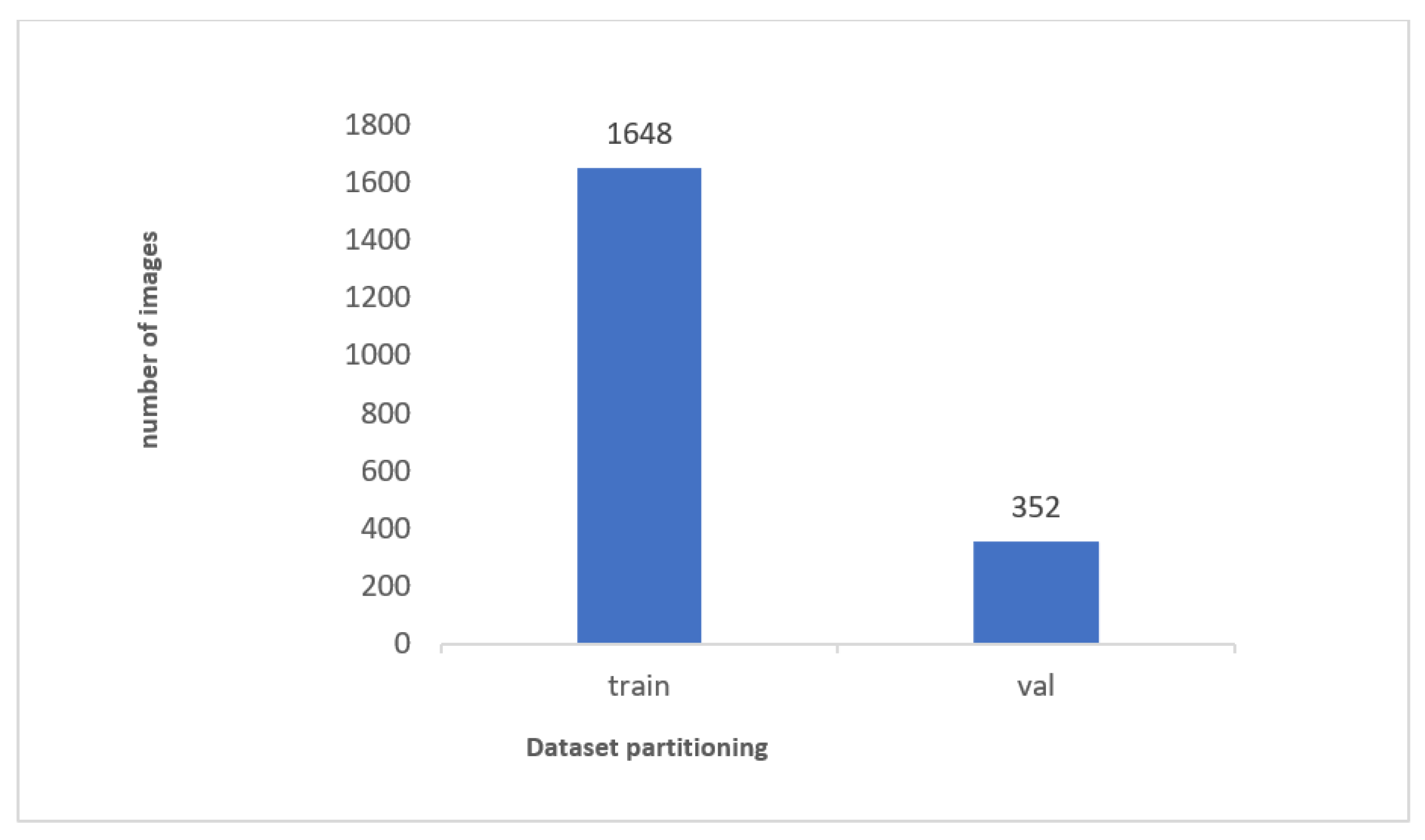
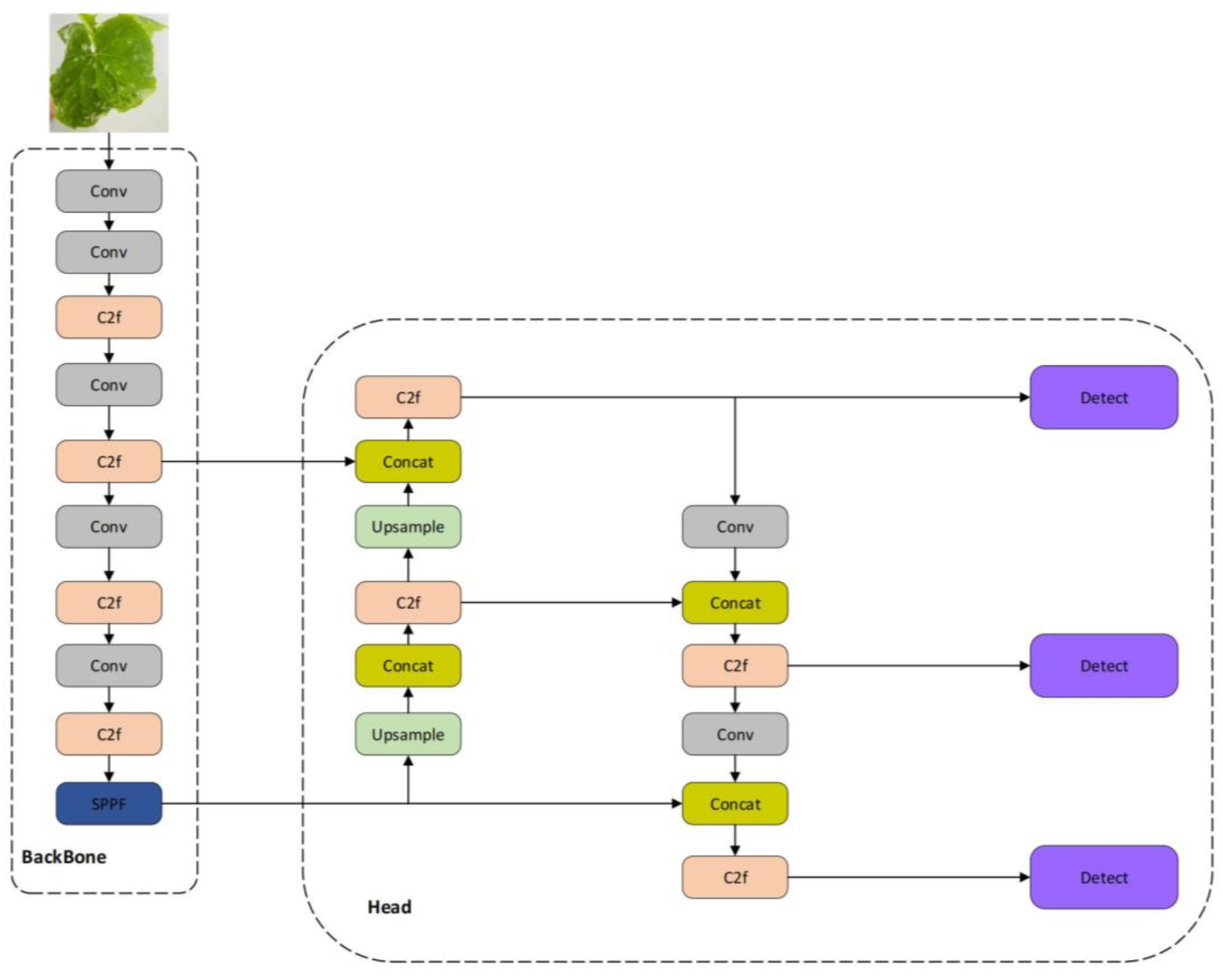
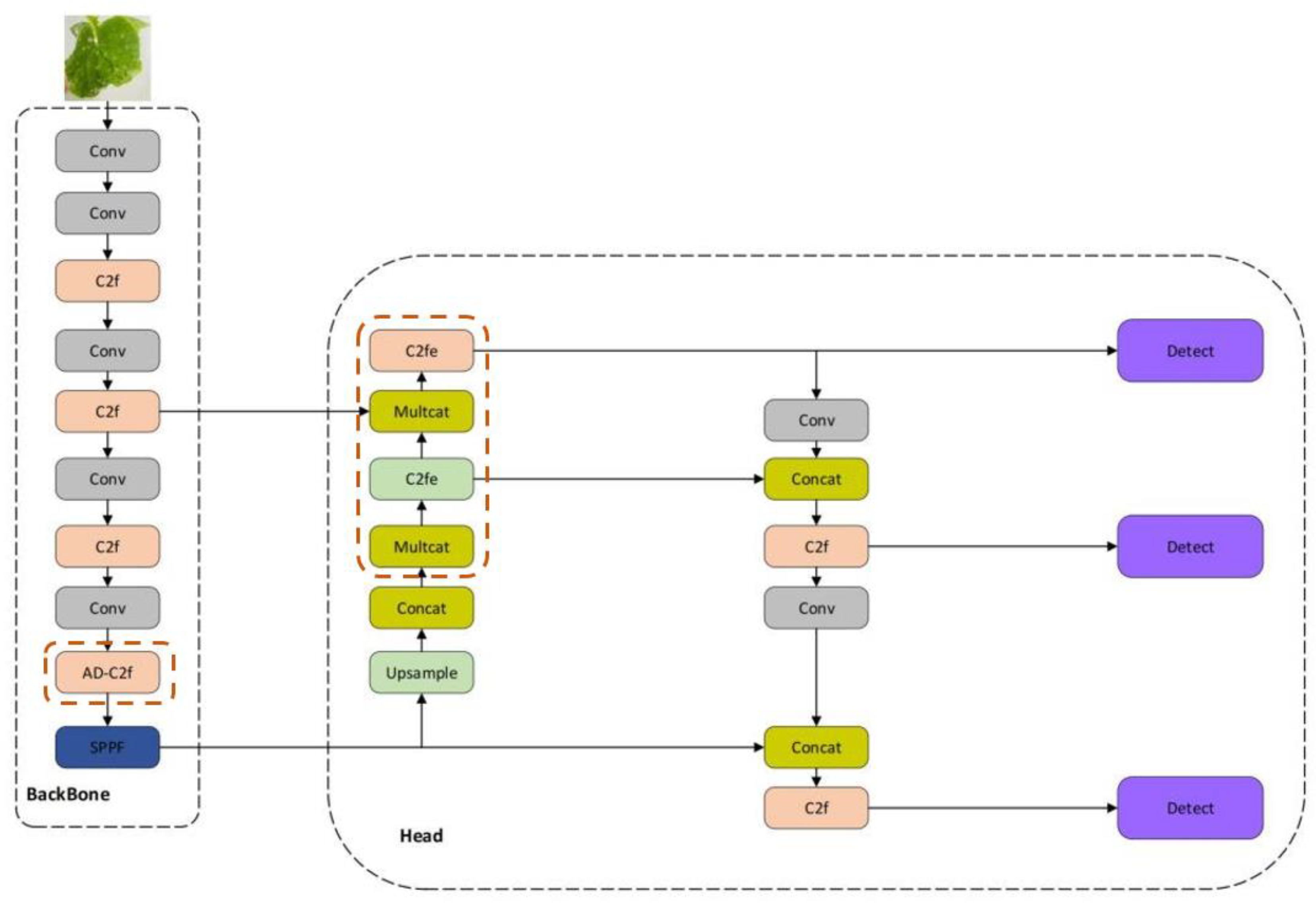
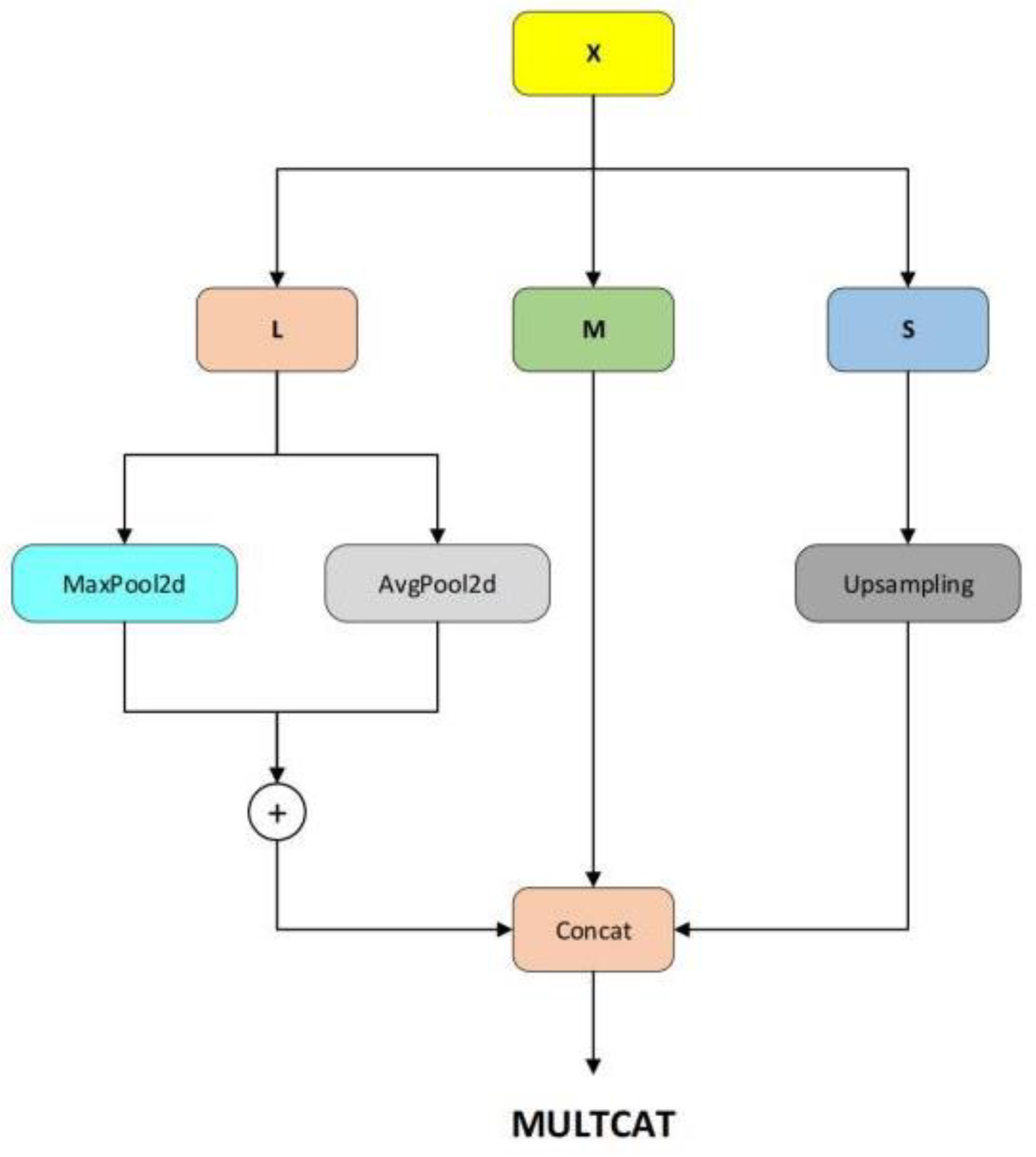
1. Introduction
In today's globalized world, ensuring food safety and high yields has become a central issue in agricultural research. Cucumber is a member of the Cucurbitaceae family, which has 90 genera and 750 species. It is one of the oldest cultivated vegetable crops grown in almost all temperate countries. It is a warm-loving, frost-tolerant plant that grows best at temperatures above 20°C [
1]. Cucumber pests and diseases are one of the main reasons for the decrease in cucumber yield [
2]. Outbreaks of agricultural pests not only affect crop production but also the use of pesticides, affecting crop production and increasing both ecological destruction and food safety risks [
3]. Therefore, it is particularly important to develop a method that can detect cucumber pests and diseases in a timely and accurate manner.
Traditional methods of detecting pests and diseases on cucumber leaves mainly rely on human visual recognition, that is, directly observing the morphology, texture, and color of leaves with the naked eye [
4]. Although this method is simple to operate, its accuracy is affected by the differences in the observer's experience and knowledge, and it is highly subjective, often leading to misdiagnosis and causing irreversible losses to farmers. Therefore, in addition to being time-consuming and costly, it is difficult to achieve the precise detection requirements for cucumber leaf pests. This is not easy to implement in large-scale agricultural production.
With the rapid development of technology, especially in the field of computer vision, deep learning provides a new direction for the identification and detection of pests and diseases in the agricultural field [
5]. Against this background, object detection has become a core topic in computer vision research, aiming to accurately identify and locate specific objects in images. Scholars have developed many innovative strategies to achieve this goal. Among them, the one-stage (One-stage) and two-stage (Two-stage) methods stand out and have become the two mainstream technologies in this field.
Representatives of one-stage object detection algorithms include SSD (Liu et al., 2016) [
6], Retina Net (Lin et al., 2017) [
7], YOLOv4 (Bochkovskiy et al., 2020) [
8], YOLOv5 (Jocher et al., 2021) [
9], DETR (Carion et al., 2020) [
10], FCOS (Tian et al., 2019) [
11], and YOLOX (Ge et al., 2021) [
12]. In contrast, two-stage object detection algorithms such as R-CNN (Girshick et al., 2014) [
13], Fast R-CNN (Girshick, 2015) [
14], Faster-RCNN (Ren et al., 2016) [
15], Mask R-CNN (He et al., 2017) [
16], and Cascade R-CNN (Cai and Vasconcelos, 2018) [
17] have a longer computational process.
Two-stage models first generate a series of candidate regions and then use a classifier to refine the classification of these regions. Although they are usually superior in accuracy, the two-step process makes them relatively slow [
18,
19]. On the other hand, one-stage models directly predict bounding boxes and categories from feature maps without generating candidate areas, providing the advantage of real-time detection. However, this speed sometimes comes at the expense of accuracy. Considering the need to make the model more practical and suitable for deployment on mobile devices, we chose the YOLO model as our object detection algorithm.
To solve the problem of low accuracy in small target detection of one-stage models, this paper proposes a new YOLOv8s model for cucumber pest identification. This study tests the new model on a specially constructed dataset. This study plans to adopt and improve the latest YOLOv8s model for the detection of cucumber pests and diseases. To compensate for the accuracy loss of lightweight models, we also adopted an attention mechanism to assign different weights to each part of the input feature layer, thereby more effectively extracting key features and improving classification performance.
The main contributions of this paper are summarized fellow:
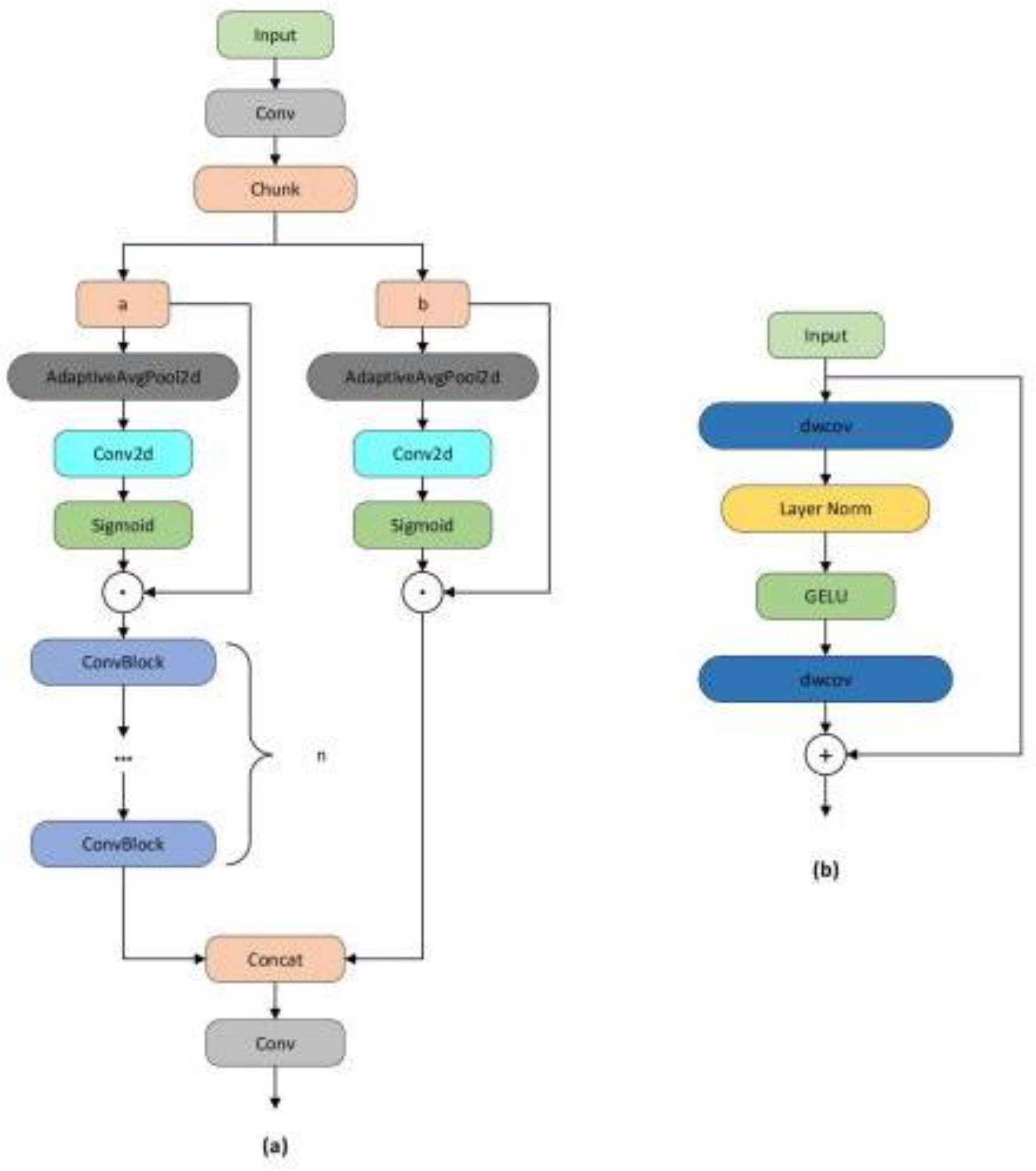
We propose a lightweight one-stage YOLOv8 model, referred to as the Detail and Multi-scale YOLO Network (DM-YOLO), built upon YOLOv8 for real-time cucumber pest and disease identification. Utilizing the MultiCat module by merging features of different scales, the model's detection capability for pests and diseases of varying sizes on cucumbers is enhanced. We introduced the C2fe module, a modification based on C2f, as a new feature fusion method aiming to more effectively combine multi-scale features. With an attention mechanism based on adaptive average pooling, we constructed a new module named AD-C2f, which intensifies the model's focus on crucial features, thereby increasing detection accuracy and overall model performance.
We extracted a portion of data from the ai-hub's " Integrated Plant Disease Induction Data " public dataset to construct a new cucumber pest and disease dataset. To ensure data quality, we manually re-annotated the leaves in each image and carefully filtered the original images. After eliminating some unorganized data, we concentrated on two primary cucumber afflictions: downy mildew and powdery mildew, culminating in this optimized new dataset.
2. Related Works
2.1. Traditional Machine Learning Methods
Traditional disease identification methods in machine learning mainly rely on extracting manually designed features from images, such as color, texture, and shape. They then utilize machine learning classifiers, such as Support Vector Machines (SVM), Decision Trees, or K-Nearest Neighbors (KNN), to differentiate between healthy and damaged plants.
Ebrahimi et al. (2017) proposed an insect detection system based on SVM, which was successfully applied to the crop canopy images in strawberry greenhouses, achieving an error rate of less than 2.25% [
20].
Mondal et al. (2017) developed a disease recognition system that combined image processing and soft computing techniques. By selecting specific morphological features, they achieved a high-accuracy classification of diseases in okra and bitter gourd leaves [
21].
Xu et al. (2020) employed BP neural networks and random forest models to detect damages caused by forest caterpillars. The random forest model performed better, emphasizing the importance of balanced sample data [
22].
Amirruddin et al. (2020) evaluated the chlorophyll sufficiency levels of mature oil palms using hyperspectral remote sensing technology and classification methods. They achieved high-accuracy chlorophyll classification through a random forest classifier, especially on younger leaves [
23].
Traditional machine learning methods might fail to capture complex and advanced patterns in high-dimensional and large-scale data, leading to subpar model performance and accuracy. Moreover, these methods often require extensive preprocessing and feature engineering, adding to the development time and cost. Compared to the adaptive learning capability of deep learning, traditional methods might perform poorly when facing data changes and uncertainties. In summary, although traditional machine learning methods may work well in certain situations, they may not be suitable for tasks requiring capturing complex patterns and relationships in high-dimensional and large-scale data.
2.1. Deep Learning Methods
Deep learning has shown significant advantages in the identification and classification of crop pests and diseases. Unlike traditional machine learning methods, deep learning can automatically learn and extract features from data, alleviating the burden of manual feature design and handling more complex and high-dimensional data. Some advanced deep learning models, like variants of Convolutional Neural Networks and pre-trained models, have been successfully applied to various crop pest and disease identification tasks.
Sethy et al. (2020) successfully identified four rice leaf diseases by combining deep convolutional neural networks and SVM, demonstrating that integrating deep features with SVM can achieve excellent classification, achieving an F1 score of 0.9838 [
24].
Yin et al. (2022) successfully developed a grape leaf disease identification method using an improved MobileNetV3 model and deep transfer learning. This method achieved recognition accuracy of up to 99.84% with limited computational resources and dataset size, and the model size was only 30 MB [
25].
Sankareshwaran et al. (2023) proposed a new method named Cross Enhanced Artificial Hummingbird Algorithm based on AX-RetinaNet (CAHA-AXRNet) for optimizing rice plant disease detection. This method proved more effective than other existing rice plant disease detection methods, achieving an accuracy of 98.1% [
26].
Liu Shiyi et al. (2023) introduced a deep learning algorithm called DCNSE-YOLOv7 based on an improved YOLOv7 algorithm. This significantly enhanced the accuracy of the detection of cucumber leaf pests and diseases, especially in detecting minute features on early-stage diseased leaves. This algorithm showed significant improvement over several mainstream object detection models, providing effective technical support for the precise detection of cucumber leaf pests and diseases [
27].
Yang et al. (2023) introduced a tomato automatic detection method based on an improved YOLOv8s model. This method achieved an mAP of 93.4%, meeting the requirements of real-time detection and providing technical support for tomato-picking robots to ensure fast and accurate operations [
28].
These studies indicate that through continuous optimization and improvement, deep learning models can achieve remarkable results in crop pest and disease detection and identification.
This section may be divided by subheadings. It should provide a concise and precise description of the experimental results, their interpretation, and the experimental conclusions that can be drawn.
4. Experiments
4.1. Equipment and Parameter Settings
The experimental operating system used in this study is Windows 10, with PyTorch serving as the framework for developing the deep learning model. The
Table 1 is provides specific details regarding the experimental environment.
Training Parameter Settings: The image input size is 640×640, batch size is 20, multi-threading is set to 4, the initial learning rate is 0.01, with a total of 120 training iterations (Epochs). The specific parameter settings are shown in
Table 2.
4.2. Evaluation Metrics
In cucumber pest and disease detection, to measure the ability and performance evaluation of the YoloV8 model in correctly detecting pests and diseases, several key metrics were primarily selected: accuracy, recall, precision, and mAP.
Among them, TP, FP, FN, and TN respectively represent True Positive, False Positive, False Negative, and True Negative. C represents the total number of classes, and AP_i represents the AP value for class i.
4.3. Experimental Results
We compared the improved model with the original YOLOv8 model to evaluate whether our improvements could enhance the performance of the model. To showcase the detection results of the algorithm proposed in this study, we randomly selected images from the test subset for comparison. The specific comparison results are shown in
Table 3, and the visual outcomes of the selected images are illustrated in
Figure 8.
The DM-YoloV8 model demonstrates superior performance over the original YoloV8 model in key metrics crucial for cucumber pest and disease detection. With a higher recall of 0.81 versus 0.76, DM-YoloV8 effectively reduces the likelihood of missing actual pest and disease instances. Its MAP50 score of 0.90 in the "A3" category, compared to YoloV8's 0.89, reflects better accuracy and precision. Despite a slightly lower frame rate, the significant improvements in recall and MAP50, along with consistent performance across various categories, underscore DM-YoloV8's enhanced suitability and reliability for cucumber pest and disease detection tasks, making it a preferable choice over the original YoloV8 model.
During the training process, we are concerned not only with the model's final performance but also with the training and validation process to ensure the model is progressing in the right direction. For this purpose, we decided to plot some critical metrics during the training and validation process so we could have a visual understanding of the model's learning situation, as shown in
Figure 8.
In
Figure 8, our investigation revealed that the enhanced DM-YOLOv8 model exhibits notable performance enhancements in the detection of foliar diseases, specifically Powdery Mildew and Downy Mildew. When benchmarked against the baseline YOLOv8s model, the DM-YOLOv8 variant demonstrated superior accuracy in bounding box delineation and augmented confidence scores, signaling a refined capability for precise pathogen feature recognition.
In particular, the DM-YOLOv8 model consistently yielded elevated confidence scores across a multitude of test instances, denoting a heightened proficiency in differentiating healthy leaf tissue from that afflicted by disease. Despite these advancements, the model occasionally generated detection boxes in healthy tissue zones, indicative of potential false positives. Furthermore, there were instances where prominent disease manifestations were not encapsulated within detection boxes, pointing to possible false negatives.
The performance of the DM-YOLOv8 model also varied when processing images characterized by intricate backgrounds and overlapping leaf structures. This variability suggests that the model's robustness in complex visual environments may require additional refinement. Notably, the model exhibited uncertainty in regions of leaf margin and vein convergence, likely attributed to the feature representation similarities between these areas and diseased segments.
Summarily, while the DM-YOLOv8 model demonstrates a distinct advantage in the domain of leaf disease detection, there is an evident imperative for enhancement in minimizing false positives and fortifying detection consistency in multifaceted scenarios. Consequently, this necessitates the development of further optimization strategies to align the model's capabilities with the pragmatic demands of accurate disease detection.
4.4. Comparative Experiments
In our research, we meticulously designed a series of comparative experiments to assess the performance of multiple advanced object detection models in the context of agricultural pest detection tasks. Our study utilized traditional models including Faster R-CNN, Retina Net, SSD, as well as the newer iterations YOLOv5, YOLOv8, and our DM-YOLOv8, testing their proficiency on a dataset of various leaf images with powdery mildew, which served as a realistic simulation of agricultural conditions. The experimental outcomes, illustrated alongside the Ground Truth, not only highlighted the differences in detection accuracy and recall rates among the models but also afforded us insightful revelations into their processing capabilities.
Figure 9.
Comparative Analysis of Object Detection Models on Leaf Disease Identification.
Figure 9.
Comparative Analysis of Object Detection Models on Leaf Disease Identification.
We observed that Faster R-CNN excelled in detecting a significant number of lesions, indicating its potential for high-confidence detection. However, this model also exhibited a relatively high false positive rate, notably underperforming in the detection of small-scale lesions. This specific limitation could lead to significant errors in sensitive pest detection tasks where precision is of the utmost importance. In contrast, RetinaNet produced fewer detection boxes than Faster R-CNN. However, it was significantly more focused and precise in accurately delineating the actual lesions, albeit with a problem of missed detections. The SSD model, while demonstrating rapid detection capabilities in specific contexts, showed a marked decrease in performance when tasked with detecting more diminutive and more nuanced features. This decrease in performance could become a hindrance in agricultural applications where the accurate identification of subtle lesions is crucial.
The latest iterations of the YOLO series, namely YOLOv5 and YOLOv8, showcased a relatively high number of detection boxes and improved precision. YOLOv8, while reducing false positives, maintained a high number of detection boxes, further enhancing the accuracy of the model. As an improved version, DM-YOLOv8's results closely matched the Ground Truth, showing significant improvements in precision and recall rates for the identification of small-scale targets.
DM-YOLOv8 stood out in the precise localization of multiple targets, offering a viable solution for agricultural applications requiring high-precision real-time detection with limited computational resources. Nevertheless, the experimental results underscore the necessity for further research to optimize the models' overall performance and validate their robustness under a broader range of real-world conditions.
When conducting a comprehensive comparative analysis of these models, data from
Table 4 further corroborated the significant advantage of DM-YOLOv8 in real-time processing capabilities. With a mean Average Precision (mAP) of 88.1%, it surpassed YOLOv5s 87.6%, and its frame rate (FPS) increased significantly to 178.57, much higher than YOLOv5's 153.84, which is particularly crucial for time-sensitive real-time pest monitoring applications. DM-YOLOv8 not only reached new heights in accuracy and speed but also showed significant advantages in model size. Its smaller size, especially compared to the larger model size of Faster R-CNN, makes it an ideal choice for environments with limited computational resources.
Through these comparative experiments, the DM-YOLOv8 model demonstrated tremendous potential for real-time agricultural pest detection applications due to its advantages in accuracy, speed, and robust performance in tasks involving small-scale and overlapping targets. While these preliminary results are encouraging, we recognize the need for further validation on a larger scale dataset and under more complex environmental conditions to ensure the efficacy and reliability of the DM-YOLOv8 model in practical agricultural applications.
4.5. Ablation Experiment and result
To systematically discern the contribution of each component in our enhanced YOLOv8 model, we conducted an ablation study. This study aimed to isolate and assess the individual and combined impacts of the C2fe, AD-C2f, and MULT modules on the model's performance. We evaluated the variants of the model on the same dataset to ensure consistency in comparisons. This was achieved by employing uniform training and validation splits, maintaining consistency across all hyperparameters and training conditions. Such methodological rigor ensured that the observed performance variations were exclusively attributable to architectural changes.
This approach allowed for a nuanced understanding of how each module influences the model's effectiveness, particularly in terms of detection accuracy, processing speed, and the ability to handle diverse image conditions. The systematic analysis provided insights into the synergistic effects of the modules when used in combination, offering valuable information on optimizing the model for specific use cases or computational constraints.
Table 5.
Ablation experiment results.
Table 5.
Ablation experiment results.
| Network |
Precision/% |
Recall/% |
mAP50 |
mAP50-95 |
FPS |
| YOLOV8s |
83.4% |
79.4% |
87.5% |
52.0% |
181.8 |
| DM-YOLOV8 |
84.2% |
80.8% |
88.2% |
53.0% |
178.5 |
| C2fe |
84.2% |
78.7% |
88.1% |
53.0% |
156.3 |
| ADC2F |
82.3% |
79.8% |
87.8% |
51.8% |
192.3 |
| MULTCAT |
83.7% |
79.4% |
88.1% |
52.9% |
172.4 |
| C2fe+ADC2F |
83.2% |
78.2% |
87.4% |
52.6% |
178.6 |
| C2fe+MULTCAT |
83.8% |
80.3% |
88.0% |
53.1% |
172.4 |
| ADC2+MULTCAT |
83.7% |
79.0% |
87.5% |
52.4% |
178.6 |
As a baseline model, YOLOv8s provided a solid foundation with a precision (P) of 83.4% and a recall (R) of 79.4%. With an IoU threshold of 0.5, its mean Average Precision (mAP50) was 87.5%, while mAP50-95 stood at 52%. The model processed images at a speed of 181.8 FPS, establishing a high-performance benchmark for real-time detection.
Upon the integration of our enhanced modules (DM-YOLOv8), enhancements in both precision and recall were observed. Notably, mAP50-95 increased to 53%, indicating more consistent performance across diverse IoU thresholds. This improvement likely resulted from the addition of the MultiCat module, which enhances the model's ability to detect features at varying resolutions through adaptive pooling and interpolation operations on different-scale feature maps. However, this gain in precision was achieved at the expense of reduced processing speed, with FPS decreasing to 178.5.
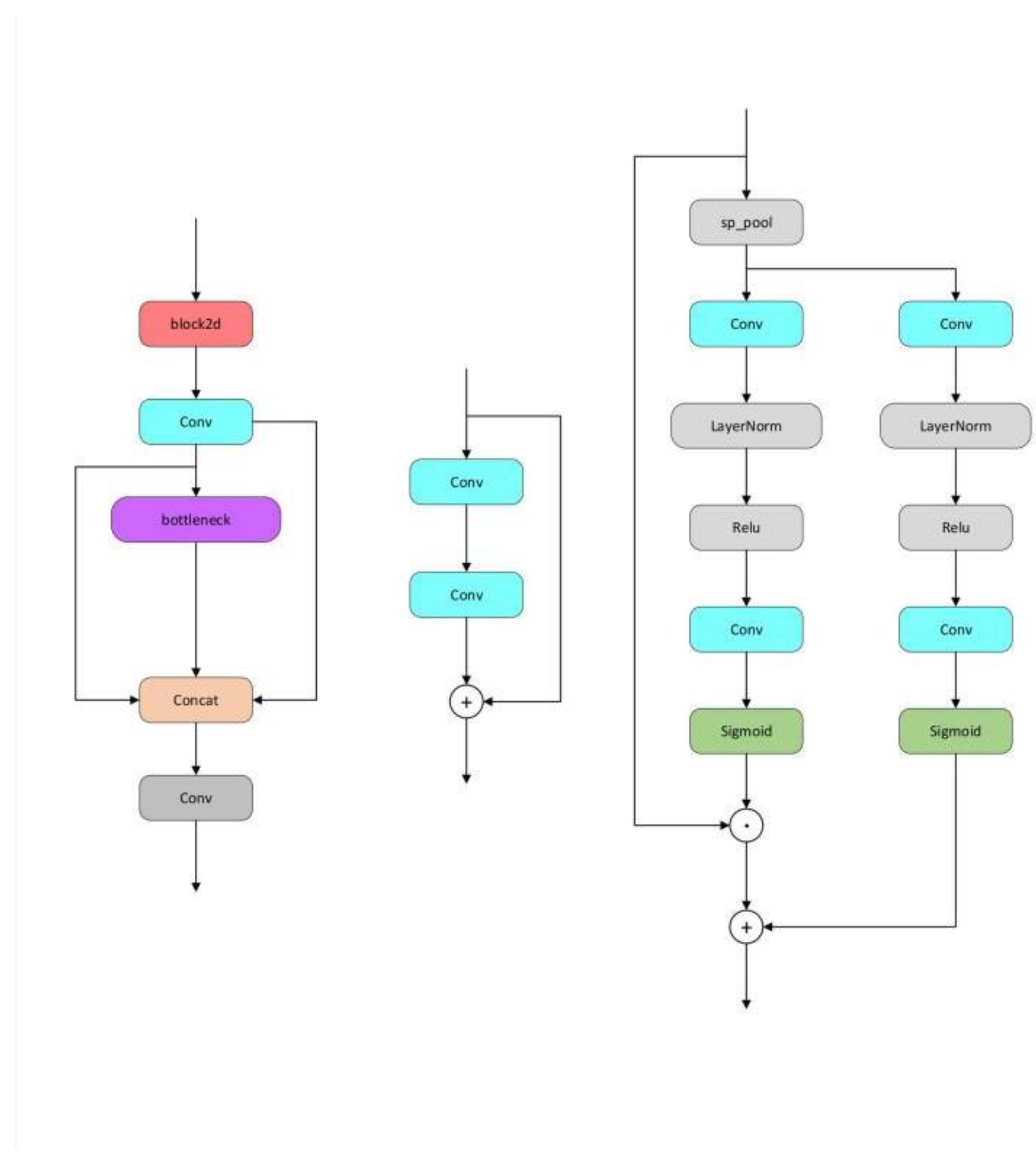
The AD-C2f module, utilizing channel-wise separation, adaptive pooling, and a Sigmoid activation function, established an attention mechanism. This method of processing channels helps emphasize relevant features while suppressing less important ones, which may explain the increase in recall noted during ablation studies. This module's efficiency in computation is also mirrored in its high FPS of 192.3, consistent with the model's performance when the AD-C2f module is employed.
The Block2d module provides spatial attention and channel feature enhancement. Its operation involving channel multiplication and addition after spatial pooling may assist in improving the model's precision. However, this increased complexity might also be the reason for the decrease in FPS when implemented in configurations.
The C2fe module merges Block2d's spatial attention with a series of bottleneck layers for feature processing. It seems to be a more intricate and robust feature extractor, leading to higher scores in mAP50-95 for the enhanced model. However, this added complexity results in a decrease in FPS.
When these modules are combined (e.g., C2fe+AD-C2f, C2fe+MULT, AD-C2f+MULT), the interaction between attention mechanisms and feature processing strategies may either complement or counteract each other, leading to observed shifts in precision, recall, mAP50, mAP50-95, and FPS. Careful balancing of these module combinations is crucial to optimize the model's overall performance.
Our ablation study reveals that the MultiCat and AD-C2f modules are crucial for improving the YOLOv8 model's detection capabilities while maintaining high computational efficiency. Conversely, the Block2d and C2fe modules contribute to increased precision and mAP50-95 but at the expense of higher computational costs. The real-world effectiveness of these modules will hinge on their impact on the precision-speed trade-off, a critical aspect in real-time application deployments.
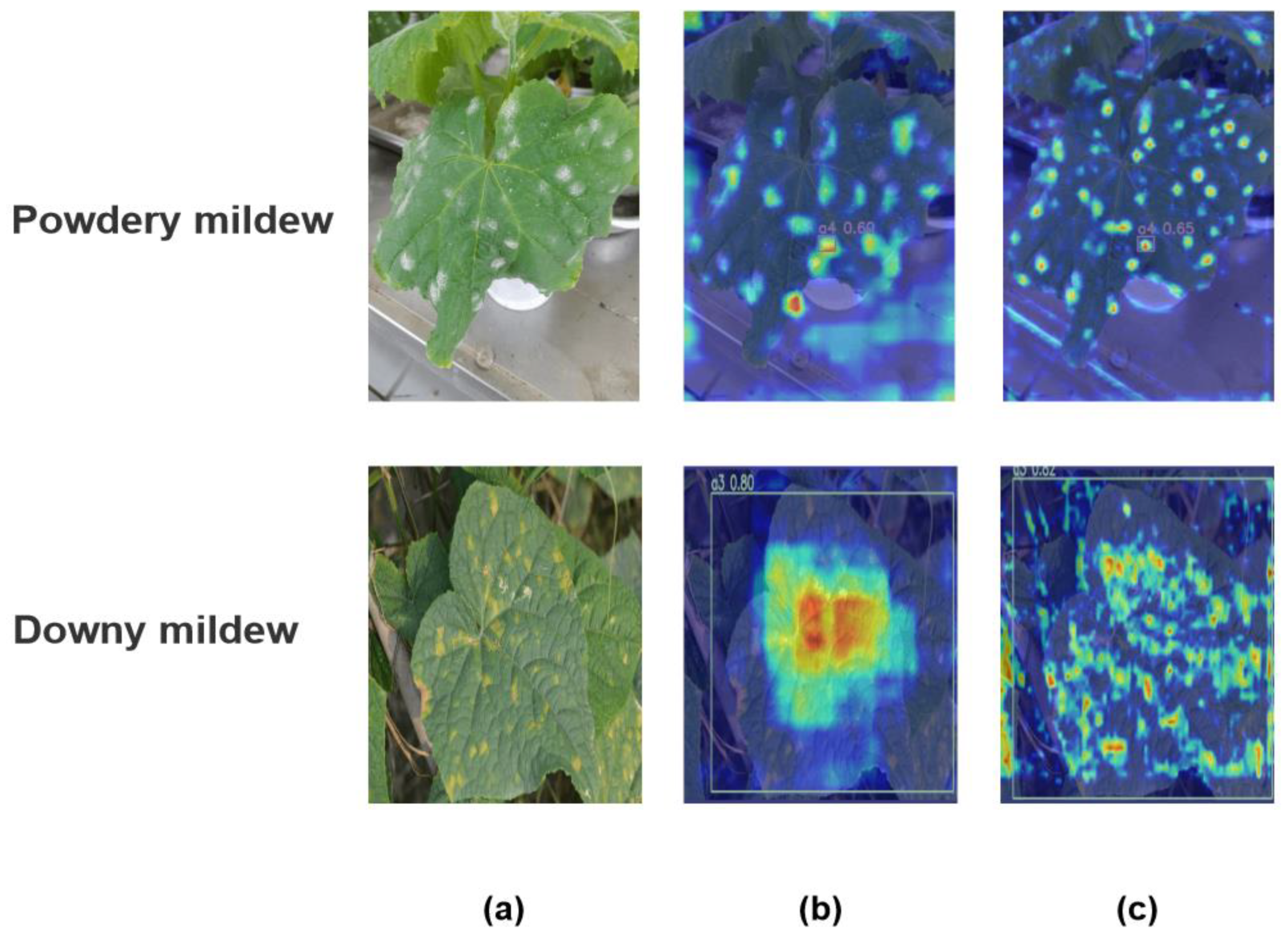
The
Figure 10. presents a visual analysis using Grad-CAM heatmaps to identify two prevalent plant diseases: Powdery Mildew (in the upper images) and Downy Mildew (in the lower images). The first column showcases original leaf images with distinct disease symptoms. Subsequent columns display Grad-CAM heatmaps, where we have specifically selected the C2fe and C2f layers of the network for comparative observation to highlight the network's focal points on features.
In Figure 11. Grad-CAM images, the integration of the C2fe layer exhibits a more concentrated activation pattern compared to the C2f layer, particularly around disease-specific features. This indicates that the C2FE module contributes to refining the convolutional neural network's focus on key attributes associated with each disease, potentially enhancing diagnostic accuracy. The heatmaps provide visual evidence of the network’s ability to localize the most informative features for disease detection, demonstrating the effectiveness of the C2fe layer in honing the model’s discriminative power for disease-specific characteristics.
5. Conclusions
This study delved into the detection of diseases and pests in cucumbers within greenhouse environments, successfully proposing an optimized YOLOv8 algorithm tailored for this purpose. Despite the reduced detection accuracy due to interference from complex backgrounds, the introduction of specific modules significantly enhanced the algorithm's feature extraction and representation capabilities. The integration of the C2fE and AD-C2f modules, in particular, collectively strengthened the network's feature extraction prowess, markedly boosting the model's detection capabilities. Additionally, in cucumber disease and pest detection tasks, this model demonstrated a higher recall rate and mean Average Precision (mAP) with an extremely high processing speed while maintaining a relatively small model size compared to other algorithms. These attributes make it an ideal choice for fast and accurate real-time object detection tasks.
In future research, we plan to enhance feature extraction accuracy and detection robustness by introducing more advanced network structures, increasing inter-layer connections, and utilizing deeper networks. We aim to expand the training dataset and adjust and test the algorithm to cater to different types of crops. Integrating the algorithm with hardware platforms such as drones and automated mobile robots, we intend to develop an automated and intelligent disease monitoring system for on-site testing and to optimize the model's real-time application performance. Through these research and development plans, we anticipate not only scientific progress but also significant technological transformation and industrial advancement in practical applications.