Submitted:
21 March 2024
Posted:
22 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
2.1. Rat Model
2.2. Mass Spectrometry Analysis of Proteins
2.2.1. Protein Extraction, Digestion and TMT-Labelling
2.2.2. Nano Liquid Chromatography Tandem Mass Spectrometry (nLC-MS/MS)
2.2.3. Proteomics Data Analysis
2.3. Statistical Analysis of Protein Expression Data
2.4. Ingenuity Pathway Analysis
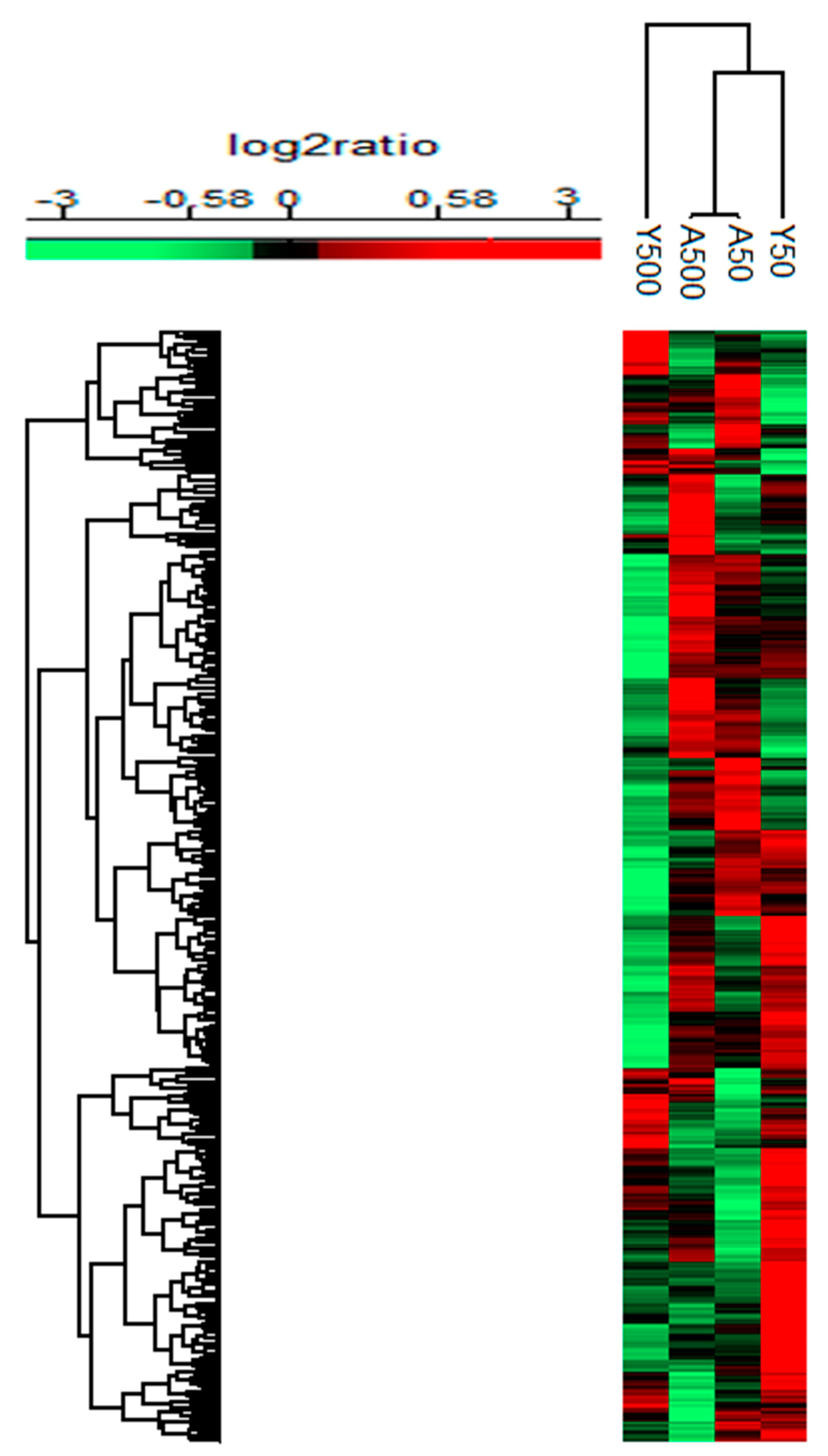
3. Results
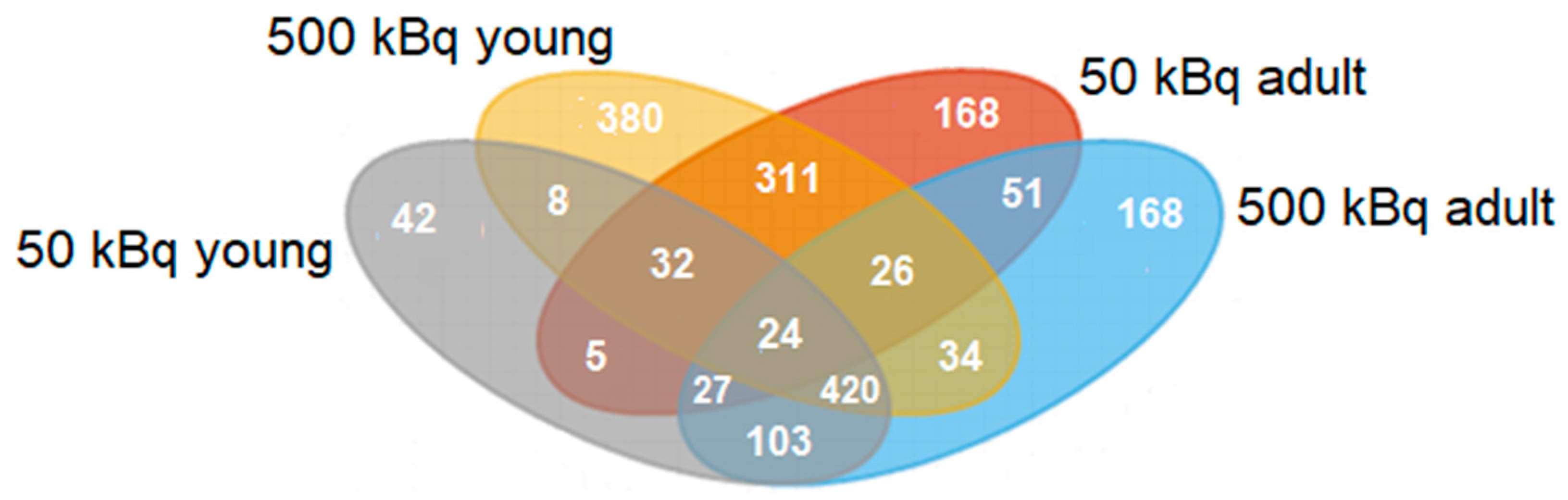
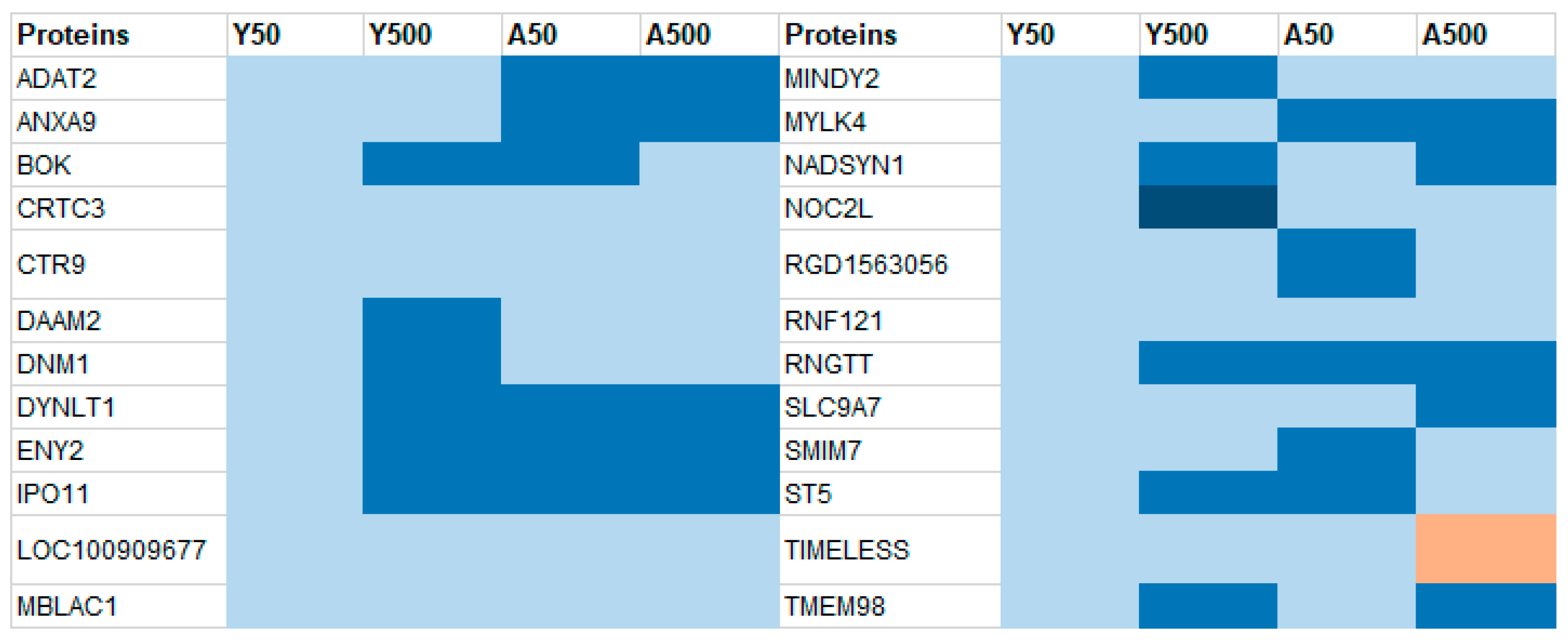
3.1. Group-Specific (Unique) Proteins
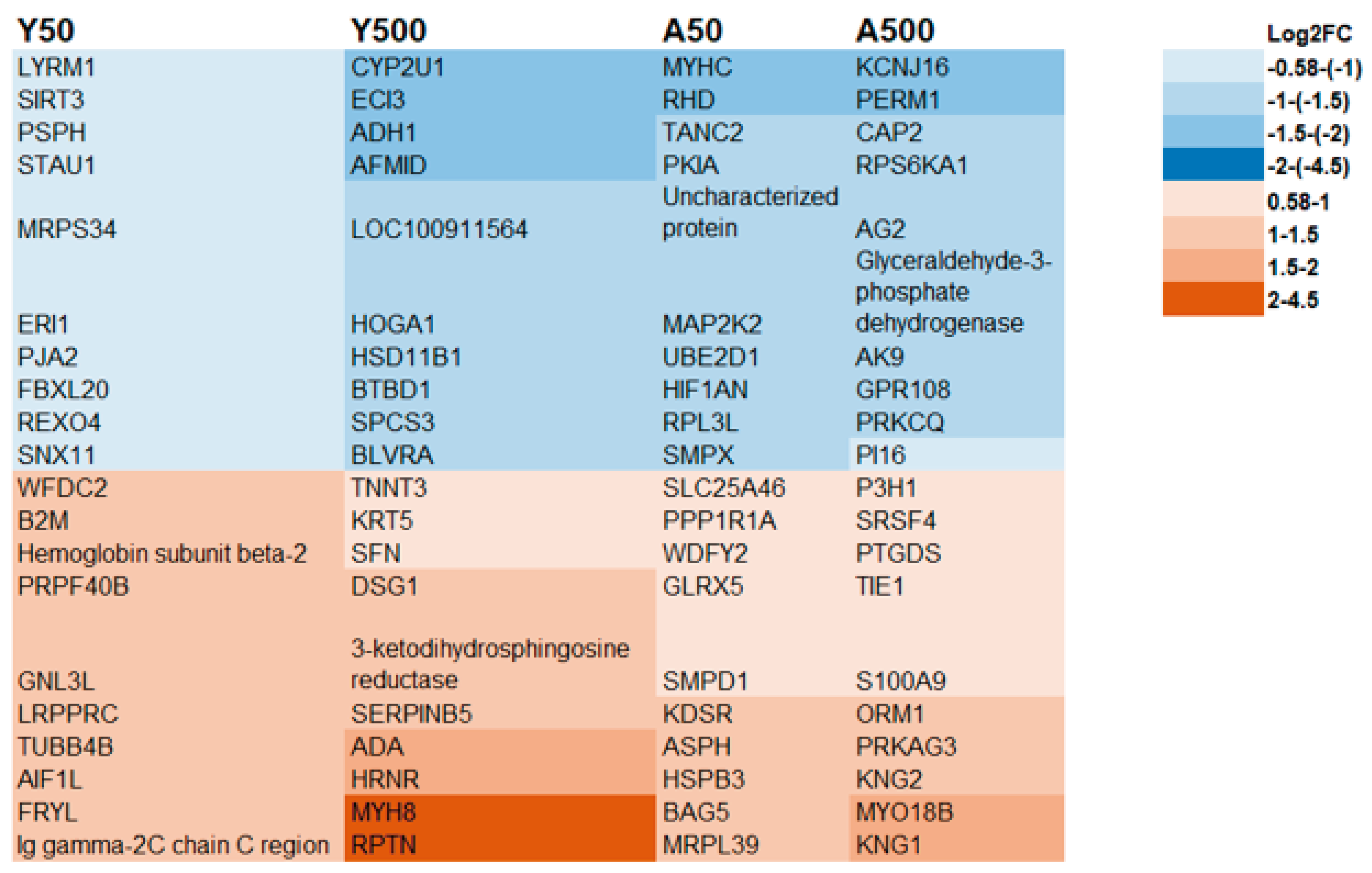
3.2. Age-Related Proteins
3.3. Dose-Related Proteins
3.4. IPA Analysis
3.5. Histological Evaluation of Rat Thyroid Tissue
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Permission
References
- Nussey S, Whitehead S. Endocrinology: An Integrated Approach. Oxford2001.
- Raymond, J.; LaFranchi, S.H. Fetal and neonatal thyroid function: review and summary of significant new findings. Curr Opin Endocrinol Diabetes Obes. 2010, 17, 1–7. [Google Scholar] [CrossRef]
- Mullur, R.; Liu, Y.Y.; Brent, G.A. Thyroid hormone regulation of metabolism. Physiol Rev. 2014, 94, 355–82. [Google Scholar] [CrossRef]
- Srikantia, N.; Rishi, K.S.; Janaki, M.G.; Bilimagga, R.S.; Ponni, A.; Rajeev, A.G.; et al. How common is hypothyroidism after external radiotherapy to neck in head and neck cancer patients? Indian journal of medical and paediatric oncology : official journal of Indian Society of Medical & Paediatric Oncology. 2011, 32, 143–8. [Google Scholar]
- Holm, L.E. Thyroid cancer after exposure to radioiodine. Strahlenschutz Forsch Prax. 1985, 25, 36–56. [Google Scholar]
- DeGroot, L.J. Effects of irradiation on the thyroid gland. Endocrinol Metab Clin North Am. 1993, 22, 607–15. [Google Scholar] [CrossRef]
- Inskip, P.D. Thyroid cancer after radiotherapy for childhood cancer. Med Pediatr Oncol. 2001, 36, 568–73. [Google Scholar] [CrossRef]
- Robbins, J.; Schneider, A.B. Radioiodine-induced thyroid cancer: Studies in the aftermath of the accident at Chernobyl. Trends Endocrinol Metab. 1998, 9, 87–94. [Google Scholar] [CrossRef]
- Ron, E.; Lubin, J.H.; Shore, R.E.; Mabuchi, K.; Modan, B.; Pottern, L.M.; et al. Thyroid cancer after exposure to external radiation: a pooled analysis of seven studies. Radiat Res. 1995, 141, 259–77. [Google Scholar] [CrossRef]
- Weiss, W. Chernobyl Thyroid Cancer: 30 Years of Follow-up Overview. Radiat Prot Dosimetry. 2018, 182, 58–61. [Google Scholar] [CrossRef]
- IAEA CF. Chernobyl’s legacy: health, environmental and socio-economic impacts and recommendations to the governments of Belarus, the Russian Federation and Ukraine. 2005.
- Zablotska, L.B.; Ron, E.; Rozhko, A.V.; Hatch, M.; Polyanskaya, O.N.; Brenner, A.V.; et al. Thyroid cancer risk in Belarus among children and adolescents exposed to radioiodine after the Chornobyl accident. Br J Cancer. 2011, 104, 181–7. [Google Scholar] [CrossRef]
- Tronko, M.D.; Howe, G.R.; Bogdanova, T.I.; Bouville, A.C.; Epstein, O.V.; Brill, A.B.; et al. A cohort study of thyroid cancer and other thyroid diseases after the chornobyl accident: thyroid cancer in Ukraine detected during first screening. J Natl Cancer Inst. 2006, 98, 897–903. [Google Scholar] [CrossRef]
- Amundson, S.A.; Fornace, A.J., Jr. Gene expression profiles for monitoring radiation exposure. Radiat Prot Dosimetry. 2001, 97, 11–6. [Google Scholar] [CrossRef]
- Chin, R.I.; Wu, F.S.; Menda, Y.; Kim, H. Radiopharmaceuticals for Neuroendocrine Tumors. Semin Radiat Oncol. 2021, 31, 60–70. [Google Scholar] [CrossRef]
- Chaudhry, M.A. Biomarkers for human radiation exposure. J Biomed Sci. 2008, 15, 557–63. [Google Scholar] [CrossRef]
- Rudqvist, N.; Spetz, J.; Schuler, E.; Parris, T.Z.; Langen, B.; Helou, K.; et al. Transcriptional response to 131I exposure of rat thyroid gland. PLoS One. 2017, 12, e0171797. [Google Scholar] [CrossRef]
- Rudqvist, N.; Schuler, E.; Parris, T.Z.; Langen, B.; Helou, K.; Forssell-Aronsson, E. Dose-specific transcriptional responses in thyroid tissue in mice after (131)I administration. Nucl Med Biol. 2015, 42, 263–8. [Google Scholar] [CrossRef]
- Langen, B.; Rudqvist, N.; Parris, T.Z.; Helou, K.; Forssell-Aronsson, E. Circadian rhythm influences genome-wide transcriptional responses to (131)I in a tissue-specific manner in mice. EJNMMI Res. 2015, 5, 75. [Google Scholar] [CrossRef]
- Larsson, M.; Rudqvist, N.; Spetz, J.; Shubbar, E.; Parris, T.Z.; Langen, B.; et al. Long-term transcriptomic and proteomic effects in Sprague Dawley rat thyroid and plasma after internal low dose 131I exposure. PLoS One. 2020, 15, e0244098. [Google Scholar] [CrossRef]
- Spetz, J.; Rudqvist, N.; Forssell-Aronsson, E. Biodistribution and dosimetry of free 211At, 125I- and 131I- in rats. Cancer Biother Radiopharm. 2013, 28, 657–64. [Google Scholar] [CrossRef]
- Rao-Rupanagudi, S.; Heywood, R.; Gopinath, C. Age-related changes in thyroid structure and function in Sprague-Dawley rats. Vet Pathol. 1992, 29, 278–87. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat Methods. 2009, 6, 359–62. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D50. [Google Scholar] [CrossRef]
- Langen, B.; Rudqvist, N.; Parris, T.Z.; Schuler, E.; Helou, K.; Forssell-Aronsson, E. Comparative analysis of transcriptional gene regulation indicates similar physiologic response in mouse tissues at low absorbed doses from intravenously administered 211At. J Nucl Med. 2013, 54, 990–8. [Google Scholar] [CrossRef]
- Celestino, R.; Nome, T.; Pestana, A.; Hoff, A.M.; Goncalves, A.P.; Pereira, L. CRABP1, C1QL1 and LCN2 are biomarkers of differentiated thyroid carcinoma, and predict extrathyroidal extension. BMC Cancer. 2018, 18, 68. [Google Scholar] [CrossRef]
- Tai, J.; Wang, S.; Zhang, J.; Ge, W.; Liu, Y.; Li, X.; et al. Up-regulated lipocalin-2 in pediatric thyroid cancer correlated with poor clinical characteristics. Eur Arch Otorhinolaryngol. 2018, 275, 2823–8. [Google Scholar] [CrossRef]
- Rosignolo, F.; Sponziello, M.; Durante, C.; Puppin, C.; Mio, C.; Baldan, F.; et al. Expression of PAX8 Target Genes in Papillary Thyroid Carcinoma. PLoS One. 2016, 11, e0156658. [Google Scholar] [CrossRef]
- Ma, H.; Xu, S.; Yan, J.; Zhang, C.; Qin, S.; Wang, X.; et al. The value of tumor markers in the diagnosis of papillary thyroid carcinoma alone and in combination. Pol J Pathol. 2014, 65, 202–9. [Google Scholar] [CrossRef]
- Ghoshal, A.; Garmo, H.; Arthur, R.; Carroll, P.; Holmberg, L.; Hammar, N.; et al. Thyroid cancer risk in the Swedish AMORIS study: the role of inflammatory biomarkers in serum. Oncotarget. 2018, 9, 774–82. [Google Scholar] [CrossRef]
- Fan, Y.; Shi, L.; Liu, Q.; Dong, R.; Zhang, Q.; Yang, S.; et al. Discovery and identification of potential biomarkers of papillary thyroid carcinoma. Mol Cancer. 2009, 8, 79. [Google Scholar] [CrossRef]
- Mitteldorf, C.A.; de Sousa-Canavez, J.M.; Massumoto, C.; da Camara-Lopes, L.H. Fine-needle aspiration biopsy of thyroid nodules as a possible source for molecular studies: analysis of RNA obtained from routine cases. Diagn Cytopathol. 2008, 36, 899–903. [Google Scholar] [CrossRef]
- Weber, R.; Bertoni, A.P.; Bessestil, L.W.; Brasil, B.M.; Brum, L.S.; Furlanetto, T.W. Validation of reference genes for normalization gene expression in reverse transcription quantitative PCR in human normal thyroid and goiter tissue. Biomed Res Int. 2014, 2014, 198582. [Google Scholar] [CrossRef]
- Razavi, S.A.; Afsharpad, M.; Modarressi, M.H.; Zarkesh, M.; Yaghmaei, P.; Nasiri, S.; et al. Validation of Reference Genes for Normalization of Relative qRT-PCR Studies in Papillary Thyroid Carcinoma. Sci Rep. 2019, 9, 15241. [Google Scholar] [CrossRef]
- Goldberg, D.M.; Goudie, R.B. Nucleases and adenosine deaminase in malignant and non-malignant lesions of the human thyroid. Br J Cancer. 1968, 22, 220–36. [Google Scholar] [CrossRef]
- Stephen, J.K.; Chen, K.M.; Merritt, J.; Chitale, D.; Divine, G.; Worsham, M.J. Methylation markers differentiate thyroid cancer from benign nodules. J Endocrinol Invest. 2018, 41, 163–70. [Google Scholar] [CrossRef]
- Traina, G.; Cataldi, S.; Siccu, P.; Loreti, E.; Ferri, I.; Sidoni, A.; et al. Mouse Thyroid Gland Changes in Aging: Implication of Galectin-3 and Sphingomyelinase. Mediators Inflamm. 2017, 2017, 8102170. [Google Scholar] [CrossRef]
- Zhang, D.L.; Wang, J.M.; Wu, T.; Du, X.; Yan, J.; Du, Z.X.; et al. BAG5 promotes invasion of papillary thyroid cancer cells via upregulation of fibronectin 1 at the translational level. Biochim Biophys Acta Mol Cell Res. 2020, 1867, 118715. [Google Scholar] [CrossRef]
- Ito, Y.; Yoshida, H.; Uruno, T.; Nakano, K.; Takamura, Y.; Miya, A.; et al. Tie-1 tyrosine kinase expression in human thyroid neoplasms. Histopathology. 2004, 44, 318–22. [Google Scholar] [CrossRef]
- Korkmaz, H.; Tabur, S.; Savas, E.; Ozkaya, M.; Aksoy, S.N.; Aksoy, N.; et al. Evaluation of Serum S100A8/S100A9 Levels in Patients with Autoimmune Thyroid Diseases. Balkan Med J. 2016, 33, 547–51. [Google Scholar] [CrossRef]
- Ito, Y.; Arai, K.; Nozawa, R.; Yoshida, H.; Hirokawa, M.; Fukushima, M.; et al. S100A8 and S100A9 expression is a crucial factor for dedifferentiation in thyroid carcinoma. Anticancer Res. 2009, 29, 4157–61. [Google Scholar]
- Ito, Y.; Arai, K.; Ryushi Nozawa Yoshida, H.; Tomoda, C.; et al. S100A9 expression is significantly linked to dedifferentiation of thyroid carcinoma. Pathol Res Pract. 2005, 201, 551–6. [Google Scholar] [CrossRef]
- Rudqvist, N. Radiobiological Effects of the Thyroid Gland. https://gupea.ub.gu.se/bitstream/2077/38006/1/gupea_2077_38006_1.pdf: University of Gothenburg; 2015.
- Larsson M. RN, Spetz J., Shubbar E., Parris TZ., Langen B., Helou K., Forssell-Aronsson E. Age related long-term response in rat thyroid tissue and plasma after internal low dose exposure to 131I 2021.
- Marchetti, F.; Coleman, M.A.; Jones, I.M.; Wyrobek, A.J. Candidate protein biodosimeters of human exposure to ionizing radiation. Int J Radiat Biol. 2006, 82, 605–39. [Google Scholar] [CrossRef] [PubMed]
- Qatanani, M.; Zhang, J.; Moore, D.D. Role of the constitutive androstane receptor in xenobiotic-induced thyroid hormone metabolism. Endocrinology. 2005, 146, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016, 54, 1 30 1-1 3. [Google Scholar] [CrossRef]
- Yen, C.F.; Wang, H.S.; Lee, C.L.; Liao, S.K. Roles of integrin-linked kinase in cell signaling and its perspectives as a therapeutic target. Gynecology and Minimally Invasive Therapy. 2014, 3, 67–72. [Google Scholar] [CrossRef]
- Perdas, E.; Stawski, R.; Nowak, D.; Zubrzycka, M. The Role of miRNA in Papillary Thyroid Cancer in the Context of miRNA Let-7 Family. Int J Mol Sci. 2016, 17. [Google Scholar] [CrossRef]
- Nieminen, T.T.; Walker, C.J.; Olkinuora, A.; Genutis, L.K.; O'Malley, M.; Wakely, P.E.; et al. Thyroid Carcinomas That Occur in Familial Adenomatous Polyposis Patients Recurrently Harbor Somatic Variants in APC, BRAF, and KTM2D. Thyroid. 2020, 30, 380–8. [Google Scholar] [CrossRef] [PubMed]
- Akaishi, J.; Kondo, T.; Sugino, K.; Ogimi, Y.; Masaki, C.; Hames, K.Y.; et al. Cribriform-Morular Variant of Papillary Thyroid Carcinoma: Clinical and Pathological Features of 30 Cases. World J Surg. 2018, 42, 3616–23. [Google Scholar] [CrossRef] [PubMed]
- Cetta, F.; Montalto, G.; Gori, M.; Curia, M.C.; Cama, A.; Olschwang, S. Germline mutations of the APC gene in patients with familial adenomatous polyposis-associated thyroid carcinoma: results from a European cooperative study. J Clin Endocrinol Metab. 2000, 85, 286–92. [Google Scholar] [PubMed]
- Li, C.; Peng, S.; Liu, X.; Han, C.; Wang, X.; Jin, T.; et al. Glycyrrhizin, a Direct HMGB1 Antagonist, Ameliorates Inflammatory Infiltration in a Model of Autoimmune Thyroiditis via Inhibition of TLR2-HMGB1 Signaling. Thyroid. 2017, 27, 722–31. [Google Scholar] [CrossRef]
- Peng, S.; Li, C.; Wang, X.; Liu, X.; Han, C.; Jin, T.; et al. Increased Toll-Like Receptors Activity and TLR Ligands in Patients with Autoimmune Thyroid Diseases. Front Immunol. 2016, 7, 578. [Google Scholar] [CrossRef]
- Jacques, C.; Guillotin, D.; Fontaine, J.F.; Franc, B.; Mirebeau-Prunier, D.; Fleury, A.; et al. DNA microarray and miRNA analyses reinforce the classification of follicular thyroid tumors. J Clin Endocrinol Metab. 2013, 98, E981–9. [Google Scholar] [CrossRef]
- Rocchi, R.; Kimura, H.; Tzou, S.C.; Suzuki, K.; Rose, N.R.; Pinchera, A.; et al. Toll-like receptor-MyD88 and Fc receptor pathways of mast cells mediate the thyroid dysfunctions observed during nonthyroidal illness. Proc Natl Acad Sci U S A. 2007, 104, 6019–24. [Google Scholar] [CrossRef]
- Fan, X.; Zhao, Y. miR-451a inhibits cancer growth, epithelial-mesenchymal transition and induces apoptosis in papillary thyroid cancer by targeting PSMB8. J Cell Mol Med. 2019, 23, 8067–75. [Google Scholar] [CrossRef]
- Lawnicka, H.; Motylewska, E.; Borkowska, M.; Kuzdak, K.; Siejka, A.; Swietoslawski, J.; et al. Elevated serum concentrations of IGF-1 and IGF-1R in patients with thyroid cancers. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2020, 164, 77–83. [Google Scholar] [CrossRef]
- He, L.; Zhang, S.; Zhang, X.; Liu, R.; Guan, H.; Zhang, H. Effects of insulin analogs and glucagon-like peptide-1 receptor agonists on proliferation and cellular energy metabolism in papillary thyroid cancer. Onco Targets Ther. 2017, 10, 5621–31. [Google Scholar] [CrossRef]
- Huang, Y.; Chang, A.; Zhou, W.; Zhao, H.; Zhuo, X. IGFBP3 as an indicator of lymph node metastasis and unfavorable prognosis for papillary thyroid carcinoma. Clin Exp Med. 2020, 20, 515–25. [Google Scholar] [CrossRef]
- Iglesias, P.; Bayon, C.; Mendez, J.; Gancedo, P.G.; Grande, C.; Diez, J.J. Serum insulin-like growth factor type 1, insulin-like growth factor-binding protein-1, and insulin-like growth factor-binding protein-3 concentrations in patients with thyroid dysfunction. Thyroid. 2001, 11, 1043–8. [Google Scholar] [CrossRef]
- Lakatos, P.; Foldes, J.; Nagy, Z.; Takacs, I.; Speer, G.; Horvath, C.; et al. Serum insulin-like growth factor-I, insulin-like growth factor binding proteins, and bone mineral content in hyperthyroidism. Thyroid. 2000, 10, 417–23. [Google Scholar] [CrossRef]
- Wescombe, L.; Lahooti, H.; Gopinath, B.; Wall, J.R. The cardiac calsequestrin gene (CASQ2) is up-regulated in the thyroid in patients with Graves' ophthalmopathy--support for a role of autoimmunity against calsequestrin as the triggering event. Clin Endocrinol (Oxf). 2010, 73, 522–8. [Google Scholar] [CrossRef]
- Liang, W.; Sun, F. Identification of key genes of papillary thyroid cancer using integrated bioinformatics analysis. J Endocrinol Invest. 2018, 41, 1237–45. [Google Scholar] [CrossRef]
- Huang, C.; Yang, X.; Han, L.; Fan, Z.; Liu, B.; Zhang, C.; et al. The prognostic potential of alpha-1 type I collagen expression in papillary thyroid cancer. Biochem Biophys Res Commun. 2019, 515, 125–32. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Suzuki, S.; Suzuki, S.; Shimura, H.; Saenko, V. Lessons from Fukushima: Latest Findings of Thyroid Cancer After the Fukushima Nuclear Power Plant Accident. Thyroid. 2018, 28, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, N.; Imaizumi, M.; Shimura, H.; Okubo, N.; Asari, Y.; Nigawara, T.; et al. Thyroid ultrasound findings in children from three Japanese prefectures: Aomori, Yamanashi and Nagasaki. PLoS One. 2013, 8, e83220. [Google Scholar] [CrossRef]
| Young | Adult | ||||
|---|---|---|---|---|---|
| Protein | Log2 ratio (Y50; Y500) |
Protein | Log2 ratio (A50; A500) |
Protein | Log2 ratio (A50; A500) |
| FDPS | -0.60; -0.72 | AHNAK 1 (Fragment) | -0.66; -0.72 | LCN2 | 0.98; 0.69 |
| GNPTAB | -0.63; -0.66 | ALDH1A7 | 1.3; 1.0 | LGALS5 | 0.89; 2.0 |
| JAK1 | -0.63; -0.69 | APOBEC2 | 1.1; 0.88 | NCAPG2 | -1.1; -0.69 |
| LSAMP | -0.59; -0.82 | APOD | --1.3; -0.81 | NHSL1 | -0.85; -1.1 |
| MAOB | -0.61; -0.73 | ATPSCKMT | 0.81; 0.63 | NR2F2 | -0.93; -0.75 |
| REEP6 | -0.58; -0.96 | BLOC1S6 | -0.82; -0.97 | PATZ1 | -0.64; -0.92 |
| RGD1566265 | -0.60; -1.0 | BUD23 | -1.1; -1.4 | PDLIM4 | -0.73; -0.61 |
| TBC1D10A | -0.66; -0.65 | CACNG1 | -1.8; -1.2 | PHKG2 | -0.59; -0.63 |
| COX3 | -0.85; -1.1 | PISD | -0.68; -0.65 | ||
| CSTF1 | -0.73; -0.74 | PLSCR1 | -0.86; -0.73 | ||
| DHRS7B | -1.2; -0.61 | PPP1R10 | -0.96; -0.99 | ||
| DNAH6 | -0.59; -1.6 | PRR33 | 1.9; 1.4 | ||
| DPH6 | -1.4; -2.1 | PSMB8 | 1.8; 1.2 | ||
| DVL1 | -0.62; -0.93 | RAC2 | -0.68; -0.61 | ||
| ECI1 | -1.3; 0.90 | RB1 | -1.1; -0.62 | ||
| FBXW17 | -0.65; -1.0 | RT1-A1B | 4.4; 1.4 | ||
| FCGBP | -0.83; -0.78 | SNPH | -1.0; -0.82 | ||
| FXYD1 | --0.99; -0.59 | TAP2 | -0.59; -1.0 | ||
| HAT1 | -0.67; -0.63 | TAP2C | 1.5; 1.3 | ||
| IGFBP6 | --0.69; -0.65 | TFE3 | -0.59; 0.99 | ||
| IGHG | 0.67; 0.71 | Titin protein homolog (Fragment) | -1.3; -0.60 | ||
| KRT1 | -1.0; -0.97 | TMEM47 | -0.63; -0.58 | ||
| KRT10 | -1.1; -0.83 | TPCR12 | -0.94; -1.2 | ||
| KRT16 | -1.6; -1.4 | TXLNB | -2.0; -1.9 | ||
| KRT80 | -0.65; -0.70 | Uncharacterized protein | -1.9; -1.7 | ||
| LAMC2 | -0.76; -0.69 | ||||
| 50 kBq | 500 kBq | ||||
|---|---|---|---|---|---|
| Protein | Log2 ratio (young; adult) |
Protein | Log2 ratio (young; adult) |
Protein | Log2 ratio (young; adult) |
| BICD2 |
-0.62; -0.59 |
ABCC8 |
-1.1; -0.90 |
MEF2D |
-0.67; -0.70 |
| HABP4 |
-0.61; -0.63 |
ADGRG2 |
-0.98; -0.78 |
MOCOS |
-1.1; -0.68 |
| HBB-B1 |
0.65; -0.71 |
ALDH1A2 |
-0.65; -0.66 |
MTM1 |
-0.85; -0.88 |
| NME3 |
0.60; 2.0 |
BHMT |
-2.7; -0.77 |
MYL3 |
1.6; -0.73 |
| PALM2 |
0.63; 0.70 |
CDC40 |
-1.1; -0.84 |
PKP1 |
1.2; -1.1 |
| CLCC1 |
0.84; 1.0 |
PRKCZ |
-0.79; -0.62 |
||
| CNST |
-0.62; -1.3 |
PSMF1 |
-0.95; -0.66 |
||
| CPT2 |
1.1; 0.90 |
RBM3 |
-0.64; -1.86 |
||
| CSRP3 |
0.99; -0.73 |
SERPINB12 |
3.1; -0.64 |
||
| DAB2 |
-0.81; -0.74 |
SLC25A15 |
-0.89; -0.74 |
||
| DNAJC17 |
-0.60; -0.69 |
SMPD3 |
1.5; -2.9 |
||
| DUSP22 |
-1.1; -0.69 |
THBS4 |
0.72; -1.0 |
||
| FLT4 |
-0.88; -1.3 |
TMEM106B |
-1.2; -1.3 |
||
| HEG1 |
-0.72; -0.60 |
TP53I11 |
-0.77; -1.1 |
||
| HP |
0.87; 0.74 |
UBAC1 |
-0.78; -0.82 |
||
| IGSF8 |
-0.89; -0.80 |
VASN |
-1.1; -0.78 |
||
| LEAP2 |
-1.0; -0.85 |
WIZ |
-0.67; -0.94 |
||
| Ingenuity canonical pathways | p | z | Molecules |
|---|---|---|---|
| Y50 | |||
| tRNA Splicing | 1.7E-02 | 2.0 | PDE10A,PDE1B,PDE5A,TSEN2 |
| Stearate Biosynthesis I (Animals) | 2.5E-02 | -2.0 | CYP2E1,DHCR24,GNPAT,SLC27A5 |
| Superpathway of Methionine Degradation | 1.0E-02 | -2.0 | BHMT,BHMT2,CTH,MAT1A |
| Thyroid Hormone Metabolism II (via Conjugation and/or Degradation) | 7.4E-03 | -2.0 | SULT1B1,UGT1A1,Ugt2b17,UGT2B28 |
| Gluconeogenesis I | 1.4E-03 | -2.0 | ALDOB,ENO3,FBP1,PGAM2 |
| Glycolysis I | 2.6E-04 | -2.2 | ALDOB,ENO3,FBP1,PGAM2,PKLR |
| Bile Acid Biosynthesis, Neutral Pathway | 7.9E-06 | -2.2 | AKR1D1,BAAT,CYP27A1,CYP3A4,SLC27A5 |
| Xenobiotic Metabolism PXR Signaling Pathway | 3.7E-06 | -2.5 | ALDH1L1,ALDH1L2,ALDH8A1,CAMK2G,CYP2C19,CYP2C9,CYP3A4,GSTA5,MAOB,PPP1R11,PRKCE,SULT1B1, SULT1E1,UGT1A1,UGT2B28,UGT8 |
| LPS/IL-1 Mediated Inhibition of RXR Function | 1.2E-07 | 2.6 | ACOX2,ALDH1L1,ALDH1L2,ALDH8A1,CPT2,CYP2A6 (includes others),CYP2C19,CYP2C9,CYP3A4,Cyp4a14, FABP1,ABP3,FMO4,GSTA5,HMGCS2,MAOB,MYD88,SLC27A5,SULT1B1,SULT1E1 |
| Serotonin Degradation | 4.3E-04 | -2.6 | ADH1C,ADH4,MAOB,SULT1B1,UGT1A1,Ugt2b17,UGT2B28 |
| Acetone Degradation I (to Methylglyoxal) | 4.0E-06 | -2.6 | CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2U1,CYP3A4 |
| Bupropion Degradation | 9.8E-07 | -2.6 | CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2U1,CYP3A4 |
| Xenobiotic Metabolism CAR Signaling Pathway | 1.4E-05 | -2.8 | ALDH1L1,ALDH1L2,ALDH8A1,Cyp2b13/Cyp2b9,CYP2C19,CYP2C9,CYP3A4,FMO4,GSTA5,PRKCE,SULT1B1,SULT1E1, UGT1A1,UGT2B28,UGT8 |
| Nicotine Degradation II | 2.4E-08 | -2.9 | AOX1,CYP2A6 (includes others),CYP2C18,CYP2C19, CYP2C9,CYP2E1,CYP2U1,CYP3A4,FMO4,UGT1A1,Ugt2b17, UGT2B28 |
| Estrogen Biosynthesis | 2.5E-07 | -3,0 | CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2U1,CYP3A4,HSD17B13,HSD17B2 |
| Melatonin Degradation I | 4.5E-08 | -3.3 | CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2U1,CYP3A4,SULT1B1,UGT1A1,Ugt2b17,UGT2B28 |
| Nicotine Degradation III | 4.5E-08 | -3.3 | AOX1,CYP2A6 (includes others),CYP2C18,CYP2C19, CYP2C9,CYP2E1,CYP2U1,CYP3A4,UGT1A1,Ugt2b17,UGT2B28 |
| Superpathway of Melatonin Degradation | 8.9E-09 | -3.5 | CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2U1,CYP3A4,MAOB,SULT1B1,UGT1A1, Ugt2b17,UGT2B28 |
| Y500 | |||
| Thyroid Hormone Metabolism II (via Conjugation and/or Degradation) | 2.0E-03 | -2.0 | SULT1B1,UGT1A1,Ugt2b17,UGT2B28 |
| Retinoate Biosynthesis I | 1.8E-03 | -2.0 | ADH1C,ADH4,ALDH8A1,Rdh7 |
| Noradrenaline and Adrenaline Degradation | 1.6E-03 | -2.0 | ADH1C,ADH4,ADHFE1,MAOB |
| Superpathway of Cholesterol Biosynthesis | 1.2E-03 | -2.0 | CYP51A1,DHCR24,FDPS,HMGCS2 |
| Bile Acid Biosynthesis, Neutral Pathway | 4.6E-05 | -2.0 | AKR1D1,BAAT,CYP3A4,SLC27A5 |
| Citrulline Biosynthesis | 8.3E-06 | -2.0 | ARG1,GLS,LOC102724788/PRODH,OTC |
| Superpathway of Citrulline Metabolism | 3.0E-06 | -2.2 | ARG1,CPS1,GLS,LOC102724788/PRODH,OTC |
| Estrogen Biosynthesis | 7.9E-13 | -2.3 | CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2F1,CYP2S1,CYP2U1,CYP3A4,CYP51A1,HSD17B13,HSD17B2 |
| Nicotine Degradation II | 1.3E-12 | -2.7 | Aox3,CYP2A6 (includes others),CYP2C18,CYP2C19, CYP2C9,CYP2E1,CYP2F1,CYP2S1,CYP2U1,CYP3A4, CYP51A1,UGT1A1,Ugt2b17,UGT2B28 |
| Melatonin Degradation I | 1.3E-13 | -2.7 | CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2F1,CYP2S1,CYP2U1,CYP3A4,CYP51A1, SULT1B1,UGT1A1,Ugt2b17,UGT2B28 |
| Nicotine Degradation III | 1.3E-13 | -2.6 | Aox3,CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2F1,CYP2S1,CYP2U1,CYP3A4,CYP51A1, UGT1A1,Ugt2b17,UGT2B28 |
| Serotonin Degradation | 4.6E-06 | -2.8 | ADH1C,ADH4,ADHFE1,MAOB,SULT1B1,UGT1A1,Ugt2b17,UGT2B28 |
| Superpathway of Melatonin Degradation | 2.0E-14 | -2.87 | CYP2A6 (includes others),CYP2C18,CYP2C19,CYP2C9, CYP2E1,CYP2F1,CYP2S1,CYP2U1,CYP3A4,CYP51A1,MAOB, SULT1B1,UGT1A1,Ugt2b17,UGT2B28 |
| Xenobiotic Metabolism CAR Signaling Pathway | 1.3E-04 | -3.3 | ALDH1L2,ALDH8A1,Cyp2b13/Cyp2b9,CYP2C19,CYP2C9, CYP3A4,GSTA5,SULT1B1,SULT1E1,UGT1A1,UGT2B28 |
| Xenobiotic Metabolism PXR Signaling Pathway | 5.9E-06 | -3.6 | ALDH1L2,ALDH8A1,CAMK2A,CYP2C19,CYP2C9,CYP3A4, GSTA5,MAOB,Ppp1cc,SULT1B1,SULT1E1,UGT1A1,UGT2B28 |
| A50 | |||
| Xenobiotic Metabolism CAR Signaling Pathway | 3.9E-03 | 2.3 | ALDH8A1,GSTA1,GSTA5,MAP2K2,MAP2K5,SULT1B1,SULT2B1,UGT1A1,UGT2B28 |
| A500 | |||
| ILK Signaling | 1.9E-04 | -2.3 | ACTB,ACTN2,JUN,MYH1,MYH2,MYH3,MYH6,MYH7,MYH8,MYL1,MYL2,MYL3 |
| Apelin Cardiomyocyte Signaling Pathway | 2.6E-03 | -2.6 | ATP2A1,MYL1,MYL2,MYL3,MYLPF,PLCL2,SLC9A2 |
| Actin Cytoskeleton Signaling | 1.4E-05 | -2.7 | ACTB,ACTN2,APC,MPRIP,MYH1,MYH2,MYH3,MYH6, MYH7,MYH8,MYL1,MYL2,MYL3,MYLPF,TTN |
| Upstream regulator | Molecule type | p | z | Target molecules in dataset |
| Y500 | ||||
| EFNA2 | kinase | 4.5E-03 | 2.0 | KRT13,KRT4,PKP1,TGM1 |
| A50 | ||||
| let-7 | microRNA | 2.3E-02 | 2.2 | APC,BOP1,IGF1R,MYD88,STARD13 |
| IGF1 | growth factor | 2.0E-03 | -2.2 | GFAP,IGF1R,IGFBP3,PSMB8,SLC20A1 |
| A500 | ||||
| MYOCD | transcription regulator | 7.8E-10 | -2.1 | ACTN2,CASQ2,CNN1,COL1A1,HSPB7,MYH6,MYH7,MYL2,TNNC1,TNNI1,TTN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





