Submitted:
18 March 2024
Posted:
22 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Compartmental Model with Mortality
Remark I
Markovian (memoryless) case
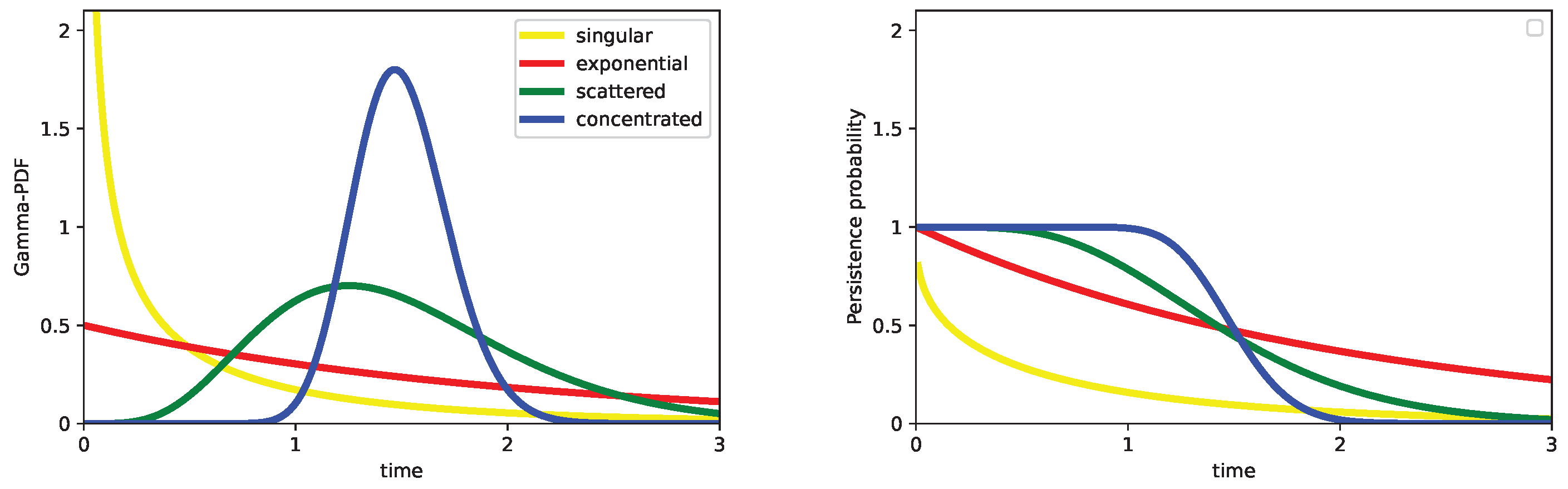
A few more words on waiting time distributions
3. Endemic Equilibrium for Zero Mortality
4. Stability Analysis of Endemic and Healthy State without Mortality
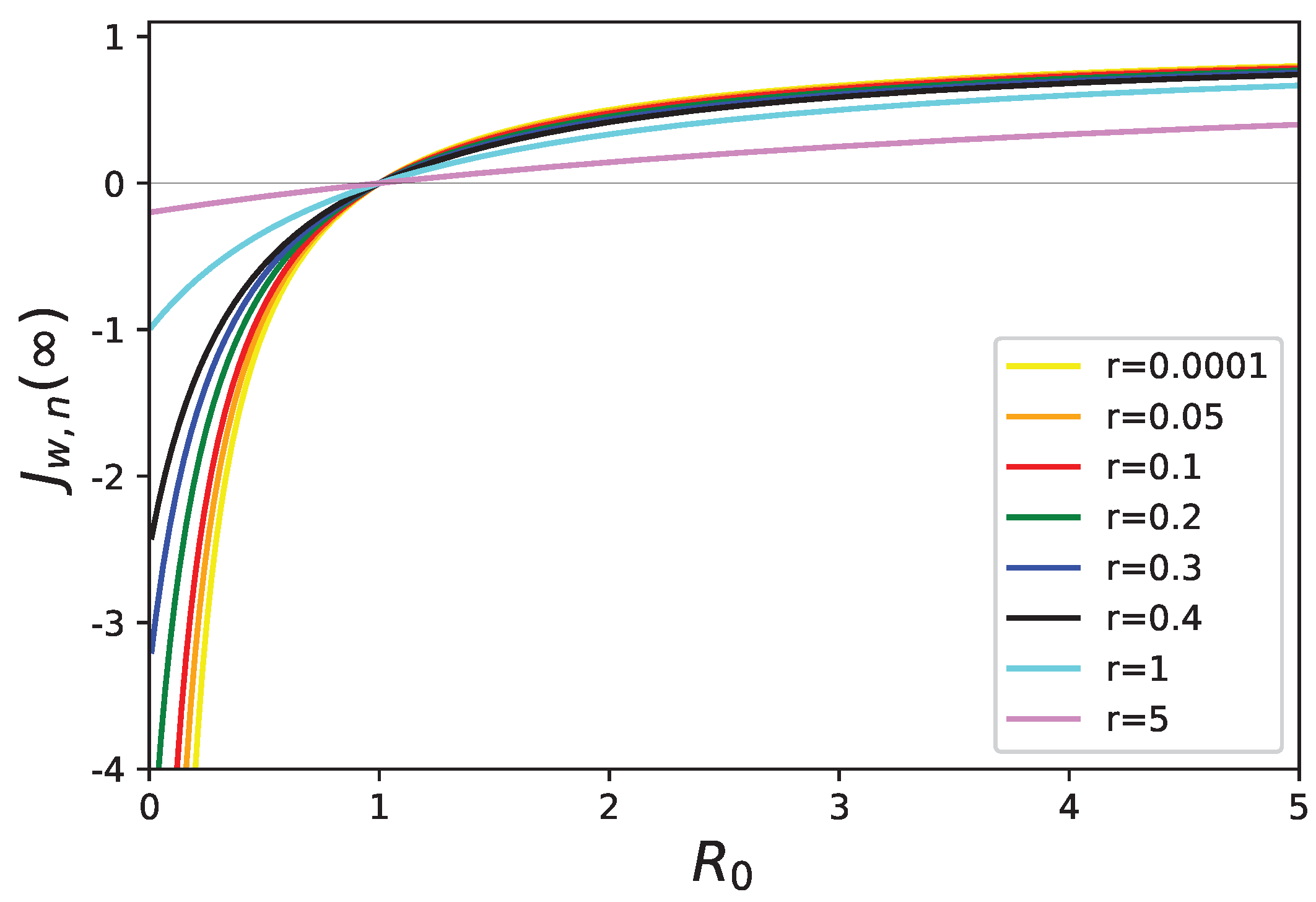
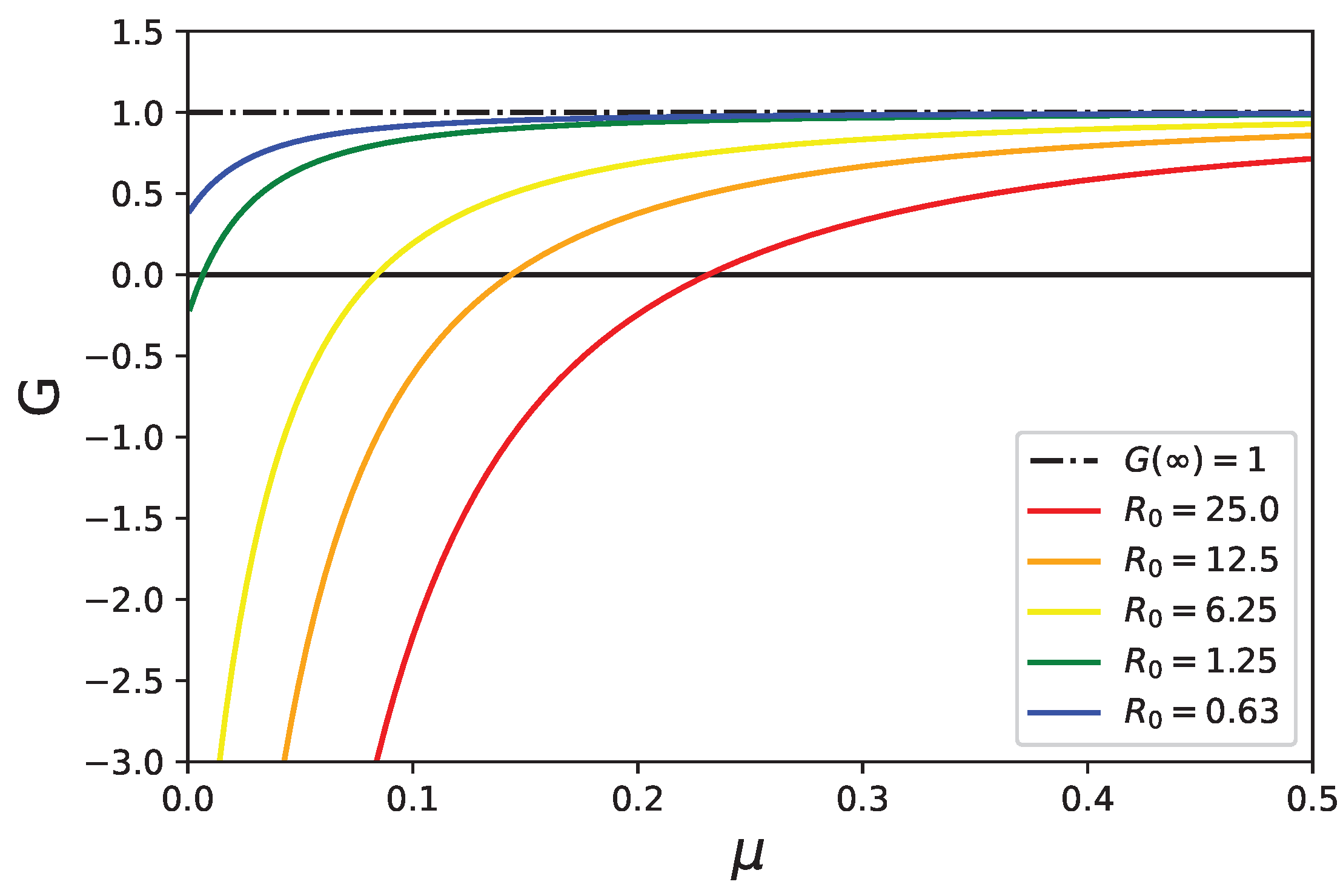
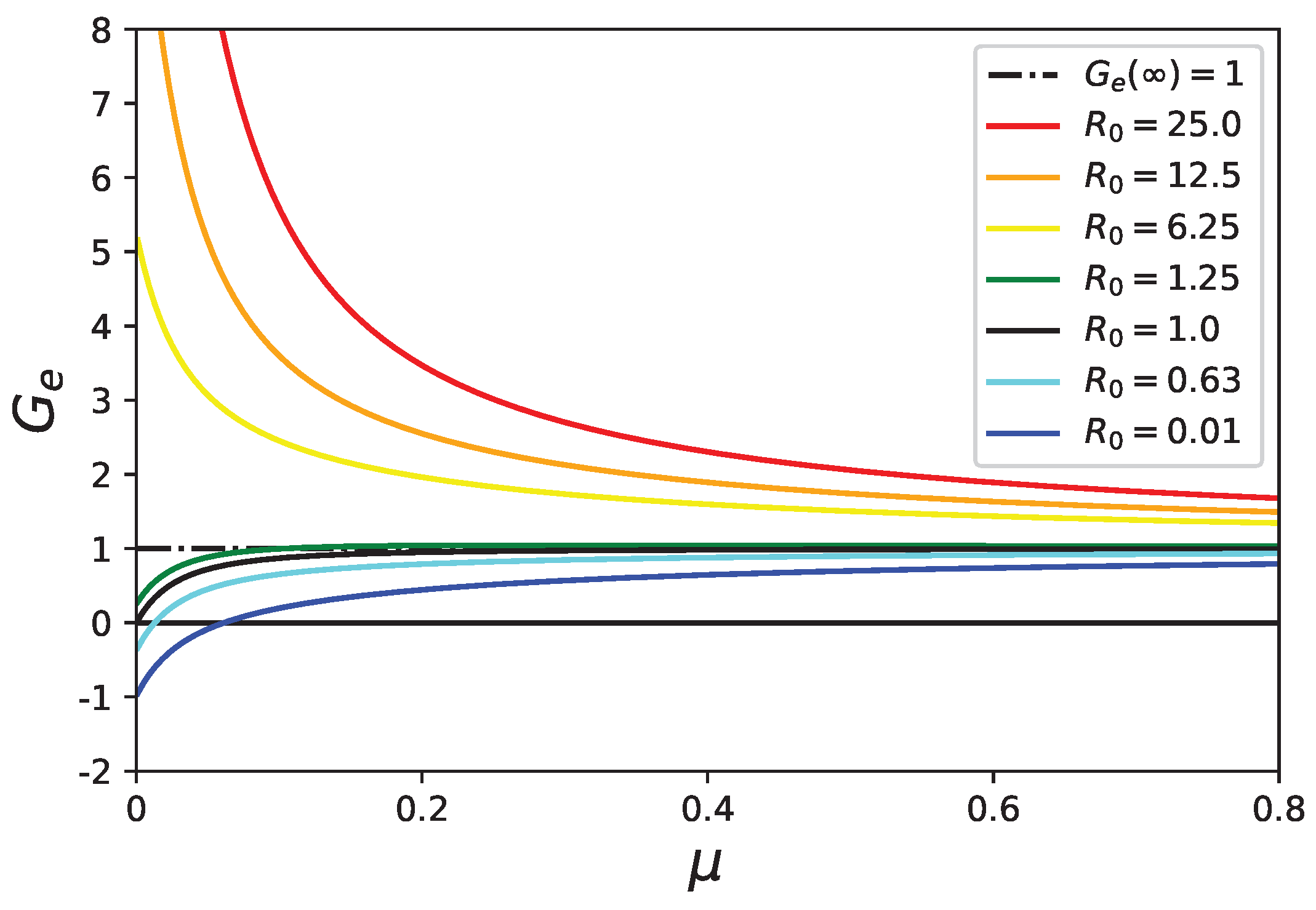
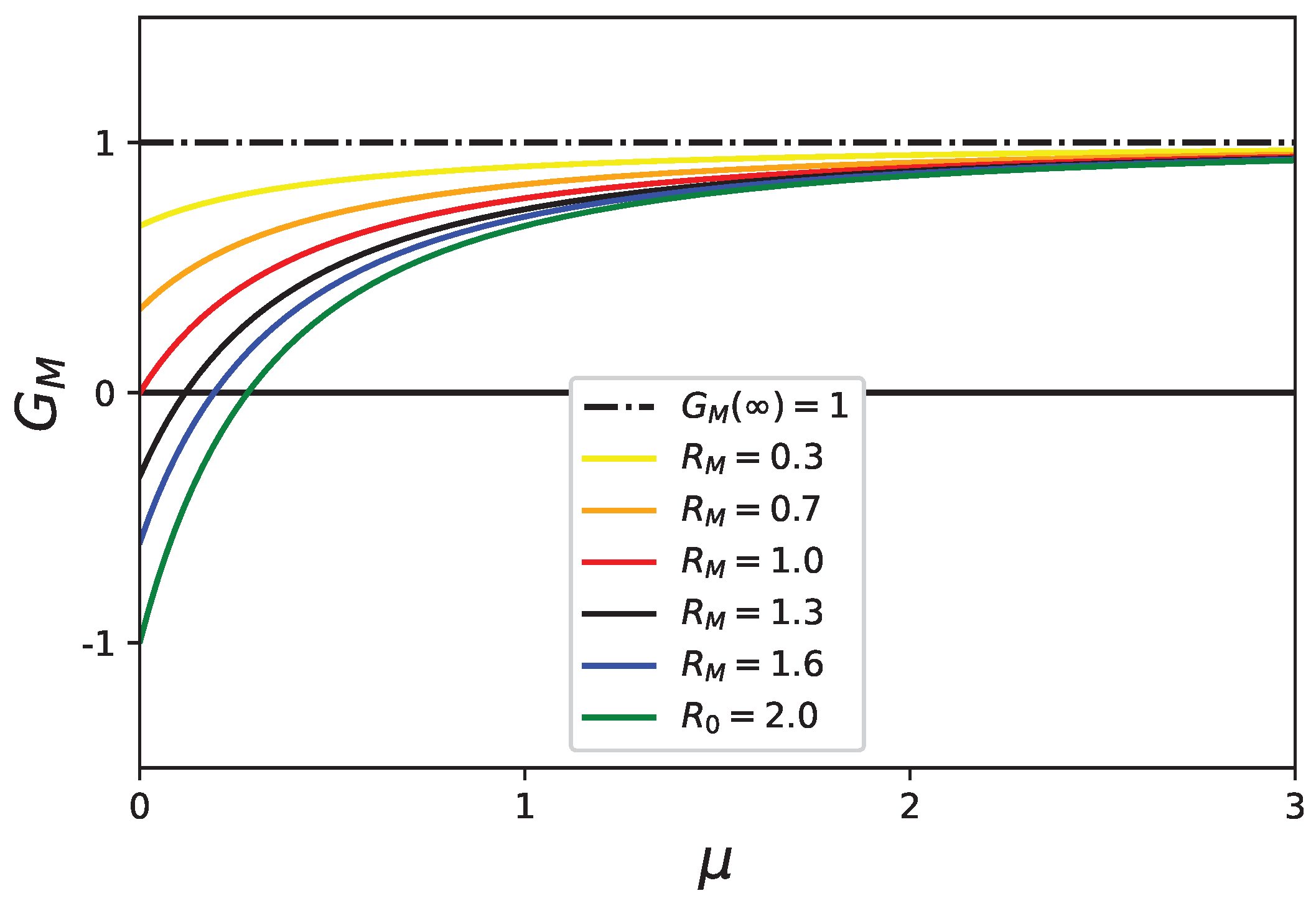
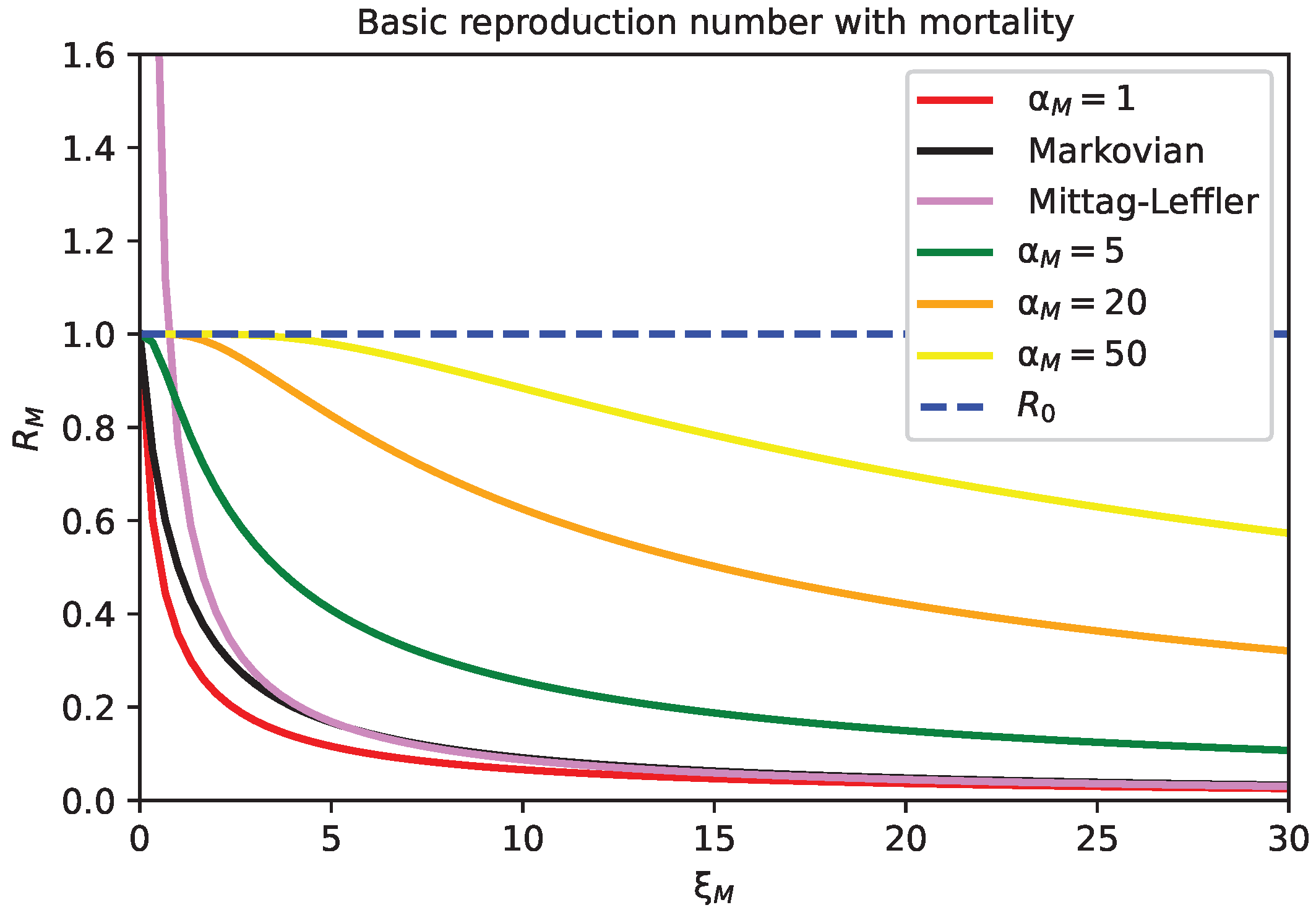
5. Stability Analysis of the Healthy State with Mortality
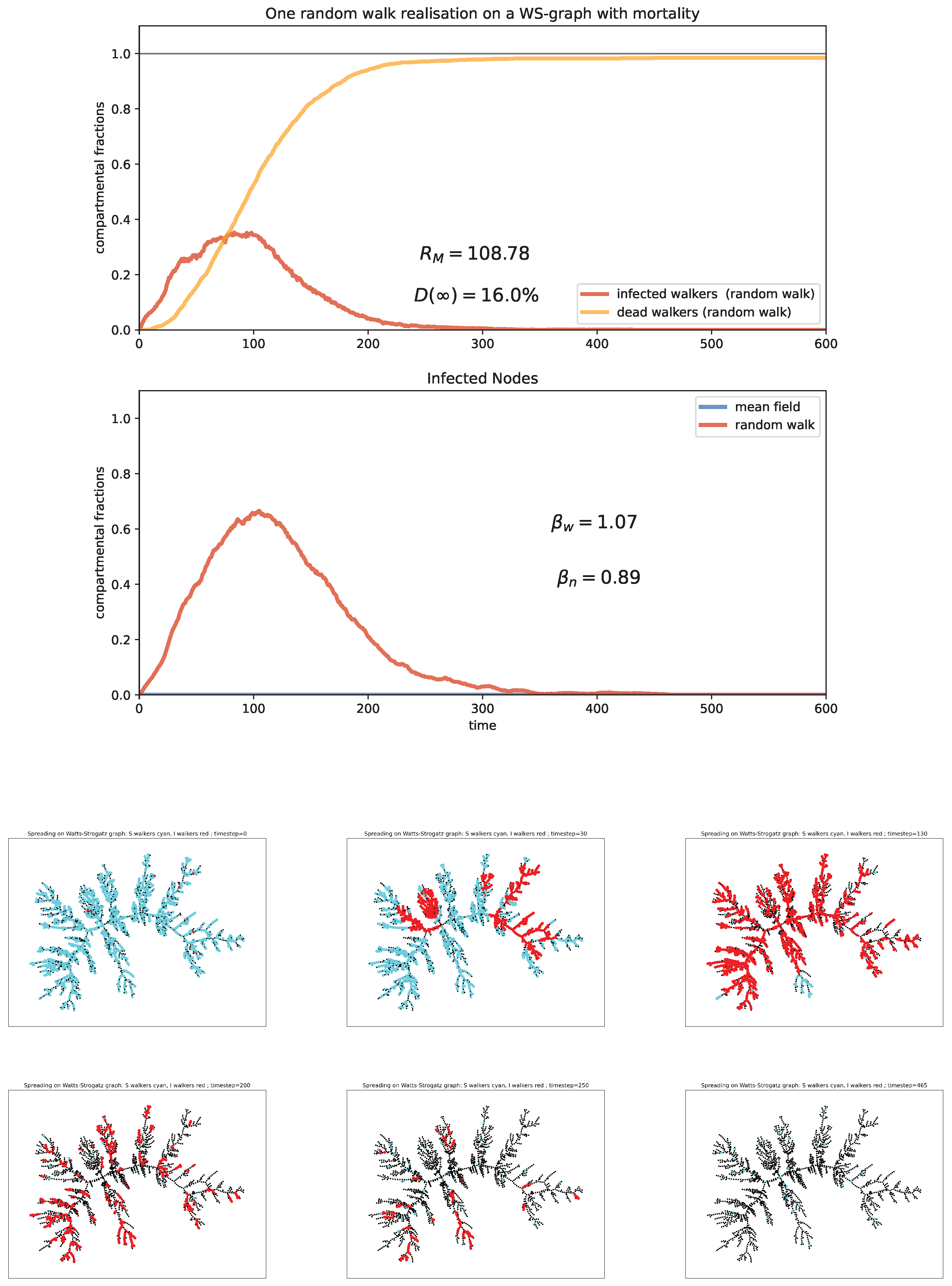
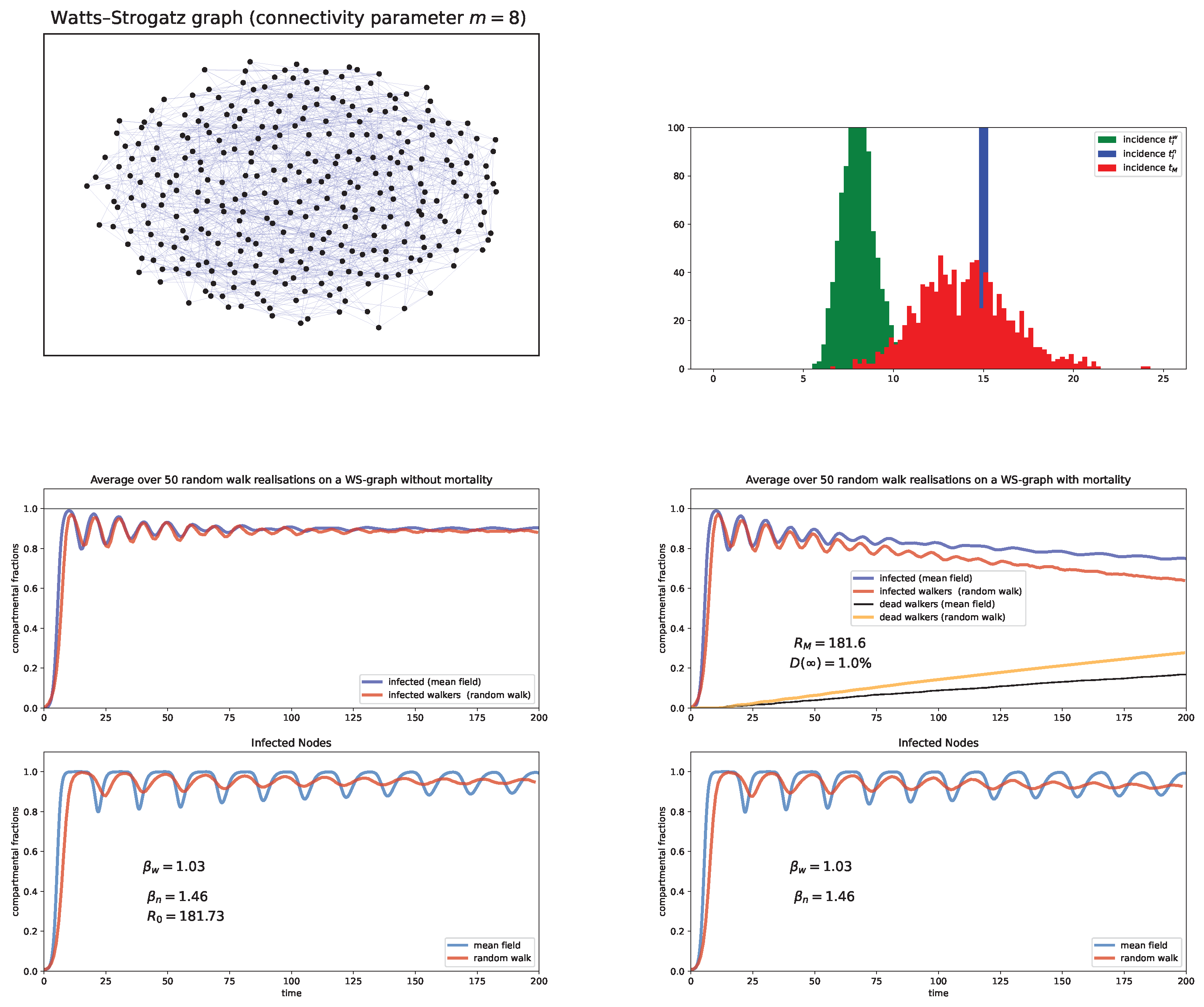
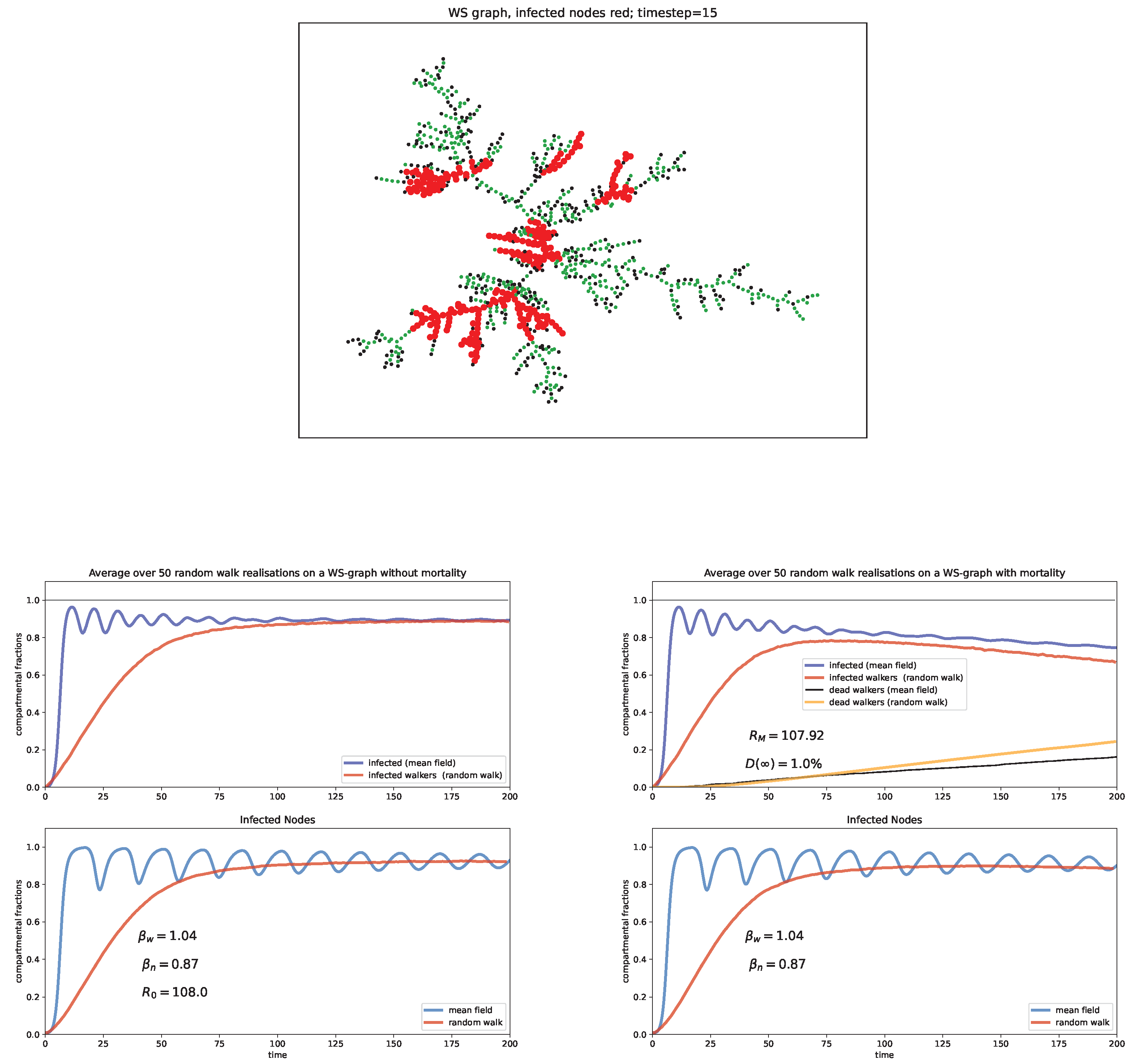
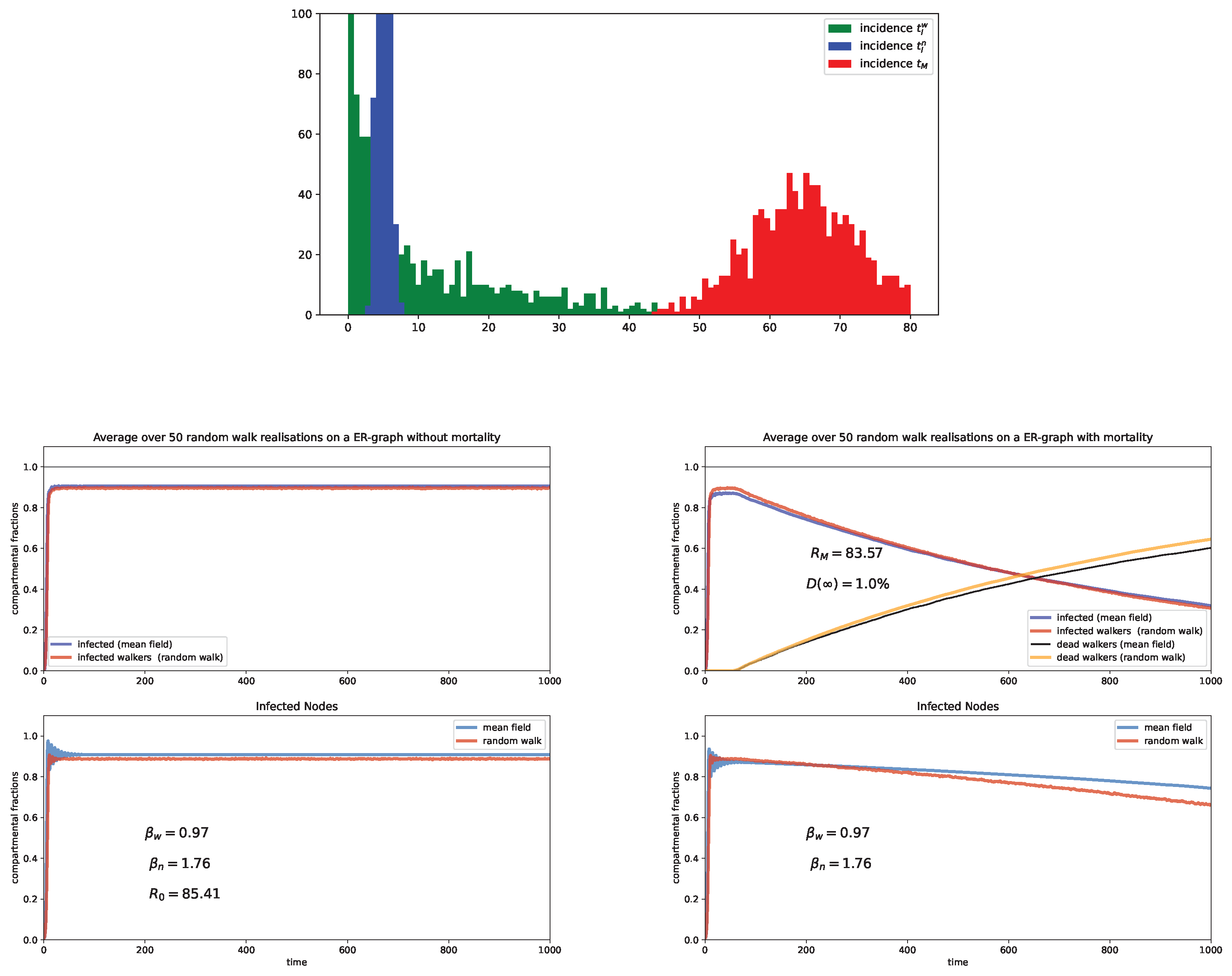
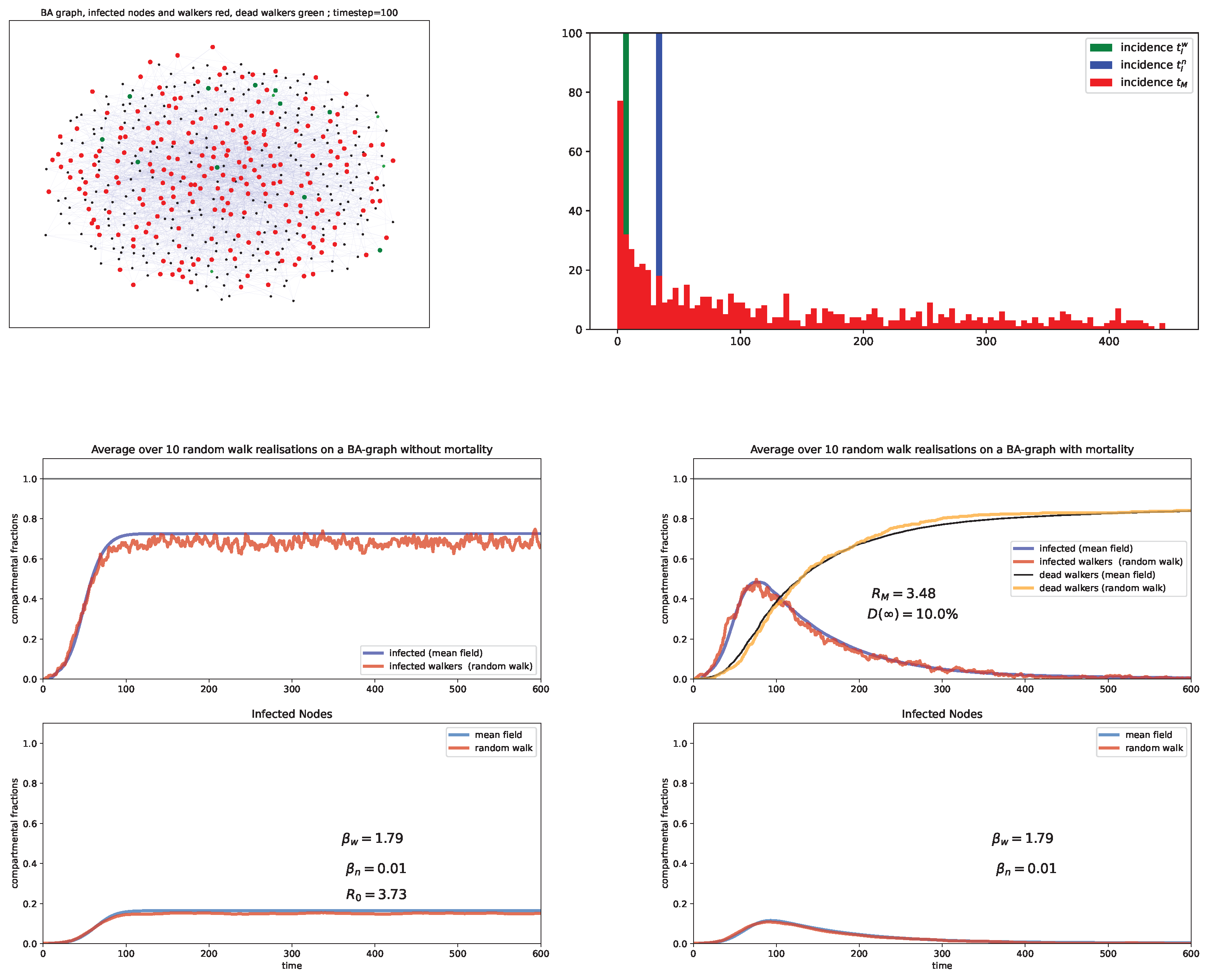
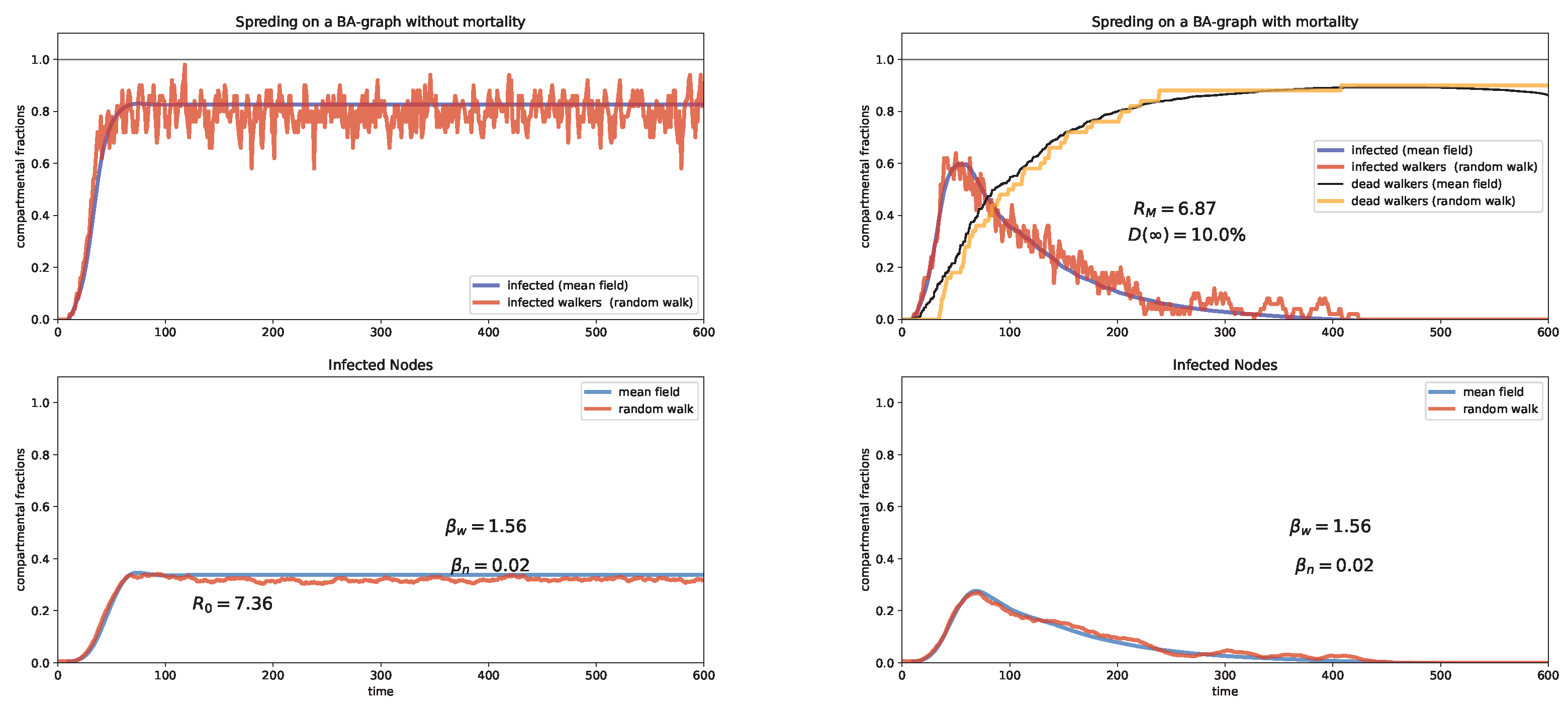
6. Random Walk Simulations
Case study and discussion
7. Conclusions
Acknowledgments
Appendix A
Appendix A.1. Some Basic Notions
Appendix A.2. Proof of Stability of the Endemic Equilibrium
A few remarks on the possibility of oscillatory (Hopf) instabilities of the endemic equilibrium
- Case (i); ():
- Case (ii); :
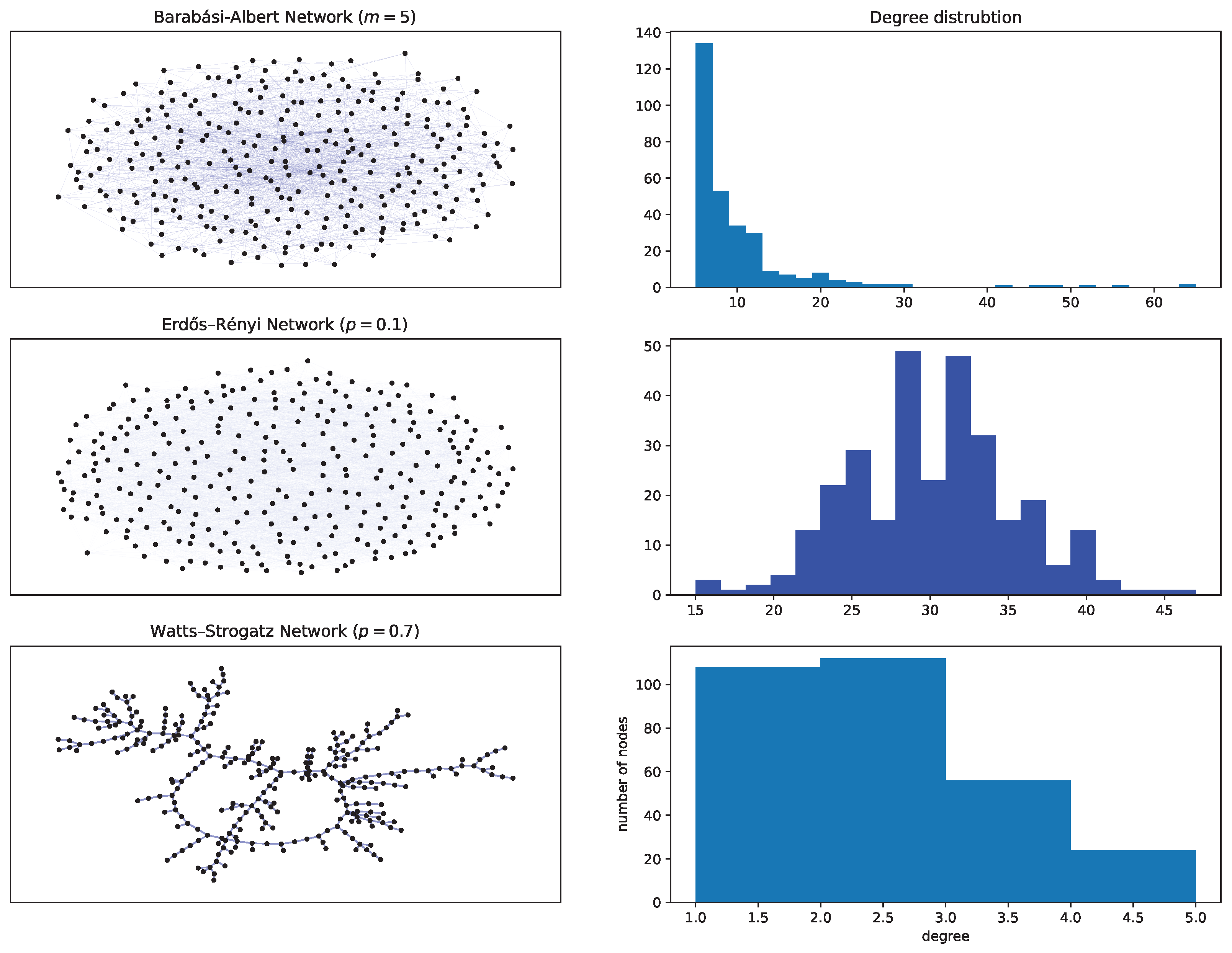
Appendix A.3. A Very Brief Recap of Random Graphs
- (i)
- Erdös and Rényi (ER) graph
- (ii)
- Watts-Strogatz (WS) network
- (iii)
- Barabási-Albert (BA) graph
References
- Rhodes, P.; Bryant, J.H. Public Health. Encycl. Br. 2024. Available online: https://www.britannica.com/topic/public-health.
- Kermack, W.O.; McKendrick, A.G. A contribution to the mathematical theory of epidemics. Proc. Roy. Soc. A 1927, 115, 700–721. [Google Scholar]
- Liu, W.M.; Levin, H.W.H.H.W.S.A. Dynamical behavior of epidemiological models with non-linear incidence rate. J. Math. Biol. 1987, 25, 359–380. [Google Scholar] [CrossRef]
- Li, M.Y.; Graef, J.R.; Wang, L.; Karsai, J. Global dynamics of a SEIR model with varying total population size. Math. Biosci. 1999, 160, 191–213. [Google Scholar] [CrossRef]
- Anderson, R.M.; May, R.M. Infectious Diseases in Humans; Oxford University Press: Oxford, 1992. [Google Scholar]
- Martcheva, M. An Introduction to Mathematical Epidemiology; Springer, 2015. [Google Scholar]
- Harris, J.E. Population-Based Model of the Fraction of Incidental COVID-19 Hospitalizations during the Omicron BA.1 Wave in the United States. COVID 2023, 3, 728–743. [Google Scholar] [CrossRef]
- Whitehead, S.S.; Blaney, J.E.; Durbin, A.P.; Murphy, B.R. Prospects for a dengue virus vaccine. Nat Rev Microbiol 2017, 5, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Satorras, R.; Castellano, C.; Mieghem, P.V.; Vespignani, A. Epidemic processes in complex networks. Rev. Mod. Phys. 2015, 87, 925–979. [Google Scholar] [CrossRef]
- Pastor-Satorras, R.; Vespignani, A. Epidemic dynamics and endemic states in complex networks. Phys. Rev. E 2001, 63, 066117. [Google Scholar] [CrossRef]
- Shudo, Y.O.Y.A. Microscopic Numerical Simulations of Epidemic Models on Networks. Mathematics 2021, 9, 932. [Google Scholar]
- Basnarkov, L.; Tomovski, I.; Sandev, T.; Kocarev, L. on-Markovian SIR epidemic spreading model of COVID-19. Chaos, Solitons and Fractals 2022, 160, 112286. [Google Scholar] [CrossRef]
- d’Onofrio, G.; Michelitsch, T.M.; Polito, F.; Riascos, A.P. On discrete-time arrival processes and related random motions. arXiv 2024, arXiv:2403.06821. [Google Scholar]
- Metzler, R.; Klafter, J. The random walk’s guide to anomalous diffusion : A fractional dynamics approach. Phys. Rep. 2000, 339, 1–77. [Google Scholar] [CrossRef]
- Mainardi, F.; Gorenflo, R.; Scalas, E. A fractional generalization of the Poisson processes. Vietnam J. Math. 2004, 32, 53–64. [Google Scholar]
- Barabási, A.-L. Network Science; Cambridge University Press: Cambridge, 2016. [Google Scholar]
- Barabási, A.-L.; Albert, R. Emergence of Scaling in Random Networks. Science 1999, 286, 509. [Google Scholar] [CrossRef] [PubMed]
- Barabási; Albert, R. ; Jeong, H. Mean-field theory for scale-free random networks. Physica A 1999, 272, 173–187. [Google Scholar] [CrossRef]
- Jeong, H.; Tombor, B.; Albert, R.; Oltvai, Z.N.; Barabási, A.L. The large-scale organization of metabolic networks. Nature 2000, 407, 651–4. [Google Scholar] [CrossRef]
- Erdös, P.; Rényi, A. On Random Graphs I. Publ. Math. 1959, 6, 290–297. [Google Scholar] [CrossRef]
- Erdös, P.; Rényi, P. On the evolution of random graphs. Publ. Math. Inst. Hung. Acad. Sci 1960, 5, 17. [Google Scholar]
- Gilbert, E.N. Random Graphs. Annals Math. Sci. 1959, 30, 1141–1144. [Google Scholar] [CrossRef]
- Ross, S.M. Stochastic Processes; John Wiley & Sons: New York, 1996. [Google Scholar]
- Newman, M.E.J. Networks: An Introduction; Oxford University Press: Oxford, 2010. [Google Scholar]
- Newman, M.E.J.; Watts, D.J.; Strogatz, S.H. Random graph models of social networks. PNAS 2002, 99, 2566–2572. [Google Scholar] [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440. [Google Scholar] [CrossRef]
- Eraso-Hernandez, L.K.; Riascos, A.P.; Michelitsch, T.M.; Wang-Michelitsch, J. Evolution of transport under cumulative damage in metro systems. Int. J. Mod. Phys. C 2023, 2450037. [Google Scholar] [CrossRef]
- Barrat, A.; Barthélemy, M.; Vespignani, A. Epidemic spreading in population networks. In Dynamic Processes on Complex Networks; Cambridge University Press, 2008; pp. 180–215. [Google Scholar] [CrossRef]
- Riascos, A.P.; Sanders, D.P. Mean encounter times for multiple random walkers on networks. Phys. Rev. E 2021, 103, 042312. [Google Scholar] [CrossRef] [PubMed]
- Bestehorn, M.; Riascos, A.P.; Michelitsch, T.M.; Collet, B.A. A Markovian random walk model of epidemic spreading. Continuum Mech. Thermodyn. 2021, 33, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- Michelitsch, T.M.; Riascos, A.P.; Collet, B.A.; Nowakowski, A.F.; Nicolleau, F.C.G.A. Fractional Dynamics on Networks and Lattices; ISTE/Wiley: London, 2019. [Google Scholar]
- Riascos, A.P.; Mateos, J.L. Long-range navigation on complex networks using Lévy random walks. Phys. Rev. E 2012, 86, 056110. [Google Scholar] [CrossRef] [PubMed]
- Bestehorn, M.; Michelitsch, T.M.; Collet, B.A.; Riascos, A.P.; Nowakowski, A.A.F. Simple model of epidemic dynamics with memory effects. Phys. Rev. E 2022, 105, 024205. [Google Scholar] [CrossRef] [PubMed]
- Granger, T.; Michelitsch, T.M.; Bestehorn, M.; Riascos, A.P.; Collet, B.A. Four-compartment epidemic model with retarded transition rates. Phys. Rev. E 2023, 107, 044207. [Google Scholar] [CrossRef]
- Bestehorn, M.; Michelitsch, T.M. Oscillating Behavior of a Compartmental Model with Retarded Noisy Dynamic Infection Rate. Int. J. Bifurcation Chaos 2023, 33, 2350056. [Google Scholar] [CrossRef]
- van Kampen, N.G. Stochastic processes in chemistry and physics; North Holland: Amsterdam, 1981. [Google Scholar]
- Mieghem, P.V. Exact Markovian SIR and SIS epidemics on networks and an upper bound for the epidemic threshold. arXiv 2014, arXiv:1402.1731. [Google Scholar]
- Zhu, Y.; Shen, R.; Dong, H.; Wang, W. Spatial heterogeneity and infection patterns on epidemic transmission disclosed by a combined contact-dependent dynamics and compartmental model. PLoS ONE 2023, 18, e0286558. [Google Scholar] [CrossRef]
- Gostiaux, L.; Bos, W.J.T.; Bertoglio, J.-P. Periodic epidemic outbursts explained by local saturation of clusters. Phys. Rev. E 2023, 107, L012201. [Google Scholar] [CrossRef]
- Peyrard, M. What can we learn from the dynamics of the Covid-19 epidemic? arXiv 2023, arXiv:2308.14090. [Google Scholar] [CrossRef] [PubMed]
- Soper, H.E. The interpretation of periodicity in disease prevalence. J. Royal Statistical Society 1929, 92, 34–61. [Google Scholar] [CrossRef]
- Noh, J.-D.; Rieger, H. Random walks on complex networks. Phys. Rev. Lett. 2004, 92. [Google Scholar] [CrossRef] [PubMed]
- Michelitsch, T.; Riascos, A.P.; Collet, B.A.; Nowakowski, A.; Nicolleau, F. Fractional Dynamics on Networks and Lattices; ISTE-Wiley: London, 2019. [Google Scholar]
- Schilling, R.; Song, R.; Vondraček, Z. Bernstein functions. In Theory and Applications, Studies in Mathematics, 37; de Gruyter: Berlin, 2010. [Google Scholar]
- Simon, C.M. The SIR dynamic model of infectious disease transmission and its analogy with chemical kinetics. PeerJ Physical Chemistry 2020. [Google Scholar] [CrossRef]
- Supplementary materials: PYTHON codes (© Téo Granger 2023) and animated films. Available online: https://sites.google.com/view/scirs-model-supplementaries/accueil.
| 1 | The network `distance’ of two nodes is the number of edges of the shortest path connecting them. |
| 2 | Free to download and non-commercial use. |
| 3 | A similar consideration of function of (39) shows as well that the healthy state for does not exhibit an oscillatory instability. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




