Introduction
Fecal Microbial transplant (FMT) is now-a-days considered as a well-established technique used to treat gastro-intestinal diseases worldwide, especially the
Clostridium dif9icile infection (CDI) in the western world [Ref]. Faecal microbiota transplantation (FMT) refers to the infusion of a faecal suspension from a healthy person into the gastrointestinal (GI) tract of another person to cure a speciCic disease [Ref]. It is a procedure in which fecal matter, or stool, is collected from a tested donor, mixed with a saline or other solution, strained, and placed in a patient, by colonoscopy, endoscopy, sigmoidoscopy, or enema [Ref]. Faecal transplant was Cirst documented in 4th century China, known as “yellow soup” [Ref]. However, there is a similar concept in ayurveda “
panchagavya prasahan”, which is being practiced since ancient times, but have not been explored using modern scientiCic tools.
Panchagavya is a system of medicine just like as homeopathy, allopathy or naturopathy [
1,
2]. The ancient ayurvedic literature (
Charak Samhita,
Sushrut, Gada nigrah) suggests a number of pharmacological applications of the substances obtained from
Panchgavya. Religious ritual of practice of ‘
Panchagavya prashan’ has been in existence for ages in our country. This practice was expected to deliver a person from all the sins (
papma in Sanskrit), which is also a synonym for the word ‘disease’. This ritual is practiced once every year during July and August at which time the Vaccinia viraemia is at its peak in cows [
3]. A systematic work needs to be carried out on chemical nature, biological activity, microbiology and pharmaceutical aspects and mechanism of bioactive compounds in
panchagavya. In the recent past due emphasis has not been given to this therapy by scientiCic community. Our aim is to decode the basis of this practice in modern scientiCic language.
Panchagavya involves the use of Cive by-products of cow viz. Cow dung, urine, milk, curd, ghee mixed in a copper vessel [
3,
4,
6]. Copper vessel is used because it is capable of detoxifying and destroying disease causing bacteria (potential pathogens) through ionization [
3]. Fresh urine being acidic in nature is antitoxic and helps to attenuate bacteria [
3]. Fresh milk and curd are also probiotic in nature and act as neutral nutritional medium to protect attenuated organisms [
3].
Ghee helps in enteric coating the organisms thereby preventing their destruction by gastric acid. Cow dung is expected to harbor all the enteric organisms. Since it’s a season when the vaccinia is rampant in cows it is expected the vaccinia virus is secreted in to all the secretions and bodily excretions of cow. Previous studies suggest that when the dung is treated with acidic urine especially in a copper container, which ionizes, the organisms undergo deCinite attenuation [
7]. When these attenuated organisms reach the intestinal villi and Payer’s patches, they stimulate activate general immunity and as well the speciCic immunity [
4,
5]. Secondly, there will be new bacterial colonies of cow origin introduced into the gut. This is also expected to change the gut microbiota composition and thereby causing deCinite changes that may be desirable in certain medical conditions, e.g. IBS, Ulcerative colitis, diabetes mellitus, obesity etc. Diseases have been traditionally studied under a paradigm of “one microbe, one disease.” However, a new understanding is emerging on how disease phenotypes are actually a result of complex interactions between bacteria, viruses, and eukaryotes, as well as their interactions with the host or with certain drugs [
8]. Virulence of some eukaryotes is, for instance, linked to the presence of certain bacteria, such as in the case of
E. histolytica and
E. coli or
S. dysenteriae [
8]. Interestingly, studies have proven that the susceptibility of the host to viral infections is conditioned by the particular conCiguration of the microbiota, whereas herpesvirus infection can confer resistance to certain bacterial infections [
8,
9]. As a clear correlation has been observed between many diseases and dysbiosis, restoring a healthy microbial community by administration of
panchagavya can be proven to be a valuable tool in the treatment of these diseases.
Further, use of antibiotics has certainly proven to be ineffective in treating many gastrointestinal diseases [
10]. Considering the rapid emergence of these lifestyle associated disease, there is an urge of developing novel tools to modulate the gut microbiota and eventually improve the health [
11].
Panchagavya prashan being a well-established technique in ayurveda may beneCit by improving the gut microbial balance and may also improve the immunological proCile of the individual. Thus, understanding the mechanism through the gut microbiome perspective is necessary. In this study, we present a preliminary investigation on the microbial composition of the fresh ‘
panchagavya’ preparation and the effect of process of using copper vessel on the observed microbial diversity. This study provides a basis for future investigations on ‘
panchagavya prashan’ and to understand its inCluence on the gut microbial composition and eventually on the human health.
Material and Methods
Panchagavya preparation
Protocol for Fresh Panchagavya Preparation
The Cive or panch ingredients of Panchagavya are cow urine, fresh cow dung, cow milk, cow curd and cow ghee.
The products with their quantities are:-
Fresh cow dung: ½, Cow urine: 1, Cow milk: 7, Cow curd: 1, Cow ghee: 1. Method of Preparation:
Step 1: Mix fresh cow dung and cow urine thoroughly and keep it for 30 minutes, Cilter this mixture using muslin cloth transfer the Ciltrate to copper vessel.
Step 2: Add cow milk and cow curd to the Ciltrate and mix well for 10-15 minutes.
Step 3: To the above mixture add liqueCied cow ghee and centrifuge the mixture at lowest possible rpm for 10 minutes.
Step 4: Transfer the Fresh Panchagavya mixture to a sterile container.
DNA Extraction and PCR Ampli5ication
Aliquots of ~1 gram of freshly prepared panchagavya preparation (DK1) and panchagavya preparation after processing in the copper vessel (DK2) were taken for total DNA extraction using the QiAmp mini stool DNA extraction kit (Qiagen, USA) as per the manufacturer’s protocol. The DNA extraction was done in replicates (in duplicate) and all the replicates were then pooled together in 1:1 ratio. The total DNA extracted DNA was then quantiCied using the NanoDrop spectrophotometer ND-1000 (Thermo ScientiCics, USA) and stored at -20° C until further use. The DNA samples were used further for the 16S rRNA based amplicon library for high throughput sequencing. BrieCly, the V3 region of the 16S rRNA gene was ampliCied using the region speciCic bacterial universal primers: forward primer 341F (5′ -C CTA C GGG A GGCA GCA G-3′ ) a nd r e verse primer 518 R (5′ -ATTACCGCGGCTGCTGG-3′; Bartram et al., 2011) and methodology as described earlier [Bhute et al., 2016].
Ion Torrent Based Amplicon Sequencing
The library generation and sequencing was carried out as previously described by Bhute et al., 2016. BrieCly, PCR products were puriCied using Agencourt AMPure XP DNA puriCication Bead (Beckman Coulter, USA). The puriCied products were end repaired and ligated with speciCic barcode adaptor as explained in IonXpressTM Plus gDNA Fragment Library Preparation user guide. Fragment size distribution and molar concentrations of amplicon were assessed on a Bioanalyzer 2100 (Agilent Technologies, USA) using High Sensitivity DNA Analysis Kit as per manufacturer’s instructions. Emulsion PCR was carried out on diluted and pooled amplicon using the Ion One TouchTM 200 Template Kit v2 DL (Life Technologies). The sequencing of the amplicon library was carried out on Ion Torrent sequencing platform using the Ion 316 chip with Ion Sequencing 200 kit, as per the manufacturer’s instructions (Life Technologies, USA).
Bacterial Diversity Analysis
The raw reads obtained from the sequencing run were assessed for quality using the FASTQC tool [
12]. The adapter sequences were trimmed and the quality based trimming of the reads was done using Mothur [
13]. Further, these quality processed sequences were used for OTU (Operational Taxonomic Sequencing) clustering using the SILVA database. The representative set of these sequences per OTU cluster were then picked and assigned taxonomy using the same SILVA taxonomy database using QIIME (Quantitative Insights In Microbial Ecology) pipeline [
14]. The BIOM Cile thus generated using the standard QIIME pipeline was then further used to carry out composition and statistical analysis using the tools such as STAMP [
15]. Appropriate statistical tests such as Wilcoxon signed-rank match paired t-test were used to determine the statistical signiCicance between the diversity in pre and post processing
panchagavya samples.
Results
Sequencing and Bacterial Composition of the Panchagavya Preparation
The high throughput sequencing carried out using Ion Torrent single end sequencing technology generated 14,642 reads for sample DK1, while 13,094 reads for sample DK2. The quality Ciltering using Mothur (>QV20) was employed to get quality sequences to be used for further analysis. It was observed that the more than 50 % erroneous reads were removed, yielding around 5.5K reads per sample. The OTU clustering using QIIME, using SILVA databases (SILVA_99) reference dataset, yielded 1,694 OTUs for DK1 and 1,756 OTUs for DK2 sample.
The analysis carried out to understand the overall bacterial composition of the
panchagavya preparation revealed that the bacterial diversity was dominated by phyla
Firmicutes (mean=81.94%), followed by
Proteobacteria (mean=9.27%) and
Bacteroidetes (mean=6.82%). Other rare taxa such as
Fibrobacteres,
Actinobacteria,
Cyanobacteria and
Saccharibacteria were also observed (<0.2%) in the
panchagavya preparation in both pre and post processing in the copper vessel (See
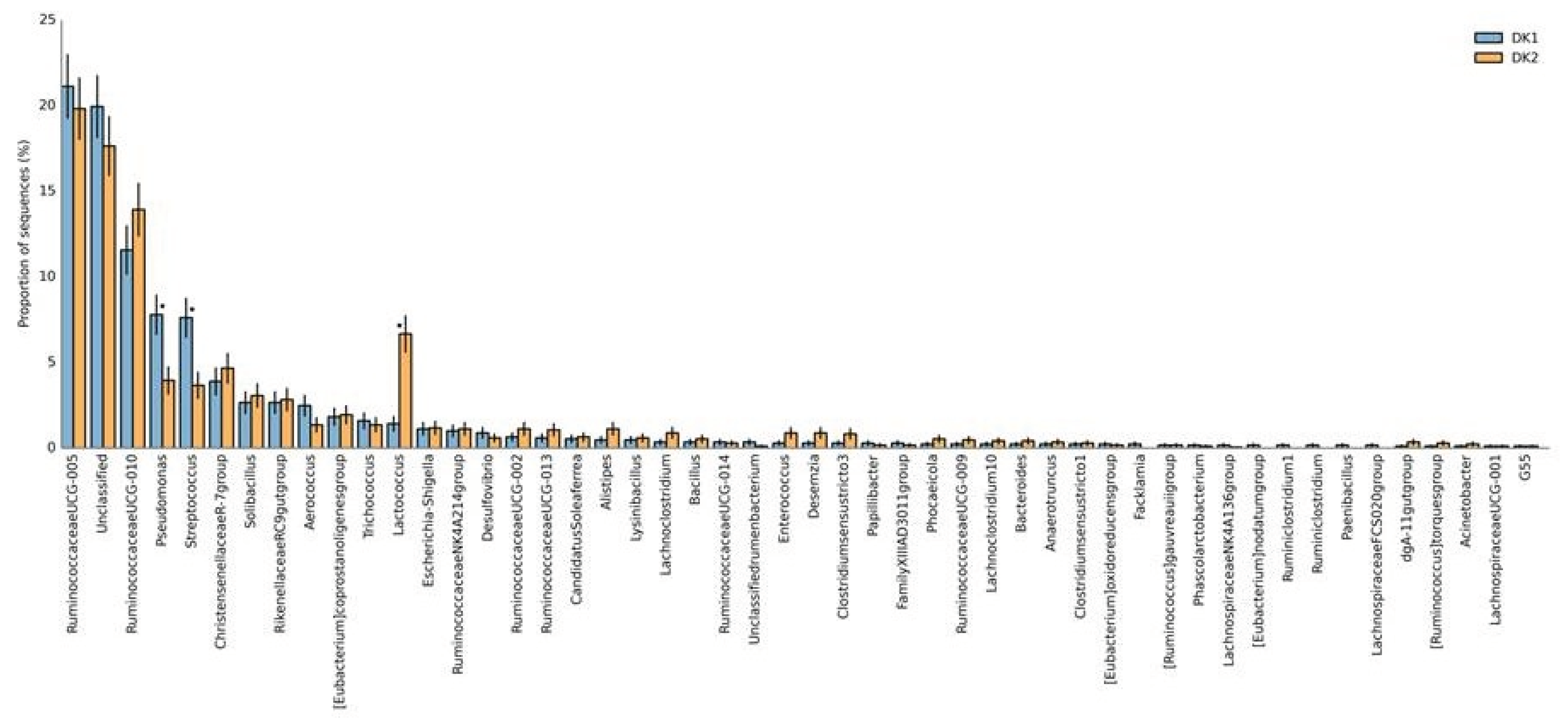
Figure 1). Further we also analysed the diversity at the deeper taxonomic level i.e. at OTU (Operational Taxonomic Unit) level. It was observed that the taxa
Ruminococcaceae UCG-005 and
Ruminococcaceae UCG-010 were the most dominant taxa (mean=17.19%) seen followed by
Streptococcus (mean=0.56%)
Christensenellaceae R-7 group (mean=0.48%),
Pseudomonas and
Lactococcus (mean=0.42% and 0.40% respectively) along with other rare taxa (See
Figure 2).
Differences in the Diversity between Pre and Post Processing
We investigated the differences in the bacterial diversity between pre- (DK1) and post- (DK2) processing samples of
panchagavya. Our analysis showed that the overall bacterial diversity underwent few compositional changes in the terms of the abundance of genera, out of which 3 genera were observed to be signiCicantly different (p<0.05, Wilcoxon signed- rank match paired t-test) between two samples. SpeciCically, the OTU belonging to
Pseudomonas and
Streptococcus (See
Figure 2) were observed to be decreased signiCicantly in the post processing samples (DK2) as compared to the post processing samples (DK1). Further it was observed that the abundance of genus
Lactococcus increases signiCicantly in the DK2 sample as compared to the DK1 sample (See
Figure 2). Although not signiCicant, however marginal differences were observed in the abundance of other genera such as
Christensenellaceae R-7 group and
Ruminococcaceae UCG-010 (See
Figure 2).
Discussion
The preliminary study of the bacterial diversity of the Panchagavya preparation revealed a highly diverse bacterial composition. Our investigation further reveals differences in the bacterial diversity in the Panchagavya preparation after processing it in the copper vessel.
We used the high throughput sequencing technology to decipher the bacterial composition of the
Panchagavya sample, which is known to be a Cive component mixtures from obtained from the cow. Our analysis revealed that the traditional Ayurvedic preparation possess a diverse group of bacterial members with few highly dominating taxa such as
Firmicutes,
Proteobacteria and
Bacteroidetes at Phylum level, while was seen dominated by taxa
Ruminococcaceae UCG-010 at lower classiCication level. Previous reports suggest that these bacterial taxa constitute the majority of the gut microbial composition of ruminant mammals [
16,
17]. As described earlier, the
Panchagavya preparation also consists of the fresh milk and curd, which are known to be probiotic in nature, as they harbour beneCicial bacteria such as
Bi9idobacterium,
Lactobacillus and
Lactococcus [
18]. Although, the former two genera were observed in less abundance, OTUs belonging to genera
Lactococcus were observed to be among one of the dominant taxa in the
Panchagavya preparation.
The comparative analysis to understand the effect of processing the fresh panchagavya preparation in the copper vessel, revealed signiCicant differences in the abundances of few speciCic bacterial genera such as Streptococcus, Pseudomanas and Lactococcus. Many of the opportunistic human pathogens are reported to belong to Streptococcus and Pseudomonas genera. The abundance of these genera was observed to be declined significantly in the post processing sample as compared to the fresh panchagavya samples. These observations can be attributed to the effect of processing in the copper vessels, however further detailed investigations need to be carried out to Cind the mechanisms of the observed dynamics.
The sample size (Biological replicates) is one of the key limitations of the study, as increase in number of replicates will conCirm these observations with higher coincidence values. However, this study serves as a preliminary ‘proof of concept’ investigation which at least provides the basic bacterial diversity proCile of the Ayurvedic ‘Panchagavya’ preparation. This study also makes an attempt to understand the effect of processing the fresh ‘Panchagavya’ sample in a copper vessel, which is a standard technique followed in Ayurveda. Though the effect is observed through the microbiome perspective, it at least provides important leads that there is a possibility of selective enrichment or reduction of speciCic bacterial members after the processing. Also, to the best of our knowledge this is the Cirst ever attempt made to understand the microbial diversity of the Panchagavya preparation, the use of which in Ayurvedic therapeutics is very well established. Hence the present study is a step towards such fundamental concepts and treatments in Ayurveda, using the modern molecular techniques.
References
- Saxena Sumit, Garg Virendra and Chauhan. Cow Urine Therapy: Promising Cure for human ailments. The Indian Cow. 2004, 1, 25–30. [Google Scholar]
- Susruta Samhita - The Medical Science of the Ancient Aryans’, Tr. Susruta Samhita - The Medical Science of the Ancient Aryans’, Tr. and Ed. A.C. Bandopadhyaya, 2nd ed. Calcutta. 1885.
- Dr.H.S.Palep, “ScientiCic Foundation of Ayurveda”. The Chaukhamba Ayurvijnana Studies (64). Chaukhamba Sanskrit Pratishthan, Delhi, 48-49,170-171, 2004.
- Jirankalgikar NM, Nariya PB, Athavale AV, De S. Trividha Snehapaka of Panchagavya Ghrita: A critical comparative evaluation. J Ayurveda Integr Med. 2013, 4, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Gajbhiye SP, Padmanabhan U, Kothari S, Patil A, Palep H, Chowdhary A Immunostimulant activity of a medical preparation panchagavya. Int J Res Pharm Sci 2014, 5, 1–5.
- Chauhan, RS. Panchagavya Therapy (Cow pathy)- Current status and future directions. Indian Cow 2004, 1, 3–7. [Google Scholar]
- Shrikant, P. Gajbhiye1, Vivek S Vishwe1, Jayashree A1, Chetan R Zope1, Sunil Mansighnka2, Abhay S. Chowdhary3 and Usha Padmanabhan. Isolation of micro-organisms from components of panchagavya. Int J of Current Advanced Research 2016, 5, 1565–1569. [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Honigsbaum, M. René Dubos, tuberculosis, and the ecological facets of virulence. History and Philosophy of the Life Sciences. 2017, 39, 15. [Google Scholar] [CrossRef]
- Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Medicine. 2016, 8, 39. [Google Scholar] [CrossRef]
- Dahiya DK, Renuka, Puniya M, Shandilya UK, Dhewa T, Kumar N, Kumar S, Puniya AK, Shukla P. Gut Microbiota Modulation and Its Relationship with Obesity Using Prebiotic Fibers and Probiotics: A Review. Front Microbiol. 2017, 8, 563. [Google Scholar] [CrossRef]
- 12. Brown J, Pirrung M, McCue LA. FQC Dashboard: integrates FastQC results into a web- based, interactive, and extensible FASTQ quality control tool. Bioinformatics. [CrossRef]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community- supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- J Gregory Caporaso, Justin Kuczynski, Jesse Stombaugh, Kyle Bittinger, Frederic D Bushman, Elizabeth K Costello, Noah Fierer, Antonio Gonzalez Pena, Julia K Goodrich, Jeffrey I Gordon, Gavin A Huttley, Scott T Kelley, Dan Knights, Jeremy E Koenig, Ruth E Ley, Catherine A Lozupone, Daniel McDonald, Brian D Muegge, Meg Pirrung, Jens Reeder, Joel R Sevinsky, Peter J Turnbaugh, William A Walters, Jeremy Widmann, Tanya Yatsunenko, Jesse Zaneveld and Rob Knight; Nature Methods, 2010. [CrossRef]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional proCiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge N, Griebel PJ, Guan le L. The Gut Microbiome and Its Potential Role in the Development and Function of Newborn Calf Gastrointestinal Tract. Front Vet Sci. 2015, 2, 36. [Google Scholar] [CrossRef]
- Ma C, Zhao J, Xi X, Ding J, Wang H, Zhang H, Kwok LY. Bovine mastitis may be associated with the deprivation of gut Lactobacillus. Benef Microbes. 2016, 7, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Ljungh A, Wadström T. Lactic acid bacteria as probiotics. Curr Issues Intest Microbiol. 2006, 7, 73–89. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).