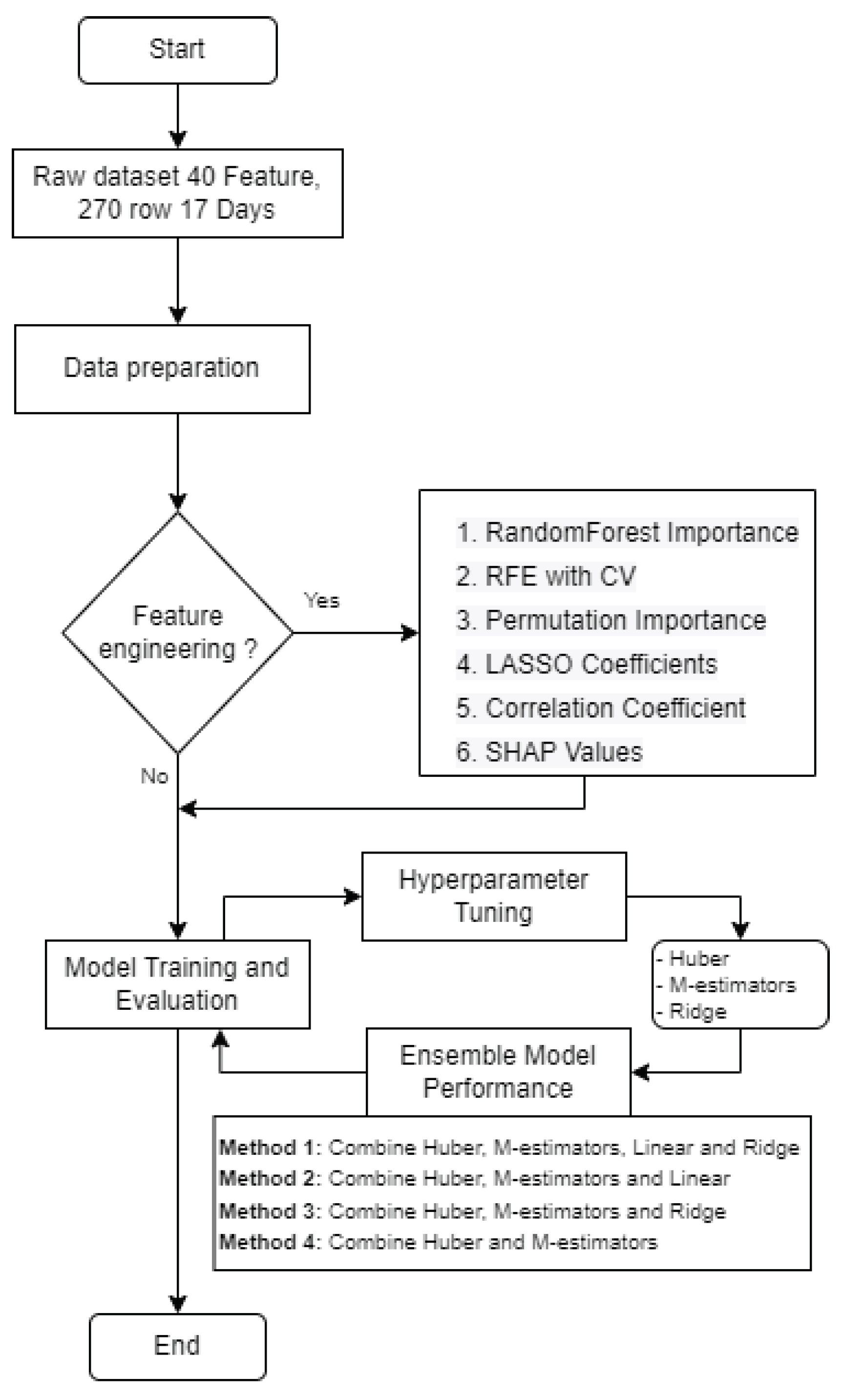
In this study, we present a comprehensive flow chart designed to predict crop yields using machine learning (ML) techniques, as depicted in
Figure 4. The flow chart comprises two primary components: the first part involves the analysis of multi-temporal images, while the second part focuses on the utilization of ML algorithms for crop yield prediction.
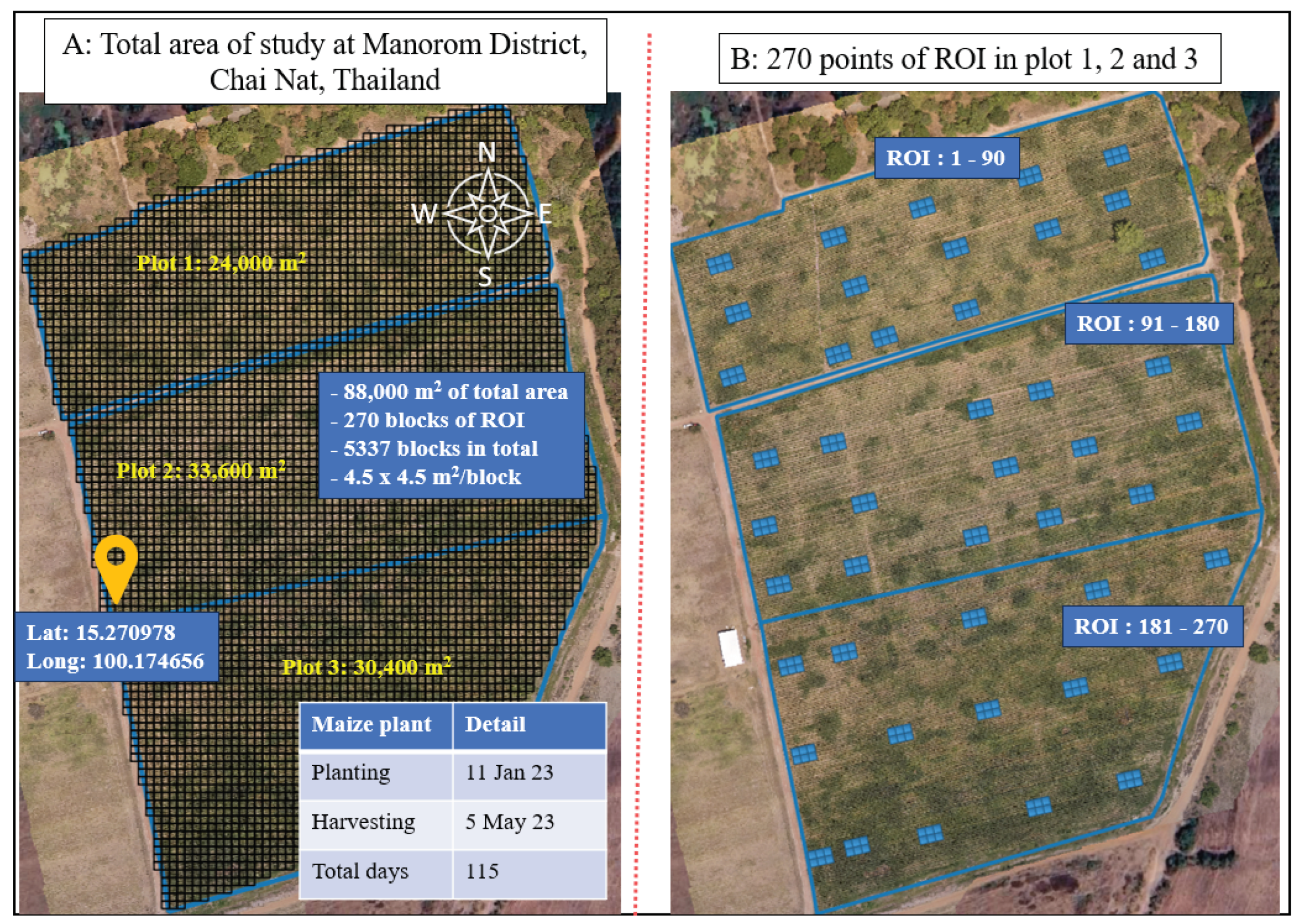
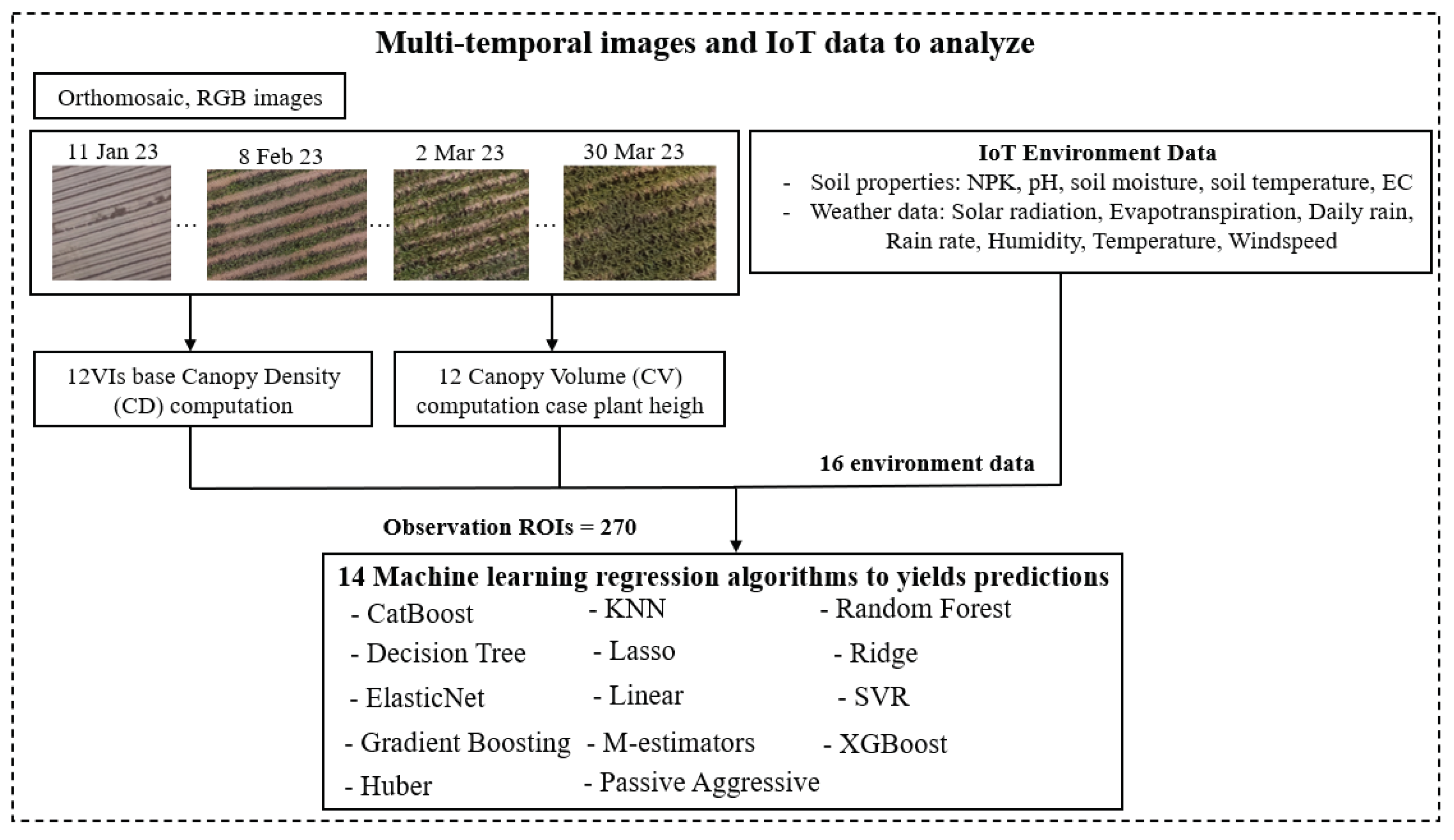
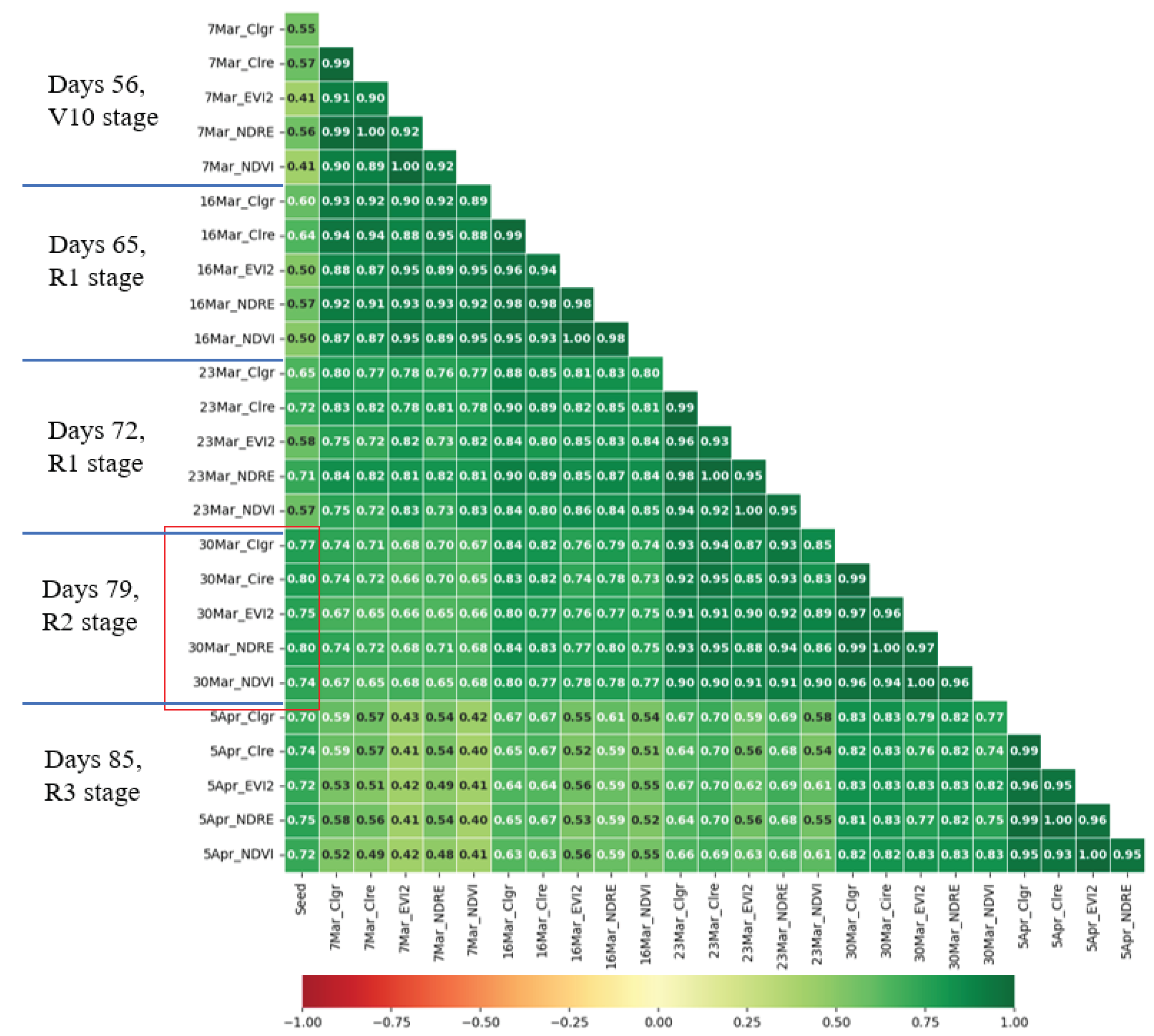
Within the first part of the diagram, multi-temporal images are subjected to thorough analysis, and various essential features are extracted. These features include vegetation indices and chlorophyll contents, among others. The imagery, spanning 17 different dates, is meticulously spliced together, incorporating precise geographic coordinates obtained through ground control points (GCPs) using standard procedures. This procedure results in the acquisition of 4,590 subsample plots (17 dates × 270 plots = 4,590) which are stitched 14,195 images from DJI Terra software as mentioned in the image processing section, each representing distinct regions of interest (ROI) within the agricultural fields. These sub-sample images of plot serve as the basis for computing VI-based canopy density, which facilitates the classification of pixels into two categories: green pixels and non-green pixels with potentially disruptive background elements, such as soil. By focusing only on green pixels, the values of twelve vegetation indices are computed for the sub-sample images. This information is then employed to establish linear relationships between chlorophyll contents, maize yields, and the vegetation indices.
To assess the performance of the various vegetation indices during critical growth stages, we obtain the R-squared (R2) values. These R2 values serve as indicators of the predictive capabilities of the different vegetation indices. By identifying the most effective indices during important growth stages, we can gain valuable insights into growth monitoring and make more accurate yield predictions.
Overall, this flow chart offers a robust methodology for predicting crop yields by leveraging the power of machine learning and in-depth analysis of multi-temporal images. The extracted vegetation indices and their associations with chlorophyll contents and maize yields are vital in understanding the growth dynamics and making informed decisions in agricultural practices.
ML algorithms outperform and show great potential in various applications such as object detection, image classification, recognition patterns, computer vision, and other domains. In supervised learning, sample data are divided into training and testing sets to build non-linear relationships between independent and dependent variables. The effectiveness of trained models is assessed using test samples. Several machine learning algorithms such as backpropagation neural network (BP), support vector machine (SVM), random forest (RF), and extreme learning machine (ELM) can be used for supervised learning tasks such as classification and regression. XGBoost is also a gradient boosting algorithm that is used for both regression and classification tasks. LASSO is a regression method that performs feature selection using L1 regularization. A hybrid approach can use XGBoost and LASSO together for feature selection and model building. Many studies have evaluated BP, SVM, RF, ELM, LASSO, and XGBoost in remote sensing domains.
In this study, the authors utilized twelve vegetation indices (VIs) based on canopy density (CD), which were calculated from images collected over a long time series for each plot. These VIs served as independent variables, and the maize yields in each plot were used as dependent variables. Additionally, canopy volume (CV) was derived by multiplying CD with plant height. We utilized a diverse set of 14 machine learning algorithms to predict the growth stage and productivity of maize. These algorithms include CatBoost Regression, decision tree Regression, ElasticNet Regression, gradient boosting Regression, Huber Regression, K-nearest neighbors Regression (KNN), Lasso Regression, linear Regression, M estimators, passive-aggressive Regression, random forest (RF) Regression, Ridge Regression, support vector Regression (SVR), and XGBoost Regression. This investigation employed a diverse array of machine learning algorithms, each meticulously chosen for its distinct characteristics to thoroughly analyze multi-temporal satellite imagery data and forecast crop yields. Each model has the following characteristics:
Model 1 (CatBoost Regression): This algorithm was chosen for its adeptness in handling categorical features prevalent in such data. By employing ordered boosting and symmetric trees, CatBoost effectively captured temporal variations and seasonality patterns, ensuring a high level of predictive accuracy.
Model 2 (Decision Tree Regression): Selected to interpret the impact of different temporal features on crop yields, Decision Tree Regression leveraged its hierarchical structure to represent complex relationships within the data.
Model 3 (ElasticNet Regression): Addressing multicollinearity and performing feature selection simultaneously, ElasticNet Regression proved crucial for high-dimensional multi-temporal data. Its combination of L1 and L2 regularization techniques played a pivotal role in achieving these objectives.
Model 4 (Gradient Boosting Regression): Demonstrating efficacy in handling complex relationships and temporal dependencies, Gradient Boosting Regression built an ensemble of decision trees sequentially, enhancing predictive accuracy in the process.
Model 5 (Huber Regression): Chosen for its robustness in accommodating outliers prevalent in agricultural datasets, Huber Regression minimized the impact of extreme values through a parameter balancing Mean Absolute Error (MAE) and Mean Squared Error (MSE).
Model 6 (K-Nearest Neighbors Regression - KNN): Predicting grain yields based on similar multi-temporal patterns observed in the past, KNN Regression relied on the assumption that analogous temporal patterns yield similar crop yields. However, consideration of an appropriate k value and computational cost for larger datasets was warranted.
Model 7 (Lasso Regression): Serving as a feature selection technique, Lasso Regression incorporated L1 regularization to encourage some feature coefficients to be exactly zero, facilitating the identification of key temporal features.
Model 8 (Linear Regression): Providing a baseline model to understand the overall trend between crop yields and multi-temporal features, Linear Regression assumed a linear relationship, offering valuable insights into the general temporal trend of grain yield.
Model 9 (M Estimators): Chosen for their robustness in handling outliers and noise in agricultural datasets, M Estimators provided flexibility in choosing the loss function, making them suitable for data with non-Gaussian noise.
Model 10 (Passive Aggressive Regression): As an online learning algorithm, Passive Aggressive Regression adapted quickly to temporal changes in crop yield patterns, making it suitable for real-time predictions, particularly in rapidly evolving agricultural conditions.
Model 11 (Random Forest Regression - RF): Handling high-dimensional multi-temporal data, Random Forest Regression provided robust predictions by building multiple decision trees on different data subsets and averaging their predictions.
Model 12 (Ridge Regression): Beneficial when dealing with multicollinear features in multi-temporal data, Ridge Regression applied L2 regularization to stabilize the model and reduce the impact of multicollinearity.
Model 13 (Support Vector Regression - SVR): Chosen for its capability to capture non-linear relationships between multi-temporal features and crop yields, SVR mapped data into a higher-dimensional space, enabling the identification of non-linear patterns.
Model 14 (XGBoost Regression): Selected for its effectiveness in various regression tasks, XGBoost Regression constructed an ensemble of decision trees using gradient boosting and regularization techniques, offering high predictive accuracy.
Collectively, these machine learning algorithms formed a comprehensive toolkit for analyzing multi-temporal satellite imagery data and predicting crop yields, effectively addressing challenges such as handling outliers, feature selection, non-linearity, and temporal dependencies.
Selection of Best-suited Models:
The selection of the best-suited models depends on the specific characteristics of the multi-temporal imagery and the nature of the grain yield data. For instance, CatBoost Regression and Gradient Boosting Regression are well-suited to capture temporal patterns and seasonality effects. ElasticNet and Lasso Regression can be useful for feature selection when dealing with high-dimensional data. Huber Regression and M Estimators are appropriate for handling outliers in crop yield data. Given the variety of scenarios and combinations of variables explored in the study, it is crucial for researchers to carefully evaluate the performance of each algorithm using the specified evaluation metrics. This thorough evaluation process will enable them to identify the most effective models for predicting grain yield in their specific agricultural context, ultimately leading to more accurate and reliable predictions.
To evaluate the performance of these various algorithms, the authors used several metrics, including mean squared error (MSE), root mean square error (RMSE), mean absolute error (MAE), and R-squared (
) as
and
where
N is the number of all samples, and
M and
P are the true values and predicted values of the yields,
represents the actual values,
represents the predicted values,
represents the absolute value, and
represents the mean of the actual values, respectively. These metrics provide insights into the accuracy and predictive power of each algorithm in estimating the growth stage and productivity of maize.
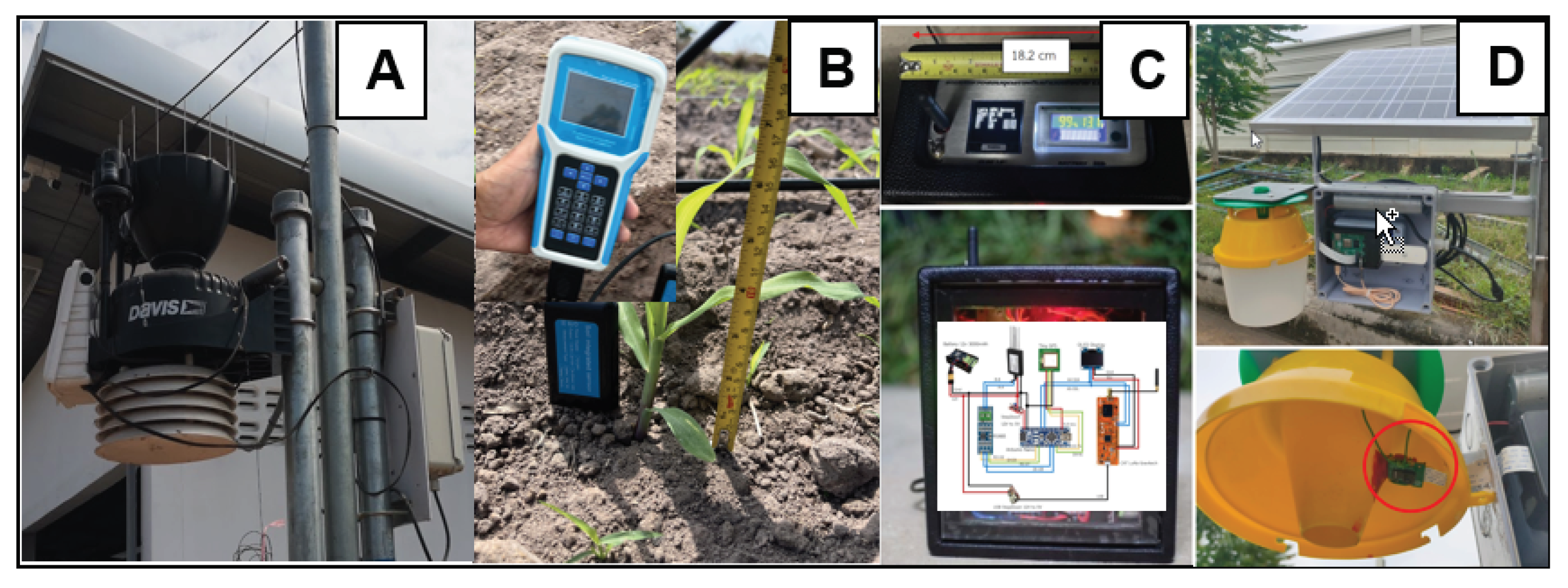
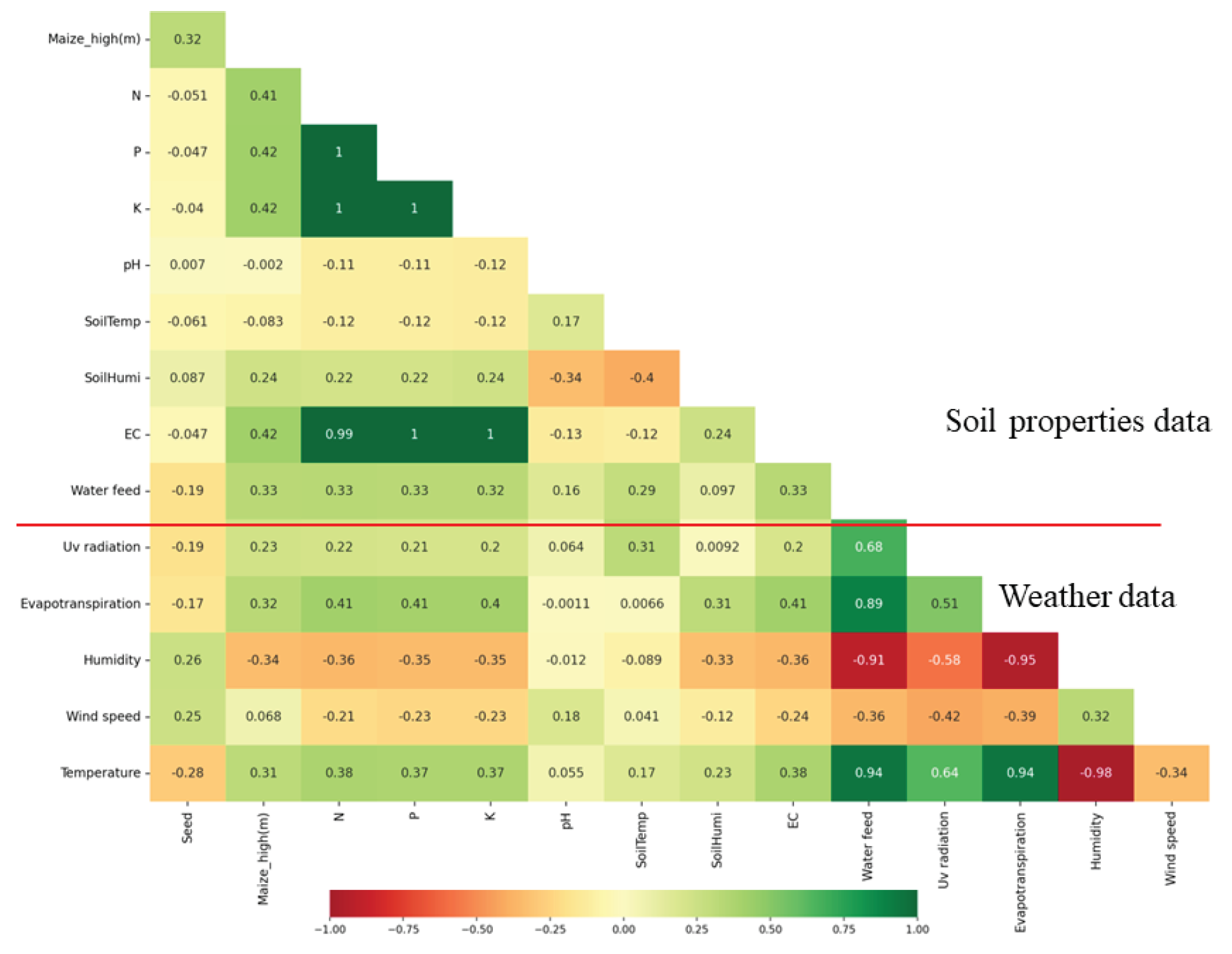
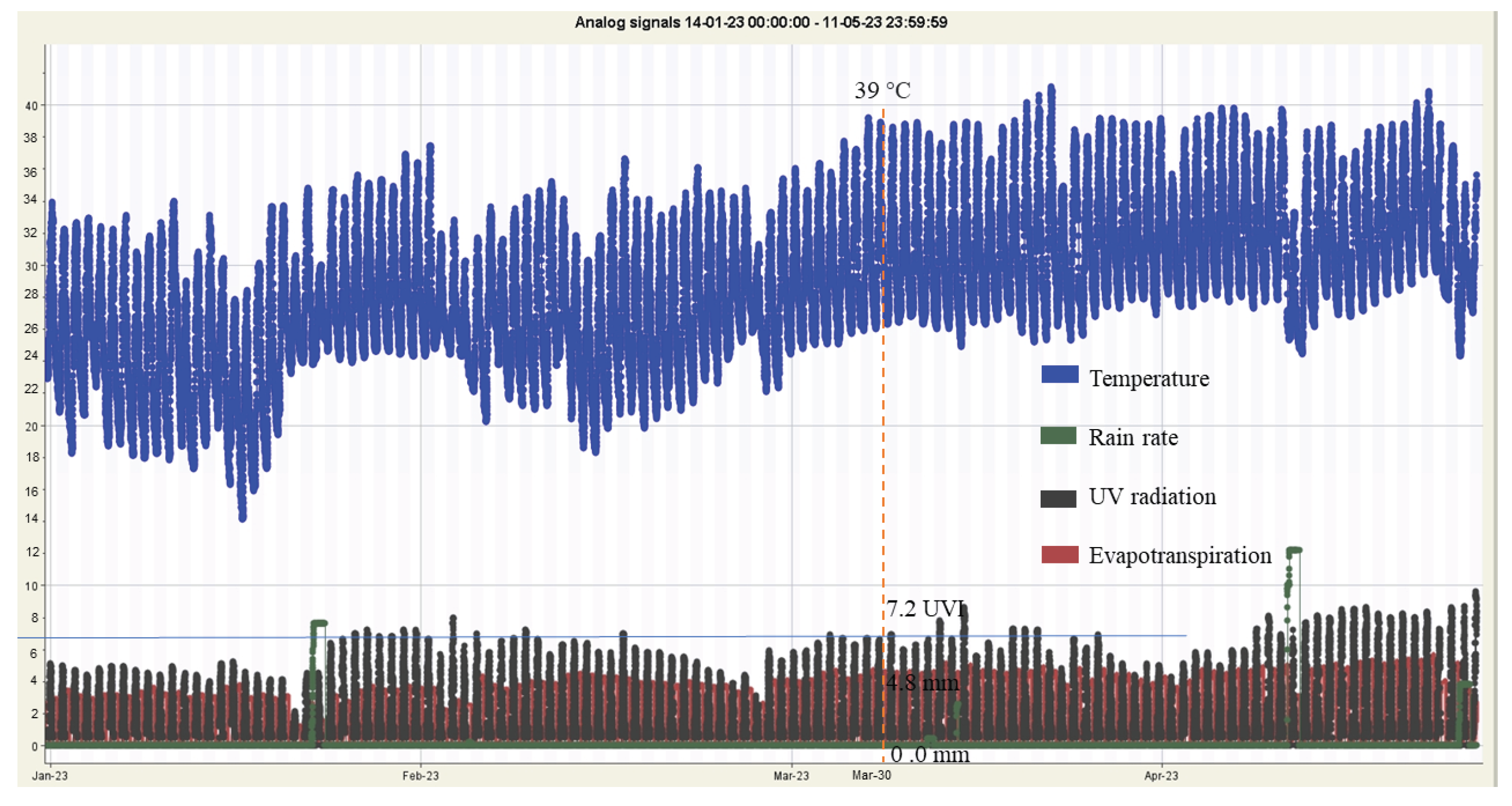
Indeed, the study encompasses a comprehensive set of forty features to feed into the machine learning models for predicting crop growth and yield. These features consist of eight factors measured from the soil, which include N, P, K, pH, soil temperature, soil humidity, EC and maize high. Additionally, there are eight parameters collected from environmental data, comprising water feed , UV radiation, evapotranspiration, daily rain, rain rate, humidity, wind speed, and temperature. Furthermore, there are twenty four feature from twelve VIs as CIgr, CIre, EVI2, GNDVI, MCARI2, MTVI2, NDRE, NDVI, NDWI, OSAVI, RDVI and RGBVI with CD and CV.
By incorporating this diverse set of forty features, the machine learning models can analyze the complex interplay between soil properties and environmental conditions that significantly influence crop health and productivity. The fusion of these factors and parameters empowers the models to make highly accurate predictions, providing valuable insights to farmers and agronomists for optimizing crop management practices, resource allocation, and decision-making in precision agriculture.
In this subsection, we introduce an ensemble machine learning model that enhances the accuracy and reliability of grain yield predictions by leveraging the strengths of multiple individual models. The ensemble approach combines the predictive power of various machine learning algorithms, optimizing their collective performance for more robust yield forecasts.
Ensemble models have gained prominence in the field of agricultural data analysis due to their ability to mitigate the limitations of individual models. By combining the outputs of multiple models, an ensemble can capture diverse patterns and relationships present in the data, leading to more accurate predictions.
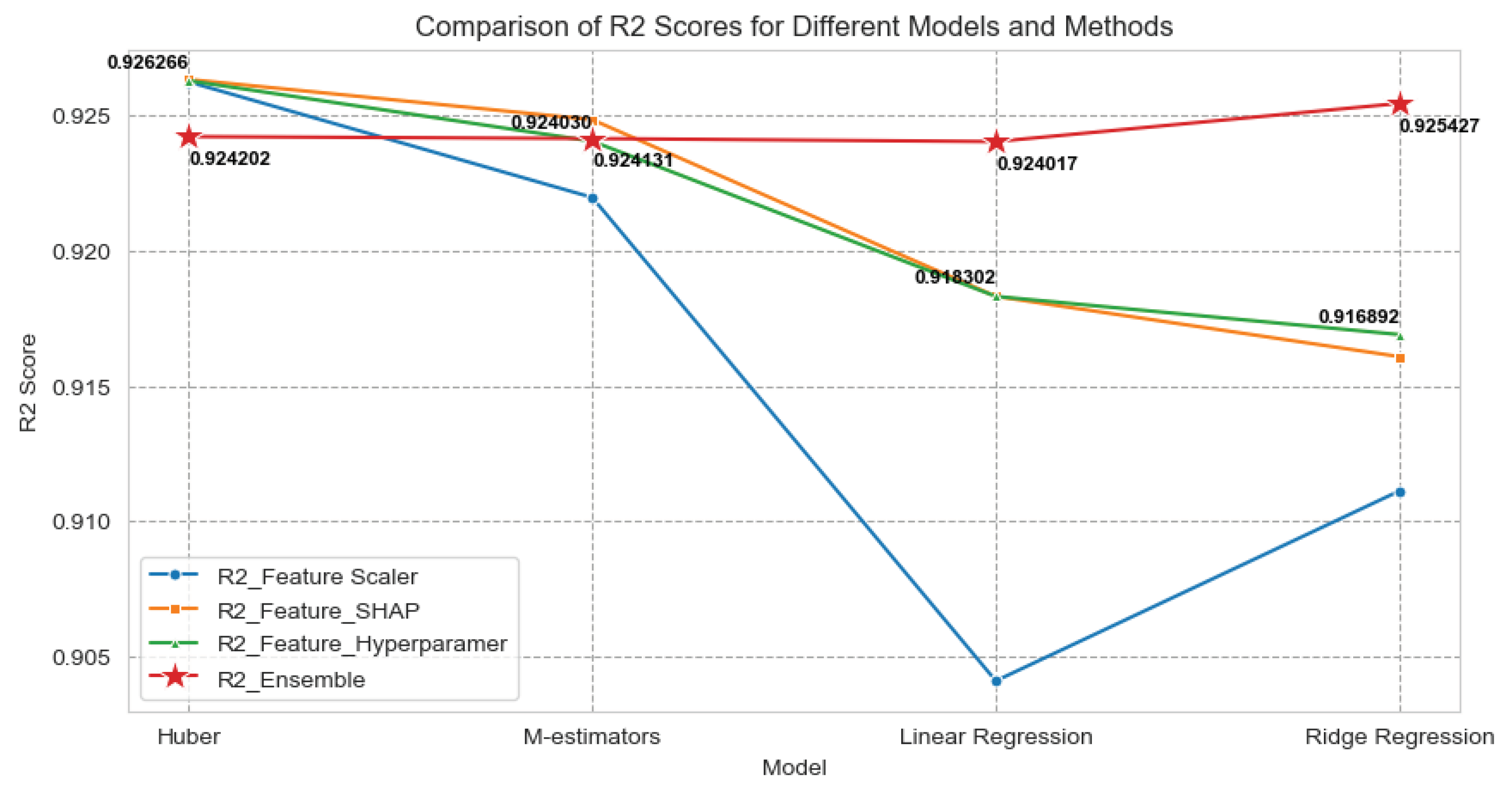
Our ensemble model comprises an arbitrary number of top-performing machine learning algorithms meticulously chosen from a pool of options, including Huber Regression, M-estimators, Linear Regression, Ridge Regression, and others. These algorithms have consistently demonstrated their effectiveness in capturing temporal variations, handling multicollinearity, and accommodating outliers within the dataset.
The ensemble model is designed to work as follows:
-
Data Preparation: To combine the data from various sources, we collect the following
- -
Twenty four VIs with CD and CV (CIgr, CIre, EVI2, GNDVI, MCARI2, MTVI2, NDRE, NDVI, NDWI, OSAVI, RDVI and RGBVI).
- -
Six teen environmental data following (maize high, N, P, K, pH, soil temperature, soil humidity, EC, water feed, uv radiation, evapotranspiration, daily rain, rain rate, humidity, wind speed, temperature).
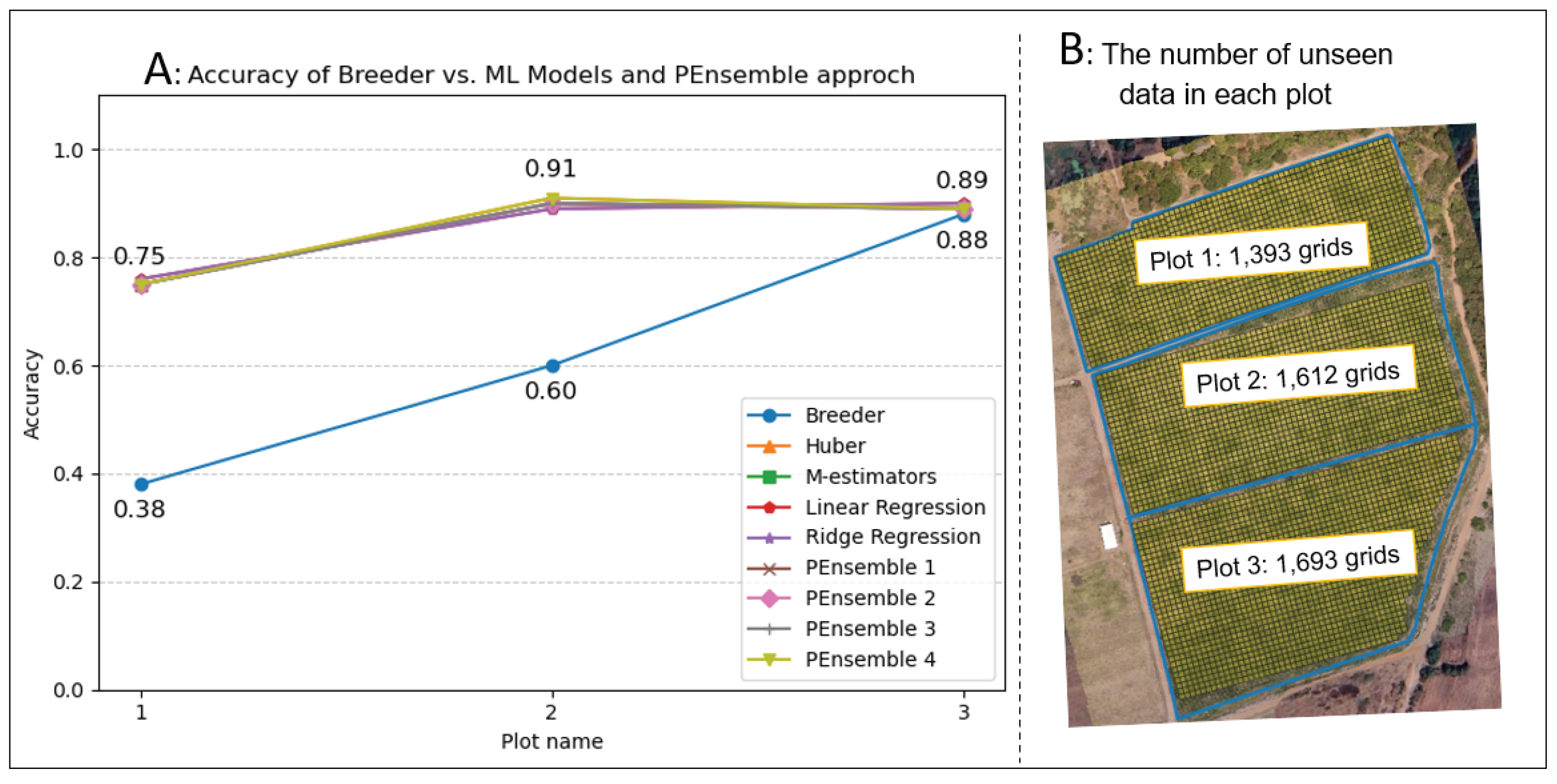
In summary, we have forty features with two hundred seventy data plots on each seventeen dates to process, with in total 137,700 records from the dataset. In addition, data cleaning, missing values handling, and null entries removal are also included.
-
Feature Engineering: Use six different feature importance techniques to extract significant features.
- -
RandomForest importance consist of a large number of individual decision trees that operate as an ensemble. Each individual tree in the random forest outputs a class prediction, and the class with the most votes becomes the model prediction. The importance of each feature is determined by the average impurity decrease calculated from all decision trees in the forest.
- -
Recursive feature elimination (RFE) with cross-validation is a method that fits the model multiple times and at each step, it removes the weakest feature (or features). RFE with cross-validation uses the cross-validation approach to find the optimal number of features.
- -
Permutation Importance can be used with any model. After a model is trained, the values in a single column of the validation data are randomly shuffled. Then the model performance metric (such as accuracy or ) is re-evaluated with the shuffled data. Features that have a significant impact on performance are considered important.
- -
LASSO Regression coefficients is a regression analysis method that performs both variable selection and regularization to enhance the prediction accuracy and interpretability of the statistical model. Features with non-zero coefficients are selected by LASSO.
- -
Correlation coefficient to measure the linear relationship between the target and the numerical features. Features that have a higher correlation with the target variable are considered important.
- -
Shapley additive explanations (SHAP) values is a unified measure of feature importance. It assigns each feature an importance value for a particular prediction.
Model Selection: We meticulously evaluate the performance of various machine learning algorithms on our grain yield prediction task. The top arbitrary number models, based on evaluation metrics such as mean squared error (MSE), root mean square error (RMSE), mean absolute error (MAE), and R-squared (), are chosen for ensemble construction.
-
Training: Each of the selected models is trained on our dataset, utilizing the same training, testing and validation splits with 70%, 20%, and 10%, respectively, to ensure consistency and fairness in the comparison. Fourteen different ML models are trained and evaluated the data. The models include:
- -
CatBoost, Decision Tree, ElasticNet, Gradient Boosting, Huber, KNN, Lasso Regression, Linear Regression, M-estimators, Passive Aggressive, RF, Ridge Regression, SVR and XGBoost.
-
Ensemble Building: We create the ensemble by combining the predictions of the top arbitrary number models using weighted averaging. The weights assigned to each model are determined based on their individual performance during training. This process optimizes the ensemble’s predictive accuracy.
The weighted ensemble prediction
is given by
where
,
, and
represent the weights assigned to each model, and
,
, and
are the predictions made by each individual model.
We fine-tune the weights (denoted as , , and ) based on the preliminary evaluation results, exploring various weight combinations to optimize the ensemble model’s performance.
Evaluation: The ensemble model’s performance is evaluated using the same evaluation metrics employed for individual models. We compare its results to those of individual models to gauge the improvement achieved through ensemble modeling.
Hyperparameter Tuning: If necessary, we fine-tune the hyperparameters of our ensemble model to further enhance its predictive capabilities. Techniques such as grid search or random search are employed for this purpose.
By incorporating the ensemble model into our grain yield prediction framework, we aim to provide more accurate and reliable forecasts, ultimately assisting farmers and agronomists in optimizing crop management practices and decision-making in precision agriculture. The fusion of multiple machine learning algorithms within the ensemble harnesses the strengths of each model, resulting in a robust tool for agricultural yield predictions.