1. Introduction
At the end of 2019, many cases of human infection in Wuhan, China, were associated with a novel coronavirus termed (Severe Acute Respiratory Syndrome-Coronavirus) SARS-CoV-2. The pandemic that followed immediately challenged the whole medical and scientific community because of the extreme seriousness of the clinical picture associated with it. Initially, the clinical presentation of Coronavirus disease (COVID)-19 ranged from asymptomatic to mild upper respiratory illness, severe bilateral pneumonia, acute respiratory distress syndrome (ARDS), disseminated thrombosis, multi-organ failure, and death [
1,
2,
3,
4]. There is no doubt that the lung was the preferred target especially in the first waves of the pandemic. The clinical presentation of the disease has changed over time with the emergence of genetic variants and following the vaccination campaigns. Clinical and scientific evidence overall suggests that SARS-CoV-2 infection is a systemic disorder whose outcome is strongly conditioned by the state of the immune system of the host.
To date, much useful information has been obtained on the mechanisms of infection since the first observations that the virus entered the target cells through the expression of a characteristic spike protein (S protein) binding to the ACE-2 receptor [
5,
6,
7]. The resulting exuberant activation of the inflammatory response, the so-called cytokine storm, has represented the most critical piece of a pathogenesis puzzle that still remains to be fully understood. Monocytes and macrophages are natural immune cells that together with dendritic cells (DCs), which are professional antigen presenting cells (APC), constitute the monocyte-phagocyte system (MPS). There is wide evidence that the MPS plays a key role in protecting against virus infections, including SARS-CoV-2 [
2,
8,
9,
10,
11]
Monocytes and monocyte-derived macrophages were reported to be significantly increased in the peripheral blood and broncho-alveolar lavage (BAL), respectively, of hospitalized patients with COVID-19 pneumonia [
12]. In particular, monocytes and macrophages expressing the type I interferon-inducible receptor CD169 have been associated with severe COVID-19 [
13,
14]. Engagement of these cells upon SARS-CoV-2 infection was found to actively contribute to the expression of pro-inflammatory cytokines and tissue damage [
9,
12,
15,
16,
17,
18,
19,
20]. DCs are essential in initiating and regulating adaptive immune responses. Several studies have investigated the interaction between SARS-CoV-2 and DCs, showing that SARS-CoV-2 can directly infect them and/or impair their number and function [
21,
22]. As for monocytes and macrophages, any dysregulation of the DC compartment resulted in an uncontrolled inflammatory reaction along with tissue damage, all suggestive of an host failure to defend itself [
21].
Given these observations, our study aimed to analyzing by multi-parametric flow cytometry the frequency distribution and absolute number of peripheral blood CD169-expressing monocytes and those of conventional CD1c+ and CD141+ (namely cDC2 and cDC1) and plasmacytoid CD303+ DCs in a cohort of hospitalized patients affected by COVID-19 pneumonia. The prognostic impact of these immune-inflammatory cell players on in-hospital mortality was investigated. A gene profile analysis focused on 770 immune-inflammatory related human genes and 20 SARS-CoV-2 genes was performed in a sub-group of cases to further address the host-SARS-CoV-2 interplay.
2. Materials and Methods
2.1. Study Population
Fifty-eight patients affected by COVID-19 pneumonia were prospectively enrolled between March and May 2021 on hospital admission in our ward. Inclusion criteria were age ≥18 years, infection by SARS-CoV-2 confirmed by a positive reverse transcriptase polymerase chain reaction (RT-PCR) assay on the nasopharyngeal swab, and evidence of lung parenchyma involvement on high resolution chest computed tomography (CT). Patients with concomitant pulmonary embolism or ARDS were excluded. The study population’s demographic and clinical and laboratory data are shown in
Table 1. Twenty-two sex- and age-matched SARS-CoV-2 negative healthy volunteers (M=15; mean age ± SD=63 ± 9.8 years), recruited among blood donors and patient family members, were enrolled as a control group for comparison. The study was conducted following the amended Declaration of Helsinki and approved by the local Ethics committee (protocol number AOC/0000524/2020). At enrolment, two peripheral blood samples (as specified below) were collected by venipuncture from all study participants once written informed consent was obtained. In addition, data of interest for the study were anonymously collected into a dedicated database. Variables were socio-demographic characteristics, smoking habit, comorbidities, laboratory data, chest CT scoring of pneumonia according to Chung et al. [
23], any device used for oxygen supply, and pharmacological therapies. To date, oxygen supplementation was administered to all patients (either via nasal cannula or vent mask). High flow nasal cannula and/or continuous positive pressure (CPAP) and/or non-invasive mechanical ventilation were required in 45 of them. All patients were treated with systemic steroids (either 0.5-0.75 mg/kg/day of methylprednisolone or 6 mg/day of dexamethasone) and low molecular weight heparin at a prophylactic dosage. Anti-viral therapies (namely remdesevir) were used in 15% of cases.
2.2. Frequency Distribution and Absolute Numbers of Circulating CD169-Expressing Monocytes
Briefly, 50 μL of the EDTA-treated whole venous blood was incubated for 20 min in the dark at room temperature with a cocktail of monoclonal antibodies (mAbs), including the anti-CD64 (Pacific Blue), anti-CD169 (PE), anti-DR (APC), and the anti-CD4 (APC-CY7). Matched isotype mAbs were used as negative controls. All reagents were purchased from Beckman Coulter (CA, USA). After erythrocyte lysis with ammonium chloride, 5×10
4 events/sample were acquired by Navios Cytometer (Beckman Coulter, CA, USA) at the Clinical Biochemistry Unit of Monaldi Hospital. The analysis was performed by Kaluza Software (Beckman Coulter, CA, USA). The morphological selection of monocytes was first based on forward and side light scatters (FSC-A and SSC-A, respectively). Gated cells were then plotted according to CD64 expression/SSC. In this way, enumeration of CD169-expressing cells was restricted to CD64
+ monocytes. The gating strategy for identifying CD169
+ monocytes is shown in Supplementary
Figure 1A. Data were expressed as percentages and absolute numbers. Absolute numbers were calculated as follows: percent of CD169
+ monocytes x total number of white blood cells (WBCs) per mm
3/100. The white blood cell (WBC) count was determined using a Sysmex XT-1800i hemocytometer (Sysmex Europe, Norderstedt, Germany).
2.3. Frequency Distribution and Absolute Numbers of Circulating Dendritic Cell Subsets
A flow cytometry DC enumeration kit (Blood Dendritic Cell Enumeration Kit, Milteny Biotech, Bergisch Gladbach, Germany) was used to identify DC subtypes according to the manufacturer’s instructions. Reference median frequencies of blood DC subsets were 0.27% (range: 0.09-0.42), 0.02% (range: 0-0.04), and 0.19% (range: 0.09-0.37) for cDC2, cDC1, and pDC, respectively. Briefly, 300 μL of the EDTA-treated whole blood was incubated for 10 min with a cocktail of mAbs, including the anti-CD1c (PE), anti-CD141(APC), and the anti-CD303 (FITC). Mouse IgG2a-PE, IgG1-FITC, and IgG1-APC mAbs were used as isotype controls. In addition, samples were co-stained with the anti-CD19-PE-Cy-5 and the anti-CD14-PE-Cy5 to exclude B cells and monocytes from the analysis. All specimens were treated with a dead-cell discriminator. Following erythrocyte lysis, washing, and fixation, 5×10
5 event per sample were acquired by Navios Cytometer (Beckman Coulter, CA, USA) at the Clinical Biochemistry Unit of Monaldi Hospital and analyzed by Kaluza Software (Beckman Coulter, CA, USA). The gating strategy for identifying each DC subtype is shown in Supplementary
Figure 1B. Results were reported as absolute numbers of cDC2, cDC1, or pDC among total WBCs. Data were expressed as percentages and absolute numbers. Absolute numbers were calculated as follows: percent of a given DC subset x total number of WBCs per mm
3/100.
2.4. Measurement of Serum Levels of Interleukin-6
Serum levels of interleukin-6 (IL-6) were evaluated by a chemiluminescence immunoassay (Immulite 2000 System, Siemens Healthcare Diagnostics) at the Clinical Biochemistry Unit of Monaldi Hospital, immediately after blood sampling in all subjects, according to the manufacturer’s instruction. The analytical sensitivity of the assay was 2 pg/mL and the upper reference value in adults was 5.9 pg/mL.
2.5. Human Immune-Inflammatory and SARS-CoV-2 Gene Profile Analysis
For gene profile analysis, 2.5 mL of peripheral blood were collected in PAXgene blood RNA tube. RNA was extracted using the RNA blood mini Kit (Qiagen), according to the manufacturer’s instructions. Purified RNA was used for hybridization and subjected to gene profiling analysis on a NanoString nCounter. Autoimmune Profiling panel of 770 human genes involved in immune-inflammatory processes were tested as target. The Coronavirus Panel Plus containing 20 probes for the SARS-CoV-2 genes (ACE2_Hs; HCoV-229E_N; HCoV-229E_S; HCoV-HKU1_N; HCoV-HKU1_S; HCoV-NL63_N; HCoV-NL63_S; HCoV-OC43_N; HCoV-OC43_S; SARS-CoV_N; SARS-CoV_S; SARS-CoV-2_E; SARS-CoV-2_M; SARS-CoV-2_N; SARS-CoV-2_orf1ab; SARS-CoV-2_orf1ab_REV; SARS-CoV-2_ORF3a; SARS-CoV-2_ORF7a; SARS-CoV-2_ORF8; SARS-CoV-2_S) was analyzed as well. Gene expression data were normalized using the nSolver Version 4.0 Software with reference to internal ERCC (External RNA Controls Consortium) technical controls and 30 housekeeping genes. Statistical analysis was performed via Benjamini-Hochberg We conducted heatmap analyses to correlate the most important covariate.
2.6. Statistical Analysis
Quantitative variables were characterized using mean ± standard deviation (SD) or median with quartile ranges, where appropriate. Categorical factors were described as absolute numbers and percentages. Comparisons between study groups were analyzed with the Mann-Whitney U test. Correlations among quantitative variables were based on the non-parametric Spearman rank correlation coefficient. Survival was calculated using Kaplan-Meier curves and compared by the log-rank test. In-hospital mortality was calculated from the date of hospital admission to the date of death. All tests were two-tailed and statistical significance was set at p< 0.05. Statistical analyses were made using the Qlucore software, GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA) or the MedCalc (MedCalcSoftware, Ostend, Belgium) platforms.
3. Results
3.1. Differential Distribution of Circulating CD169+ Monocytes and Dendritic Cell Subsets in COVID-19 Pneumonia
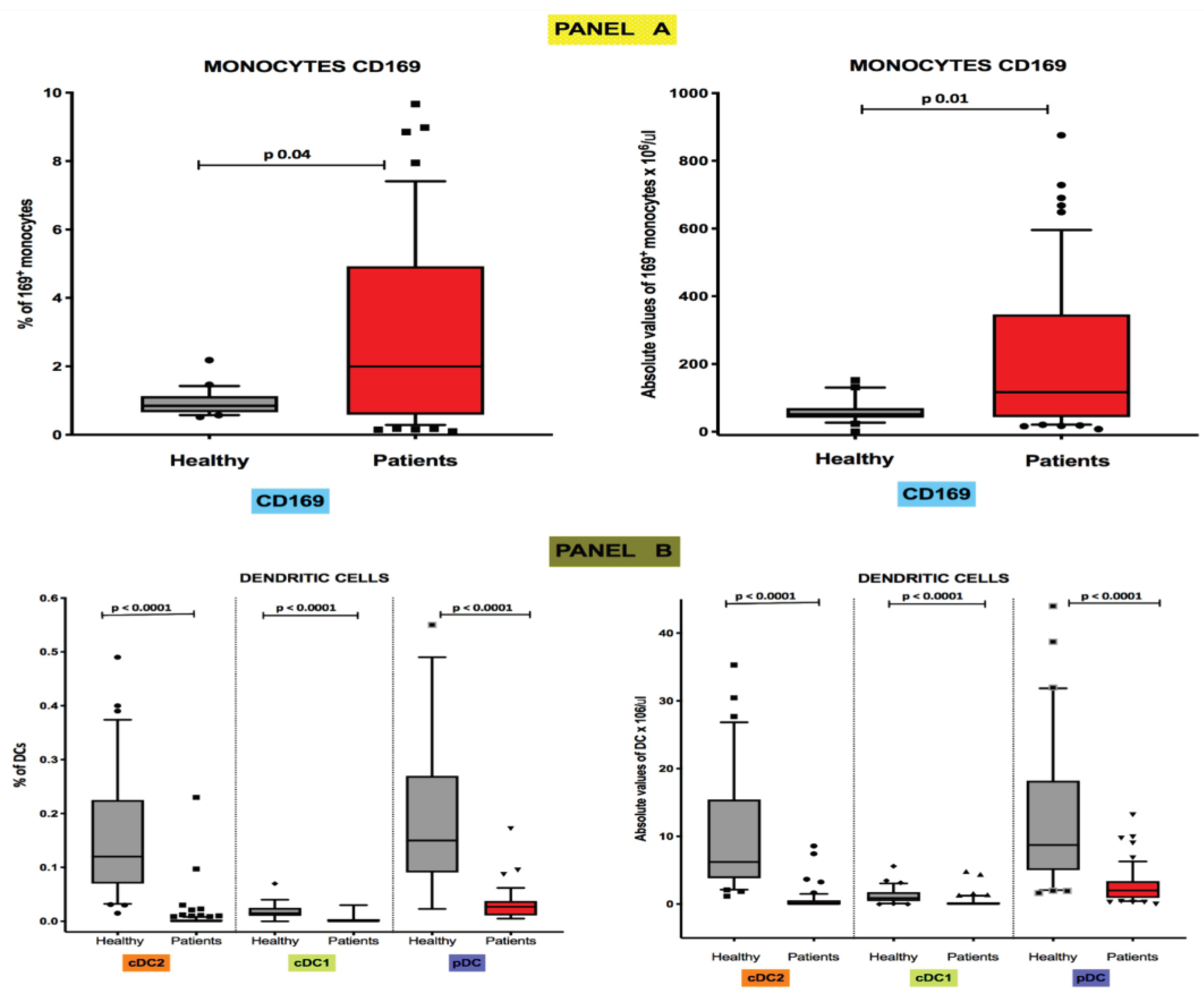
Flow cytometry estimates of median frequencies and absolute levels of peripheral blood CD169-expressing monocytes were significantly higher in COVID-19 patients than in healthy controls. In particular, the median frequency values [25th; 75th percentile] were 0.85% [0.65; 1.13] in healthy individuals and 1.99% [0.58; 4.92] in COVID-19 patients (p 0.04) (
Figure 1, Panel A), while the median absolute levels were 51.4 cells/μL [41.3; 69.7] and 116.9 cells/μL [43.4; 346.1], respectively (p 0.01) (
Figure 1, Panel A). Conversely, as reported in
Figure 1 (Panel B), both frequencies and absolute median counts of all DC subsets were severely depleted in COVID-19 patients. Specifically, the median frequencies in healthy controls were respectively 0.120% [0.070;0.225] for CD1c
+ cDC2s, 0.0150% [0.010;0.025] for CD141
+cDC1s and 0.150% [0.090; 0.270] for CD303
+ pDCs (
Figure 3, Panel B), while in COVID-19 patients were 0.000% [0.000; 0.003] for CD1c
+ cDC2s, 0.000% [0.000; 0.004] for CD141
+cDC1s, and 0.0270% [0.011; 0.037] for CD303
+ pDCs (
Figure 1, Panel B). The median absolute numbers of DC subsets in healthy individuals were 6.226 cells/μL [3.85; 15.43] for CD1c
+ cDC2s, 0.870 cells/μL [0.87; 1.78] for CD141
+cDC1s, and 8.721 cells/μL [5.02; 18.21] for CD303
+ pDCs (
Figure 3, Panel B), while DC absolute counts in COVID-19 patients were 0.00 cells/μL [0.00; 0.58] for CD1c
+ cDC2s, 0.00 cells/μL [0.00; 0.28] for CD141
+cDC1s, and 2.01 cells/μL [0.91; 3.38] for CD303
+ pDCs (
Figure 1, Panel B). For all the DC subsets differences with the control group were equally statistically significant in all comparisons (p <0.0001).
Figure 1.
Highs levels of CD169+ monocytes and low values of circulating DC subsets are present in peripheral blood of COVID-19 patients Boxplot showing the percentage distribution and absolute counts of monocytes CD169+ (Panel A) and circulating cDC2, cDC1, and pDC (Panel B) subsets among total leukocytes in healthy subjects and COVID-19 patients.
Figure 1.
Highs levels of CD169+ monocytes and low values of circulating DC subsets are present in peripheral blood of COVID-19 patients Boxplot showing the percentage distribution and absolute counts of monocytes CD169+ (Panel A) and circulating cDC2, cDC1, and pDC (Panel B) subsets among total leukocytes in healthy subjects and COVID-19 patients.
3.2. Inverse Correlation of Absolute Levels of CD169-Positive Monocytes with CD141+ cDC1s Numbers in COVID-19 Pneumonia Patients
Perturbations at baseline of cDC1s and pDCs were inversely correlated with CD169-positive monocytes (r=-0.29; p=0.0309; r=-0.31; p=0.0347 respectively).
Moreover, pDCs absolute values were significantly correlated with lymphocytes (r=0.46; p=0.0001) and monocytes (r=0.44; p=0.0003). No further correlations were found between the peripheral cell subtypes analyzed and the patients’ clinical and laboratory data.
3.3. Short-Term Distribution of Circulating CD169+ Monocytes DC Subsets Is Not Modified by Systemic Steroids
To address the impact of systemic steroids on the peripheral distribution of CD169-expressing monocytes and DC subsets, a sub-cohort of 20 COVID-19 patients treated with systemic steroids were tested at 7 and 14 days from hospital admission. Absolute numbers of both CD169+monocytes and DC subsets remained unchanged at the time points analyzed (data not shown).
3.4. Circulating CD169+ Monocytes Are Associated with In-Hospital Mortality of COVID-19 Pneumonia
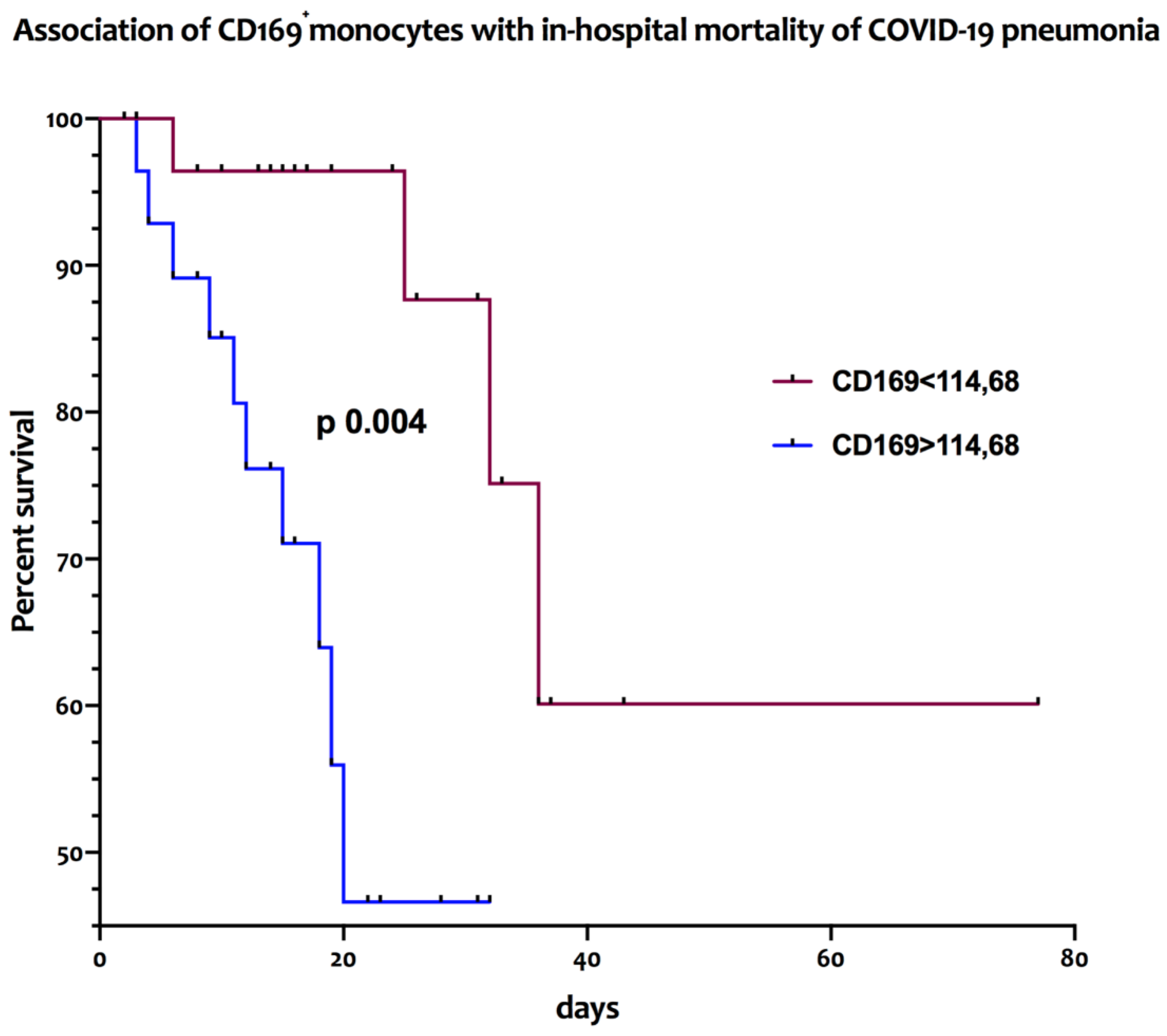
The association between the frequencies and absolute numbers of circulating CD169-positive monocytes and of DC subsets with in-hospital mortality was assessed with a log rank test. As reported in
Figure 2, patients affected by COVID-19 pneumonia with absolute counts of peripheral CD169
+ monocytes above the median value of 114.68/μL had a significantly higher in-hospital mortality (median survival of 20 days) [HR 4.33; 95% CI: 1.46-12.88] than those with lower values (median survival >75 days) [HR 0.23; 95% CI: 0.07-0.68; p=0.004].
Figure 2.
High numbers of peripheral CD169+ monocytes are associated with increased in-hospital mortality in COVID-19 patients Kaplan Meier curves representative of the in-hospital mortality of COVID-19 patients according to the high (>cut-off) or low (<cut-off) baseline levels (T0) of circulating CD169+ monocytes.
Figure 2.
High numbers of peripheral CD169+ monocytes are associated with increased in-hospital mortality in COVID-19 patients Kaplan Meier curves representative of the in-hospital mortality of COVID-19 patients according to the high (>cut-off) or low (<cut-off) baseline levels (T0) of circulating CD169+ monocytes.
Conversely, no significant difference of in-hospital mortality was observed between patients when looking at frequencies of CD169+ monocytes or to percentages and absolute levels of any circulating DC subset (data not shown).
3.5. Serum Levels of the Pro-Inflammatory Cytokine IL-6 Are Increased in COVID-19 Patients and Inversely Correlated with the Absolute Numbers of cDC1s and pDCs
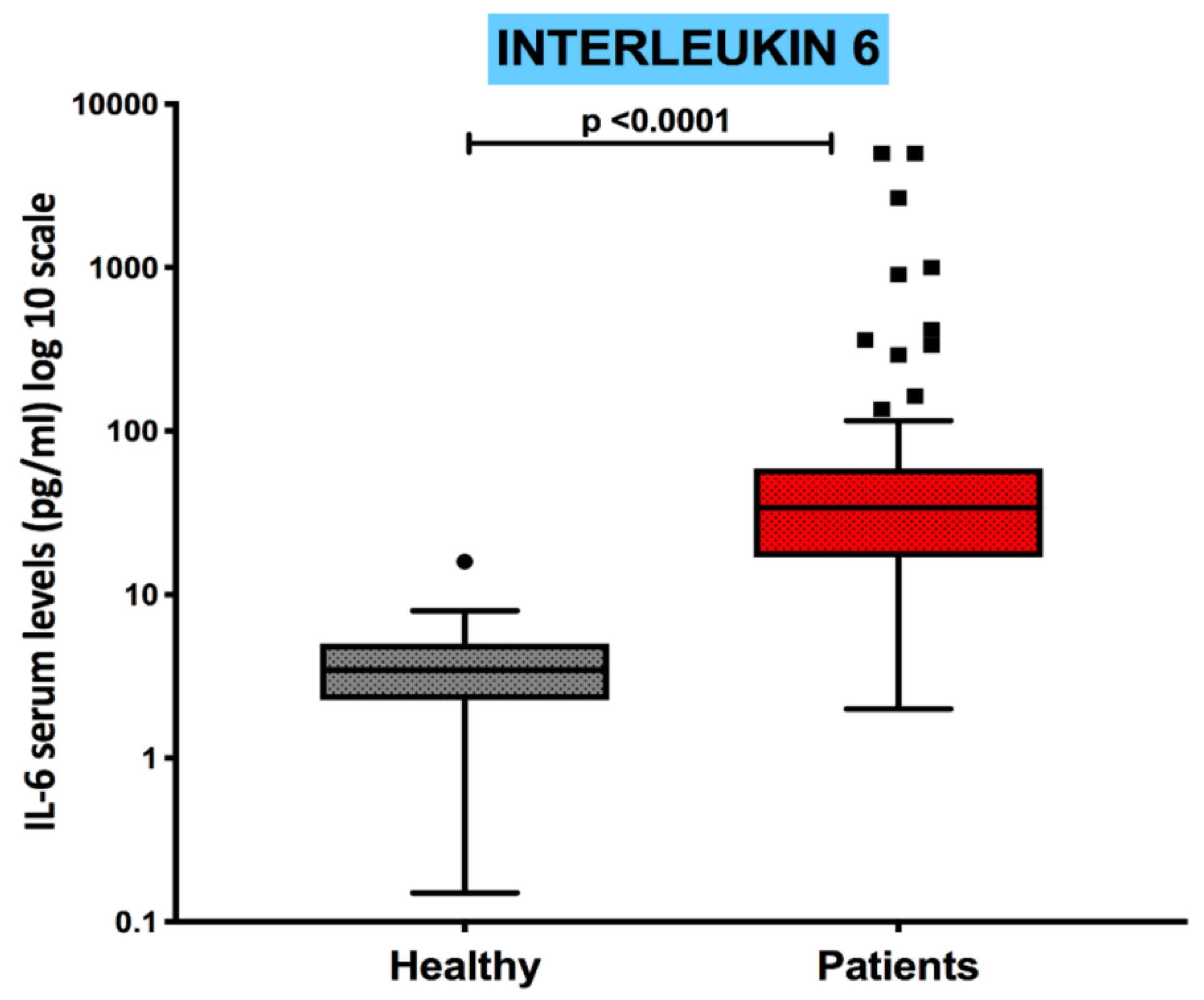
Serum levels of the pro-inflammatory cytokine IL-6 were measured in all patients to assess the COVID-19 related peripheral immune-inflammatory milieu. IL-6 concentrations were significantly increased in COVID-19 patients as compared to the control group (p<0.0001), as shown in
Figure 3, ranging from a minimum of 2.00 pg/mL to a maximum of 5000 pg/mL.
Figure 3.
Serum concentrations of interleukin-6 are increased in COVID-19 patients Boxplot showing the distribution of Interleukin-6 analyzed in healthy subjects and COVID-19 patients.
Figure 3.
Serum concentrations of interleukin-6 are increased in COVID-19 patients Boxplot showing the distribution of Interleukin-6 analyzed in healthy subjects and COVID-19 patients.
Correlation analysis between IL-6 serum levels and the distribution of all DC subtypes further showed that in COVID-19 patients IL-6 was negatively related with the circulating absolute levels of CD1c+ cDC2 subpopulation (r=-0.29, p=0.034) and CD303+ pDC subsets (r=-0.29, p=0.036). No significant correlation was observed with the CD141+cDC1 and the expression of CD169-expressing monocytes.
3.6. Interplay of Host Immune-Inflammatory Genes and SARS-CoV-2 Gene Signature in COVID-19 Pneumonia
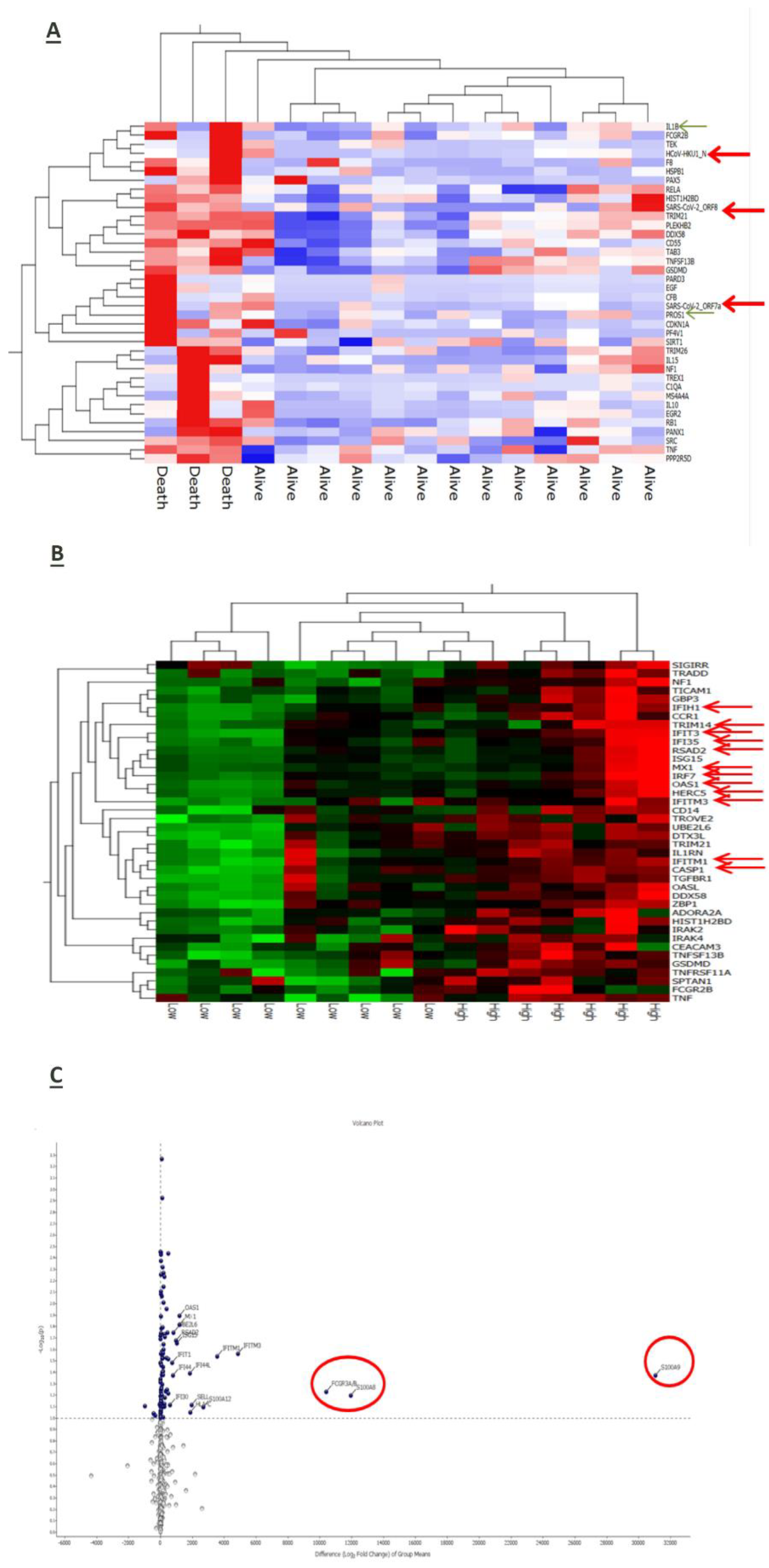
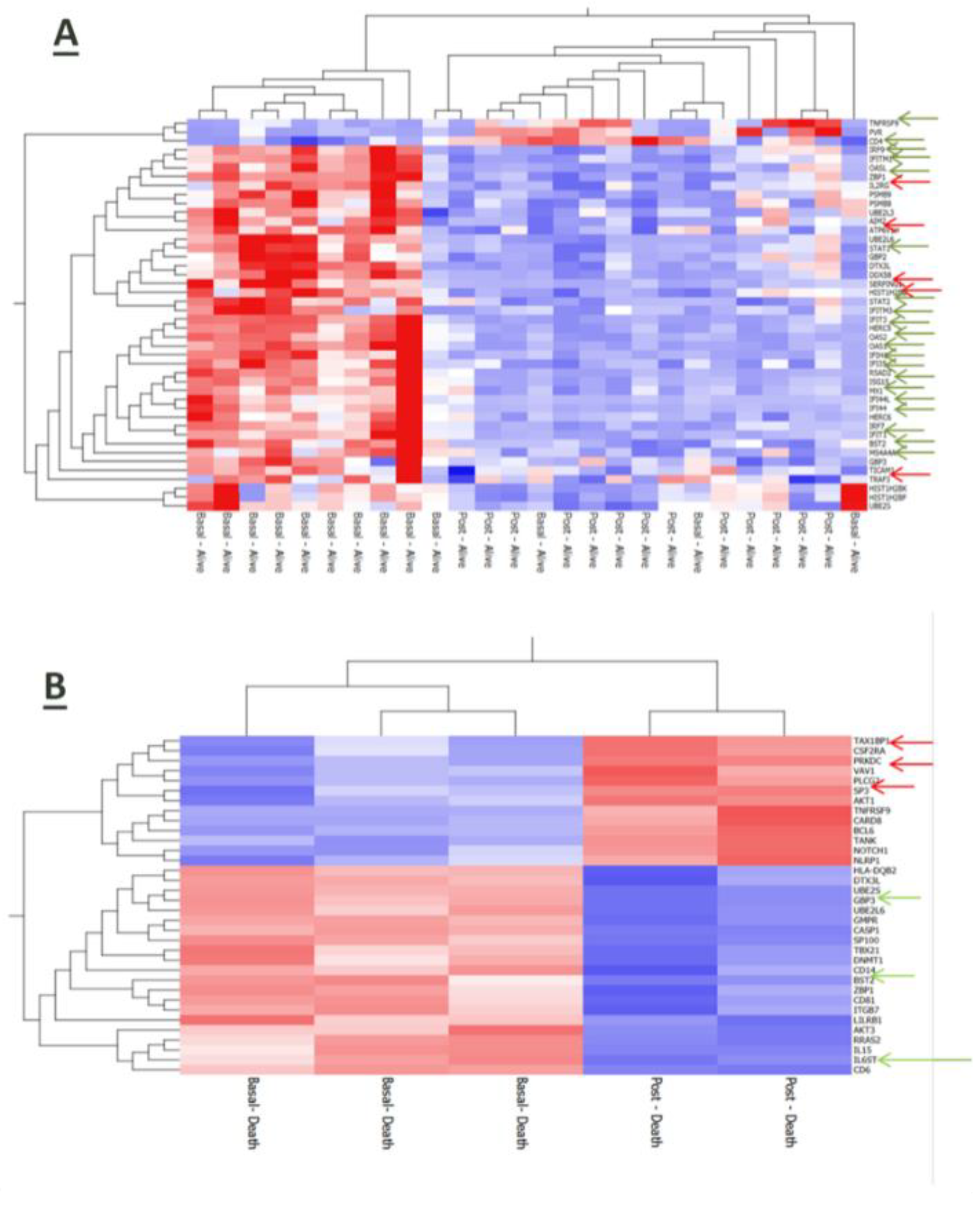
RNA-based gene profiling analysis of a panel of 770 human genes involved in immune-inflammatory processes was available for a sub-cohort of 16 patients at enrollment on hospital admission. Three of them died during the observation period due to a sudden deterioration of their clinical conditions. Interestingly, the expression of some viral genes (HCoV-HKU1_N, SARS-CoV2_ORF8, and SARS-CoV2_ORF7a) was up-regulated in patients with worse outcome along with inflammatory mediators such as interleukin (IL)-1 beta, tumor necrosis-α (TNF-α) and the anticoagulant protein (PROS1) (
Figure 4A). Also, stratification of the whole patient sub-group according to the median reference value of IL-6 serum levels (31.46 pg/mL) allowed to identify 7 cases in whom higher IL-6 expression was associated with the upregulation of genes involved in the anti-viral response induced by interferon, such as IFIH1, IFIT3, IFI35, IRF7, IFITM3, IFITM1, and of the inflammatory cytokines S100A9 and S100A8 (
Figure 4B) and (
Figure 4C).
Even after 7 days from hospitalization, immune-inflammatory gene expression was modulated according to disease outcome of COVID-19 patients. In particular, surviving patients upregulated genes related to inflammatory and anti-viral-related pathways along with the T cell membrane molecule CD4 (
Figure 5A). Conversely, patients who died had a reduced expression of genes involved in anti-viral defense (
Figure 5B).
4. Discussion
The pathogenesis of COVID-19 remains a not fully clarified puzzle which involves the contribution of multiple host-related immune factors. Aim of our study was to assess the interplay between circulating CD169
+ monocytes and DCs, which are part of the monocyte-phagocyte system and key players in the modulation of the immune-inflammatory process against viral infections [
12,
21], in a cohort of patients hospitalized for COVID-19 pneumonia. In agreement with previous data, our study confirms the involvement of the MPS in COVID-19 pneumonia through the finding of a lack of balance between CD169
+ monocytes (increased) and dendritic cell subsets (severely depleted) at the peripheral level. This observation was significantly associated with a worse disease outcome (in-hospital mortality). Interestingly, in line with cellular findings, gene profiling analysis revealed that a panel of inflammation-related host products, including interleukin-6 which notably exerts a suppressive effect on DC maturation, along with viral genes were over-expressed in patients with bad prognosis.
CD169-expressing monocytes are present in high numbers in the spleen and lymph nodes where they are involved in autoimmunity and defense against viral infections. This happens upon stimulation with interferons (IFNs) that typically increase their constitutive low expression of CD169 [
24,
25,
26]. Therefore, the overexpression of CD169 may be considered as a promising host marker of viral infections. Within a pandemic context, this possibility has ignited interest on CD169-expressing monocytes as a rapid marker for the triage of patients with suspected COVID-19. Also, there is evidence that CD169
+ monocytes display a distinct gene expression profile and are functionally different in COVID-19 patients [
27]. We found that both the proportion and the absolute number of CD169
+ monocytes were increased in our patient cohort. Interestingly, in-hospital mortality was increased in cases with circulating CD169
+ monocytes above the median reference value. Similarly to our setting, previous studies have shown that blood CD169
+ monocytes were significantly increased in COVID-19 cases, correlating with disease severity [
12,
28]. In this direction, Ortillon
et al. have shown that CD169-expressing monocytes were more activated in bad prognosis COVID-19 patients requiring mechanical ventilation [
29]. Despite this, the relationship of CD169
+ monocytes and patient outcome still remains not univocal in COVID-19. Indeed, lower numbers of CD169
+ monocytes have been equally related with a poorer prognosis in COVID-19 patients [
30,
31], while other observations have reported no association [
18].
Herein we also show that the increase of CD169
+ monocytes was associated with a severe depletion of all circulating DC subsets, either analyzed as frequencies or absolute numbers. This combined observation is the first time to be reported to emphasize the close complementarity between these two different cell types. Previous studies have reported that the numbers of cDCs and pDCs were lower in COVID-19 subjects than in healthy individuals [
32,
33,
34,
35], and that DCs impairment was restored in convalescent phases [
32]. Among the different DC subsets, pDCs display high antiviral activities due to their ability to produce IFN-I and are thought to be involved in immune tolerance [
36]. By virtue of these properties, pDCs, similarly to CD169-expressing monocytes, have been suggested as a biomarker of COVID-19, strongly related with disease severity [
30]. The so-called cytokine-storm that characterized the most severe forms of COVID-19 was an exuberant inflammatory response associated with high levels of serum IL-6 [
19,
37]. Cumulative evidence suggests that IL-6 inhibits the differentiation of DCs by affecting the transition from the resting/immature phenotype to the activated/mature one through the IL-6/JAK2/STAT3 axis, which is emerging as a major player in inflammation [
38,
39]. In our series we found that serum concentrations of the pro-inflammatory cytokine IL-6 were significantly higher in COVID-19 patients than in healthy controls. Also, IL-6 levels negatively correlated with the absolute number of both pDCs and cDCs suggesting the close relationship of inflammation with the impairment of anti-viral responses and initiation of adaptive immunity. According to these data, the ancillary human gene profiling analysis performed in a sub-cohort of our patient population also revealed that inflammation-related gene products, such as IL-1 beta, TNF-α and the anticoagulant protein (PROS1), were over-expressed in COVID-19 patients along with some viral genes (HCoV-HKU1_N, SARS-CoV2_ORF8, and SARS-CoV2_ORF7a). In addition, as far as our study was a qualitative assessment, the increased expression of IL-6 was found to be associated with the upregulation of genes involved in the innate antiviral response, such as IFIH1, IFIT3, IFI35, IRF7, IFITM3, IFITM1, and of the inflammatory cytokines S100A9 and S100A8. Of note, patients who died had a reduced expression of genes involved in anti-viral defense.
Our study has some limitations. First, it has been realized in a small cohort of patients. Unfortunately, the restrictions imposed by the pandemic have strongly penalized experimental studies on potentially contagious samples. Also the short period of study was representative of a clinical picture that has changed over time as a result of the virus gene variability. However, our data are noteworthy due to the fact that enumeration and phenotypic analysis of CD169-expressing cells and DC subtypes suffers from their low values at the peripheral level, even in healthy individuals. In addition, the use of high-cost molecular analytical procedures was a further constraint on the extension of the study to a larger case study in an extremely difficult context. In this issue, our effort represents a precious piece of the intriguing puzzle of the immuno-pathology of SARS-CoV-2 infection.
In conclusion, our findings suggest that the interplay between the different components of the MPS is dysregulated in COVID-19 patients presenting with acute pneumonia. An excessive inflammatory response together with the recruitment of cells of the natural immunity (that is increased availability of CD169-expressing monocytes) is at the same time cause of disease progression but also an attempt to compensate for the impairment of the anti-viral and adaptive responses (that is depletion of DC subsets). This may explain, at least in part, the imbalance between innate and adaptive immunity and its impact on disease outcome. Despite the clinical manifestations of COVID-19 have significantly changed, even in the face of a tight vaccination campaign, the deepening of the pathogenesis mechanisms underlying a condition that has devastated the planet remains a priority of clinical and experimental research.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
study conception and design: MB and DG; collection and interpretation of data: MB, DG, SZ, CN, DM; statistical analysis: MB, DG, SZ; manuscript drafting: MB, DG; manuscript editing: MB, DG, SZ, DM, PAA; approval to submit: MB, DG, DM, SZ, CN, SS, FL, EC, BB, RB, LA, CL, LC, PAA.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was approved by local Ethics committee of Aziends Ospedaliera dei Colli (AOC) (protocol number AOC/0000524/2020).
Informed Consent Statement
Informed consent was obtained from all participants at the time of data collection.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
No acknowledgements.
Conflicts of Interest
PAA has/had a consultant/advisory role for Bristol Myers Squibb, Roche-Genentech, Merck Sharp & Dohme, Novartis, Merck Serono, Pierre-Fabre, AstraZeneca, Sun Pharma, Sanofi, Idera, Sandoz, Immunocore, 4SC, Italfarmaco, Nektar, Boehringer-Ingelheim, Eisai, Regeneron, Daiichi Sankyo, Pfizer, Oncosec, Nouscom, Lunaphore, Seagen, iTeos, Medicenna, Bio-Al Health, ValoTX, Replimmune, Bayer. He also received research funding from Bristol Myers Squibb, Roche-Genentech, Pfizer, and Sanofi. Travel support by Pfizer, Bio-Al Health, and Replimmune. All other authors have declared no conflicts of interest.
References
- M.F. Osuchowski, M.S. Winkler, T. Skirecki, S. Cajander, M. Shankar-Hari, G. Lachmann, G. Monneret, F. Venet, M. Bauer, F.M. Brunkhorst, S. Weis, A. Garcia-Salido, M. Kox, J.M. Cavaillon, F. Uhle, M.A. Weigand, S.B. Flohe, W.J. Wiersinga, R. Almansa, A. de la Fuente, I. Martin-Loeches, C. Meisel, T. Spinetti, J.C. Schefold, C. Cilloniz, A. Torres, E.J. Giamarellos-Bourboulis, R. Ferrer, M. Girardis, A. Cossarizza, M.G. Netea, T. van der Poll, J.F. Bermejo-Martin, I. Rubio, The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity, Lancet Respir Med (2021). [CrossRef]
- Y. Shi, Y. Wang, C. Shao, J. Huang, J. Gan, X. Huang, E. Bucci, M. Piacentini, G. Ippolito, G. Melino, COVID-19 infection: the perspectives on immune responses, Cell Death Differ (2020). [CrossRef]
- Z. Wu, J.M. McGoogan, Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention, JAMA 323(13) (2020) 1239-1242. [CrossRef]
- Q. Zhang, Z. Wang, Y. Lv, J. Zhao, Q. Dang, D. Xu, D. Zhao, H. Liu, Z. Wang, X. Zhao, Z. Xu, X. Zhang, Clinical features and prognostic factors of patients with COVID-19 in Henan Province, China, Hum Cell 34(2) (2021) 419-435. [CrossRef]
- L. Casalino, Z. Gaieb, J.A. Goldsmith, C.K. Hjorth, A.C. Dommer, A.M. Harbison, C.A. Fogarty, E.P. Barros, B.C. Taylor, J.S. McLellan, E. Fadda, R.E. Amaro, Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein, ACS Cent Sci 6(10) (2020) 1722-1734. [CrossRef]
- L.K. Gadanec, K.R. McSweeney, T. Qaradakhi, B. Ali, A. Zulli, V. Apostolopoulos, Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells?, Int J Mol Sci 22(3) (2021). [CrossRef]
- A.C. Walls, Y.J. Park, M.A. Tortorici, A. Wall, A.T. McGuire, D. Veesler, Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell 183(6) (2020) 1735.
- W.J. Guan, Z.Y. Ni, Y. Hu, W.H. Liang, C.Q. Ou, J.X. He, L. Liu, H. Shan, C.L. Lei, D.S.C. Hui, B. Du, L.J. Li, G. Zeng, K.Y. Yuen, R.C. Chen, C.L. Tang, T. Wang, P.Y. Chen, J. Xiang, S.Y. Li, J.L. Wang, Z.J. Liang, Y.X. Peng, L. Wei, Y. Liu, Y.H. Hu, P. Peng, J.M. Wang, J.Y. Liu, Z. Chen, G. Li, Z.J. Zheng, S.Q. Qiu, J. Luo, C.J. Ye, S.Y. Zhu, N.S. Zhong, C. China Medical Treatment Expert Group for, Clinical Characteristics of Coronavirus Disease 2019 in China, N Engl J Med (2020). [CrossRef]
- S.F. Pedersen, Y.C. Ho, SARS-CoV-2: a storm is raging, J Clin Invest (2020).
- B. Vellingiri, K. Jayaramayya, M. Iyer, A. Narayanasamy, V. Govindasamy, B. Giridharan, S. Ganesan, A. Venugopal, D. Venkatesan, H. Ganesan, K. Rajagopalan, P. Rahman, S.G. Cho, N.S. Kumar, M.D. Subramaniam, COVID-19: A promising cure for the global panic, Sci Total Environ 725 (2020) 138277. [CrossRef]
- Z. Zhou, L. Ren, L. Zhang, J. Zhong, Y. Xiao, Z. Jia, L. Guo, J. Yang, C. Wang, S. Jiang, D. Yang, G. Zhang, H. Li, F. Chen, Y. Xu, M. Chen, Z. Gao, J. Yang, J. Dong, B. Liu, X. Zhang, W. Wang, K. He, Q. Jin, M. Li, J. Wang, Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients, Cell Host Microbe 27(6) (2020) 883-890 e2. [CrossRef]
- M. Merad, J.C. Martin, Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages, Nat Rev Immunol 20(6) (2020) 355-362. [CrossRef]
- A.J. Affandi, K. Olesek, J. Grabowska, M.K. Nijen Twilhaar, E. Rodriguez, A. Saris, E.S. Zwart, E.J. Nossent, H. Kalay, M. de Kok, G. Kazemier, J. Stockl, A.J.M. van den Eertwegh, T.D. de Gruijl, J.J. Garcia-Vallejo, G. Storm, Y. van Kooyk, J.M.M. den Haan, CD169 Defines Activated CD14(+) Monocytes With Enhanced CD8(+) T Cell Activation Capacity, Front Immunol 12 (2021) 697840. [CrossRef]
- C. Lucas, P. Wong, J. Klein, T.B.R. Castro, J. Silva, M. Sundaram, M.K. Ellingson, T. Mao, J.E. Oh, B. Israelow, T. Takahashi, M. Tokuyama, P. Lu, A. Venkataraman, A. Park, S. Mohanty, H. Wang, A.L. Wyllie, C.B.F. Vogels, R. Earnest, S. Lapidus, I.M. Ott, A.J. Moore, M.C. Muenker, J.B. Fournier, M. Campbell, C.D. Odio, A. Casanovas-Massana, I.T. Yale, R. Herbst, A.C. Shaw, R. Medzhitov, W.L. Schulz, N.D. Grubaugh, C. Dela Cruz, S. Farhadian, A.I. Ko, S.B. Omer, A. Iwasaki, Longitudinal analyses reveal immunological misfiring in severe COVID-19, Nature 584(7821) (2020) 463-469. [CrossRef]
- H. Chu, J.F. Chan, Y. Wang, T.T. Yuen, Y. Chai, Y. Hou, H. Shuai, D. Yang, B. Hu, X. Huang, X. Zhang, J.P. Cai, J. Zhou, S. Yuan, K.H. Kok, K.K. To, I.H. Chan, A.J. Zhang, K.Y. Sit, W.K. Au, K.Y. Yuen, Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19, Clin Infect Dis (2020). [CrossRef]
- Z. Parackova, I. Zentsova, M. Bloomfield, P. Vrabcova, J. Smetanova, A. Klocperk, G. Meseznikov, L.F. Casas Mendez, T. Vymazal, A. Sediva, Disharmonic Inflammatory Signatures in COVID-19: Augmented Neutrophils' but Impaired Monocytes' and Dendritic Cells' Responsiveness, Cells 9(10) (2020). [CrossRef]
- D. Ragab, H. Salah Eldin, M. Taeimah, R. Khattab, R. Salem, The COVID-19 Cytokine Storm; What We Know So Far, Front Immunol 11 (2020) 1446. [CrossRef]
- G. Chen, D. Wu, W. Guo, Y. Cao, D. Huang, H. Wang, T. Wang, X. Zhang, H. Chen, H. Yu, X. Zhang, M. Zhang, S. Wu, J. Song, T. Chen, M. Han, S. Li, X. Luo, J. Zhao, Q. Ning, Clinical and immunological features of severe and moderate coronavirus disease 2019, J Clin Invest 130(5) (2020) 2620-2629. [CrossRef]
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang, B. Cao, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet 395(10223) (2020) 497-506. [CrossRef]
- J. Schulte-Schrepping, N. Reusch, D. Paclik, K. Bassler, S. Schlickeiser, B. Zhang, B. Kramer, T. Krammer, S. Brumhard, L. Bonaguro, E. De Domenico, D. Wendisch, M. Grasshoff, T.S. Kapellos, M. Beckstette, T. Pecht, A. Saglam, O. Dietrich, H.E. Mei, A.R. Schulz, C. Conrad, D. Kunkel, E. Vafadarnejad, C.J. Xu, A. Horne, M. Herbert, A. Drews, C. Thibeault, M. Pfeiffer, S. Hippenstiel, A. Hocke, H. Muller-Redetzky, K.M. Heim, F. Machleidt, A. Uhrig, L. Bosquillon de Jarcy, L. Jurgens, M. Stegemann, C.R. Glosenkamp, H.D. Volk, C. Goffinet, M. Landthaler, E. Wyler, P. Georg, M. Schneider, C. Dang-Heine, N. Neuwinger, K. Kappert, R. Tauber, V. Corman, J. Raabe, K.M. Kaiser, M.T. Vinh, G. Rieke, C. Meisel, T. Ulas, M. Becker, R. Geffers, M. Witzenrath, C. Drosten, N. Suttorp, C. von Kalle, F. Kurth, K. Handler, J.L. Schultze, A.C. Aschenbrenner, Y. Li, J. Nattermann, B. Sawitzki, A.E. Saliba, L.E. Sander, C.-O.I. Deutsche, Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment, Cell 182(6) (2020) 1419-1440 e23. [CrossRef]
- D. Galati, S. Zanotta, L. Capitelli, M. Bocchino, A bird's eye view on the role of dendritic cells in SARS-CoV-2 infection: Perspectives for immune-based vaccines, Allergy 77(1) (2022) 100-110. [CrossRef]
- Vanderheiden, P. Ralfs, T. Chirkova, A.A. Upadhyay, M.G. Zimmerman, S. Bedoya, H. Aoued, G.M. Tharp, K.L. Pellegrini, C. Manfredi, E. Sorscher, B. Mainou, J.L. Lobby, J.E. Kohlmeier, A.C. Lowen, P.Y. Shi, V.D. Menachery, L.J. Anderson, A. Grakoui, S.E. Bosinger, M.S. Suthar, Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures, J Virol 94(19) (2020). [CrossRef]
- M. Chung, A. Bernheim, X. Mei, N. Zhang, M. Huang, X. Zeng, J. Cui, W. Xu, Y. Yang, Z.A. Fayad, A. Jacobi, K. Li, S. Li, H. Shan, CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV), Radiology 295(1) (2020) 202-207. [CrossRef]
- P. Bourgoin, G. Biechele, I. Ait Belkacem, P.E. Morange, F. Malergue, Role of the interferons in CD64 and CD169 expressions in whole blood: Relevance in the balance between viral- or bacterial-oriented immune responses, Immun Inflamm Dis 8(1) (2020) 106-123. [CrossRef]
- H. Rempel, C. Calosing, B. Sun, L. Pulliam, Sialoadhesin expressed on IFN-induced monocytes binds HIV-1 and enhances infectivity, PLoS One 3(4) (2008) e1967. [CrossRef]
- Y.S. Xiong, Y. Cheng, Q.S. Lin, A.L. Wu, J. Yu, C. Li, Y. Sun, R.Q. Zhong, L.J. Wu, Increased expression of Siglec-1 on peripheral blood monocytes and its role in mononuclear cell reactivity to autoantigen in rheumatoid arthritis, Rheumatology (Oxford) 53(2) (2014) 250-9. [CrossRef]
- A.J. Wilk, A. Rustagi, N.Q. Zhao, J. Roque, G.J. Martinez-Colon, J.L. McKechnie, G.T. Ivison, T. Ranganath, R. Vergara, T. Hollis, L.J. Simpson, P. Grant, A. Subramanian, A.J. Rogers, C.A. Blish, A single-cell atlas of the peripheral immune response in patients with severe COVID-19, Nat Med 26(7) (2020) 1070-1076. [CrossRef]
- W. Wen, W. Su, H. Tang, W. Le, X. Zhang, Y. Zheng, X. Liu, L. Xie, J. Li, J. Ye, L. Dong, X. Cui, Y. Miao, D. Wang, J. Dong, C. Xiao, W. Chen, H. Wang, Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing, Cell Discov 6(1) (2020) 31.
- M. Ortillon, R. Coudereau, M. Cour, T. Rimmele, M. Godignon, M. Gossez, H. Yonis, L. Argaud, A.C. Lukaszewicz, F. Venet, G. Monneret, Monocyte CD169 expression in COVID-19 patients upon intensive care unit admission, Cytometry A 99(5) (2021) 466-471. [CrossRef]
- A.G. Laing, A. Lorenc, I. Del Molino Del Barrio, A. Das, M. Fish, L. Monin, M. Munoz-Ruiz, D.R. McKenzie, T.S. Hayday, I. Francos-Quijorna, S. Kamdar, M. Joseph, D. Davies, R. Davis, A. Jennings, I. Zlatareva, P. Vantourout, Y. Wu, V. Sofra, F. Cano, M. Greco, E. Theodoridis, J.D. Freedman, S. Gee, J.N.E. Chan, S. Ryan, E. Bugallo-Blanco, P. Peterson, K. Kisand, L. Haljasmagi, L. Chadli, P. Moingeon, L. Martinez, B. Merrick, K. Bisnauthsing, K. Brooks, M.A.A. Ibrahim, J. Mason, F. Lopez Gomez, K. Babalola, S. Abdul-Jawad, J. Cason, C. Mant, J. Seow, C. Graham, K.J. Doores, F. Di Rosa, J. Edgeworth, M. Shankar-Hari, A.C. Hayday, A dynamic COVID-19 immune signature includes associations with poor prognosis, Nat Med 26(10) (2020) 1623-1635. [CrossRef]
- R. Zhou, K.K. To, Y.C. Wong, L. Liu, B. Zhou, X. Li, H. Huang, Y. Mo, T.Y. Luk, T.T. Lau, P. Yeung, W.M. Chan, A.K. Wu, K.C. Lung, O.T. Tsang, W.S. Leung, I.F. Hung, K.Y. Yuen, Z. Chen, Acute SARS-CoV-2 Infection Impairs Dendritic Cell and T Cell Responses, Immunity 53(4) (2020) 864-877 e5.
- Q. Chen, B. Yu, Y. Yang, J. Huang, Y. Liang, J. Zhou, L. Li, X. Peng, B. Cheng, Y. Lin, Immunological and inflammatory profiles during acute and convalescent phases of severe/ critically ill COVID-19 patients, Int Immunopharmacol 97 (2021) 107685. [CrossRef]
- J. Hadjadj, N. Yatim, L. Barnabei, A. Corneau, J. Boussier, N. Smith, H. Pere, B. Charbit, V. Bondet, C. Chenevier-Gobeaux, P. Breillat, N. Carlier, R. Gauzit, C. Morbieu, F. Pene, N. Marin, N. Roche, T.A. Szwebel, S.H. Merkling, J.M. Treluyer, D. Veyer, L. Mouthon, C. Blanc, P.L. Tharaux, F. Rozenberg, A. Fischer, D. Duffy, F. Rieux-Laucat, S. Kerneis, B. Terrier, Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients, Science 369(6504) (2020) 718-724. [CrossRef]
- S. Matic, S. Popovic, P. Djurdjevic, D. Todorovic, N. Djordjevic, Z. Mijailovic, P. Sazdanovic, D. Milovanovic, D. Ruzic Zecevic, M. Petrovic, M. Sazdanovic, N. Zornic, V. Vukicevic, I. Petrovic, S. Matic, M. Karic Vukicevic, D. Baskic, SARS-CoV-2 infection induces mixed M1/M2 phenotype in circulating monocytes and alterations in both dendritic cell and monocyte subsets, PLoS One 15(12) (2020) e0241097. [CrossRef]
- M.A. Zingaropoli, P. Nijhawan, A. Carraro, P. Pasculli, P. Zuccala, V. Perri, R. Marocco, B. Kertusha, G. Siccardi, C. Del Borgo, A. Curtolo, C. Ajassa, M. Iannetta, M.R. Ciardi, C.M. Mastroianni, M. Lichtner, Increased sCD163 and sCD14 Plasmatic Levels and Depletion of Peripheral Blood Pro-Inflammatory Monocytes, Myeloid and Plasmacytoid Dendritic Cells in Patients With Severe COVID-19 Pneumonia, Front Immunol 12 (2021) 627548. [CrossRef]
- E. Mantlo, N. Bukreyeva, J. Maruyama, S. Paessler, C. Huang, Antiviral activities of type I interferons to SARS-CoV-2 infection, Antiviral Res 179 (2020) 104811. [CrossRef]
- E. Prompetchara, C. Ketloy, T. Palaga, Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic, Asian Pac J Allergy Immunol 38(1) (2020) 1-9. [CrossRef]
- D. Oosterhoff, S. Lougheed, R. van de Ven, J. Lindenberg, H. van Cruijsen, L. Hiddingh, J. Kroon, A.J. van den Eertwegh, B. Hangalapura, R.J. Scheper, T.D. de Gruijl, Tumor-mediated inhibition of human dendritic cell differentiation and function is consistently counteracted by combined p38 MAPK and STAT3 inhibition, Oncoimmunology 1(5) (2012) 649-658. [CrossRef]
- H. Yu, D. Pardoll, R. Jove, STATs in cancer inflammation and immunity: a leading role for STAT3, Nat Rev Cancer 9(11) (2009) 798-809. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).