Submitted:
19 March 2024
Posted:
22 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Replication
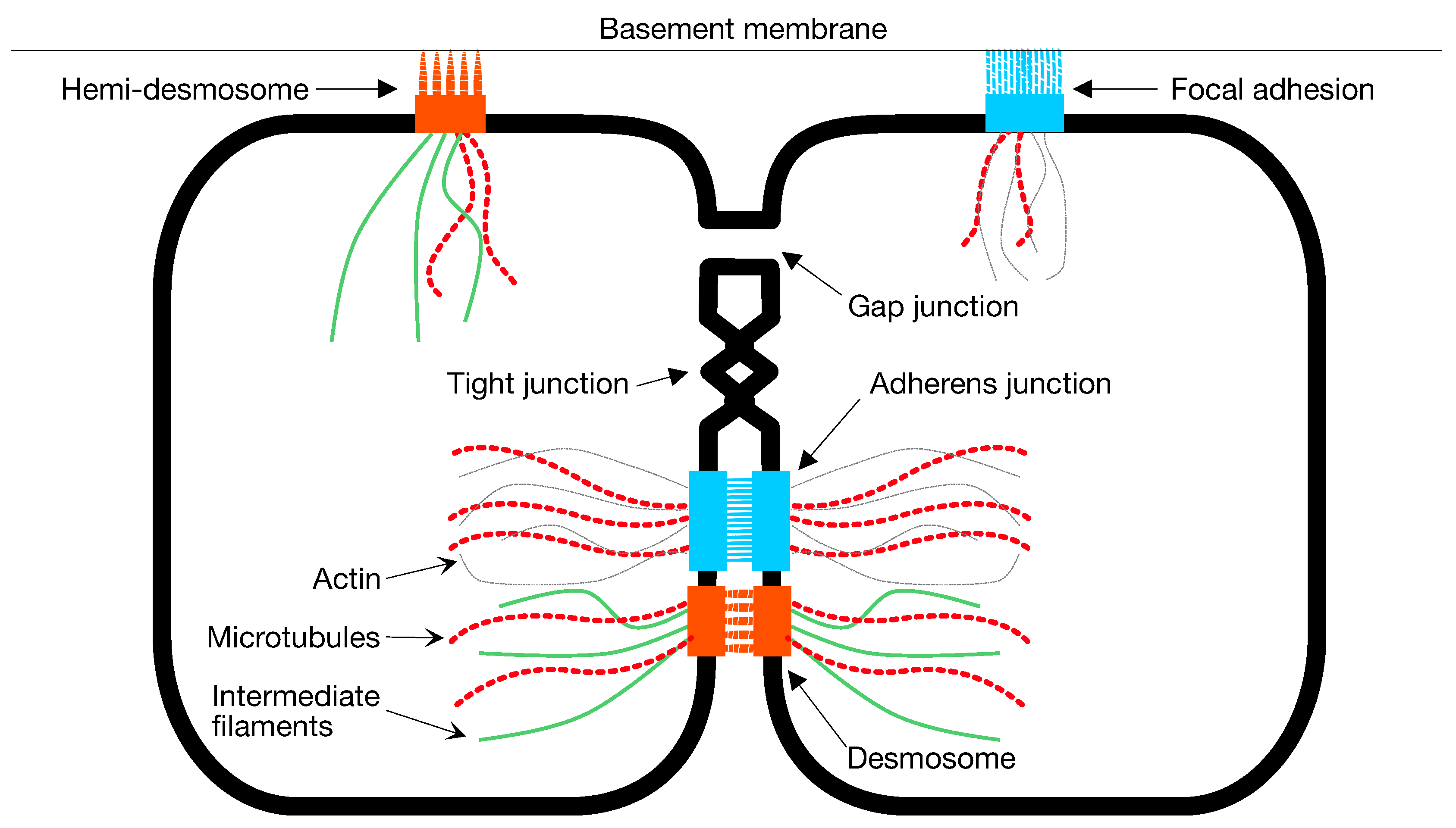
1.2. Adhesion
2. Cellular Adhesion and Replication Patterns that May Produce Fibonacci Population Counts
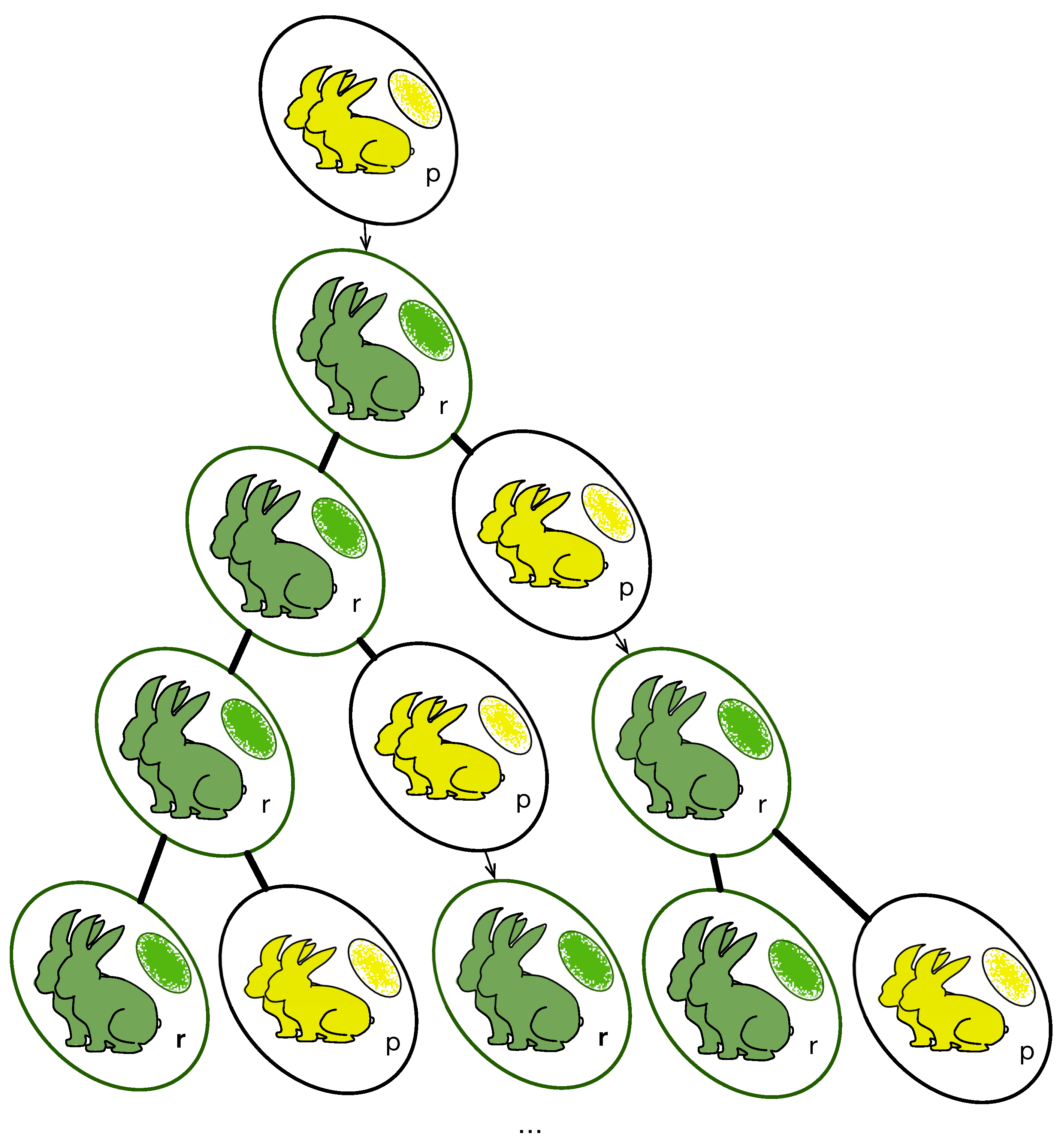
2.1. Differential Mitosis Timing between Progeny
2.1.1. Epigenomic Divergence
2.2. Contact Inhibition
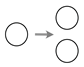
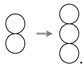
2.3. Cooperative Mitosis and Adhesion
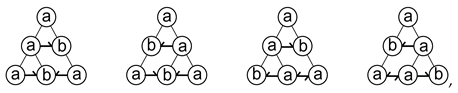
3. Candidate Molecular Mechanisms
4. Simplicial Steps by Replication and Adhesion
- Increase the space dimensions from to .
- This increases the vector length of every point. Let be the first points. Let each be a copy of with a 0 placed in the last position.
- Let be a copy of the point to be replicated, , with a 1 placed in the last position.
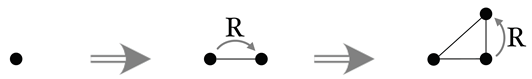
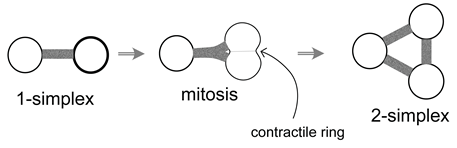
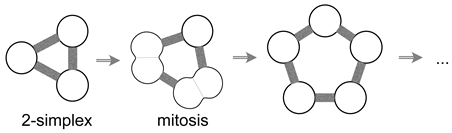
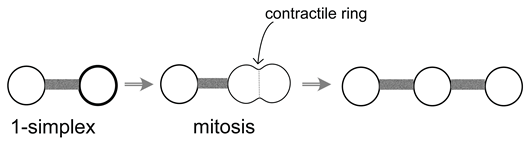
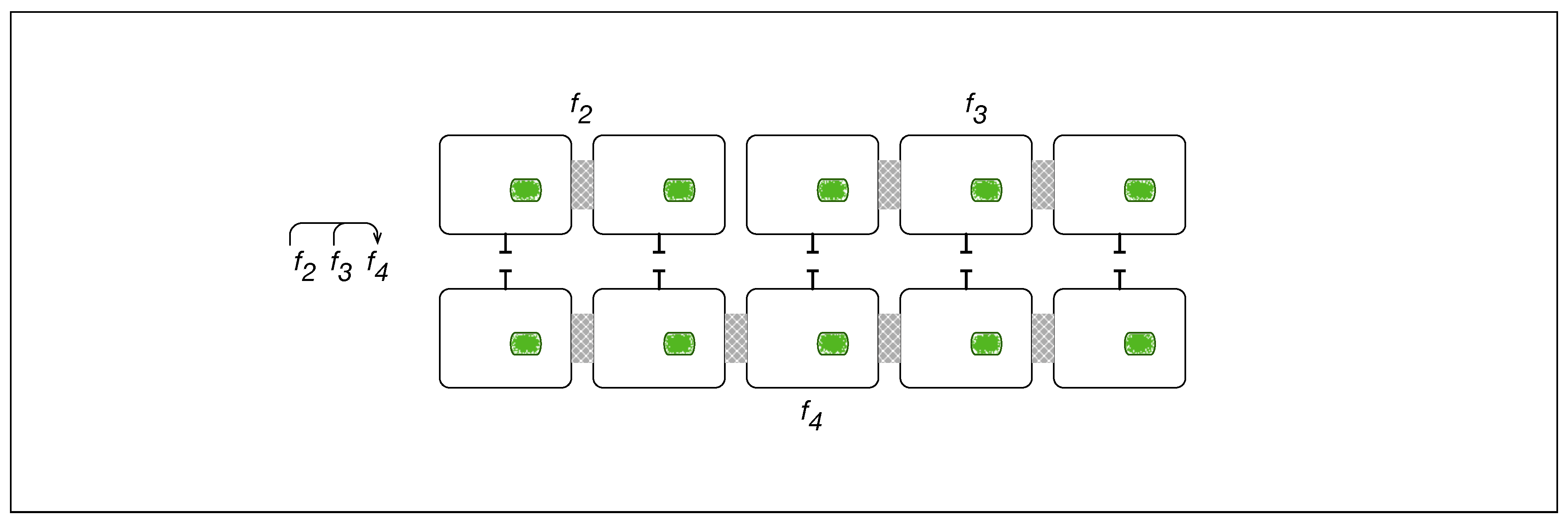
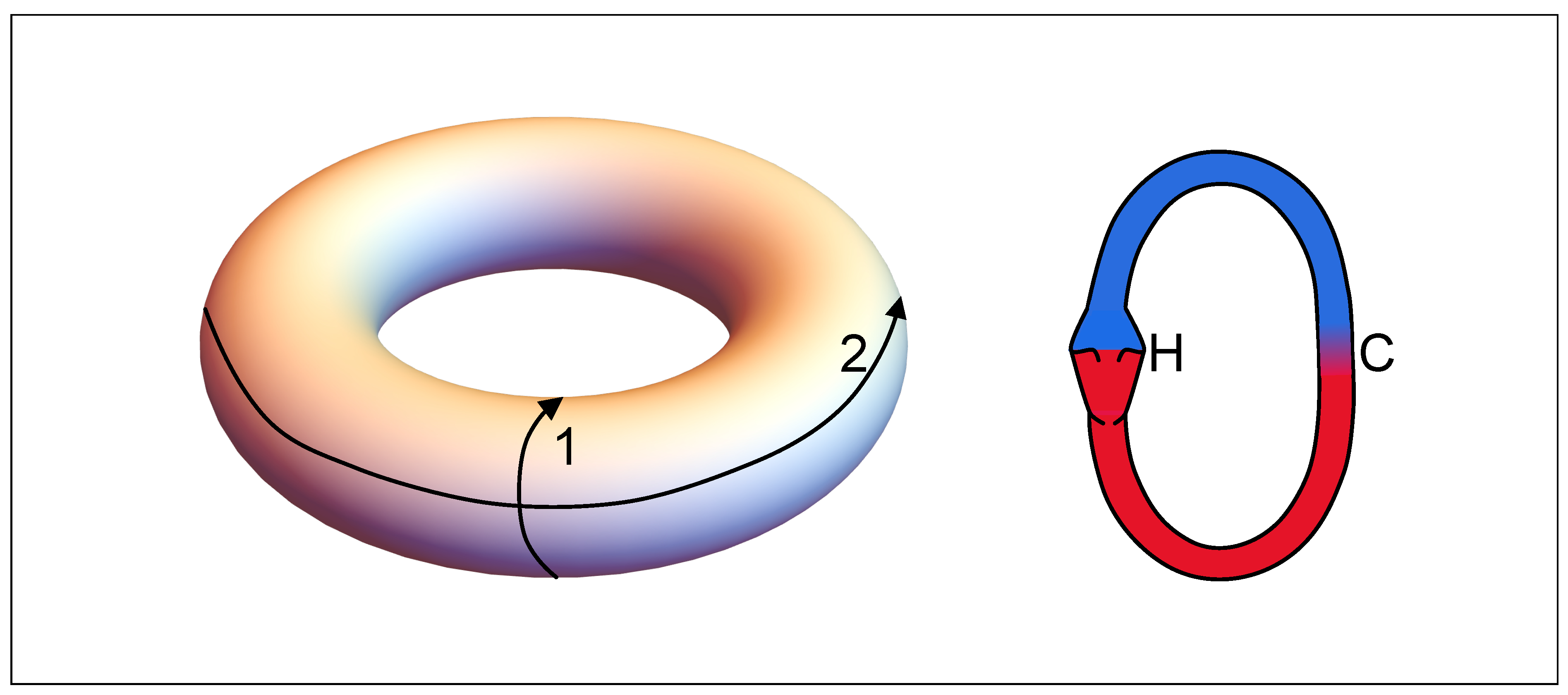
5. A Visual Representation for Algebraic Topology of Replication and Adhesion
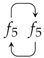
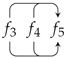
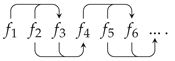
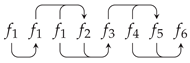
6. Non-Simplicial Topological Transformation
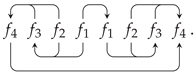
7. Combinatorial Properties of Circular Forms
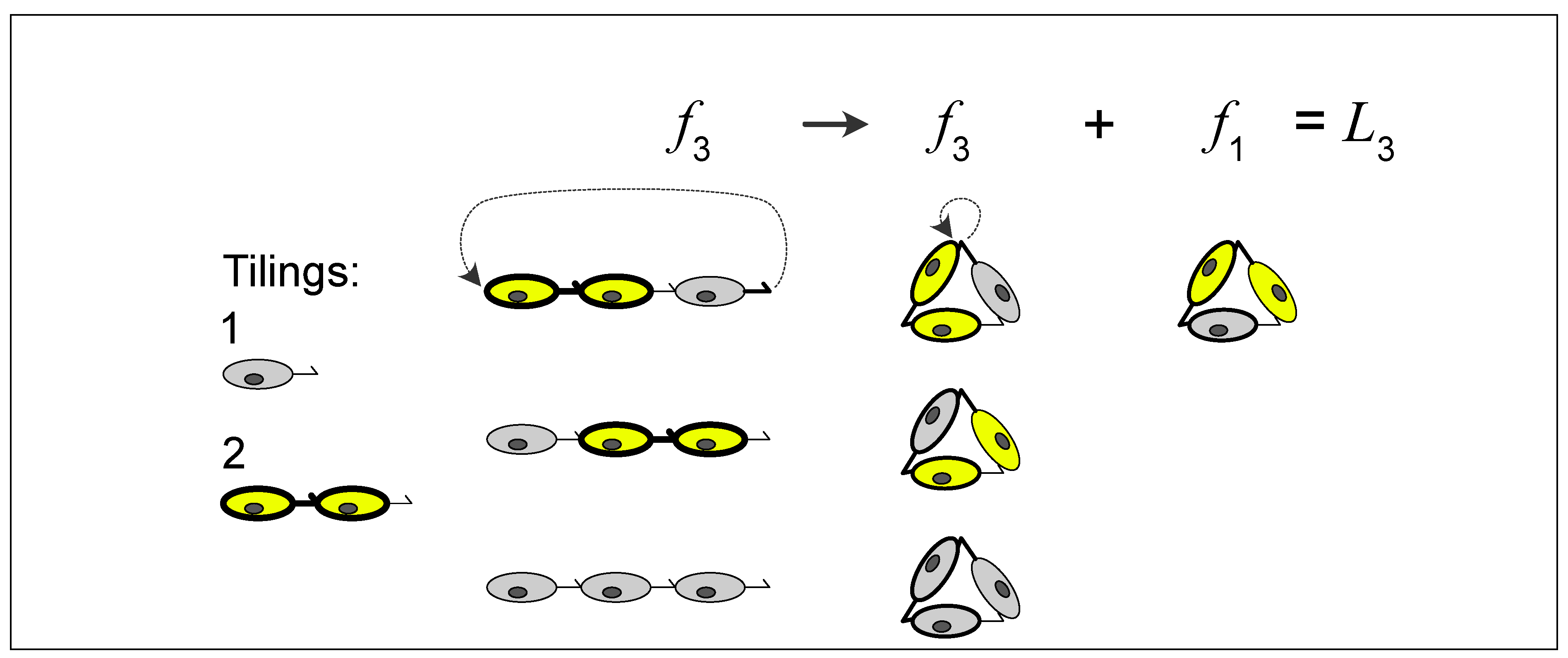
8. Molecular Enablement of Simplicial Topological Steps
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
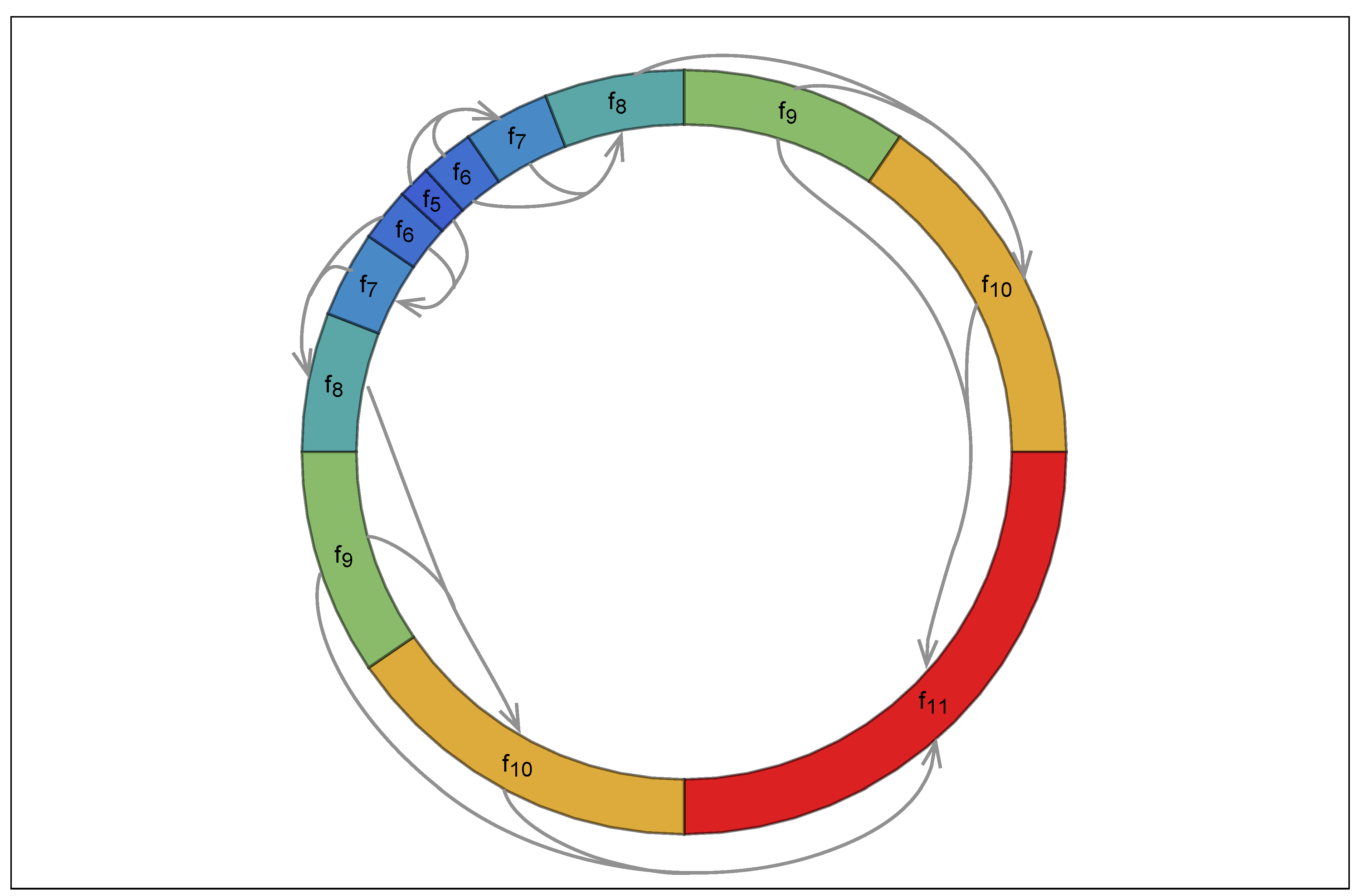
Appendix A. Random Ring Patterns
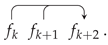

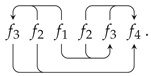
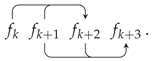
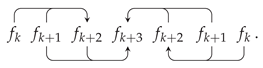
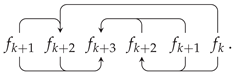
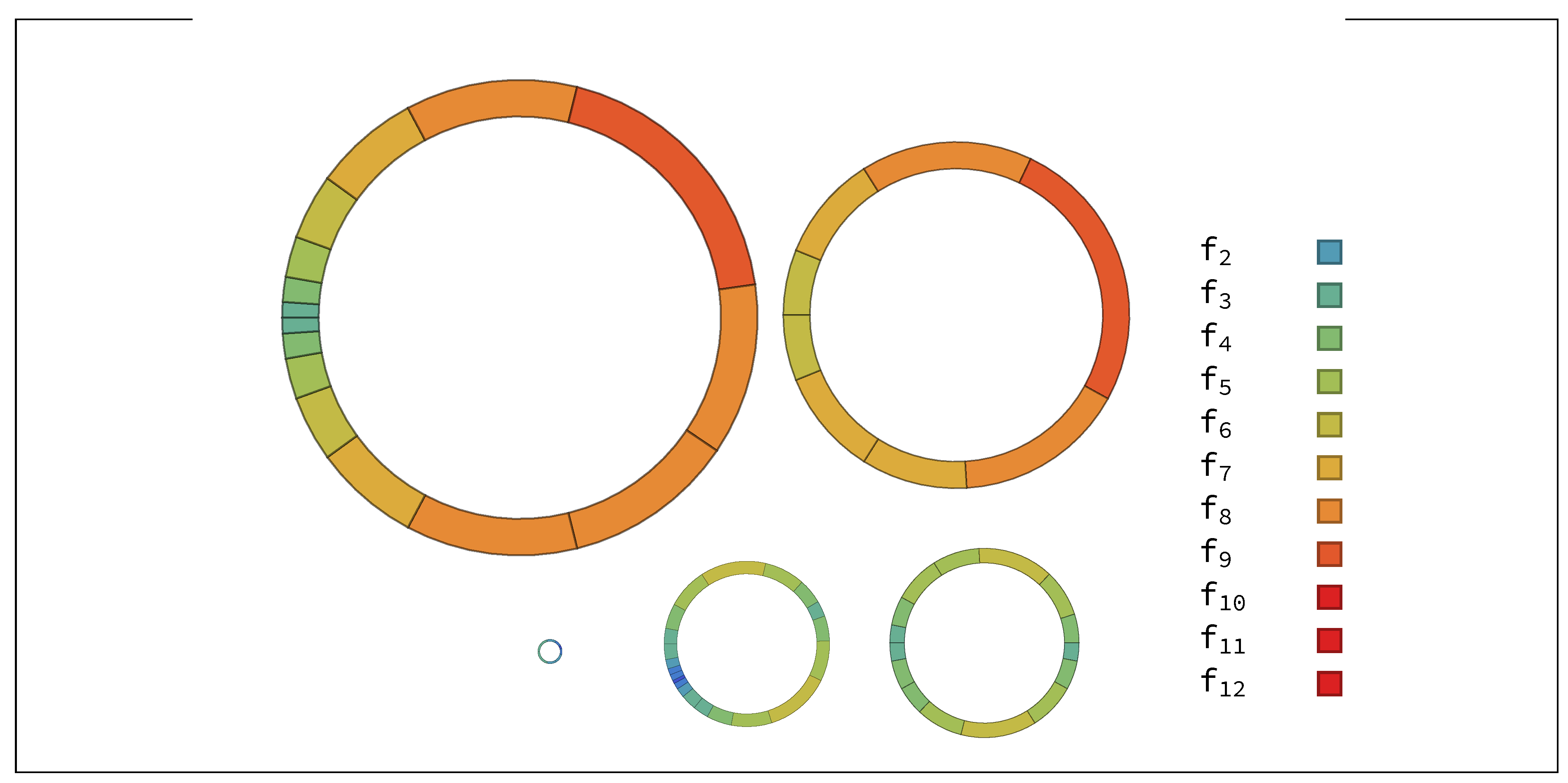
References
- Meisner, G.B. The Golden Ratio; Race Point Publishing: New York, 2018. [Google Scholar]
- Livio, M. The Golden Ratio; Broadway Books: New York, 2008; p. 304. [Google Scholar]
- de Campos, D.; Malysz, T.; Bonatto-Costa, J.A.; Pereira Jotz, G.; Pinto de Oliveira Junior, L.; Oxley da Rocha, A. Michelangelo, the Last Judgment fresco, Saint Bartholomew and the Golden Ratio. Clin Anat 2015. [Google Scholar] [CrossRef]
- Iosa, M.; Morone, G.; Paolucci, S. Phi in physiology, psychology and biomechanics: The golden ratio between myth and science. 2018. [Google Scholar] [CrossRef]
- Sebé-Pedrós, A.; Degnan, B.; Ruiz-Trillo, I. The origin of Metazoa: a unicellular perspective. Nature Reviews Genetics 2017, 18, 498–512. [Google Scholar] [CrossRef]
- Wille, J.J. Occurrence of Fibonacci numbers in development and structure of animal forms: Phylogenetic observations and epigenetic significance. NS 2012, 04, 216–232. [Google Scholar] [CrossRef]
- Peaucelle, A.; Couder, Y. Fibonacci spirals in a brown alga [Sargassum muticum (Yendo) Fensholt] and in a land plant [Arabidopsis thaliana (L.) Heynh.]: a case of morphogenetic convergence. Acta Societatis Botanicorum Poloniae 2016, 85. [Google Scholar] [CrossRef]
- Steurer, W.; Deloudi, S. Cluster packing from a higher dimensional perspective. Struct Chem 2011, 23, 1115–1120. [Google Scholar] [CrossRef]
- Malygin, A.G. Morphodynamics of phyllotaxis. International Journal of Developmental Biology 2006, 50, 277–287. [Google Scholar] [CrossRef]
- Mughal, A.; Weaire, D. Phyllotaxis, disk packing, and Fibonacci numbers. Physical Review E 2017, 95, 22401. [Google Scholar] [CrossRef] [PubMed]
- Bozdag, G.O.; Zamani-Dahaj, S.A.; Kahn, P.C.; Day, T.C.; Tong, K.; Balwani, A.H.; Dyer, E.L.; Yunker, P.J.; Ratcliff, W.C. De novo evolution of macroscopic multicellularity. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Singh, P. The so-called fibonacci numbers in ancient and medieval India. Historia Mathematica 1985, 12, 229–244. [Google Scholar] [CrossRef]
- Sigler, L. Fibonacci’s Liber Abaci: A Translation into Modern English of Leonardo Pisano’s Book of Calculation; Springer Science and Business Media: New York, 2003. [Google Scholar]
- Benjamin, A.T.; Plott, S.S. A combinatorial approach to fibonomial coefficients. Fibonacci Quarterly 2008, 46-47, 7–9. [Google Scholar]
- Schnyder, S.; Molina, J.; Yamamoto, R. Control of cell colony growth by contact inhibition. Sci Rep 2020, 10, 6713. [Google Scholar] [CrossRef] [PubMed]
- Lujan, P.; Rubio, T.; Varsano, G.; Köhn, M. Keep it on the edge: The post-mitotic midbody as a polarity signal unit. Commun Integr Biol 2017, 10, e1338990. [Google Scholar] [CrossRef]
- Juanes, M.; Piatti, S. The final cut: cell polarity meets cytokinesis at the bud neck in S. cerevisiae. Cell Mol Life Sci 2016, 73, 3115–3136. [Google Scholar] [CrossRef]
- Johnson, A.; McCollum, D.; Gould, K. Polar opposites: Fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton (Hoboken) 2012, 69, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Fleming, E.; Zajac, M.; Moschenross, D.; Montrose, D.; Rosenberg, D.; Cowan, A.; Tirnauer, J. Planar spindle orientation and asymmetric cytokinesis in the mouse small intestine. J Histochem Cytochem 2007, 55, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Caydasi, A.; Lohel, M.; Grünert, G.; Dittrich, P.; Pereira, G.; Ibrahim, B. A dynamical model of the spindle position checkpoint. Mol Syst Biol 2012, 8, 582. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, J.; Gondaliya, P.; Patel, T. Tunneling Nanotube-Mediated Communication: A Mechanism of Intercellular Nucleic Acid Transfer. Int J Mol Sci 2022, 23, 5487. [Google Scholar] [CrossRef] [PubMed]
- Gilberto, S.; Peter, M. Dynamic ubiquitin signaling in cell cycle regulation. The Journal of cell biology 2017, 216, 2259–2271. [Google Scholar] [CrossRef]
- Ogura, Y.; Sasakura, Y. Emerging mechanisms regulating mitotic synchrony during animal embryogenesis. Development, growth & differentiation 2017, 59, 565–579. [Google Scholar]
- Üretmen Kagıalı, Z.C.; Şentürk, A.; Özkan Küçük, N.E.; Qureshi, M.H.; Özlü, N. Proteomics in Cell Division. Proteomics 2017, 17, 1600100. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Gerlach, B. The roles and regulation of the actin cytoskeleton, intermediate filaments and microtubules in smooth muscle cell migration. Respir Res 2017, 18, 54. [Google Scholar] [CrossRef] [PubMed]
- Rizzelli, F.; Malabarba, M.; Sigismund, S.; Mapelli, M. The crosstalk between microtubules, actin and membranes shapes cell division. Open Biol 2020, 10, 190314. [Google Scholar] [CrossRef] [PubMed]
- Dyson, R.J.; Green, J.E.F.; Whiteley, J.P.; Byrne, H.M. An investigation of the influence of extracellular matrix anisotropy and cell-matrix interactions on tissue architecture. Journal of Mathematical Biology 2016, 72, 1775–1809. [Google Scholar] [CrossRef] [PubMed]
- Motealleh, A.; Hermes, H.; Jose, J.; Kehr, N.S. Chirality-dependent cell adhesion and enrichment in Janus nanocomposite hydrogels. Nanomedicine: Nanotechnology, Biology, and Medicine 2018, 14, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Walma, D.A.; Yamada, K.M. The extracellular matrix in development. Development 2020, 147. [Google Scholar] [CrossRef]
- Ostrander, E.A. Central Dogma of Molecular Biology, 2022.
- Gould, H.W. A history of the Fibonacci Q-matrix and a higher-dimensional problem. Fibonacci Quarterly 1981, 19, 250–257. [Google Scholar]
- Silvester, J.R. Fibonacci Properties by Matrix Methods. The Mathematical Gazette 1979, 63, 188–191. [Google Scholar] [CrossRef]
- Spears, C.; Bicknell-johnson, M.; Yan, J.J. Fibonacci phyllotaxis by asymmetric cell division : Zeckendorf and Wythoff trees. Congressus Numerantium 2016, 201, 257–257. [Google Scholar]
- Butler, W.; Kinane, T. Ring shape Golden Ratio multicellular structures are algebraically afforded by asymmetric mitosis and one to one cell adhesion. bioRxiv 2018. [Google Scholar]
- Lindenmayer, A. Mathematical models for cellular interactions in development I. Filaments with one-sided inputs. Journal of theoretical biology 1968, 18, 280–299. [Google Scholar] [CrossRef]
- Lindenmayer, A. Developmental algorithms for multicellular organisms: A survey of L-systems. Journal of Theoretical Biology 1975, 54, 3–22. [Google Scholar] [CrossRef]
- Kelly-Sacks, C. The biological and mathematical basis of L systems. PhD thesis, Rochester Institute of Technology, 1981.
- Owens, N.; Stepney, S. Investigations of Game of Life cellular automata rules on Penrose Tilings: lifetime and ash statistics. Automata 2008, 1–35. [Google Scholar]
- Giavitto, J.L.; Michel, O. Modeling the topological organization of cellular processes. BioSystems 2003, 70, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Nehaniv, C.L.; Rhodes, J.L. The evolution and understanding of hierarchical complexity in biology from an algebraic perspective. Artificial Life 2000, 6, 45–67. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Samaranayake, M.; Pradhan, S. Epigenetic mechanisms in mammals. Cell Mol Life Sci 2009, 66, 596–612. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R. Epigenetics: a historical overview. Epigenetics 2006. [Google Scholar] [CrossRef]
- Fuentealba, L.; Eivers, E.; Geissert, D.; Taelman, V.; De Robertis, E. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc Natl Acad Sci U S A 2008, 105, 7732–7737. [Google Scholar] [CrossRef] [PubMed]
- Charlton, J.; Downing, T.; Smith, Z.; Gu, H.; Clement, K.; Pop, R.; Akopian, V.; Klages, S.; Santos, D.; Tsankov, A.; Timmermann, B.; Ziller, M.; Kiskinis, E.; Gnirke, A.; Meissner, A. Global delay in nascent strand DNA methylation. Nat Struct Mol Biol 2018, 25, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef]
- Brero, A.; Leonhardt, H.; Cardoso, M.C. Replication and Translation of Epigenetic Information. In DNA Methylation: Basic Mechanisms; 2006. [Google Scholar]
- Podobinska, M.; Szablowska-Gadomska, I.; Augustyniak, J.; Sandvig, I.; Sandvig, A.; Buzanska, L. Epigenetic Modulation of Stem Cells in Neurodevelopment: The Role of Methylation and Acetylation. Frontiers in cellular neuroscience 2017. [Google Scholar] [CrossRef]
- Laugesen, A.; Helin, K. Chromatin repressive complexes in stem cells, development, and cancer. Cell stem cell 2014. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.J.; Nelson, W.J.; Weis, W.I. An epithelial tissue in Dictyostelium challenges the traditional origin of metazoan multicellularity. BioEssays 2012, 34, 833–840. [Google Scholar] [CrossRef]
- Yin, T.; Green, K. Regulation of desmosome assembly and adhesion. Semin Cell Dev Biol 2004, 15, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Coránguez, M.; Liu, X.; Antonetti, D. Tight Junctions in Cell Proliferation. Int J Mol Sci 2019, 20, E5972. [Google Scholar] [CrossRef] [PubMed]
- Balda, M.; Matter, K. Tight junctions and the regulation of gene expression. Biochim Biophys Acta 2009, 1788, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Le Bras, S.; Le Borgne, R. Epithelial cell division - multiplying without losing touch. J Cell Sci 2014, 127, 5127–5137. [Google Scholar] [CrossRef]
- Hatte, G.; Prigent, C.; Tassan, J. Adherens junctions are involved in polarized contractile ring formation in dividing epithelial cells of Xenopus laevis embryos. Exp Cell Res 2021, 402, 112525. [Google Scholar] [CrossRef]
- Leithe, E. Regulation of connexins by the ubiquitin system: Implications for intercellular communication and cancer. Biochim Biophys Acta 2016, 1865, 133–146. [Google Scholar] [CrossRef]
- Herszterg, S.; Pinheiro, D.; Bellaïche, Y. A multicellular view of cytokinesis in epithelial tissue. Trends Cell Biol 2014, 24, 285–293. [Google Scholar] [CrossRef]
- Dong, L.; Gong, J.; Wang, Y.; He, J.; You, D.; Zhou, Y.; Li, Q.; Liu, Y.; Cheng, K.; Qian, J.; Weng, W.; Wang, H.; Yu, M. Chiral geometry regulates stem cell fate and activity. Biomaterials 2019, 222, 119456. [Google Scholar] [CrossRef]
- Farmer, T.; Prekeris. New signaling kid on the block: the role of the postmitotic midbody in polarity, stemness, and proliferation. Mol Biol Cell 2022, 33. [Google Scholar] [CrossRef] [PubMed]
- Peterman, E.; Prekeris, R. The postmitotic midbody: Regulating polarity, stemness, and proliferation. J Cell Biol 2019, 218, 3903–3911. [Google Scholar] [CrossRef] [PubMed]
- Novak, M.; Polak, B.; Simunić, J.; Boban, Z.; Kuzmić, B.; Thomae, A.W.; Tolić, I.M.; Pavin, N. The mitotic spindle is chiral due to torques within microtubule bundles. Nature Communications 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Satir, P. Chirality of the cytoskeleton in the origins of cellular asymmetry. Philosophical transactions of the Royal Society of London. Series B, Biological sciences 2016, 371, 20150408. [Google Scholar] [CrossRef]
- Schuchert, P. Phylogenetic analysis of the Cnidaria. Journal of Zoological Systematics and Evolutionary Research 2009, 31, 161–173. [Google Scholar] [CrossRef]
- Berzins, I.K.; Yanong, R.P.E.; LaDouceur, E.E.; Peters, E.C. Cnidaria. In Invertebrate Histology; LaDouceur, E.E., Ed.; Wiley, 2021; pp. 55–86. [Google Scholar]
- Weisstein, E.W. Torus. From MathWorld—A Wolfram Web Resource. Accessed 4 Oct 2022.
- Basic three-dimensional objects constructed with simplex meshes, Vol. (ICEEE). 1st International Conference on Electrical and Electronics Engineering, 2004., New York, 2004. IEEE.
- Nicolson, S.J.S.; L., G. The Fluid Mosaic Model of the Structure of Cell Membranes. Science, New Series, 175, No. 4023, 720–731.
- Fletcher, A. The cell membrane and receptors. Anaesthesia & Intensive Care Medicine 2007, 8, 443–446. [Google Scholar]
- Parfrey, L.W.; Lahr, D.J.G. Multicellularity arose several times in the evolution of eukaryotes (Response to DOI 10.1002/bies.201100187). BioEssays 2013, 35, 339–347. [Google Scholar] [CrossRef]
- Meng, W.; Takeichi, M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol 2009, 1, a002899. [Google Scholar] [CrossRef]
- Matsuzawa, K.; Himoto, T.; Mochizuki, Y.; Ikenouchi, J. alpha-Catenin Controls the Anisotropy of Force Distribution at Cell-Cell Junctions during Collective Cell Migration. Cell Rep 2018, 23, 3447–3456. [Google Scholar] [CrossRef]
- Knopfmacher, A.; Tichy, R.F.; Wagner, S.; Ziegler, V. Graphs, partitions and Fibonacci numbers. Discrete Applied Mathematics 2007, 155, 1175–1187. [Google Scholar] [CrossRef]
- Benjamin, A.; Quinn, J. Proofs that Really Count; The Mathematical Association of America: Washington, D.C, 2009. [Google Scholar]
- Gessel, I.M.; Li, J. Compositions and Fibonacci Identities. arXivJournal of Integer Sequences 2013, arXiv:1303.136616. [Google Scholar]
- Koken, F.; Bozkurt, D. On Lucas numbers by the matrix method. Hacettepe Journal of Mathematics and Statistics 2010. [Google Scholar]
- Stephenson, A.; Adams, J.W.; Vaccarezza, M. The vertebrate heart: an evolutionary perspective. Journal of anatomy 2017, 231, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Schwille, P. Jump-starting life? Fundamental aspects of synthetic biology. J Cell Biol 2015, 210, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Szathmáry, E.; Smith, J.M. The major evolutionary transitions. Nature 1995, 374, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Grosberg, R.K.; Strathmann, R.R. The Evolution of Multicellularity: A Minor Major Transition? Annual Review of Ecology, Evolution, and Systematics 2007, 38, 621–654. [Google Scholar] [CrossRef]
- Fisher, R.M.; Cornwallis, C.K.; West, S.A. Group formation, relatedness, and the evolution of multicellularity. Current Biology 2013, 23, 1120–1125. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




