1. Introduction
In recent years, there has been a significant growth in electronic scientific publications, driven by technological advances, the rapid expansion of the World Wide Web (WWW), and largely many of these publications emerge directly in digital format, considerably accelerating their availability [
1,
2,
3].
Digital libraries have become crucial resources for scientific and academic communities, serving not just as repositories for publications but also as platforms for information classification and analysis. This enhances the ability to group and retrieve relevant data. Accurate recording and analysis of citations and bibliographic references are particularly important.
In the digital era, the surge in scientific publications has necessitated advanced solutions for managing and processing large volumes of bibliographic data. As libraries and information repositories move towards comprehensive digitization, the need for efficient and accurate bibliographic reference segmentation has become paramount.
The recording and analysis of citations and bibliographic references not only allow measuring the impact of a publication in the scientific field but also extracting valuable information, such as the disciplines citing a specific work, the geographic regions where it is most read, or the language in which it is most cited. This enables libraries to identify needs and opportunities in their activities of material acquisition and building special collections [
4].
Given the exponential growth of scientific and academic literature, automated processes for tasks such as storage, consultation, classification, and information analysis become essential. The first step to achieve this, is the correct detection, extraction, and segmentation of bibliographic references (also known as "reference mining") within academic documents [
5].
In this paper, we address the task of bibliographic reference segmentation by comparing models based on three distinct natural language processing architectures: Conditional Random Fields (CRF), Bidirectional Long Short-Term Memory with CRF (BiLSTM+CRF), and Transformer Encoder with CRF (Transformer+CRF). These models are evaluated using the Giant corpus for training and the Cora corpus for further assessment, highlighting the capabilities and differences of each architecture in handling the complexities of bibliographic data.
The problem of bibliographic reference segmentation has been tackled using various approaches, ranging from heuristic methods to machine learning (ML) and deep learning (DL) techniques. Conditional Random Fields (CRF) stand out as the most prominent representative of ML approaches. However, DL-based approaches exhibit notable variability, as they employ diverse types of embeddings and context-capturing architectures such as LSTM or Transformers [
6,
7]. The purpose of this study is to evaluate, under uniform conditions, the three most representative architectures for segmenting bibliographic references. Despite advances in natural language processing techniques, bibliographic reference segmentation continues to present unique challenges, especially due to the variety of formats and styles, as well as the presence of specialized terminology and proper names. In this context, our study focuses on identifying the most effective architecture for this task, considering factors such as accuracy and efficiency.
We present a comparative evaluation of three natural language processing architectures and an analysis under uniform conditions, emphasizing the BiLSTM+CRF model’s superiority. This model’s ability to handle complex syntactic loads highlights the importance of selecting architecture based on specific task demands, contributing valuable insights for digital library management and automated bibliographic reference processing.
The rest of the paper is organized as follows:
Section 2 presents an overview of the various approaches that have been used to address reference segmentation.
Section 3 not only describes the data sets used but also details the preprocessing steps applied to prepare the data for model training. In
Section 4, we detail the implemented architectures, each encapsulated in a different model, and their respective training processes and evaluation.
Section 5 addresses the experimentation carried out and discusses the results. Finally,
Section 6 presents the conclusions, highlighting the effectiveness of the BiLSTM+CRF model in comparison with other techniques and discussing the implications of these findings for managing digital libraries and the automated processing of bibliographic references.
2. State-of-the-Art
The challenge of reference segmentation has persisted as an open research problem for decades, with numerous attempts made towards its efficient resolution. Each effort has approached the problem from a unique perspective. Hence, it is crucial to understand the primary function of a citation parser, which is to take an input string (formatted in a specific style such as APA, ISO, Chicago, etc.), extract the metadata, and then generate a labeled output [
6].
In 2000, the first attempts to automate the segmentation of bibliographic references emerged [
8], focusing on the syntactic analysis (parser) of online documents based on HyperText Markup Language (HTML) and simultaneously proposing the transformation of other formats like Portable Document Format (PDF) to HTML or Extensible HyperText Markup Language (XHTML). Many of the approaches consist of using different proposals for the syntactic analysis of information, using techniques similar to web scraping (a set of techniques that use software programs to extract information from websites), the use of character pattern identification, also known as regular expressions, for their use as labels (for example, the ’pp’ associated with the number of pages, etc.) that allow establishing analysis contexts for the identification and extraction of these.
Other works employ machine learning-based models for the syntactic analysis of strings containing references, such as the case of Hidden Markov Models [
9]; the clustering proposal through the TOP-K algorithm [
10]; or the Conditional Random Fields model, implemented in the GROBID and CERMINE systems [
11,
12,
13], which seems to be the technique that has given the best results.
From 2018 onwards, works based on deep learning began to emerge, improving the precision of the results obtained with machine learning. These works usually use a Long Short-Term Memory neural network architecture (LSTM), which combines its output with the Conditional Random Fields (CRF) technique [
14,
15].
It is important to mention the particular case of the ParsRec system, which has the peculiarity of being an approach based on recommendation and meta-learning. The main premise of ParsRec is "Although numerous reference parsers address the problem from different approaches, offers optimal results in all scenarios". ParsRec recommends the best analyzer, out of the ten contemplated, based on the reference string in turn [
16].
A detail worth highlighting is the fact that all the main proposals based on Deep Learning [
12,
14,
15] make use of vector representations based on Word2Vec and ELMO.
Lastly, we have the case of Choi et al. [
7], who propose a model based on transfer learning using BERT [
17] as a base, which its authors claim is the best exponent in the state-of-the-art for working with multilingual references.
4. Architectures of the Evaluated Models
For the development of each of the three models, the Flair NLP framework
4 created by [
21] was used for the following reasons:
Its ability to efficiently integrate and manage different types of embeddings.
Extensible and modular architecture makes it easy to add additional model-specific layers, such as Word Dropout and Locked Dropout.
Comprehensive documentation and practical examples available.
The models evaluated in this study share a common base architecture, which incorporates Byte Pair Encoding (BPE) and Character Embeddings for vector representation. This strategic combination is adept at capturing both the semantic essence of words and the nuanced characteristics at the character level, an approach that proves crucial for addressing variations and common errors encountered in bibliographic references. These variations and errors primarily manifest as omissions in bibliographic fields and variability in writing styles, such as the inversion of the order of names (where last names and first names may be swapped) and the inconsistent expression of volume numbers (sometimes represented in arabic numerals, other times in roman numerals). By accommodating these peculiarities, the chosen embedding strategies enhance the models’ ability to accurately process and segment bibliographic data, reflecting the complexity and diversity inherent in academic references.
In addition to these representation layers, the common architecture of the models includes several additional layers designed to optimize performance and generalization:
Word Dropout: This layer reduces overfitting by randomly "turning off" (i.e., setting to zero) some word vectors during training, which helps the model not to rely too much on specific words.
Locked Dropout: Similar to word dropout, but applied uniformly across all dimensions of a word vector at a given step. This improves the robustness of the model by preventing it from overfitting to specific patterns (token combinations) in the training data.
Embedding2NN: A layer that transforms concatenated embeddings into a representation more appropriate for processing by subsequent layers. This transformation can include non-linear operations to capture more complex relationships in the data.
Linear: A linear layer that acts as a classifier, mapping the processed representations to the target segmentation labels.
Furthermore, this study tested three models, each incorporating a specific processing layer that capitalizes on the strengths of distinct approaches. These models, set to be detailed in the following subsections, were selected based on their status as the most recently utilized and best-performing architectures in the literature for reference segmentation. The choice of these three architectures allows for an ideal comparison, as they represent the cutting edge in tackling the complexities of bibliographic data [
7,
12,
15], providing a comprehensive overview of current capabilities and identifying potential areas for innovation in reference segmentation techniques.
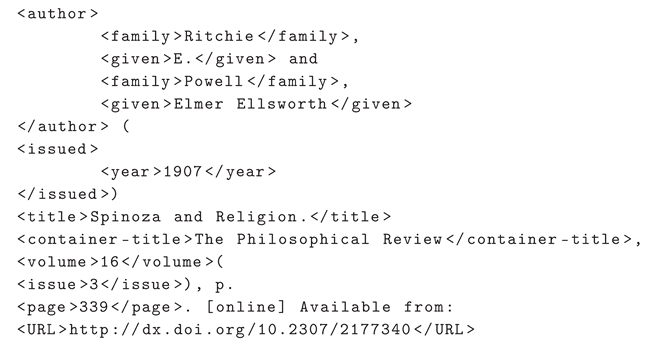
4.1. CRF Model
The CRF model focuses on using Conditional Random Fields for sequence segmentation [
22]. This technique is particularly effective in capturing dependencies and contextual patterns in sequential data (see
Figure 1).
The following outlines the layers that comprise the architecture of the CRF model:


The description of the CRF model outlines its configuration and structure in terms of components and their functionalities:
Model: "CRF": This line denotes the name of the model, the following vignettes represent the layers that compose it:
-
(embeddings): StackedEmbeddings: Refers to combining various types of word embeddings to form a rich and complex representation. It utilizes BytePairEmbeddings and CharacterEmbeddings:
- -
(list_embedding_0): BytePairEmbeddings(model=0-bpe-multi-100000-50):BytePairEmbeddings are based on Byte Pair Encoding (BPE), capturing subword-level semantics, useful for handling out-of-vocabulary (OOV) words. The specific BPE model used is indicated by "model=0-bpe-multi-100000-50", detailing its parameters.
- -
-
(list_embedding_1): CharacterEmbeddings: Uses character-level embeddings, crucial for understanding orthographic peculiarities and common errors in texts.
- *
(char_embedding): Embedding(275, 25): Defines a character embedding with a vocabulary size of 275 and 25-dimensional vectors.
- *
(char_rnn): LSTM(25, 25, bidirectional=True): A bidirectional LSTM network that processes character embeddings, with 25 units in both directions, capturing contexts before and after each character.
(word_dropout): WordDropout(p=0.05): Applies dropout at the word level with a probability of 0.05, helping prevent overfitting by randomly "turning off" words during training.
(locked_dropout): LockedDropout(p=0.5): Applies dropout uniformly across all dimensions of word vectors at a given step, with a probability of 0.5, enhancing the model’s robustness.
(embedding2nn): Linear(in_features=650, out_features=650, bias=True): A linear layer transforming concatenated embeddings representation, preparing them for processing by subsequent layers.
(linear): Linear(in_features=650, out_features=29, bias=True): Another linear layer acting as a classifier, mapping processed representations to 29 target label categories.
(loss_function): ViterbiLoss(): Employs the Viterbi loss function, suitable for sequential prediction tasks like reference segmentation.
(crf): CRF(): Indicates the use of a Conditional Random Field for sequence label prediction, optimizing the coherence and accuracy of predicted labels.
Each line succinctly summarizes a key component of the CRF model and its role in the learning and prediction process, highlighting the complexity and sophistication of the approach taken for bibliographic reference segmentation. Next, the equations describing the interactions of the components of the CRF model are presented.
The embeddings represent the combination of Byte Pair Encoding (BPE) and character embeddings. BPE captures the semantic aspects of words, while character embeddings focus on the syntactic nuances at the character level. This dual approach is crucial for processing bibliographic references, where both semantic context and specific syntactic forms (like abbreviations or special characters) play key roles.
The word dropout randomly deactivates a portion of the word embeddings during training (here, 5% as indicated by p=0.05). This method prevents the model from over-relying on particular words, encouraging it to learn more generalized patterns. This approach is particularly beneficial in bibliographic texts where certain terms, such as common author names or publication titles, might appear with significantly higher frequency than others, potentially skewing the model’s learning focus.
Locked dropout extends the dropout concept to entire embedding vectors, turning off the same set of neurons for the entire sequence. This approach helps in maintaining consistency in the representation of words across different contexts, an essential factor in processing structured bibliographic data.
This linear transformation adapts the embeddings for further processing by the neural network layers. It is a crucial step for converting the rich, but potentially unwieldy, embedding information into a more suitable format for the classification tasks ahead.
The final linear layer maps the transformed embeddings to the target classes. In this model, there are 29 classes, likely corresponding to different components of a bibliographic reference (like author, title, year, etc.). This layer is pivotal for the actual task of reference segmentation.
The CRF layer is key for capturing the dependencies between tags in the sequence. It considers not only the individual likelihood of each tag but also how likely they are in the context of neighboring tags. This sequential aspect is vital for bibliographic references, where the order and context of elements (like the sequence of authors or the structure of a citation) are crucial for accurate segmentation.
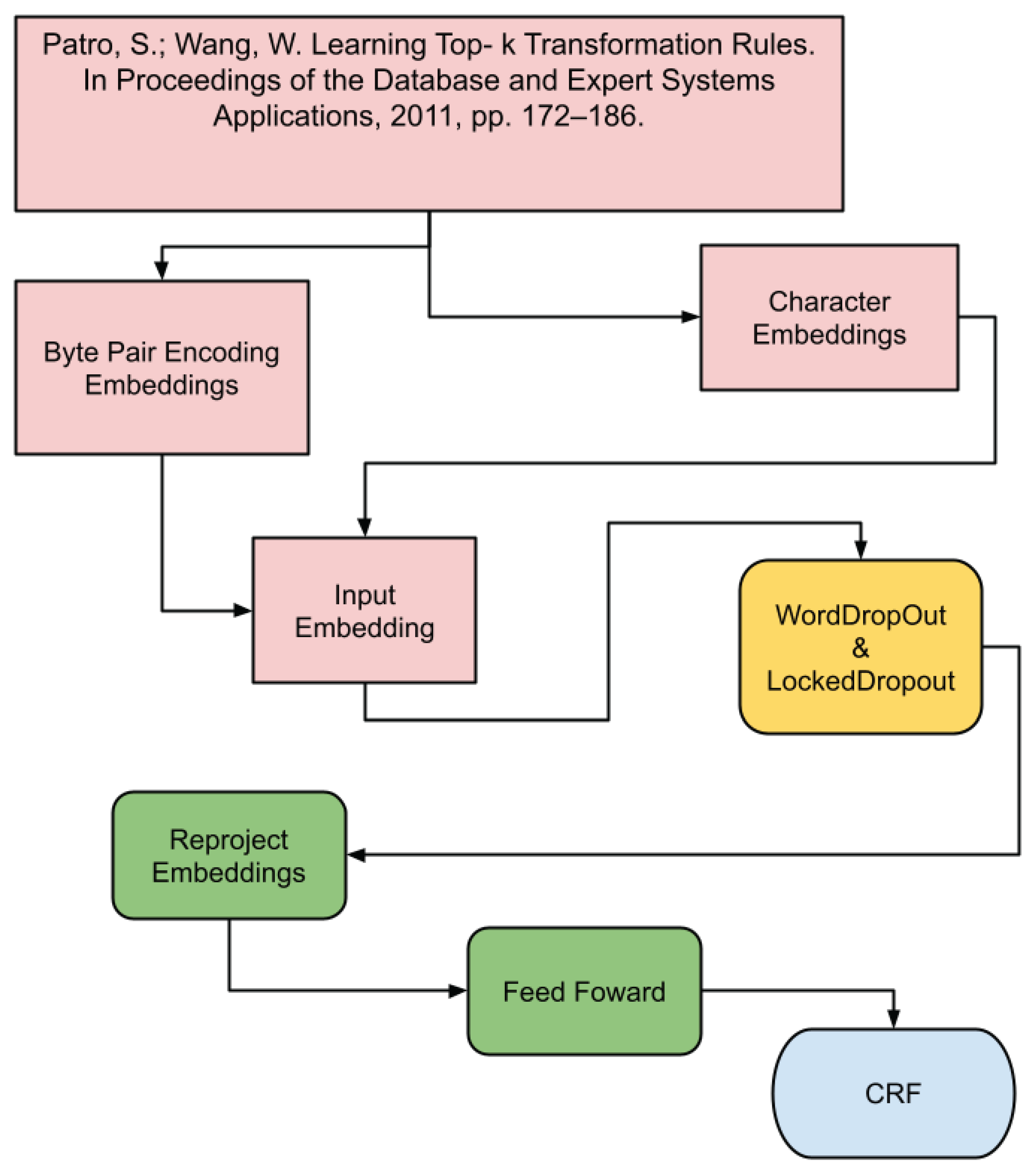
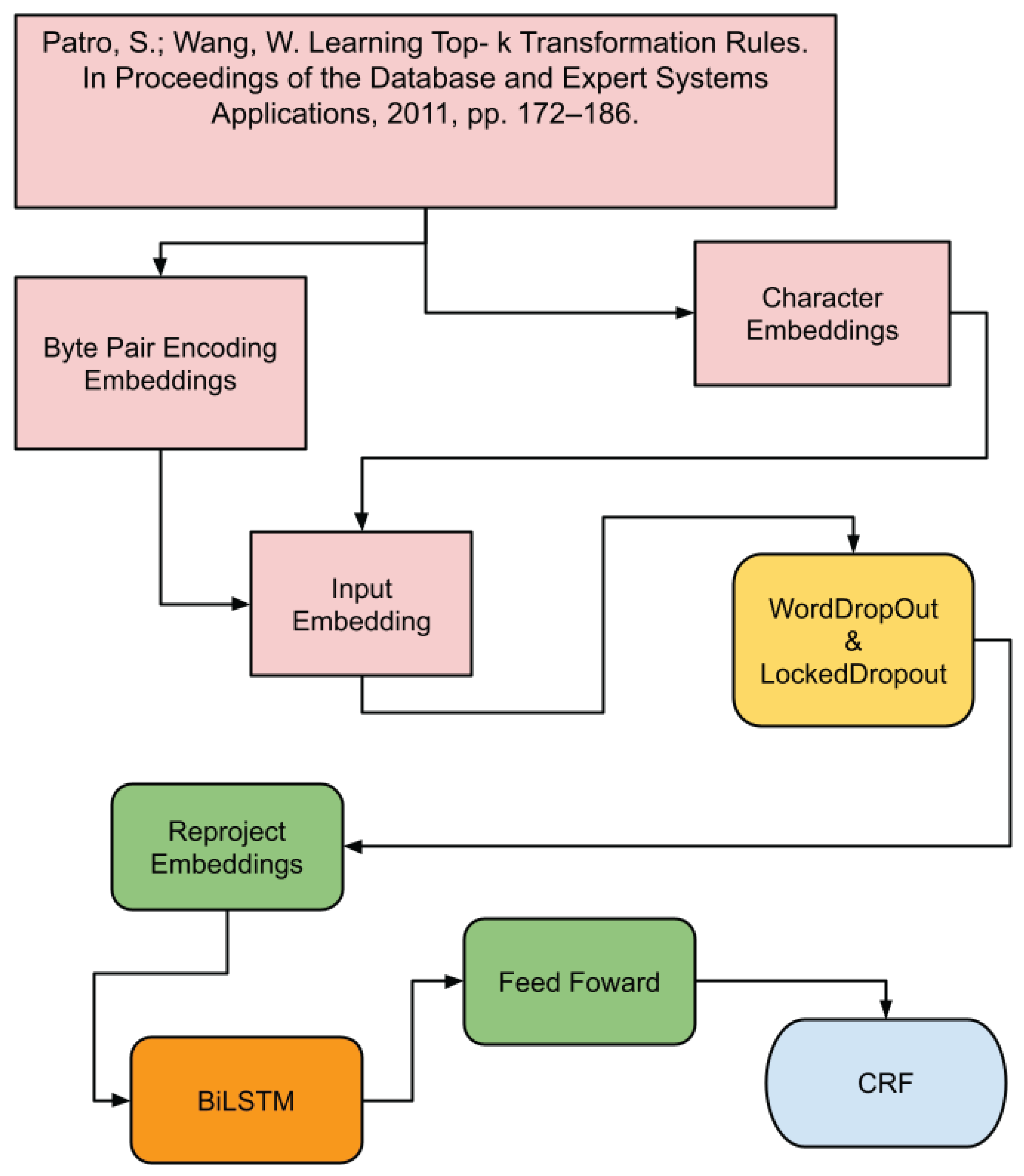
4.2. BiLSTM+CRF Model
The BiLSTM+CRF model combines Bidirectional Long Short-Term Memory (BiLSTM) networks with CRF [
23] to better capturing both past and future context in the text sequence. BiLSTMs process the sequence in both directions, offering a deeper understanding of the syntactic structure (see
Figure 2).
The following outlines the layers that comprise the architecture of the BiLSTM+CRF model:


This model is fundamentally similar to the CRF, with a key distinction being the incorporation of a Bidirectional Long Short-Term Memory ( highlighted in yellow as the rnn layer). This layer is strategically positioned between embedding2nn and the linear. The BiLSTM layer is crucial for capturing both past and future context, which is particularly beneficial for structured tasks like bibliographic reference segmentation.
The rnn layer is described below:
(rnn): LSTM(650, 256, batch_first=True, bidirectional=True): A bidirectional LSTM layer that processes sequences in both forward and backward directions. With an input size of 650 features and an output of 256 features, it captures contextual information from both past and future data points within a sequence, enhancing the model’s ability to understand complex dependencies in bibliographic reference segmentation.
Let’s delve into the mathematical aspects of this layer:
These equations represent the forward and backward passes of the BiLSTM. The forward pass processes the sequence from start to end, capturing the context up to the current word. Conversely, the backward pass processes the sequence in reverse, capturing the context from the end to the current word. The final hidden state is a concatenation of these two passes, providing a comprehensive view of the context surrounding each word.
This bidirectional context is invaluable for bibliographic data. For instance, in a list of authors, the context of surrounding names helps in accurately identifying the start and end of each author’s name. Similarly, for titles or journal names, the BiLSTM can effectively use the context to delineate these components accurately.
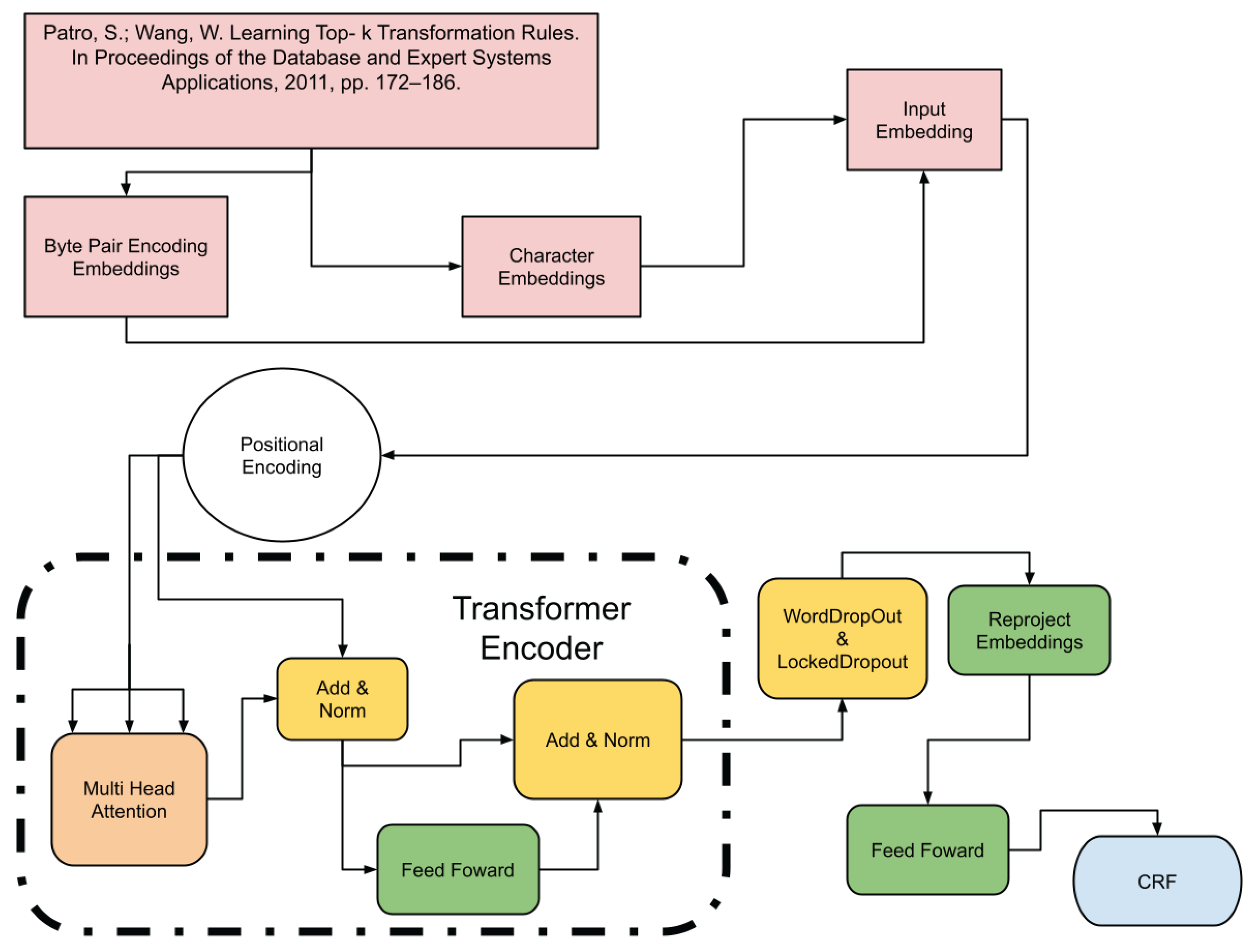
4.3. Transformer+CRF Model
Finally, the model incorporating the Transformer Encoder [
24] with CRF leverages the architecture of transformers for global attention processing of the sequence. This approach allows capturing complex and non-linear relationships in the data (see
Figure 3).
The following outlines the layers that comprise the architecture of the Transformer+CRF model:


The positional_enconder, transformer_encoder_layer and transformer_encoder layers are described below:
(positional_encoding): PositionalEncoding(dropout=0.1, inplace=False): This layer adds positional information to the input embeddings, allowing the model to capture the sequence of the text. The addition of positional encodings is crucial for attention-based models, such as transformers, as it enables them to distinguish the order of words in a sequence. The use of dropout with a probability of 0.1 helps to prevent overfitting by randomly "turning off" parts of the positional embeddings during training to enhance the model’s robustness.
(transformer_encoder_layer): CustomTransformerEncoderLayer(...) and (transformer_encoder): TransformerEncoder(...): These two layers represent the heart of the Transformer model, where the first defines the structure of a single layer of the transformer encoder, including multi-head attention, residual connections, and layer normalization, while the second stacks multiple of these layers to construct the complete encoder. The apparent redundancy between these layers is because the first specifies the architecture and configuration of an individual layer within the encoder, including specific operations such as attention and normalization, and the second encapsulates the repetition of these layers to form the complete encoder, allowing the model to process and learn from sequences with greater depth and complexity. The inclusion of BatchNorm1d in the custom layer suggests an adaptation to stabilize and accelerate training by normalizing activations across mini-batches, which is not typical in standard transformers but can offer benefits in terms of convergence and performance in specific tasks like bibliographic reference segmentation.
It should be mentioned that in the case of the Transformer+CRF model, placing the Transformer Encoder layer before the embedding2nn layer is due to the following reasons.
First, in bibliographic references, context is of vital importance. The position of a word or phrase can significantly alter its interpretation, such as distinguishing between an author’s name and the title of a work. By processing the embeddings through the positional encoder and the Transformer from the beginning, the model can more effectively capture the contextual and structural relationships specific to bibliographic references.
Second, the early inclusion of the Transformer allows for early capture of these contextual relationships. This is crucial in bibliographic references, where the structure and order of elements (authors, title, publication year, etc.) follow patterns that can be complex and varied. The Transformer, known for its ability to handle long-distance dependencies, is ideal for detecting and learning these patterns.
Finally, once contextualized representations are generated, the embedding2nn layer acts as a fine-tuner, making specific adjustments and improvements to these representations. This makes the representations even more suitable for the Named Entity Recognition (NER) task, optimally adapting them for the accurate identification of the different components within bibliographic references.
The model being analyzed here, while structurally similar to the CRF model, introduces a significant variation with the addition of positional_encoding and a transformer_encoder_layer between the embedding and word_dropout layers. This inclusion is a key differentiator, enhancing the model’s ability to process and understand the sequence data more effectively. Let’s explore these additional layers:
positional_encodingPositional encoding is added to the embeddings to provide context about the position of each word in the sequence. This is particularly crucial in transformer-based models, as they do not inherently capture sequential information. By incorporating positional data, the model can better understand the order and structure of elements in bibliographic references, such as distinguishing between the start and end of titles or authors’ lists.
The transformer encoder layer employs multi-head attention mechanisms, enabling the model to focus on different parts of the sequence simultaneously. This multi-faceted approach is beneficial for bibliographic references, as it allows the model to capture complex relationships and dependencies between different parts of a reference, such as correlating authors with titles or publication years. The layer also includes several normalization and dropout steps, ensuring stable and efficient training.
Each of these models was carefully designed and optimized for reference segmentation, considering both accuracy in identifying reference components and computational efficiency.
4.4. Training
From the Giant corpus, described in
Section 3, the training and validation sets were generated, using a Python script that transforms the XML-tagged reference into CONLL-BIO format. It is important to emphasize that each token representing punctuation will be marked with the PUNC class, as these elements are key to distinguishing the different components of a reference string. Below is an example of a reference tagged with a class.
Raw text string citation:
Ritchie, E. and Powell, Elmer Ellsworth (1907) Spinoza and Religion. The Philosophical Review, 16(3), p. 339. [online] Available from: http://dx.doi.org/10.2307/2177340
Citation in CONLL-BIO format:
This dataset comprises 125,801 records, representing all citation styles and document types. These were divided proportionally and randomly using the SciKit-Learn library
5 in Python, as follows:
In the process of developing and evaluating machine learning models, the partitioning of data into distinct sets for training, hyperparameter tuning, and performance evaluation plays a pivotal role in ensuring the model’s effectiveness and generalizability. For this study, the dataset was divided into three subsets: 80% allocated for training, 10% for hyperparameter tuning, and the remaining 10% for performance evaluation. This distribution was chosen to provide a substantial amount of data for the model to learn from during the training phase, ensuring a deep and robust understanding of the task. The allocation of 10% for hyperparameter tuning allows for sufficient experimentation with model configurations to find the optimal set of parameters that yield the best performance. Similarly, reserving 10% for the evaluation set ensures that the model’s effectiveness is tested on unseen data, offering a reliable estimate of its real-world performance. This balanced approach facilitates a comprehensive model development cycle, from learning and tuning to a fair and unbiased evaluation, critical for achieving high accuracy and generalizability in bibliographic reference segmentation tasks.
The hyperparameters used for the three models (CRF, BiLSTM+CRF, Transformer+CRF) are shown in
Table 1:
The selection of hyperparameters for the three models (CRF, BiLSTM+CRF, Transformer+CRF) was a meticulous process aimed at optimizing performance while efficiently utilizing available computational resources. As detailed in
Table 1, critical parameters such as learning rate, batch size, maximum epochs, the optimizer used, and patience were carefully adjusted through experimental iteration until the models achieved the lowest possible loss per epoch. This iterative approach ensured that each model could learn effectively from the training data, adapting its parameters to minimize error rates progressively. The chosen learning rate of 0.003 and the batch size of 1024 were particularly instrumental in this process, striking a balance between rapid learning and the capacity to process a substantial amount of data in each training iteration. Additionally, the AdamW optimizer facilitated a more nuanced adjustment of the learning rate throughout the training process, further contributing to the models’ ability to converge towards optimal solutions. The patience parameter, set at 2, allowed for early stopping to prevent overfitting, ensuring the models remained generalizable to new, unseen data. This strategic selection and tuning of hyperparameters reflect a deliberate effort to maximize the models’ learning efficiency and performance, capitalizing on the computational resources to achieve the best possible outcomes in bibliographic reference segmentation tasks.
4.5. Model Evaluation
The evaluation of the Transformer+CRF, BiLSTM+CRF, and CRF models, was conducted using a subset of data specifically allocated for performance assessment, which constituted 10% of the dataset extracted from the Giant corpus, revealing differences in their performance. These differences manifest in general metrics and in specific class performance, providing a deep understanding of each model’s effectiveness in bibliographic reference segmentation.
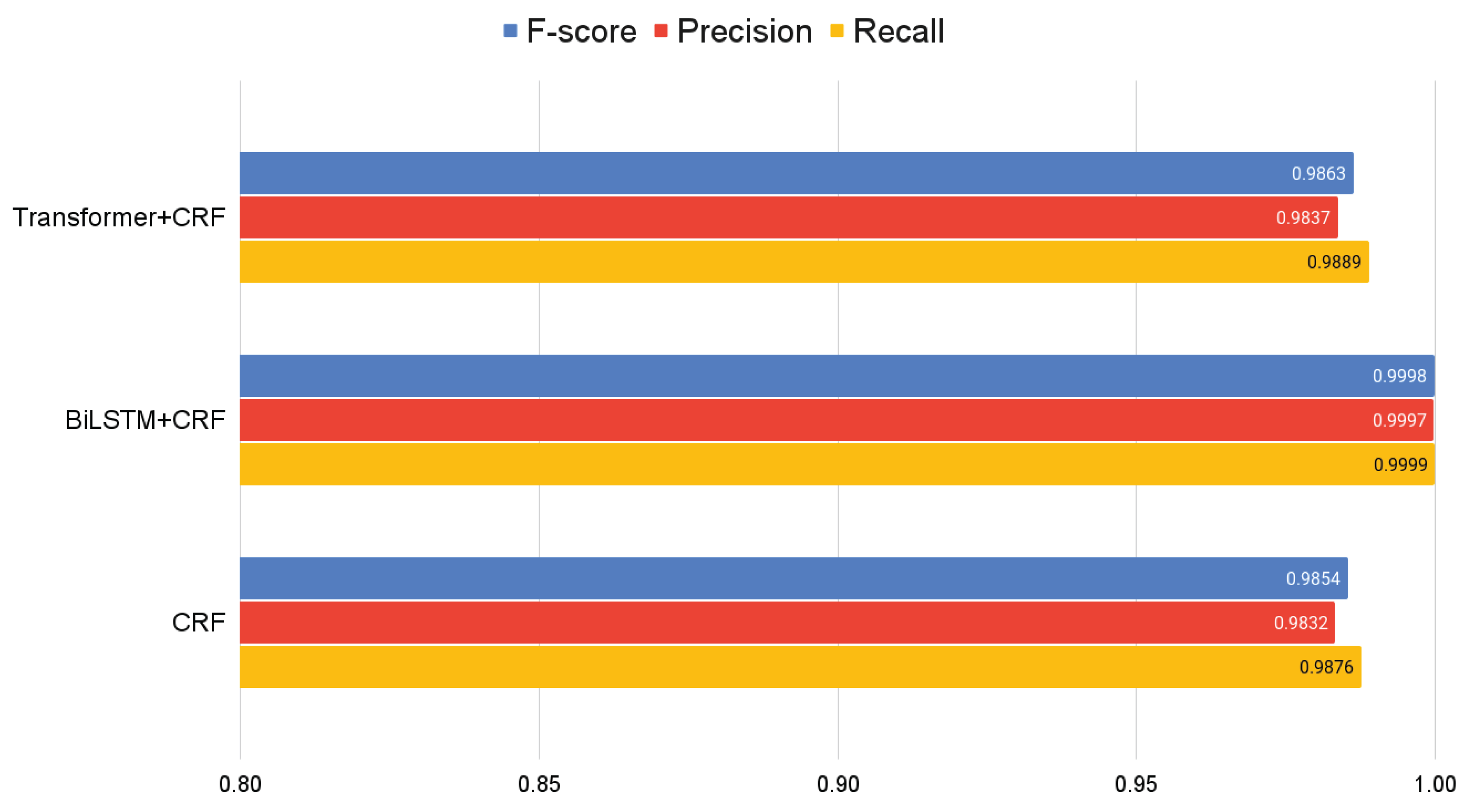
The selection of F-score, Precision, and Recall as evaluation metrics for our models, particularly in Named Entity Recognition (NER) tasks and bibliographic reference segmentation, follows established best practices within the field. These metrics are widely recognized in the state-of-the-art for their ability to provide a comprehensive assessment of a model’s performance. In terms of these metrics, the BiLSTM+CRF model demonstrated high performance, achieving nearly perfect scores with an F-score of 0.9998, a Precision of 0.9997, and a Recall of 0.9999. This level of accuracy signifies an almost flawless capability of the model to correctly identify and classify the components of bibliographic references, underlining the effectiveness of the chosen metrics in capturing the nuanced performance of NER models in the specialized task of bibliographic reference segmentation.
On the other hand, both the Transformer+CRF and traditional CRF models showed equally but slightly lower results compared to BiLSTM+CRF, with F-scores of 0.9863 and 0.9854, respectively. These results suggest that, although effective, these models may not be as precise in capturing certain fine details in references as the BiLSTM+CRF, see
Figure 4.
When examining the performance by class, interesting trends are observed. In categories like PUNC, URL, and ISSN, all three models demonstrated high effectiveness, with BiLSTM+CRF and Transformer+CRF even achieving perfect precision in several classes.
However, in categories like VOLUME and ISSUE, which may present greater challenges due to their lower frequency or greater variability in references, there is a noticeable decrease in the performance of Transformer+CRF and CRF, while BiLSTM+CRF maintains relatively high efficacy, see
Table 2.
Notably, certain categories like VOLUME and ISSUE present a greater challenge for the models, with the BiLSTM+CRF showing a significant improvement over the other two models. This could reflect the contextual variability and complexity of these categories within bibliographic references.
5. Experiments and Results
An experiment was carried out on a test corpus that was totally different from the training corpus in order to assess the generalization and robustness of the produced models (CRF, BiLSTM+CRF, and Trasnformer+CRF). This corpus, known as CORA (described in
Section 3.1.2), consists of a wide range of bibliographic references and represents a significant challenge due to its diversity and differences from the training dataset.
The CORA corpus, with its distinctive feature of containing references with missing components, offers an ideal test scenario to evaluate the adaptability of the trained models to previously unseen data [
13,
14,
20]. This unique characteristic of the corpus underscores the importance of model resilience in handling incomplete data, providing a rigorous test bed to assess the models’ capability to adjust to new contexts and data structures. Such an evaluation is crucial for real-world applications, where bibliographic references often exhibit significant variability in format, style, and completeness.
In this section, we present the results obtained by applying the trained models to the CORA corpus. The same metrics used in the training evaluation - F-score, Precision, and Recall - were employed to maintain consistency and allow direct comparisons. Additionally, the performance by class for each model was analyzed, offering a view of their effectiveness in specific categories of bibliographic references.
The results obtained in this experimental scenario provide a thorough evaluation of each model’s ability to generalize and adapt to new datasets. The following subsection details these results, offering a comparison of the performance of the models in the CORA corpus.
5.1. Results on the CORA Corpus
Evaluating the CRF, BiLSTM+CRF, and Transformer+CRF models on the CORA corpus provides insights into their ability to adapt and perform on a dataset different from the one used for their training. It is important to note that there are classes in CORA that are different or non-existent in the Giant dataset, so adjustments (described in
Section 3.2.2) were made to reduce variability of them, see
Table 3.
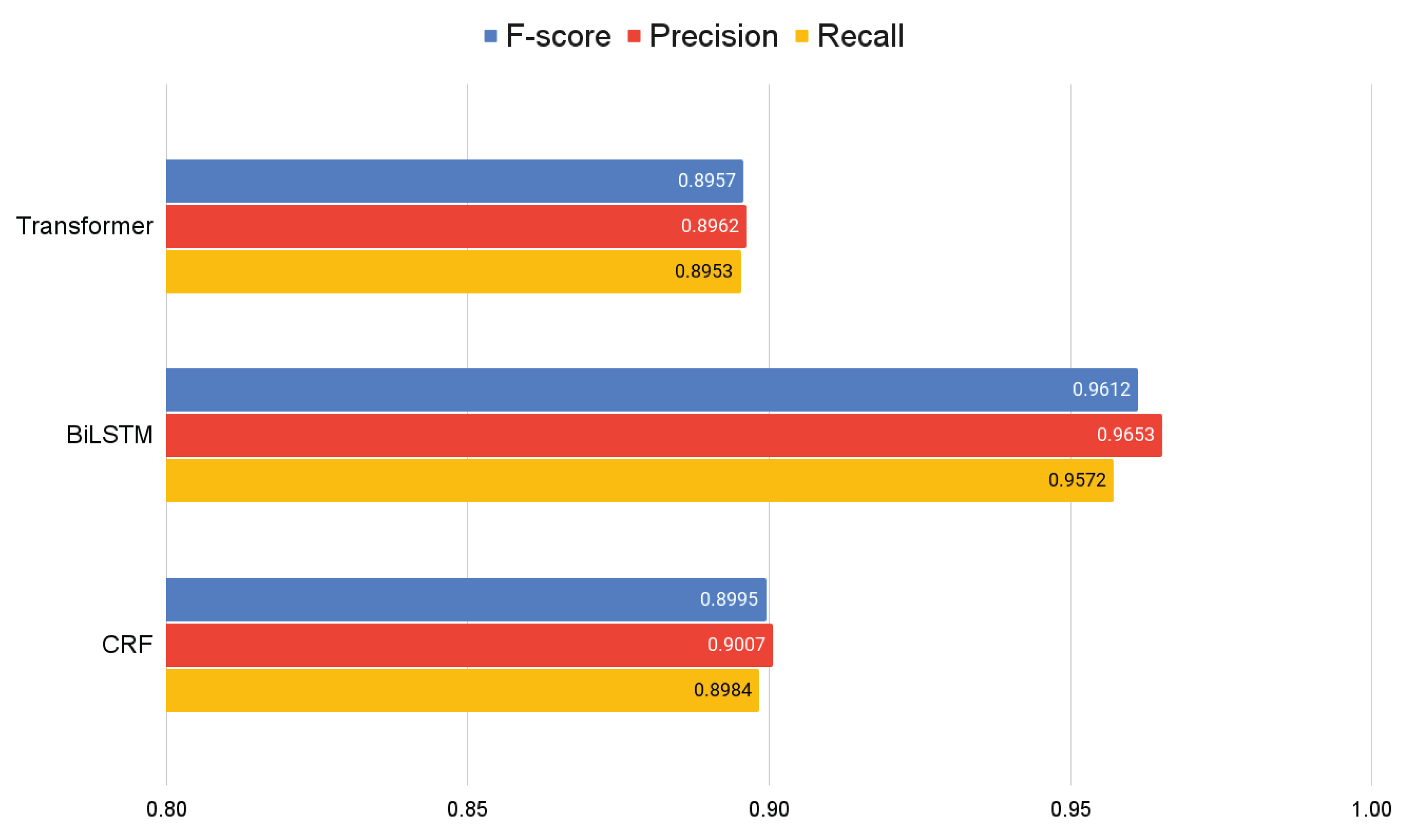
Regarding general metrics, the results show similar trends to those observed during the training phase. The BiLSTM+CRF model continues to demonstrate superior performance, with an F-score of 0.9612, a precision of 0.9653, and a recall of 0.9572, see
Figure 5.
These results reaffirm the robustness of the BiLSTM+CRF in terms of accuracy and its ability to capture relevant contexts. Meanwhile, the Transformer+CRF and the CRF alone show slightly lower performance, with F-scores of 0.8957 and 0.8995, respectively.
These findings highlight the BiLSTM+CRF’s superior adaptability and accuracy in dealing with a diverse set of bibliographic references, a critical aspect for real-world applications in digital libraries and information management systems.
The analysis by category reveals notable variations in performance among the models. While all variants achieve a perfect F-score in the PUNC category, there are significant differences in other categories. For instance, in the TITLE category, BiLSTM+CRF considerably outperforms its counterparts, with an F-score of 0.838 compared to 0.6096 (CRF) and 0.5441 (Transformer). Similarly, in categories like CONTAINER-TITLE, PUBLISHER, and VOLUME, BiLSTM+CRF shows a notably greater ability to correctly identify these elements, which could be attributed to its better handling of the variability and complexity of these categories, see
Table 4.
On the other hand, it is interesting to note that in categories like YEAR and PAGE, all models show relatively high performance, indicating a certain uniformity in the structure of these categories that the models can effectively capture.
In summary, the results on the CORA corpus suggest that while the BiLSTM+CRF model is consistently superior in several categories, the differences in performance between the models become more pronounced in more complex and varied categories. This underscores the importance of choosing the right model based on the specific characteristics of the task and dataset. The superiority of the BiLSTM+CRF on CORA Corpus reinforces its potential as a reliable tool for bibliographic reference segmentation tasks, especially in environments where data diversity and complexity are high.
5.2. Considerations about the CORA Corpus
It is crucial to consider certain specific characteristics of the CORA corpus when evaluating the performance of the models. One of the most significant peculiarities is the presence of references with missing components, which represents a unique challenge for reference segmentation models. A representative example of this type of case is as follows:
<author> M. Ahlskog, J. Paloheimo, H. Stubb, P. Dyreklev, M. Fahlman,
O. Inganas and M.R. Andersson, </author> <container-title> J Appl.
Phys., </container-title> <volume> 76, </volume><pages>893, </pages>
<year> (1994). </year>
In this example, the reference lacks the TITLE label, leading the model to erroneously inferring that the first tokens of CONTAINER-TITLE belong to TITLE. This situation affects the scores of both classes and highlights the challenge of handling incomplete references, which are common in the CORA corpus.
5.3. Discussion
The detailed performance analysis of Transformer+CRF, BiLSTM+CRF, and CRF models on the CORA corpus provides valuable insights into their efficacy in addressing the segmentation of bibliographic references across various classes. While the BiLSTM+CRF model exhibits a clear advantage in handling diverse and incomplete data sets, as evidenced by its superior performance across almost all categories, the results also shed light on the challenges and limitations faced by the other models.
The Transformer+CRF model, despite its innovative architecture designed to capture long-range dependencies and contextual nuances, struggles significantly with certain classes such as CONTAINER-TITLE and PUBLISHER, where it scores remarkably lower than its counterparts. This suggests a potential limitation in its ability to handle instances where contextual cues are sparse or irregular, common in real-world bibliographic data.
Conversely, the CRF model, while not achieving the high performance of the BiLSTM+CRF model, demonstrates a degree of resilience, outperforming the Transformer+CRF model in several classes. This indicates that traditional CRF models, despite their simpler architecture, can still be competitive, particularly in scenarios where the data structure benefits from their sequence modeling capabilities. However, its performance in critical categories such as TITLE and CONTAINER-TITLE remains suboptimal, highlighting the necessity for more sophisticated sequence modeling techniques to capture the complex patterns present in bibliographic references effectively.
The comparative analysis underscores the BiLSTM+CRF model’s robustness and its capacity to adapt to the CORA dataset’s irregularities, making it a potent tool for bibliographic reference segmentation tasks. Meanwhile, the observed performance disparities among the models underscore the imperative of selecting a modeling approach that is not only theoretically sound but also practically attuned to the specific challenges posed by the data. This entails a nuanced consideration of each model’s architectural strengths and weaknesses, ensuring the chosen method aligns with the inherent complexities of bibliographic data encountered in digital libraries and databases.
Furthermore, these findings align with recent observations reported [
25], which suggest that transformer models may not always excel in tasks such as Named Entity Recognition (NER) where understanding the specific structure and immediate context is crucial. This study reinforces the notion that while transformers offer advanced capabilities for capturing long-range dependencies, their performance in structured prediction tasks like bibliographic reference segmentation might not match that of models designed to navigate syntactic complexities more effectively, such as BiLSTM+CRF.
6. Conclusions
This study embarked on a comparative analysis of three models—Conditional Random Fields (CRF), Bidirectional Long Short-Term Memory with CRF (BiLSTM+CRF), and Transformer Encoder with CRF (Transformer+CRF)—for the task of bibliographic reference segmentation. Conducted with the Giant corpus for model training and the CORA corpus for evaluation, this research aimed to identify the most effective model for parsing the intricate structure of bibliographic references. The BiLSTM+CRF model demonstrated superior performance, particularly excelling in the precise delineation of bibliographic components. This model’s success can be attributed to its effective management of the syntactic relationships within bibliographic data, which is essential given the structured nature of these references.
The findings of this research not only emphasize the efficacy of the BiLSTM+CRF model in addressing data imperfections but also underscore its potential utility in digital library systems and automated bibliographic processing tools. By ensuring high accuracy across diverse datasets, the BiLSTM+CRF model emerges as a robust solution for the management and processing of bibliographic information in academic libraries.
Moreover, the insights gained from this study highlight the importance of selecting a model that aligns with the specific requirements of the task and the dataset. This investigation advances our understanding of bibliographic reference segmentation and paves the way for future research to explore and refine these models further, enhancing their applicability in real-world scenarios.
Looking to the future, the research pathway in this domain is rich with potential. An immediate direction involves delving into the bio-inspired mechanisms, particularly focusing on lateral inhibition mechanisms, given that the BiLSTM architecture, closely mirroring biological neural networks more than transformers, shows promise in reference segmentation tasks. The exploration of reference segmentation through lateral inhibition mechanisms[
26] could provide a novel approach, building on the bio-inspired foundations established by the BiLSTM architecture. This could open avenues for enhancing the model’s ability to manage the wide variety of bibliographic formats and styles more effectively. Assessing the model’s performance across a broader spectrum of languages and bibliographic traditions is also essential for ensuring its global applicability and utility in digital library systems. These future endeavors promise not only to refine the capabilities of bibliographic reference segmentation models but also to broaden their practical applications, significantly contributing to advancements in digital library services and scholarly communication.