1. Introduction
Date palm sap collection is a widespread and common practice during the winter season in South Asia. Most commonly it is consumed raw or processed into other products, such as molasses or tari. There are several food safety concerns during these processes from the circumstances of collection to storage and transport and later processes. Nipah virus is a well-known and highly lethal viral pathogen in relation to this food product. All Nipah cases in Bangladesh were epidemiologically linked to raw date palm sap consumption to date. These index cases presumably consumed the sap, contaminated by Pteropus bats during collection [
1]. Even though the consumption has known health risks of infectious diseases it is still a widely common practice. In a survey-based study, conducted in Bangladesh, more than one-third of the responders reported drinking raw palm sap at least once per month. Ten percent of the responders related the consumption of unprocessed palm sap with mild diseases, such as diarrhea, vomiting or indigestion [
1,
2]. Nevertheless, the bacterial components of this product are less studied and there is not enough knowledge about the fermentation processes or common pathogenic bacterial species and their origin.
Considering the importance of this product as a nutrient during the winter season, increased food safety awareness is required both from the general consumers and from respective authorities. Novel techniques may help to conduct non targeted and rapid surveillance of pathogens. Bacterial metabarcoding offers a solution for the discovery and the examination of bacterial communities.
Oxford Nanopore Technologies (ONT) stands out as the sole platform currently facilitating off-lab and in-field (point-of-care) library preparation and sequencing, coupled with online real-time data analysis. This feature is particularly advantageous for diagnostic applications conducted in the field. Moreover, ONT is an emerging technology in food safety applications where it was already developed for different foodborne pathogen screening applications with reduced time, material and expenses needed, compared to other technologies [
3].
In our study, we demonstrated a proof-of-concept bacterial metabarcoding analysis on raw date palm sap samples, focusing on the demonstration of the advantages of ONT fast bacterial metabarcoding and its mobility. This was carried out as an initial step towards establishing a reference for future food safety interventions. By leveraging the portability of Nanopore-based sequencing techniques, we are able to conduct this analysis in remote locations. The primary aim of this study was to demonstrate that nanopore sequencing-based bacterial metabarcoding is an effective and rapid method for identifying bacterial communities in food products, offering a significantly shorter diagnostic timeframe compared to conventional methods. With our study we targeted a common, but under investigated food type, the raw date palm sap, usually collected in remote places.
2. Materials and Methods
Sample Collection
We collected 15 samples from separate collection pots with the help of a gachi, a local palm sap collector who admitted not using protective coverage during collection of the sap, therefore animals and contamination could potentially reach the product. Samples were taken into 50 mL falcon tubes and were frozen until laboratory processing.
Sequencing and Analysis
Nucleic acid extraction was performed with the Zymo Quick DNA Miniprep Kit, following the manufacturer’s protocol with a final elution to 50 uL. Prior to this we sedimented the samples with centrifugation for 30 minutes at 15 000 g, thereafter the sediment was subjected to nucleic acid extraction.
For quantification during the library preparation, we used a Qubit 4 Fluorometer, with the relevant data detailed in the accompanying table. The amplification targeted approximately 1500bp fragments, encompassing the entire 16S gene, and both this step and the library preparation were conducted according to the Nanopore protocol (SQK-16S024). Both the amplification step and library preparation were done following the manufacturer’s protocol of Nanopore 16S Barcoding Kit. The sequencing took place on a MinION Mk1B, lasting for 42 hours and employing only fast base-calling techniques. Analysis was subsequently performed using EPI2ME’s “Fastq 16S’’ workflow. In the classified reads, those achieving a minimum of 1% are represented in the figures.
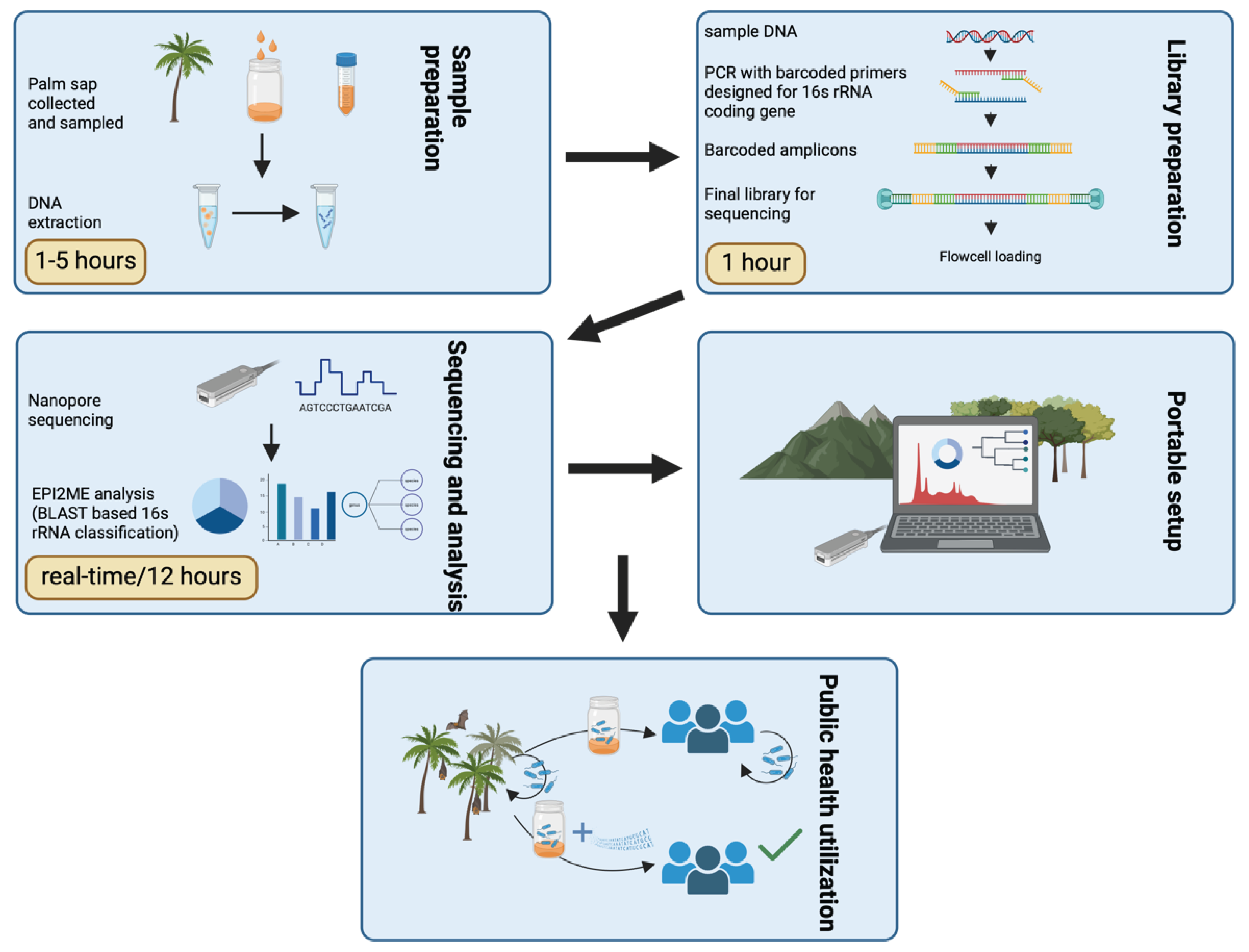
Figure 1.
Representative scheme of the study workflow with highlights on approximate processing times for each step.
Figure 1.
Representative scheme of the study workflow with highlights on approximate processing times for each step.
3. Results and Discussion
The study successfully identified several bacterial genera, important in the natural fermentation process of raw date palm sap (Leuconostoc spp., Lactococcus spp.). Also some additional genera was identified with low abundance with the potential for opportunistic infections in humans (Citrobacter spp., Enterobacter spp.) (
Figure 2).
This demonstrates the feasibility of nanopore sequencing for monitoring such products and also its ability to examine bacterial communities on the genus level. In addition, as a major advantage of the technology, Nanopore sequencing produces complete 16s rDNA sequences, resulting in a detailed taxonomic resolution of bacterial communities.
Carbohydrate concentration of palm sap usually reaches up to 94.98g/100g of dry matter basis, containing mainly sucrose, glucose and fructose [
4]. The research discovered two primary fermentation directions in the freshly collected raw date palm sap, dominated by either
Leuconostoc species or
Lactococcus species. With lower representation, only in sample 5 and 15, Lactococcus sp. was the most dominant, whilst in all of the other 13 samples the Leuconostoc sp. (
Figure 3).
Leuconostoc spp. are heterofermentative lactic acid bacteria (LAB), they ferment sugars to lactic acid and ethanol/acetic acid. Young
Leuconostoc mesenteroides cultures produce lactate, acetic acid and dextran from sugars, the stationary-growth-phase cultures produce lactic acid and ethanol.
Streptococcus and
Lactococcus genera are members of the
Lactobacillales order, similarly to
Leuconostoc genus, but members of the first two genera are homofermentative LAB, producing only lactate from glucose fermentation [
5]. LABs produce bacteriocins, hydrogen peroxide, antimicrobial peptides in addition to acids and alcohols, which have an antagonistic effect [
6]. Nallala et al. showed that Lactobacillus strains inhibit, among others, the growth of
Escherichia coli (E. coli),
Listeria monocytogenes,
Pseudomonas aeruginosa,
Staphylococcus aureus strains [
6]. Reuben et al. documented that LAB strains showed antagonistic activity against
E. coli,
Enterococcus faecalis,
Salmonella typhimurium and
Salmonella enteritidis [
7]. LABs also inhibit the growth of
Citrobacter [
8],
Enterobacter [
9] and
Serratia marcescens [
10].
Fermentation end- and by-products are the reasons why Streptococcus spp. only occur with Lactococcus spp.. In high sugar content medium, LAB strains exclude and overgrow species from the Enterobacterales order, which are plant pathogens and common soil microbes. The recycled and unwashed clay pots provide the starter culture, causing some tested samples to appear as “pure” cultures (barcodes 7, 12, 24).
Acinetobacter, a common soil bacteria, is the third most frequently detected genus among the 15 samples. A previous study showed that
Acinetobacter was also presented in 4% of grapevine sap samples [
11]. Furthermore another study confirmed the presence of
Acinetobacter in floral nectar, a medium with similarly high sugar content, of wild Mediterranean plants [
12].
Since the dominance of either of these bacteria was obvious in the pre-market stage of collection we hypothesize the natural fermentation process as a protective state of this product by out-competing potentially harmful bacteria. However, community members after consumption of unprocessed palm sap reported a range of mild diseases possibly related to enteric pathogens. This highlights the importance of rapid and generalized food safety diagnostic measures, as presented in our current study to discover these pathogens. Also this means the potential of the opportunistic pathogens with low abundance to act as the source of these negative health impacts. We verified the presence of such bacteria, however the origin is uncertain and more focused research is necessary to better understand the sources of contamination.
Overall, the species composition is similar through all 15 samples. Detailed sequencing results by barcodes with the most abundant genera is summarized in
Table S1. It is uncertain if there is any difference between regions of the collection of seasonal patterns which may lead to more optimal circumstances for pathogenic bacteria. This also needs to be addressed in future studies.
Beyond fermentation bacteria we identified a number of opportunistic bacteria from the Enterobacter, Citrobacter, Acinetobacter, Streptococcus and Serratia genera. Enterobacter was found in four samples (barcodes 4, 8, 9, 13); Citrobacter in one sample (barcode 6); Acinetobacter in seven samples (barcodes 1, 2, 3, 4, 6, 9, 11) and Serratia in four samples (barcodes 5, 6, 11, 13). Interestingly, Streptococcus was identified only in those two samples, where the Lactococcus dominance is observable. However the sequencing method could not provide species-level details of these, simply the presence of these bacteria supports the potential for causing negative health effects such as gastrointestinal problems. Particularly, Streptococcus may refer to faecal contamination which may originate from visiting animals during the collection [
13].
Interestingly, we identified multiple plant pathogenic bacteria, represented by Erwinia and Pectobacterium groups in the data. These are very important to raise awareness in regard to maintaining clean cultivation and harvesting practices to avoid the cross contamination of palm trees with pathogens during the cutting [
14]. Although these potential plant pathogens were identified in three samples, Erwinia in two samples (barcodes 9, 13); and Pectobacterium in one sample (barcode 6), the importance for safe agriculture is notable.
Most interestingly we report the presence of
Exiguobacterium in one of the palm sap samples with over 1% sequence ratio (Barcode 9). A range of isolates from diverse environments has been investigated for their potential in biotechnology and industry, such as for producing enzymes, bioremediating, and breaking down harmful substances discharged into the environment. Additionally, certain isolates with the ability to promote plant growth are under study for their capacity to enhance agricultural yields. The role of these bacteria in this food product is uncertain [
15].
4. Conclusions
Our study served as an initial step towards establishing a reference for future food safety interventions, highlighting the effectiveness of nanopore sequencing in remote locations. Effectiveness of the method includes the high mobility and finescale resolution of results as we demonstrated in this study. Also the possibility for mobile experimental and bioinformatic analysis offers a faster diagnostic timeframe compared to conventional methods.
The study reinforces earlier findings about the health risks associated with consuming raw date palm sap, particularly highlighting the risk of bacterial contamination. The samples analysed showed the presence of various opportunistic human pathogenic bacteria, albeit in low abundance. This research underscores the potential health hazards and emphasises the need for caution and further safety measures in the consumption of this product. Our research also identified the presence of plant pathogenic bacteria in the samples, underscoring the need for clean collection practices. This finding is significant as it highlights the potential adverse impact on agricultural practices, emphasising the importance of maintaining hygiene standards during the collection of raw date palm sap in relation to plant pathogens.
Overall, the study provides significant insights into the bacterial composition of raw date palm sap and demonstrates the utility of advanced sequencing techniques in food safety surveillance.
Supplementary Materials
The following supporting information can be downloaded at: Preprints.org, Table S1. Sequencing data summary per barcodes
Author Contributions
“Conceptualization, G.K. and M.N.I.; methodology, Á.Á.; validation, Á.Á. and S.A.; investigation, S.A., Á.Á., M.N.I., Z.G..; resources, G.K.; data curation, Á.Á; writing—original draft preparation, Á.Á., Z.G., G.K., S.A., M.N.I.; writing—review and editing, S.A.K., S.C., Z.G.; visualization, Á.Á.; supervision, G.K., S.A., S.A.K., S.C.; funding acquisition, G.K.. All authors have read and agreed to the published version of the manuscript.”
Funding
The work was supported by the grant from the National Research, Development and Innovation Office, under grant number 2021-4.1.2-NEMZ_KI-2022-00020.
Data Availability Statement
All related data to the manuscript are available in the paper and as a supplementary file.
Acknowledgments
We are grateful for the collaboration of local helpers and guides during the field trip and their assistance and hospitality.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Nahar, N., Sultana, R., Gurley, E. S., Hossain, M. J., & Luby, S. P. (2010). Date Palm Sap Collection: Exploring Opportunities to Prevent Nipah Transmission. EcoHealth, 7(2), 196–203. [CrossRef]
- Nahar, N., Paul, R. C., Sultana, R., Gurley, E. S., Garcia, F., Abedin, J., Sumon, S. A., Banik, K. C., Asaduzzaman, M., Rimi, N. A., Rahman, M., & Luby, S. P. (2015). Raw Sap Consumption Habits and Its Association with Knowledge of Nipah Virus in Two Endemic Districts in Bangladesh. PLOS ONE, 10(11), e0142292. [CrossRef]
- Counihan, K. L., Kanrar, S., Tilman, S., & Gehring, A. (2023). Evaluation of Long-Read Sequencing Simulators to Assess Real-World Applications for Food Safety. Foods, 13(1), 16. [CrossRef]
- Ben Thabet, I., Besbes, S., Attia, H., Deroanne, C., Francis, F., Drira, N.-E., & Blecker, C. (2009). Physicochemical Characteristics of Date Sap “ Lagmi ” from Deglet Nour Palm ( Phoenix Dactylifera L.). International Journal of Food Properties, 12(3), 659–670. [CrossRef]
- Harper, A. R., Dobson, R. C. J., Morris, V. K., & Moggré, G. (2022). Fermentation of plant-based dairy alternatives by lactic acid bacteria. Microbial Biotechnology, 15(5), 1404–1421. [CrossRef]
- Nallala, V., Sadishkumar, V., & Jeevaratnam, K. (2017). Molecular characterization of antimicrobial Lactobacillus isolates and evaluation of their probiotic characteristics in vitro for use in poultry. Food Biotechnology, 31(1), 20–41. [CrossRef]
- Reuben, R. C., Roy, P. C., Sarkar, S. L., Alam, R.-U., & Jahid, I. K. (2019). Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiology, 19(1), 253. [CrossRef]
- Pasteris, S. E., Guidoli, M. G., Otero, M. C., Bühler, M. I., & Nader-Macías, M. E. (2011). In vitro inhibition of Citrobacter freundii, a red-leg syndrome associated pathogen in raniculture, by indigenous Lactococcus lactis CRL 1584. Veterinary Microbiology, 151(3–4), 336–344. [CrossRef]
- Zhang, X., Han, J., Zheng, X., Yan, J., Chen, X., Zhou, Q., Zhao, X., Gu, Q., & Li, P. (2022). Use of Lactiplantibacillus plantarum ZJ316 as a starter culture for nitrite degradation, foodborne pathogens inhibition and microbial community modulation in pickled mustard fermentation. Food Chemistry: X, 14, 100344. [CrossRef]
- Vahedi-Shahandashti, R., Kasra-Kermanshahi, R., Shokouhfard, M., Ghadam, P., Feizabadi, M. M., & Teimourian, S. (2017). Antagonistic activities of some probiotic lactobacilli culture supernatant on Serratia marcescens swarming motility and antibiotic resistance. Iranian Journal of Microbiology, 9(6), 348–355.
- Deyett, E., & Rolshausen, P. E. (2019). Temporal Dynamics of the Sap Microbiome of Grapevine Under High Pierce’s Disease Pressure. Frontiers in Plant Science, 10, 1246. [CrossRef]
- Álvarez-Pérez, S., Lievens, B., Jacquemyn, H., & Herrera, C. M. (2013). Acinetobacter nectaris sp. Nov. And Acinetobacter boissieri sp. Nov., isolated from floral nectar of wild Mediterranean insect-pollinated plants. International Journal of Systematic and Evolutionary Microbiology, 63(Pt_4), 1532–1539. [CrossRef]
- Rossi, F., Santonicola, S., Amadoro, C., Marino, L., & Colavita, G. (2023). Recent Records on Bacterial Opportunistic Infections via the Dietary Route. Microorganisms, 12(1), 69. [CrossRef]
- Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., Dow, M., Verdier, V., Beer, S. V., Machado, M. A., Toth, I., Salmond, G., & Foster, G. D. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Molecular Plant Pathology, 13(6), 614–629. [CrossRef]
- Kasana, R. C., & Pandey, C. B. (2018). Exiguobacterium: An overview of a versatile genus with potential in industry and agriculture. Critical Reviews in Biotechnology, 38(1), 141–156. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).