Submitted:
15 March 2024
Posted:
18 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
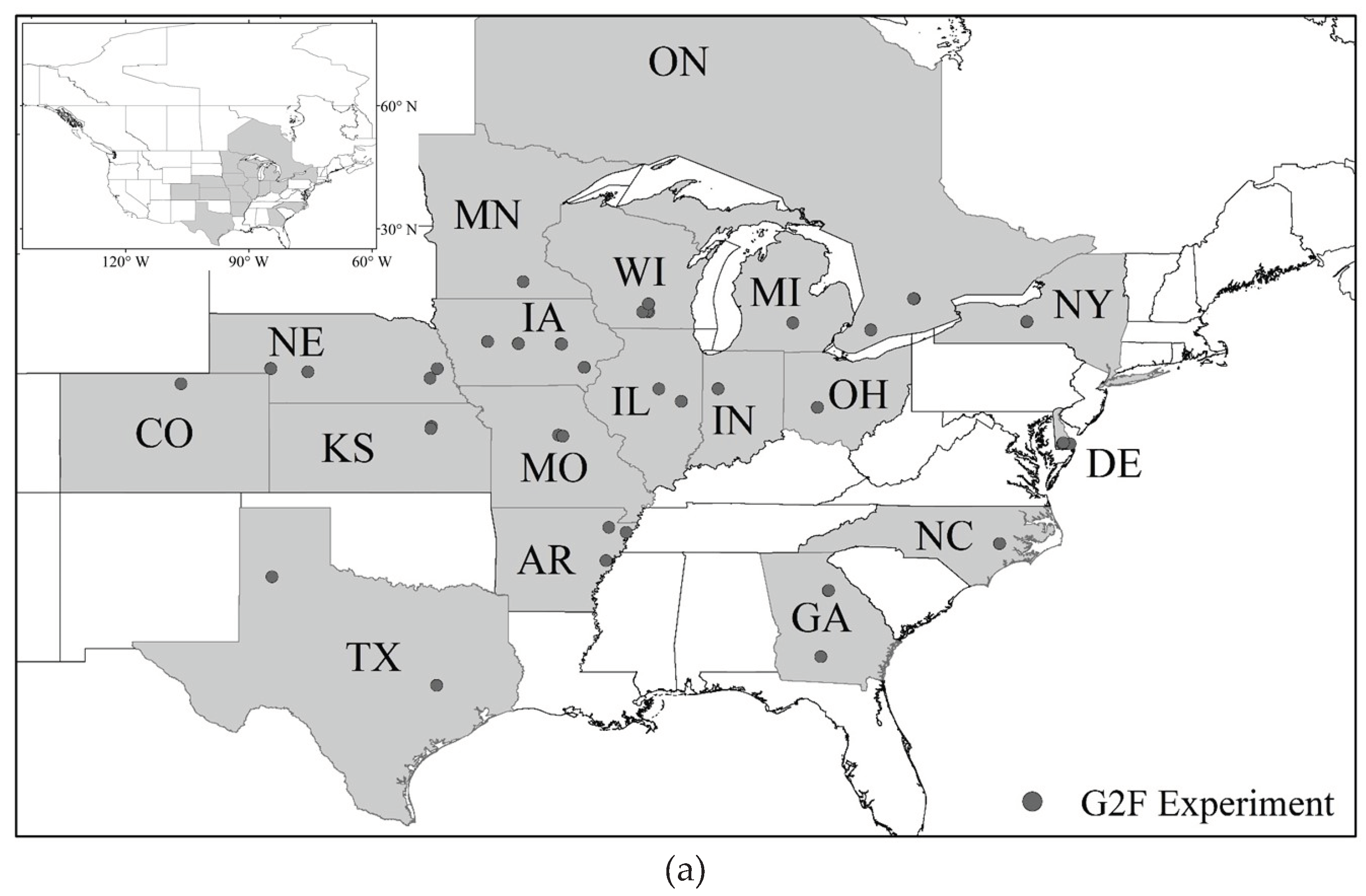
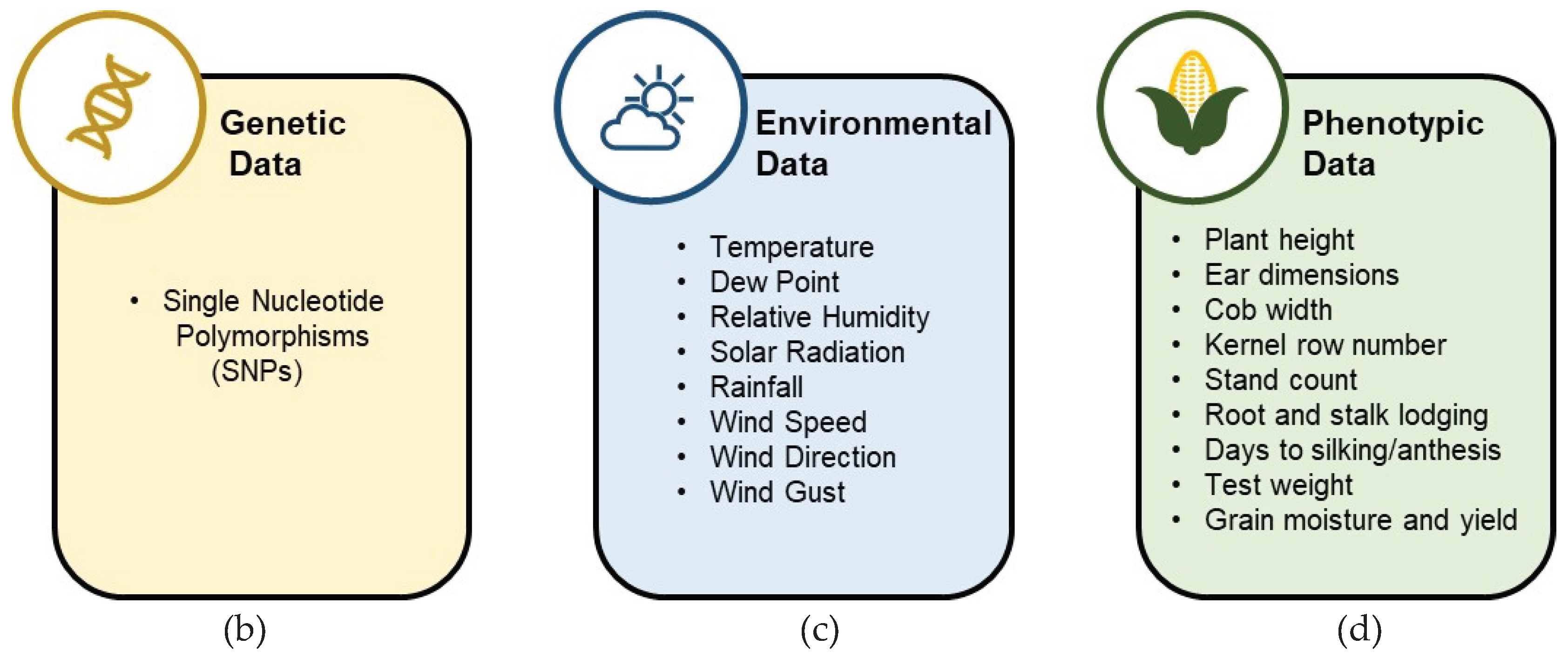
2.1. Database and Data Availability
2.1.1. Multi-Dimensional Genomes to Field (G2F) Database
2.1.2. G2F-E Pre-Processing
2.2. Modeling the GxE Interactions
2.2.1. Environmental Covariates (ECs) and Environmental Covariance Matrix Structure
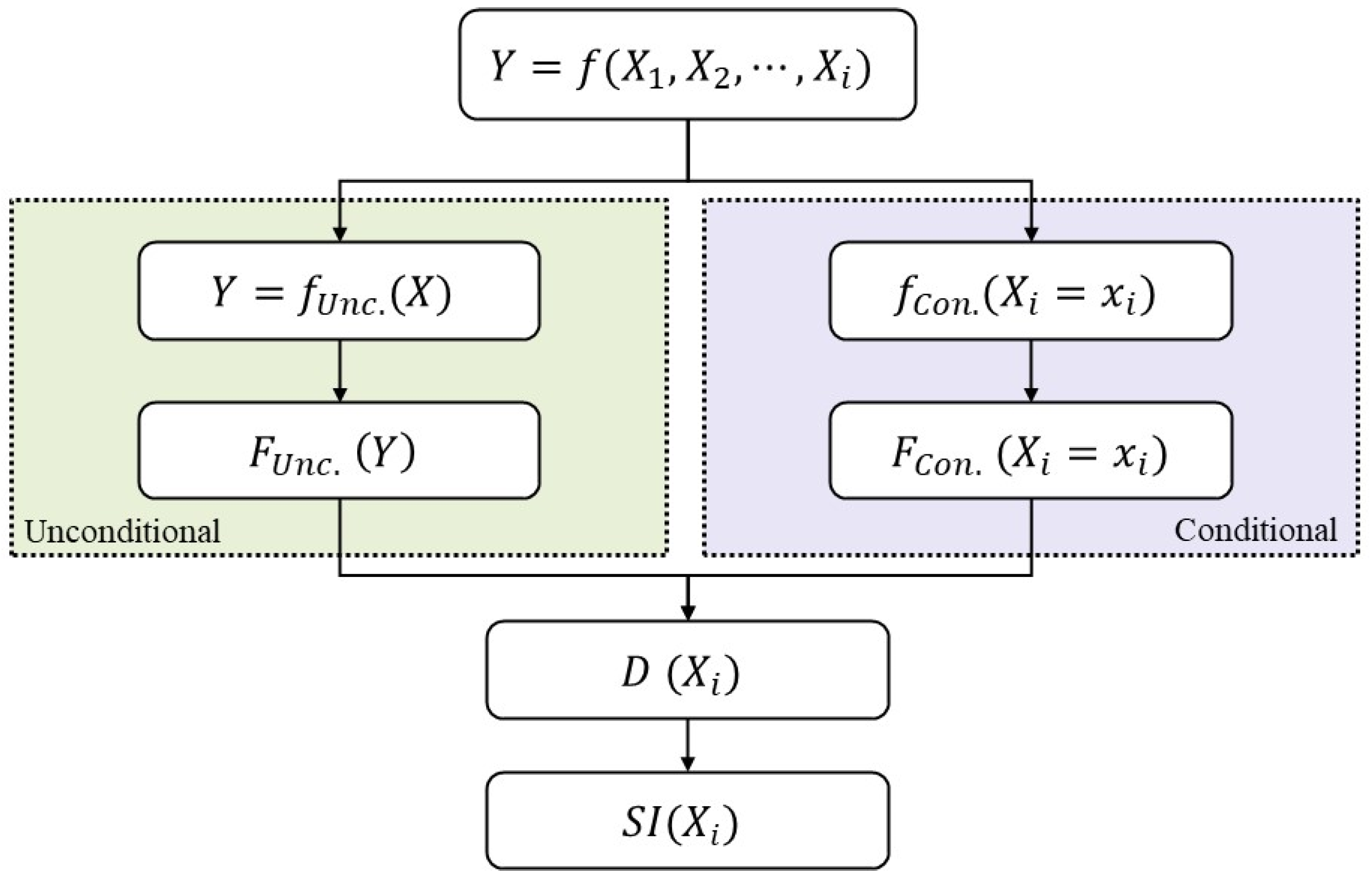
2.2.3. PAWN Global Sensitivity Analysis (GSA)
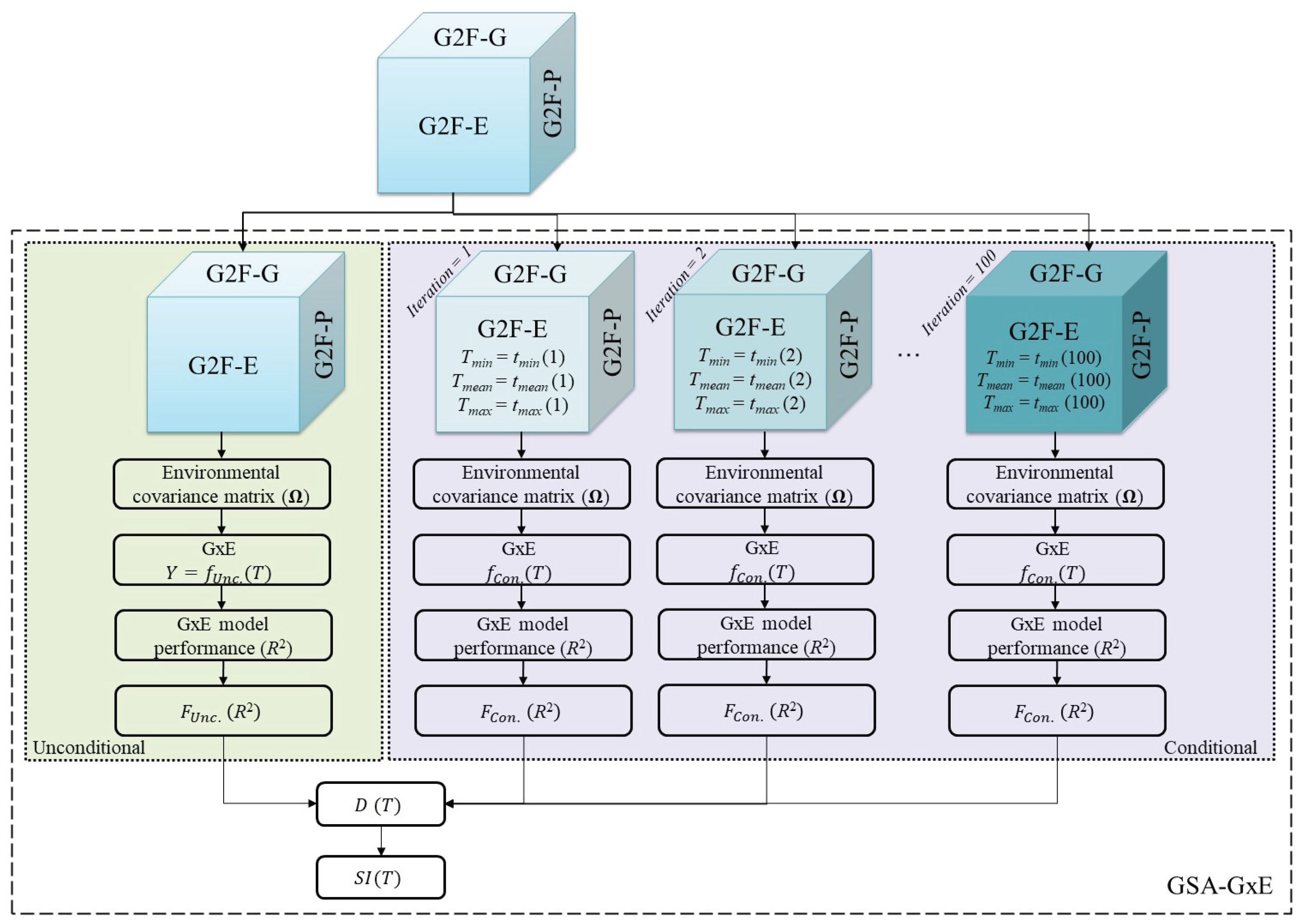
2.3. Coupling the GxE Model with PAWN Global Sensitivity Analysis
3. Results and Discussion
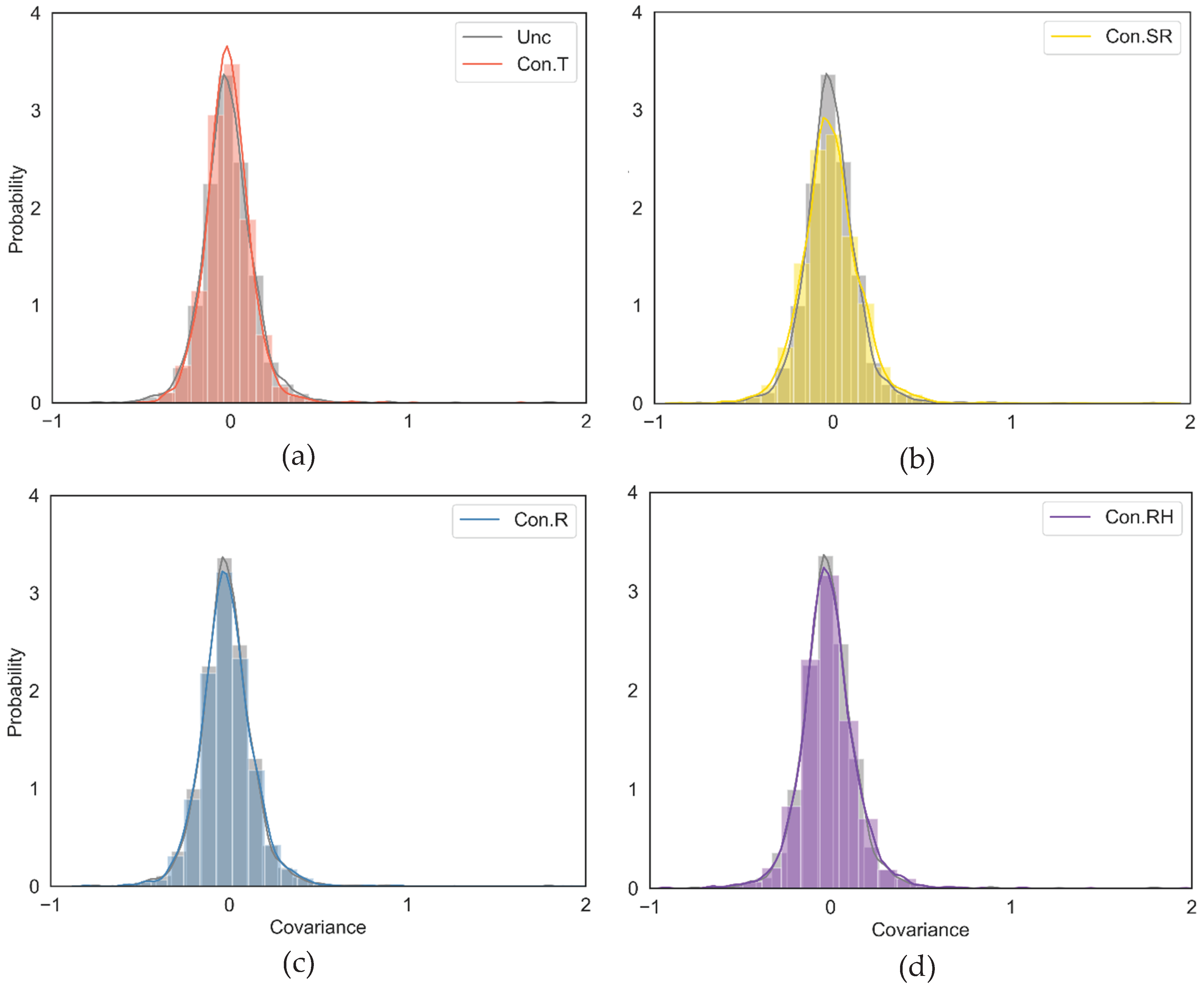
3.1. The Environmental Covariance Matrix
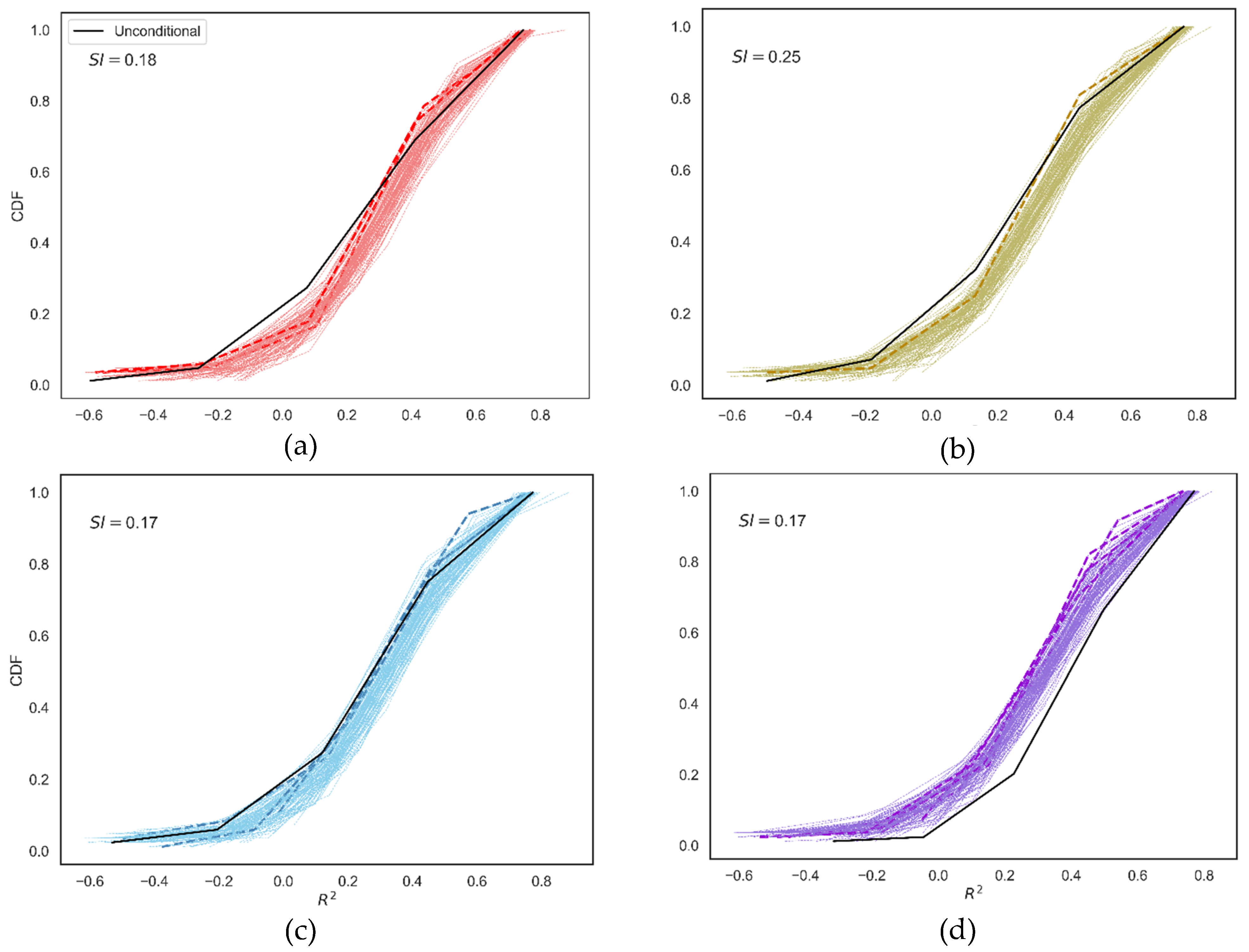
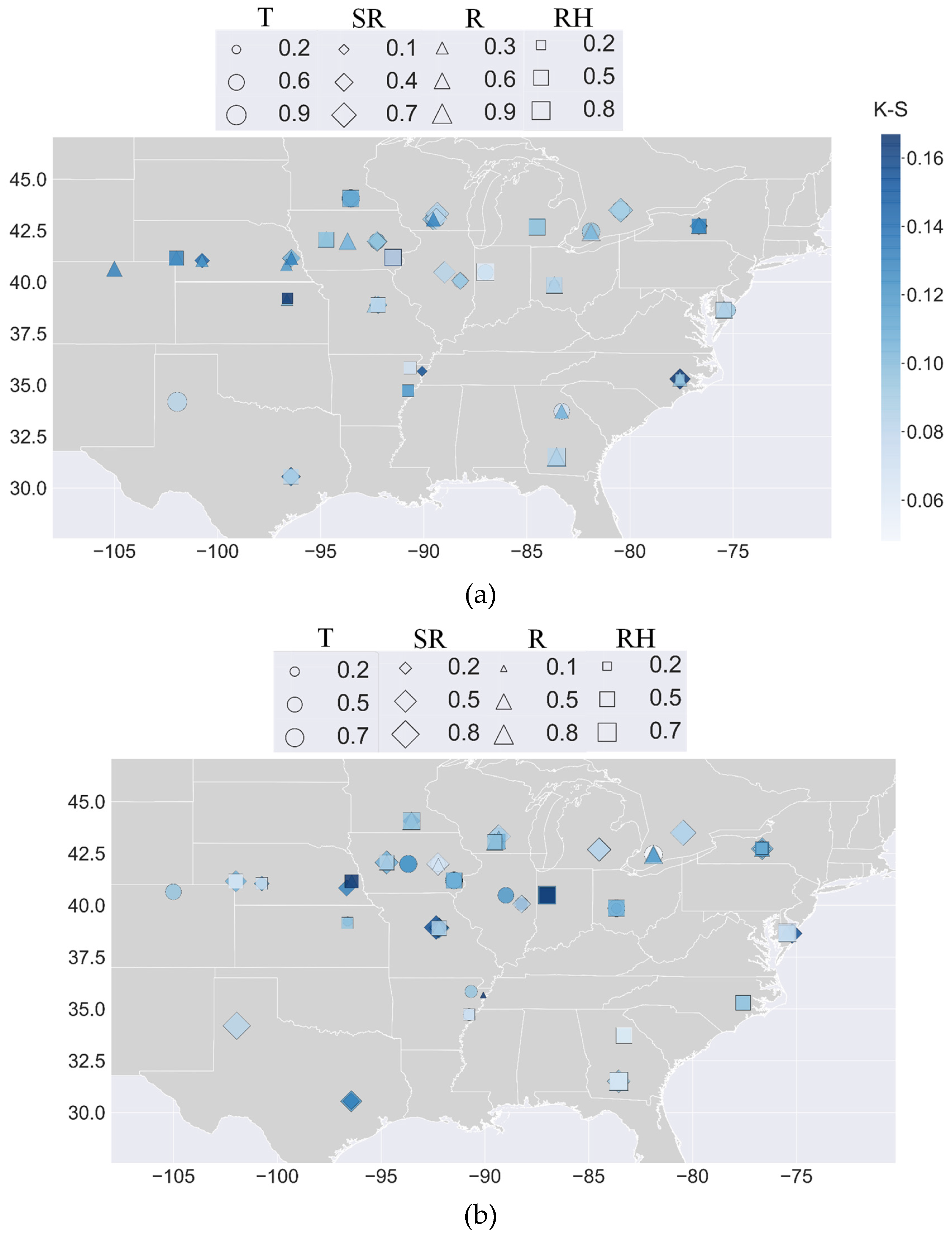
3.2. The GSA-GxE Framework
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muñoz Orozco, A. , 2003: CENTLI MAIZ. Prehistoria e historia, Diversidad, Potencial, Origen Génetico y Geográfico, Glosario Centli-Maíz. Colegio de Postgraduados en Ciencias Agrícolas. https://search.worldcat. 6514. [Google Scholar]
- Wheeler, T.; Von Braun, J. Climate Change Impacts on Global Food Security. Science 2013, 341, 508–511. [Google Scholar] [CrossRef] [PubMed]
- FAO, 2018: The Future of Food and Agriculture: Alternative Pathways to 2050, Food and Agriculture Organization of the United Nations. https://www.fao. 1157.
- Intergovernmental Panel on Climate Change (IPCC). Special Report on Climate Change and Land 2019. Available online: https://www.ipcc.ch/site/assets/uploads/2019/11/SRCCL-Full-Report-Compiled-191128.pdf.
- Stuart, L.; Hobbins, M.; Niebuhr, E.; Ruane, A.C.; Pulwarty, R.; Hoell, A.; Thiaw, W.; Rosenzweig, C.; Muñoz-Arriola, F.; Jahn, M.; et al. Enhancing Global Food Security: Opportunities for the American Meteorological Society. Bull. Am. Meteorol. Soc. [CrossRef]
- Springate, D.A.; Kover, P.X. Plant responses to elevated temperatures: a field study on phenological sensitivity and fitness responses to simulated climate warming. Glob. Chang. Biol. 2013, 20, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Zabel, F.; Müller, C.; Elliott, J.; Minoli, S.; Jägermeyr, J.; Schneider, J.M.; Franke, J.A.; Moyer, E.; Dury, M.; Francois, L.; et al. Large potential for crop production adaptation depends on available future varieties. Glob. Chang. Biol. 2021, 27, 3870–3882. [Google Scholar] [CrossRef] [PubMed]
- Fradgley, N.S.; Bacon, J.; Bentley, A.R.; Costa-Neto, G.; Cottrell, A.; Crossa, J.; Cuevas, J.; Kerton, M.; Pope, E.; Swarbreck, S.M.; et al. Prediction of near-term climate change impacts on UK wheat quality and the potential for adaptation through plant breeding. Glob. Chang. Biol. 2022, 29, 1296–1313. [Google Scholar] [CrossRef]
- Jarquín, D.; Crossa, J.; Lacaze, X.; Du Cheyron, P.; Daucourt, J.; Lorgeou, J.; Piraux, F.; Guerreiro, L.; Pérez, P.; Calus, M.; et al. A reaction norm model for genomic selection using high-dimensional genomic and environmental data. Theor. Appl. Genet. 2013, 127, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.L., D. G. Wang, K.E. Trenberth, A. Erfanian, M. Yu, M.G. Bosilovich, and D. Parr, 2017: Peak structure and future changes of the relationships between extreme precipitation and temperature. Nat. Climate Change 2017, 7, 268–274. [Google Scholar] [CrossRef]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- A Montesinos-López, O.; Montesinos-López, A.; Crossa, J.; Gianola, D.; Hernández-Suárez, C.M.; Martín-Vallejo, J. Multi-trait, Multi-environment Deep Learning Modeling for Genomic-Enabled Prediction of Plant Traits. G3 Genes|Genomes|Genetics 2018, 8, 3829–3840. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.R.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genom. 2021, 22, 1–23. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Crespo-Herrera, L.; Pierre, C.S.; Bentley, A.R.; de la Rosa-Santamaria, R.; Ascencio-Laguna, J.A.; Agbona, A.; Gerard, G.S.; Montesinos-López, A.; Crossa, J. Do feature selection methods for selecting environmental covariables enhance genomic prediction accuracy? Front. Genet. 2023, 14, 1209275. [Google Scholar] [CrossRef]
- Kick, D.R.; Wallace, J.G.; Schnable, J.C.; Kolkman, J.M.; Alaca, B.; Beissinger, T.M.; Edwards, J.; Ertl, D.; Flint-Garcia, S.; Gage, J.L.; et al. Yield prediction through integration of genetic, environment, and management data through deep learning. G3 Genes|Genomes|Genetics 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yang, M.; Mohammadi, K.; Song, D.; Bi, J.; Wang, G. Machine Learning Crop Yield Models Based on Meteorological Features and Comparison with a Process-Based Model. Artif. Intell. Earth Syst. 2022, 1, 1–34. [Google Scholar] [CrossRef]
- Sarzaeim, P.; Muñoz-Arriola, F.; Jarquín, D. Climate and genetic data enhancement using deep learning analytics to improve maize yield predictability. J. Exp. Bot. 2022, 73, 5336–5354. [Google Scholar] [CrossRef] [PubMed]
- Sarzaeim, P., F. Munoz-Arriola, and D. Jarquin, 2022b: Large-scale and Multi-dimensional Climate, Genetics, and Phenotypes Database for Maize Yield Predictability in the U.S. and Canada, Zenodo, accessed. 29 September. [CrossRef]
- Sarzaeim, P.; Muñoz-Arriola, F.; Jarquin, D.; Aslam, H.; Gatti, N.D.L. CLIM4OMICS: a geospatially comprehensive climate and multi-OMICS database for maize phenotype predictability in the United States and Canada. Earth Syst. Sci. Data 2023, 15, 3963–3990. [Google Scholar] [CrossRef]
- Lopez-Cruz, M.; Aguate, F.M.; Washburn, J.D.; de Leon, N.; Kaeppler, S.M.; Lima, D.C.; Tan, R.; Thompson, A.; De La Bretonne, L.W.; Campos, G.d.L. Leveraging data from the Genomes-to-Fields Initiative to investigate genotype-by-environment interactions in maize in North America. Nat. Commun. 2023, 14, 690. [Google Scholar] [CrossRef] [PubMed]
- van Voorn, G.A.K.; Boer, M.P.; Truong, S.H.; Friedenberg, N.A.; Gugushvili, S.; McCormick, R.; Korts, D.B.; Messina, C.D.; van Eeuwijk, F.A. A conceptual framework for the dynamic modeling of time-resolved phenotypes for sets of genotype-environment-management combinations: a model library. Front. Plant Sci. 2023, 14, 1172359. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Fritsche-Neto, R.; Montesinos-Lopez, O.A.; Costa-Neto, G.; Dreisigacker, S.; Montesinos-Lopez, A.; Bentley, A.R. The Modern Plant Breeding Triangle: Optimizing the Use of Genomics, Phenomics, and Enviromics Data. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Olesen, J.E.; Jensen, T.; Petersen, J. Sensitivity of field-scale winter wheat production in Denmark to climate variability and climate change. Clim. Res. 2000, 15, 221–238. [Google Scholar] [CrossRef]
- Southworth, J.; Randolph, J.; Habeck, M.; Doering, O.; Pfeifer, R.; Rao, D.; Johnston, J. Consequences of future climate change and changing climate variability on maize yields in the midwestern United States. Agric. Ecosyst. Environ. 2000, 82, 139–158. [Google Scholar] [CrossRef]
- Lobell, D.B.; Burke, M.B. Why are agricultural impacts of climate change so uncertain? The importance of temperature relative to precipitation. Environ. Res. Lett. 2008, 3, 034007. [Google Scholar] [CrossRef]
- Leng, G.; Zhang, X.; Huang, M.; Asrar, G.R.; Leung, L.R. The Role of Climate Covariability on Crop Yields in the Conterminous United States. Sci. Rep. 2016, 6, 33160–33160. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q. Temperature thresholds and crop production: a review. Clim. Chang. 2011, 109, 583–598. [Google Scholar] [CrossRef]
- Ray, D.K.; Gerber, J.S.; MacDonald, G.K.; West, P.C. Climate variation explains a third of global crop yield variability. Nat. Commun. 2015, 6, 5989. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, K.F.; Wang, G.; Yu, M.; Koo, J.; You, L. Potential impact of climate change on cereal crop yield in West Africa. Clim. Chang. 2015, 133, 321–334. [Google Scholar] [CrossRef]
- Iizumi, T.; Ramankutty, N. Changes in yield variability of major crops for 1981–2010 explained by climate change. Environ. Res. Lett. 2016, 11, 034003. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Ahmed, K.F.; You, L.; Yu, M.; Pal, J.; Ji, Z. Projecting regional climate and cropland changes using a linked biogeophysical-socioeconomic modeling framework: 1. Model description and an equilibrium application over West Africa. Geosci. Model Dev. 2017, 9, 354–376. [Google Scholar] [CrossRef]
- Adams, M.W. Principles of Plant Breeding. Agron. J. 1962, 54, 372–372. [Google Scholar] [CrossRef]
- Evans, L.T. , 1993: Crop Evolution, Adaptation and Yield. University of Cambridge, New York, 514 pp.
- Duvick, D.N.; Cassman, K.G. Post–Green Revolution Trends in Yield Potential of Temperate Maize in the North-Central United States. Crop. Sci. 1999, 39, 1622–1630. [Google Scholar] [CrossRef]
- Ben Miflin, B. Crop improvement in the 21st century. J. Exp. Bot. 2000, 51, 1–8. [Google Scholar] [CrossRef]
- Howden, S.M.; Soussana, J.-F.; Tubiello, F.N.; Chhetri, N.; Dunlop, M.; Meinke, H. Adapting agriculture to climate change. Proc. Natl. Acad. Sci. USA 2007, 104, 19691–19696. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.E. , and C.C. Funk, 2008: Climate: Food Security under Climate Change.
- Li, T.; Hasegawa, T.; Yin, X.; Zhu, Y.; Boote, K.; Adam, M.; Bregaglio, S.; Buis, S.; Confalonieri, R.; Fumoto, T.; et al. Uncertainties in predicting rice yield by current crop models under a wide range of climatic conditions. Glob. Chang. Biol. 2014, 21, 1328–1341. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Bohra, A.; Roorkiwal, M.; Barmukh, R.; Cowling, W.A.; Chitikineni, A.; Lam, H.-M.; Hickey, L.T.; Croser, J.S.; Bayer, P.E.; et al. Fast-forward breeding for a food-secure world. Trends Genet. 2021, 37, 1124–1136. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.P.; Lewis, J.M.; Ammar, K.; Basnet, B.R.; Crespo-Herrera, L.; Crossa, J.; Dhugga, K.S.; Dreisigacker, S.; Juliana, P.; Karwat, H.; et al. Harnessing translational research in wheat for climate resilience. J. Exp. Bot. 2021, 72, 5134–5157. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Khan, S.; Ma, X. Climate change impacts on crop yield, crop water productivity and food security – A review. Prog. Nat. Sci. 2009, 19, 1665–1674. [Google Scholar] [CrossRef]
- Asseng, S.; Ewert, F.; Rosenzweig, C.; Jones, J.W.; Hatfield, J.L.; Ruane, A.C.; Boote, K.J.; Thorburn, P.J.; Rötter, R.P.; Cammarano, D.; et al. Uncertainty in simulating wheat yields under climate change. Nat. Clim. Chang. 2013, 3, 827–832. [Google Scholar] [CrossRef]
- Cammarano, D.; Rötter, R.P.; Asseng, S.; Ewert, F.; Wallach, D.; Martre, P.; Hatfield, J.L.; Jones, J.W.; Rosenzweig, C.; Ruane, A.C.; et al. Uncertainty of wheat water use: Simulated patterns and sensitivity to temperature and CO2. Field Crop. Res. 2016, 198, 80–92. [Google Scholar] [CrossRef]
- Jones, J.W.; Antle, J.M.; Basso, B.; Boote, K.J.; Conant, R.T.; Foster, I.; Godfray, H.C.J.; Herrero, M.; Howitt, R.E.; Janssen, S.; et al. Toward a new generation of agricultural system data, models, and knowledge products: State of agricultural systems science. Agric. Syst. 2016, 155, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, S., J. Colletti, F. Muñoz-Arriola, L. Ramaswamy, C. Krintz, B.L. Varshney, and D. Richardson, 2017: Preprint Intelligent Infrastructure for Smart Agriculture: An Integrated Food, Energy and Water System. [CrossRef]
- Rosenzweig, C.; Mbow, C.; Barioni, L.G.; Benton, T.G.; Herrero, M.; Krishnapillai, M.; Liwenga, E.T.; Pradhan, P.; Rivera-Ferre, M.G.; Sapkota, T.; et al. Climate change responses benefit from a global food system approach. Nat. Food 2020, 1, 94–97. [Google Scholar] [CrossRef]
- Demaria, E.M.; Nijssen, B.; Wagener, T. Monte Carlo sensitivity analysis of land surface parameters using the Variable Infiltration Capacity model. 112. [CrossRef]
- Muñoz-Arriola, F.; Avissar, R.; Zhu, C.; Lettenmaier, D.P. Sensitivity of the water resources of Rio Yaqui Basin, Mexico, to agriculture extensification under multiscale climate conditions. Water Resour. Res. 2009, 45. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, L.; Guan, Y.; Chen, X.; Chen, X. Sensitivity analysis of Xinanjiang rainfall–runoff model parameters: a case study in Lianghui, Zhejiang province, China. Hydrol. Res. 2012, 43, 123–134. [Google Scholar] [CrossRef]
- Dzotsi, K.; Basso, B.; Jones, J. Development, uncertainty and sensitivity analysis of the simple SALUS crop model in DSSAT. Ecol. Model. 2013, 260, 62–76. [Google Scholar] [CrossRef]
- Merchant, C.J.; Paul, F.; Popp, T.; Ablain, M.; Bontemps, S.; Defourny, P.; Hollmann, R.; Lavergne, T.; Laeng, A.; de Leeuw, G.; et al. Uncertainty information in climate data records from Earth observation. Earth Syst. Sci. Data 2017, 9, 511–527. [Google Scholar] [CrossRef]
- Khatun, S.; Sahana, M.; Jain, S.K.; Jain, N. Simulation of surface runoff using semi distributed hydrological model for a part of Satluj Basin: parameterization and global sensitivity analysis using SWAT CUP. Model. Earth Syst. Environ. 2018, 4, 1111–1124. [Google Scholar] [CrossRef]
- Wang, A.; Solomatine, D.P. Practical Experience of Sensitivity Analysis: Comparing Six Methods, on Three Hydrological Models, with Three Performance Criteria. Water 2019, 11, 1062. [Google Scholar] [CrossRef]
- Jaimes-Correa, J.C.; Muñoz-Arriola, F.; Bartelt-Hunt, S. Modeling Water Quantity and Quality Nonlinearities for Watershed Adaptability to Hydroclimate Extremes in Agricultural Landscapes. Hydrology 2022, 9, 80. [Google Scholar] [CrossRef]
- Song, X.; Zhang, J.; Zhan, C.; Xuan, Y.; Ye, M.; Xu, C. Global sensitivity analysis in hydrological modeling: Review of concepts, methods, theoretical framework, and applications. J. Hydrol. 2015, 523, 739–757. [Google Scholar] [CrossRef]
- Pianosi, F.; Beven, K.; Freer, J.; Hall, J.W.; Rougier, J.; Stephenson, D.B.; Wagener, T. Sensitivity analysis of environmental models: A systematic review with practical workflow. Environ. Model. Softw. 2016, 79, 214–232. [Google Scholar] [CrossRef]
- Pianosi, F.; Wagener, T. Understanding the time-varying importance of different uncertainty sources in hydrological modelling using global sensitivity analysis. Hydrol. Process. 2016, 30, 3991–4003. [Google Scholar] [CrossRef]
- Amaranto, A.; Pianosi, F.; Solomatine, D.; Corzo, G.; Muñoz-Arriola, F. Sensitivity analysis of data-driven groundwater forecasts to hydroclimatic controls in irrigated croplands. J. Hydrol. 2020, 587. [Google Scholar] [CrossRef]
- Amaranto, A.; Juizo, D.; Castelletti, A. Disentangling sources of future uncertainties for water management in sub-Saharan river basins. Hydrol. Earth Syst. Sci. 2022, 26, 245–263. [Google Scholar] [CrossRef]
- Sarzaeim, P.; Ou, W.; de Oliveira, L.A.; Munoz-Arriola, F. Flood-Risk Analytics for Climate-Resilient Agriculture Using Remote Sensing in the Northern High Plains. Geo-Extreme 2021. LOCATION OF CONFERENCE, GeorgiaDATE OF CONFERENCE; pp. 234–244.
- Bert, F.E.; Laciana, C.E.; Podestá, G.P.; Satorre, E.H.; Menéndez, A.N. Sensitivity of CERES-Maize simulated yields to uncertainty in soil properties and daily solar radiation. Agric. Syst. 2007, 94, 141–150. [Google Scholar] [CrossRef]
- Pathak, T.B.; Fraisse, C.W.; Jones, J.W.; Messina, C.D.; Hoogenboom, G. Use of Global Sensitivity Analysis for CROPGRO Cotton Model Development. Trans. ASABE 2007, 50, 2295–2302. [Google Scholar] [CrossRef]
- Lamboni, M.; Makowski, D.; Lehuger, S.; Gabrielle, B.; Monod, H. Multivariate global sensitivity analysis for dynamic crop models. Field Crop. Res. 2009, 113, 312–320. [Google Scholar] [CrossRef]
- Casadebaig, P.; Zheng, B.; Chapman, S.; Huth, N.; Faivre, R.; Chenu, K. Assessment of the Potential Impacts of Wheat Plant Traits across Environments by Combining Crop Modeling and Global Sensitivity Analysis. PLOS ONE 2016, 11, e0146385–e0146385. [Google Scholar] [CrossRef]
- Lu, Y.; Chibarabada, T.P.; McCabe, M.F.; De Lannoy, G.J.; Sheffield, J. Global sensitivity analysis of crop yield and transpiration from the FAO-AquaCrop model for dryland environments. Field Crop. Res. 2021, 269, 108182. [Google Scholar] [CrossRef]
- Jong, S.K.; Brewbaker, J.L.; Lee, C.H. Effects of Solar Radiation on the Performance of Maize in 41 Successive Monthly Plantings in Hawaii 1. Crop. Sci. 1982, 22, 13–18. [Google Scholar] [CrossRef]
- Muchow, R.C.; Sinclair, T.R.; Bennett, J.M. Temperature and Solar Radiation Effects on Potential Maize Yield across Locations. Agron. J. 1990, 82, 338–343. [Google Scholar] [CrossRef]
- Li, X.; Takahashi, T.; Suzuki, N.; Kaiser, H.M. The impact of climate change on maize yields in the United States and China. Agric. Syst. 2011, 104, 348–353. [Google Scholar] [CrossRef]
- Lobell, D.B.; Roberts, M.J.; Schlenker, W.; Braun, N.; Little, B.B.; Rejesus, R.M.; Hammer, G.L. Greater Sensitivity to Drought Accompanies Maize Yield Increase in the U.S. Midwest. Science 2014, 344, 516–519. [Google Scholar] [CrossRef]
- Johnston, R.Z.; Sandefur, H.N.; Bandekar, P.; Matlock, M.D.; Haggard, B.E.; Thoma, G. Predicting changes in yield and water use in the production of corn in the United States under climate change scenarios. Ecol. Eng. 2015, 82, 555–565. [Google Scholar] [CrossRef]
- Sarzaeim, P.; Muñoz-Arriola, F.; Jarquin, D. Analytics for climate-uncertainty estimation and propagation in maize-phenotype predictions. 2020 ASABE Annual International Virtual Meeting, -15, 2020. LOCATION OF CONFERENCE, COUNTRYDATE OF CONFERENCE; p. 1. 13 July.
- Pianosi, F.; Wagener, T. A simple and efficient method for global sensitivity analysis based on cumulative distribution functions. Environ. Model. Softw. 2015, 67, 1–11. [Google Scholar] [CrossRef]
- Aslam, H., P. Sarzaeim, and F. Munoz-Arriola, 2023: CLImate-for-Maize-OMICS_CLIM4OMICS-Analytics-and-Database: CLImate-for-Maize-OMICS_CLIM4OMICS-Analytics-and-Database Code, version 2., Zenodo, accessed. 29 September. [CrossRef]
- The Genomes To Fields (G2F) Initiative, 2013: Project: Genomes by Environment (GxE). Accessed . https://www.genomes2fields.org/home/#project-gxe. 21 November.
- van Eeuwijk, F.A.; Bustos-Korts, D.V.; Malosetti, M. What Should Students in Plant Breeding Know About the Statistical Aspects of Genotype × Environment Interactions? Crop. Sci. 2016, 56, 2119–2140. [Google Scholar] [CrossRef]
- Bustos-Korts, D., I. Romagosa, G. Borràs-Gelonch, A.M. Casas, G.A. Slafer, and F.V. Eeuwijk, 2018: Genotype by Environment Interaction and Adaptation. -44. [CrossRef]
- dos Reis, D.M.; Flach, P.; Matwin, S.; Batista, G. Fast Unsupervised Online Drift Detection Using Incremental Kolmogorov-Smirnov Test. KDD '16: The 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. LOCATION OF CONFERENCE, United StatesDATE OF CONFERENCE;
- Moghim, S.; Bras, R.L. Bias Correction of Climate Modeled Temperature and Precipitation Using Artificial Neural Networks. J. Hydrometeorol. 2017, 18, 1867–1884. [Google Scholar] [CrossRef]
- Lobell, D.B.; Asseng, S. Comparing estimates of climate change impacts from process-based and statistical crop models. Environ. Res. Lett. 2017, 12, 015001. [Google Scholar] [CrossRef]
- Jägermeyr, J.; Müller, C.; Ruane, A.C.; Elliott, J.; Balkovic, J.; Castillo, O.; Faye, B.; Foster, I.; Folberth, C.; Franke, J.A.; et al. Climate impacts on global agriculture emerge earlier in new generation of climate and crop models. Nat. Food 2021, 2, 873–885. [Google Scholar] [CrossRef]
- Schwalbert, R.; Amado, T.; Nieto, L.; Corassa, G.; Rice, C.; Peralta, N.; Schauberger, B.; Gornott, C.; Ciampitti, I. Mid-season county-level corn yield forecast for US Corn Belt integrating satellite imagery and weather variables. Crop. Sci. 2020, 60, 739–750. [Google Scholar] [CrossRef]
- Rogers, A.R.; Holland, J.B. Environment-specific genomic prediction ability in maize using environmental covariates depends on environmental similarity to training data. G3 Genes|Genomes|Genetics 2021, 12. [Google Scholar] [CrossRef]
- Parent, B.; Tardieu, F. Temperature responses of developmental processes have not been affected by breeding in different ecological areas for 17 crop species. New Phytol. 2012, 194, 760–774. [Google Scholar] [CrossRef]
- Frey, H.C.; Patil, S.R. Identification and Review of Sensitivity Analysis Methods. Risk Anal. 2002, 22, 553–578. [Google Scholar] [CrossRef]
- Brown, R.A.; Rosenberg, N.J. Sensitivity of crop yield and water use to change in a range of climatic factors and CO2 concentrations: a simulation study applying EPIC to the central USA. Agric. For. Meteorol. 1997, 83, 171–203. [Google Scholar] [CrossRef]
- Ruane, A.C.; Cecil, L.D.; Horton, R.M.; Gordón, R.; McCollum, R.; Brown, D.; Killough, B.; Goldberg, R.; Greeley, A.P.; Rosenzweig, C. Climate change impact uncertainties for maize in Panama: Farm information, climate projections, and yield sensitivities. Agric. For. Meteorol. 2013, 170, 132–145. [Google Scholar] [CrossRef]
- Meng, Q.; Chen, X.; Lobell, D.B.; Cui, Z.; Zhang, Y.; Yang, H.; Zhang, F. Growing sensitivity of maize to water scarcity under climate change. Sci. Rep. 2016, 6, 19605–19605. [Google Scholar] [CrossRef]
- Silvestro, P.C.; Pignatti, S.; Yang, H.; Yang, G.; Pascucci, S.; Castaldi, F.; Casa, R. Sensitivity analysis of the Aquacrop and SAFYE crop models for the assessment of water limited winter wheat yield in regional scale applications. PLOS ONE 2017, 12, e0187485. [Google Scholar] [CrossRef]
- Fronzek, S.; Pirttioja, N.; Carter, T.R.; Bindi, M.; Hoffmann, H.; Palosuo, T.; Ruiz-Ramos, M.; Tao, F.; Trnka, M.; Acutis, M.; et al. Classifying multi-model wheat yield impact response surfaces showing sensitivity to temperature and precipitation change. Agric. Syst. 2018, 159, 209–224. [Google Scholar] [CrossRef]
- Jarquin, D.; de Leon, N.; Romay, C.; Bohn, M.; Buckler, E.S.; Ciampitti, I.; Edwards, J.; Ertl, D.; Flint-Garcia, S.; Gore, M.A.; et al. Utility of Climatic Information via Combining Ability Models to Improve Genomic Prediction for Yield Within the Genomes to Fields Maize Project. Front. Genet. 2021, 11. [Google Scholar] [CrossRef]
- Crossa, J. , and Coauthors, 2022: Genome and environment based prediction models and methods of complex traits incorporating genotype× environment interaction. Methods Mol. Biol., 2467, 245-283. [CrossRef]
- Tollenaar, M.; Fridgen, J.; Tyagi, P.; Jr, P.W.S.; Kumudini, S. The contribution of solar brightening to the US maize yield trend. Nat. Clim. Chang. 2017, 7, 275–278. [Google Scholar] [CrossRef]
- Chen, C.; Baethgen, W.E.; Robertson, A. Contributions of individual variation in temperature, solar radiation and precipitation to crop yield in the North China Plain, 1961–2003. Clim. Chang. 2012, 116, 767–788. [Google Scholar] [CrossRef]
- Bruce, W.B.; Edmeades, G.O.; Barker, T.C. Molecular and physiological approaches to maize improvement for drought tolerance. J. Exp. Bot. 2002, 53, 13–25. [Google Scholar] [CrossRef]
- Schlenker, W.; Roberts, M.J. Nonlinear temperature effects indicate severe damages to U.S. crop yields under climate change. Proc. Natl. Acad. Sci. 2009, 106, 15594–15598. [Google Scholar] [CrossRef]
- Maltais-Landry, G.; Lobell, D.B. Evaluating the Contribution of Weather to Maize and Wheat Yield Trends in 12 U.S. Counties. Agron. J. 2012, 104, 301–311. [Google Scholar] [CrossRef]
- Carrillo, C.M.; Muñoz-Arriola, F.; Chen, L. Multi-scale Sources of Precipitation Predictability in the Northern Great Plains, 2023120362, preprint 2023. [CrossRef]
- Priya, S.; Shibasaki, R. National spatial crop yield simulation using GIS-based crop production model. Ecol. Model. 2001, 136, 113–129. [Google Scholar] [CrossRef]
- Rogers, A.R.; Dunne, J.C.; Romay, C.; Bohn, M.; Buckler, E.S.; A Ciampitti, I.; Edwards, J.; Ertl, D.; Flint-Garcia, S.; A Gore, M.; et al. The importance of dominance and genotype-by-environment interactions on grain yield variation in a large-scale public cooperative maize experiment. G3 Genes|Genomes|Genetics 2021, 11. [Google Scholar] [CrossRef]
- Rico, D.A.; Munoz-Arriola, F.; Detweiler, C. Trajectory Selection for Power-over-Tether Atmospheric Sensing UAS. 2021 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS). LOCATION OF CONFERENCE, Czech RepublicDATE OF CONFERENCE; pp. 2321–2328.
- Rico, D.A.; Detweiler, C.J.; Muñoz-Arriola, F. Power-over-Tether UAS Leveraged for Nearly-Indefinite Meteorological Data Acquisition. 2020 ASABE Annual International Virtual Meeting, -15, 2020. LOCATION OF CONFERENCE, COUNTRYDATE OF CONFERENCE; p. 1. 13 July.
- Quiñones, R.; Samal, A.; Das Choudhury, S.; Muñoz-Arriola, F. OSC-CO2: coattention and cosegmentation framework for plant state change with multiple features. Front. Plant Sci. 2023, 14, 1211409. [Google Scholar] [CrossRef]
- Quiñones, R.; Munoz-Arriola, F.; Das Choudhury, S.; Samal, A. Multi-feature data repository development and analytics for image cosegmentation in high-throughput plant phenotyping. PLOS ONE 2021, 16, e0257001. [Google Scholar] [CrossRef]
- Volk, J.M.; Huntington, J.L.; Melton, F.S.; Allen, R.; Anderson, M.; Fisher, J.B.; Kilic, A.; Ruhoff, A.; Senay, G.B.; Minor, B.; et al. Assessing the accuracy of OpenET satellite-based evapotranspiration data to support water resource and land management applications. Nat. Water 2024, 2, 193–205. [Google Scholar] [CrossRef]
| Conditional variable | Min | Max | Range |
|---|---|---|---|
| Tmin (°C) | -10.7 (2014NYH1) | 30.5 (2017ARH2) | 41.2 |
| Tmean (°C) | -6.4 (2014NYH2) | 32.5 (2017ARH2) | 38.9 |
| Tmax (°C) | -5.7 (2014NYH2) | 51.4 (2016IAH2) | 57.1 |
| SRmin (W/m2) | 0 (All) | 0 (All) | 0 |
| SRmean (W/m2) | 0 (2017IAH4) | 1168.3 (2017TXH1) | 1168.3 |
| SRmax (W/m2) | 0 (2017IAH1, 2017IAH2, 2017IAH3, 2017IAH4) | 1507.0 (2017MOH1) | 1507.0 |
| Racc (mm) | 0 (All) | 451.8 (2016IAH1) | 451.8 |
| RHmin (%) | 0 (2016NCH1, 2017ARH2) | 99.5 (2014IAH3) | 99.5 |
| RHmean (%) | 10.2 (2017WIH1) | 99.9 (2014IAH3) | 89.7 |
| RHmax (%) | 11.2 (2017IAH3) | 100.0 (All except 2016GAH2 and 2017NYH1) | 88.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





