1. Introduction
The genus
Micromonospora (phylum:
Actinobacteria) was first described by Ørskov in 1926 and currently includes 122 validly published species (
https://lpsn.dsmz.de/genus/micromonospora). These Gram-positive, aerobic and filamentous bacteria are characterized by a well-developed substrate mycelium. Bacteria of
Micromonospora genus are widespread in the environment, e.g. soil, plant internal tissues, marine sediments, desert sands, and even marine sponges, rocks, and Antarctic sands [
1,
2,
3].
Micromonospora members are regarded as rich biosources of novel bioactive compounds with a promising use in medicine due to their ability to produce antibiotics (probably the second antibiotic producer after
Streptomyces), anti-tumor substances, enzyme inhibitors, and antioxidants [
4,
5,
6]. Moreover, the biochemical properties of
Micromonospora show potential to be used in various biotechnological processes, such as nitrogen preservation in composting [
7], degradation of organic and artificial polymers [
8,
9,
10] and even in the concrete industry as a bio-healing agent [
3]. The beneficial influence of these bacteria on soil ecology and plant growth and development has recently been elucidated [
5]. Endophytic
Micromonospora isolates, successfully recovered from different parts of various plants: roots, leaves, or root nodules of legume and actinorhizal plants [
11], have been shown to be beneficial for plants with an important role as plant growth promoting bacteria (PGPB) and biocontrol agents [
6,
12]. Many of them have been recovered and investigated in the recent decade [
13,
14]. In this study, nineteen endophytic bacterial isolates originating from root nodules of white clover were recovered and classified into the genus
Micromonospora. To evaluate their potential as biocontrol agents, their antifungal properties against common phytopathogens were examined. The ability to produce bacterial metallophores and indole-3-acetic acid was tested to assess the function of isolates as PGPB.
2. Materials and Methods
2.1. Plant Material and Isolation of Micromonospora Strains
Bacteria were isolated from root nodules of wild white clover plants (
Trifolium repens L.) collected in Poland and South Africa. Until the bacteria isolation, the nodules were stored at 4 °C. Following the procedure developed by Vincent [
15], the root nodules were washed in running tap water and sterilized by soaking in a 0.1% HgCl
2 solution for 1 min and 75% EtOH for 1 min, followed by rinsing five times in sterile deionized water. Surface sterilized nodules were crushed aseptically, streaked onto 79CA agar medium, and incubated at 28 °C for 35 days. Orange to brown
Micromonospora-like colonies were transferred and cultivated on ISP2 agar [
16] and used in further analysis. Two isolates obtained from plants collected in Poland (strains 5052 and 5056) and seventeen other isolates obtained from African plants (Supplementary Material,
Table S1) were examined. All isolates are deposited in Department of Genetic and Microbiology (UMCS, Poland) microbial culture collections and preserved in 15% glycerol (v/v) at -70 °C.
2.2. Genomic DNA Extraction
Extraction of genomic DNA from the endophytic bacterial isolates was conducted with the method proposed by Pitcher et al. [
17] with modification of the bacterial cell degradation step. The 15-minute incubation of the bacterial suspension in Tris-EDTA (TE) buffer with guanidinium thiocyanate-EDTA-sarcosyl (GES) reagent was replaced by 16-hour incubation at 37 °C. Instead of GES buffer, 20 µL of 228 U/µL lysozyme and 10 µL of 10 U/µL mutanolysin were added.
2.3. Amplification of 16S rRNA Gene and Housekeeping Genes
Amplification of 16S rRNA gene fragments (about 1500 bp) was carried out by PCR using fD1d (5'-GAGAGTTTGATCCTGGCTCAGA-3') and rPla (5'-CTACGGCTACCTTGTTACGACTT-3') primers [
18]. The reaction mixture contained 12.5 μL of Taq PCR Master Mix 2x (Eurx, Poland), 0.25 μL of each primer (100 mM), 100 ng of DNA, and nuclease free sterile water up to 25 μL. The initial denaturation step of PCR was performed at 95 °C for 4 min, followed by 34 cycles of denaturation at 95 °C for 45 sec, annealing at 55 °C for 1 min, extension at 72 °C for 2 min, and final extension at 72 °C for 7 min. Housekeeping genes (
recA,
gyrB,
rpoB, and
atpD) were amplified according to previously published PCR conditions and primers [
19]. The amplicons were analyzed by electrophoresis on 1% agarose gel. Amplified gene fragments were purified with the Clean-Up Kit (A&A Biotechnology, Poland) according to the manufacturer's instructions and sequenced by Genomed S. A. Company (Warsaw, Poland). DNA sequences of amplified genes were deposited in the GenBank under the accession numbers: OQ892262-OQ892276, OQ880495-OQ880497, and OQ874508 for the 16S rRNA gene and OQ943085-OQ943103 (
recA), OQ943104-OQ943122 (
gyrB), OQ943123-OQ943141 (
rpoB), and OQ943142-OQ943160 (
atpD) for the housekeeping genes.
2.4. Phylogenetic Analysis of Sequencing Data
The most closely related sequences to the 16S rRNA,
recA,
gyrB,
rpoB, and
atpD genes of the tested isolates were obtained from the GenBank database with the Basic Local Alignment Search Tool (BLAST) algorithm [
20]. MEGA 7 software [
21] was used to construct the alignments of the 16S rRNA gene and concatenated housekeeping genes sequences. In the next step, phylogenetic trees were constructed according to the models of evolution determined by the model test (MEGA 7 software option). Maximum-likelihood algorithm evaluated by bootstrap analysis of 1000 replicates was used.
Catellatospora citrea DSM 44097 was used as an outgroup in both phylogenetic analyses.
2.5. Evaluation of Antifungal Properties
The antifungal activity of the tested bacteria was evaluated
in vitro with the agar plug diffusion test [
22] and the dual culture plate assay according to the method developed by Trujillo et al. [
1]. Five fungal plant pathogens:
Sclerotinia sclerotiorum strain 10Ss01,
Fusarium oxysporum strain 10Fo01,
Botrytis cinerea strain 10Bc01,
Verticillium albo-atrum CBS 745.83, and a wild pathogenic
Fusarium equiseti isolate were used in both methods. Phytopathogenic fungi were obtained from Institute of Horticulture in Skierniewice, Poland and University of Life Sciences in Lublin, Poland.
2.6. Agar Plug Diffusion Assay
7-day-old cultures of the tested bacteria growing on ISP2 agar were used to inoculate different types of solid culture media dedicated to investigation of actinobacterial growth and antagonism (ISP2, ISP3, ISP4, and ISP5) [
16], minimal medium [
23] (with addition of 10 g/L of mannitol instead of glucose), and yeast extract-malt extract (YEME) agar [
24]. After 11 days of incubation at 28 °C, 0.7-cm diameter agar plugs were cut with a sterile cork borer and placed on the borders of Petri dishes (four plugs per plate) with potato dextrose agar (PDA) [
25]. In the center of the PDA plates, 0.7-cm diameter plugs of the 7-day-old phytopathogenic fungal cultures were placed simultaneously and incubated at 28 °C. Plates without agar plugs with the bacteria, inoculated only with the fungal strain were considered as a negative control. The inhibition zones of fungal growth around the agar plugs indicated bacterial abilities to suppress the phytopathogenic fungi; the measurement of inhibition range was carried out after the full colonization of the control plates by the phytopathogenic fungi. Biological triplicates were done for each treatment.
2.7. Dual Culture Plate Assay
7-day-old actinobacteria cultured on ISP2 medium were suspended in sterile saline (0.85% NaCl solution) at optical density (OD
600) = 0.2. 15 µL of bacterial suspension were inoculated on the edge of Petri dishes with SA1 medium [
1] and incubated for 7 days at 28 °C. After the incubation period, 0.7-cm diameter agar plugs of the phytopathogenic fungi were placed in the center of Petri dishes and incubated at suitable temperature to the moment of the full colonization by the fungi on the control plates (fungi without bacteria). After this period, the inhibition zones of fungal growth around the bacterial colonies were measured. In the case of
V.
albo-atrum, suppression range was measured after 28 days of fungus growth. Biological triplicates were done for each tested strain.
2.8. Production of Metallophores and Indole-3-Acetic Acid
The potential to produce iron, copper, aluminum, and arsenic sequestering metallophores was tested on chrome azurol S (CAS) agar, where iron ions were substituted by respective metal ions, according to the modification described by Mehnert et al. [
26]. The ability to produce metallophores was determined with the method proposed by Rungin et al. [
27]. 0.7-cm diameter agar plugs cut from 7-day-old bacterial cultures on ISP2 medium agar were placed on CAS medium agar with addition of respective metal compounds and incubated at 28 °C for 14 days. Biological triplicates were prepared for each treatment; sterile ISP2 agar plugs were used as a negative control. An orange halo around the agar plug was considered a positive result. Indole-3-acetic acid (IAA) production by the tested isolates was assessed using a colorimetric assay [
28]. 100 µL of the bacterial suspension (OD
600=0.2) were inoculated into 5 ml of ISP2 broth amended with 0.2% L-tryptophan. After 14 days of agitation at 28 °C and 160 rpm, 1 ml of each bacterial culture was centrifuged at 11,000 rpm for 5 min, and 100 µL of the supernatant were mixed with 200 µL of Salkowski reagent [
29]. After 25 min of incubation in the dark, the samples were measured at 530 nm with a spectrophotometer. The IAA production was estimated by comparison of the obtained results with the prepared IAA standard curve (range of 10-100 μg×mL
−1 ISP2 broth). Biological triplicates were done for each treatment.
3. Results and Discussion
3.1. Isolation of Micromonospora Strains and Genomic DNA Extraction
79CA plates inoculated with aseptically crushed root nodules slurry were incubated at 28 °C for 35 days, and the newly grown, gradually appearing small orange colonies were subcultured. Genomic DNA of pure colonies, extracted with the modified Pitcher method [
17], was used for genotyping of all isolates using the BOX-PCR method. As result, nineteen various fingerprinting profiles were obtained (data not shown). Based on the BOX-PCR genotyping, nineteen different isolates were selected for genetic analysis and further tests.
3.2. Phylogenetic Analysis of 16S rRNA Gene Sequences
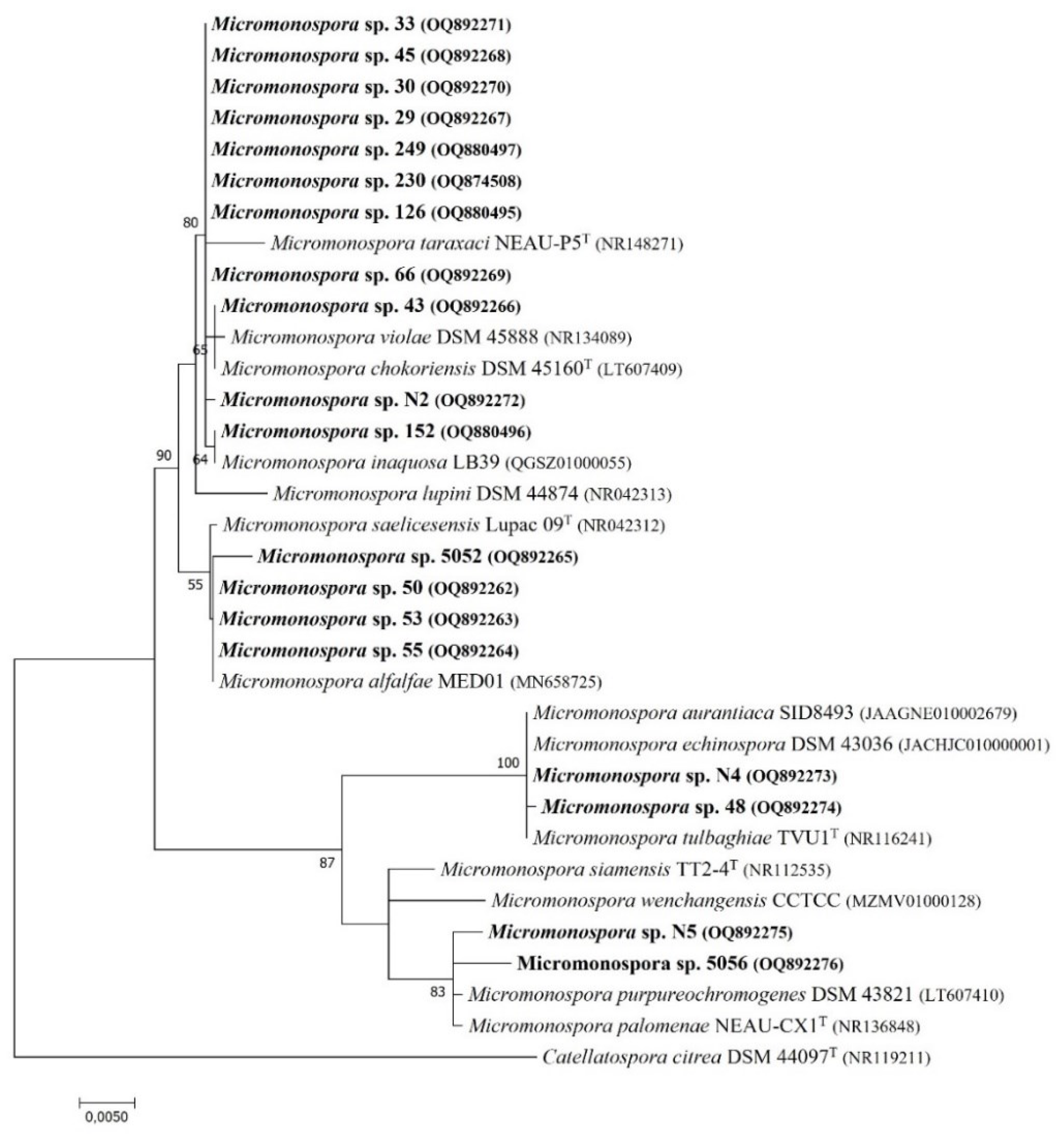
The evolutionary phylogenetic tree of the 16S rRNA gene, based on 1274 bp sequences of the tested isolates and the most closely related reference strains obtained from the GenBank database is presented (
Figure 1). The analysis showed the segregation of the taxa into two large groups comprising 15 and 4 isolates, both with two smaller subgroups. All the tested strains showed high similarities of the 16S rRNA gene sequences to various
Micromonospora species (from 99.6 to 100% similarity). These results allowed classification of the isolates to the
Micromonospora genus, but the limitation of the resolution power of this method did not allow affiliating the isolates to the species [
30,
31].
3.3. Multilocus Sequence Analysis (MLSA) of Housekeeping Genes
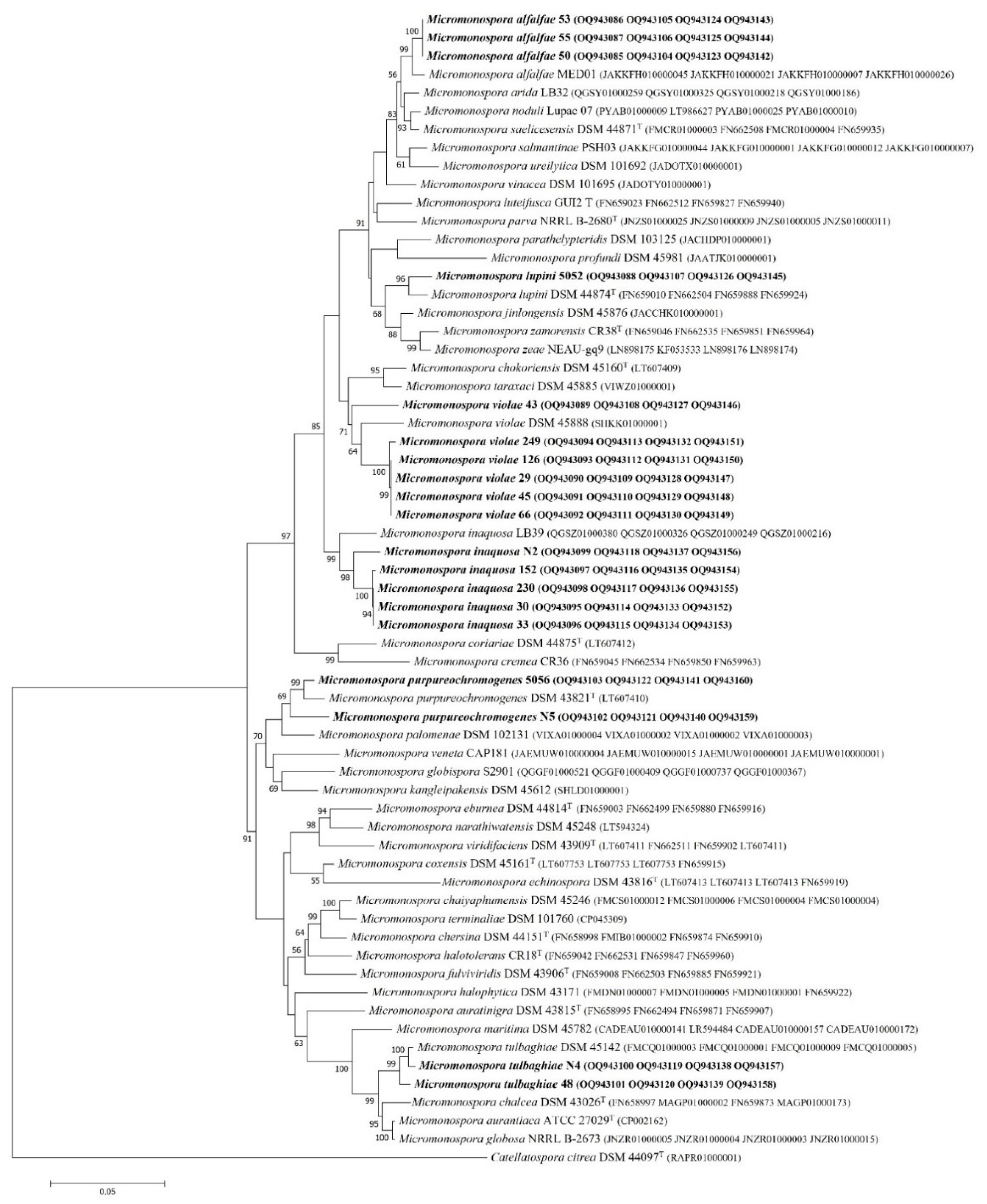
To reveal the relationship between the tested isolates and the most related reference strains at the species level, multilocus sequence analysis MLSA was carried out [
31,
33].
As a result, the maximum-likelihood phylogenetic tree of the strains examined in this study and the valid taxa based on the concatenated 2620 bp of four housekeeping gene sequence fragments is presented (
Figure 2). The tested strains are divided into two large groups (encompassing 15 and 4 strains) in a similar way as in the 16S rRNA gene phylogenetic tree. Strains 43, 249, 126, 29, 45, and 66 are grouped with the most closely related
Micromonospora violae DSM 45888 (sequence similarity 97.4%). The second phylogenetic group comprises 152, 33, N2, 230, 30, and
Micromonospora inaquosa LB39 as the most closely related reference species with the sequence similarity range from 97.3 to 97.8%.
Micromonospora purpureochromogenes DSM 43821 is the most closely related to strains 5056 and N5 (97.6-98.8% sequence similarity); strains 48 and N4 are closely related (99.1-99.5% sequence similarity) to
Micromonospora tulbaghiae DSM 45142. Strains 53, 55, and 50 show 99.1% sequence similarity to reference strain
Micromonospora alfalfae MED01. Strain 5052 exhibits 98.5% sequence similarity to type strain
Micromonospora lupini DSM 44874.
3.4. Antifungal Properties
The evaluation of endophytic
Micromonospora antifungal properties was the main object of this study. Bacterial ability to suppress phytopathogenic fungi was firstly assessed by the agar plug diffusion test with six various culture media used to bacterial growth. Despite this, suppression of plant pathogens was not observed. In the second step, the dual culture plates assay was carried out and recognized as appropriate tool to detect antifungal properties of tested endophytic strains. The suppression range of phytopathogenic fungi was measured after 4 days of incubation (
B.
cinerea and
S.
sclerotiorum), 6 days of incubation (
F.
oxysporum and
F.
equiseti) and 28 days of incubation (
V.
albo-atrum). The antimicrobial effect against phytopathogenic fungi was observed (
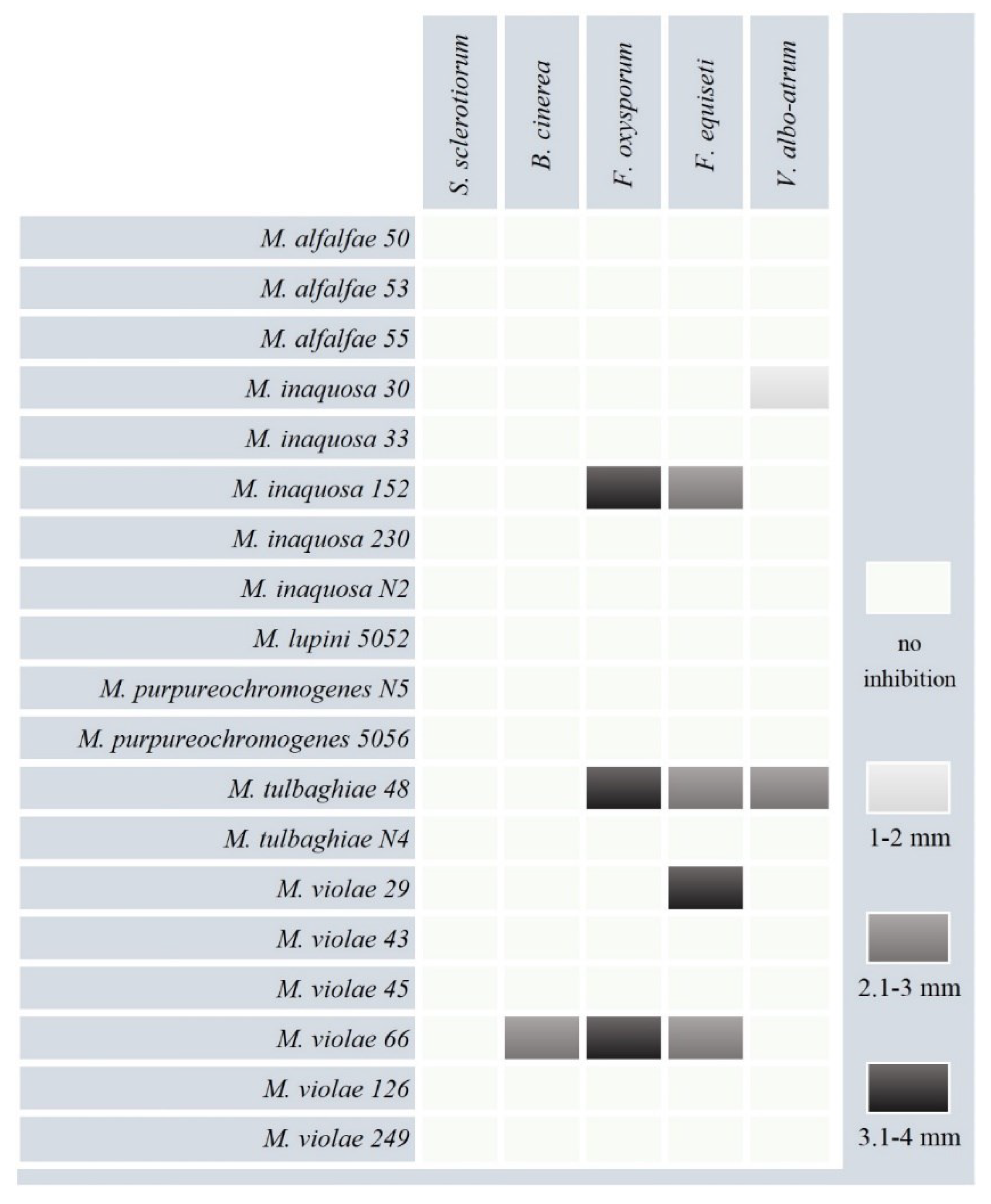
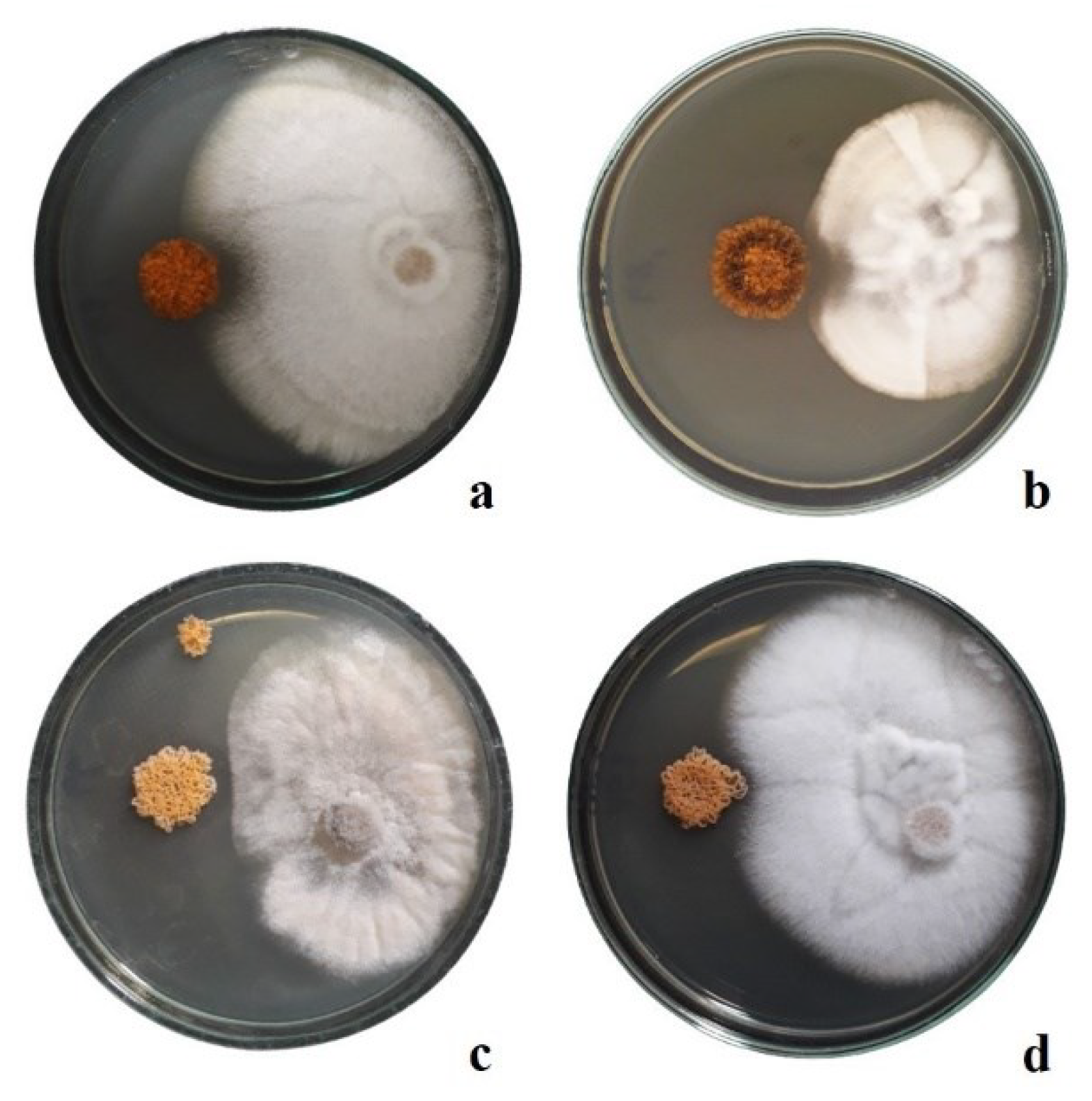
Figure 3).
Five strains (~26% of total) indicate ability to suppress phytopathogenic fungi; the diameter of inhibition zone range from 1 mm to 4 mm.
Micromonospora tulbaghiae 48 showed the antagonism against
F.
oxysporum,
F.
equiseti (
Figure 4a) and
V.
albo-atrum (
Figure 4b).
Micromonospora violae 66 was able to suppress three fungal pathogen (
B.
cinerea –
Figure 4c,
F.
oxysporum and
F.
equiseti);
Micromonospora inaquosa 152 indicate antifungal properties against
F.
oxysporum and
F.
equiseti (
Figure 4d);
Micromonospora inaquosa 30 showed the antimicrobial activity against
V.
albo-atrum. Strain
Micromonospora violae 29 was able to inhibit growth of
F.
equiseti. Members of the
Micromonospora genus are considered as great candidates for novel bioactive compounds producers, including secondary metabolites [
35]. The results confirm previous reports, where
Micromonospora strains had antagonistic activity against other microorganisms, including fungi [
36,
37,
38].
However, in our study the antifungal activity was observed only in the dual culture assay, where SA1 agar was used as a growth medium. It is worth emphasizing that the production of antimicrobial compounds by actinobacteria is highly dependent on growth conditions, including carbon and nitrogen sources [
39], and various strains of
Micromonospora may require differently composed media to produce antimicrobials [
40]. The SA1 agar was the only accurate medium to indicate the antimicrobial activity of the tested strains. Similarly, SA1 agar was also successfully used by Martínez-Hidalgo et al. [
41], who evaluated the antimicrobial activity of
Micromonospora strains against fungal plant pathogens, including
Sclerotinia sclerotiorum and
Botrytis cinerea. As reported by Amin et al. [
39], the addition of glucose (the main carbon source in SA1 medium) resulted in an increase in the antimicrobial agent yields in the case of
Micromonospora. In turn, Mark et al. [
42] reported antimicrobial activity of
Micromonospora bacteria with properties inducible also by culturing on International Streptomyces Project (ISP) media, which proved to be unhelpful in our investigations.
3.5. Production of Metallophores
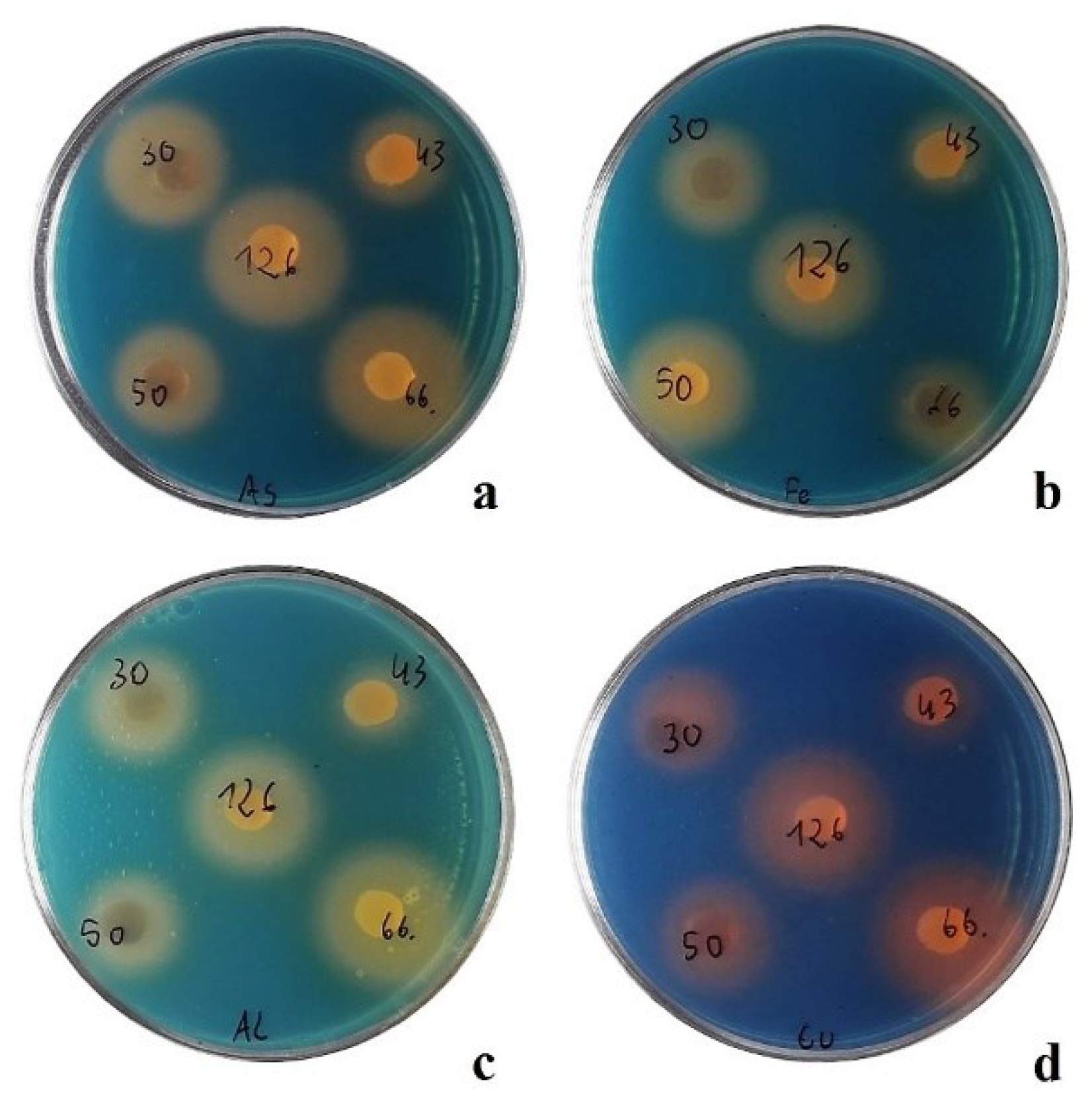
Orange halos around agar plugs (
Figure 5) were observed in the samples with all the nineteen tested strains. This indicate their ability to produce metallophores and sequester ions of iron (Fe), copper (Cu), aluminum (Al), and arsenic (As). The halo zone diameter ranged from 16 mm to 40 mm (
Figure 6). The largest siderophore diffusion zones (over 30 mm for three different metal ions added to the agar) were observed for the strains
M.
inaquosa 152,
M.
violae 126,
M.
violae 66, and
M.
violae 45.
The smallest zones were observed for strains
M.
violae 43,
M.
tulbaghiae 48,
M.
tulbaghiae N4 and
Micromonospora purpureochromogenes N5 (under 20 mm for three of four tested metals). The halo zones on the As-CAS medium had the largest diameters. Due to the high toxicity of hexadecyltrimethylammonium bromide (HDTMA), it was not possible to grow the tested actinomycetes on CAS agar. Metallophores, i.e. various ion-chelating molecules, facilitate metal uptake as nutrient elements and are useful as metal stress management agents. This study reported that all the tested
Micromonospora strains exhibit the ability to produce various metallophores. The same conclusion was presented by previous
in vitro and
in vivo studies conducted by Ortúzar et al. [
43]. Moreover, the genome mining of the complete genome sequence of
Micromonospora craniellae LHW63014T [
44] showed biosynthetic gene clusters indicating the ability to produce ion-chelating agents.
3.6. Production of Indole-3-Acetic Acid
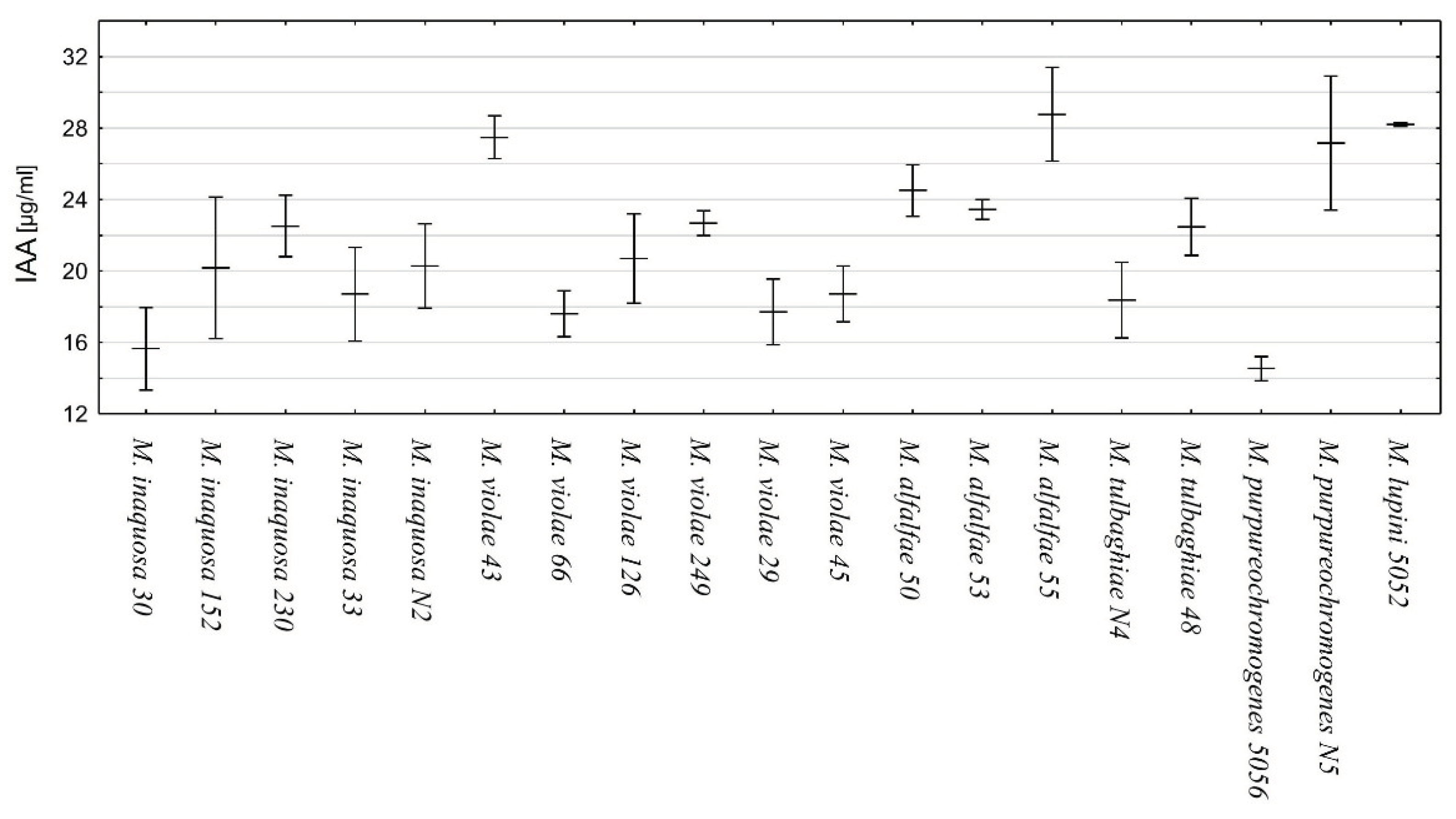
The spectrophotometric comparison of the standard curve of IAA and that of the samples of bacterial supernatant and Salkowsky reagent indicated the ability of all the studied strains of
Micromonospora to produce IAA as a result of tryptophan metabolism (
Figure 7).
M.
alfalfae strain 55,
M.
purpureochromogenes strain N5,
M.
lupini strain 5052, and
M.
violae strain 43 showed the highest productivity (above 27 µg/ml). The lowest amount of IAA was detected in the case of
M.
purpureochromogenes strain 5056.
Production of phytohormones plays a significant role in plant growth promotion by plant beneficial bacteria [
45]. As demonstrated by the widely used spectrophotometric measurement of IAA production (despite the limitation of this method described by Goswami et al. [
46]), numerous
Micromonospora isolates have been reported as auxin-producing PGPB [
47,
48,
49].
4. Conclusions
Nineteen endophytic Micromonospora strains isolated from the root nodules of white clover (Trifolium repens L.) were classified as M. inaquosa, M. violae, M. alfalfae, M. tulbaghiae, M. purpureochromogenes, and M. lupini. The antifungal properties and two mechanisms of plant growth promotion (production of metallophores and auxin) by these strains were assessed. The strains M. tulbaghiae 48, M. inaquosa 30, M. inaquosa 152, M. violae 66, and M. violae 29 showed antimicrobial properties against phytopathogenic fungi, especially against Fusarium equiseti and Fusarium oxysporum. Further comprehensive analyzes of these phenomena are necessary, especially the characterization of antifungal bacterial metabolites. Moreover, strains with the ability to produce metallophores and auxin are candidates for use in agriculture to promote plant growth, which should be confirmed by in planta studies. The most promising strains seem to be M. alfalfae 55 and M. lupini 5052. The results indicate the in vitro potential of selected strains as biocontrol factors and plant growth promotion.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Table S1: Micromonospora strains used in this study.
Author Contributions
[W.S.], [S.W.-W.], [M.M.-K.], [M.K.] participated in the conceptualization, design, and modification of methods, data analysis, and manuscript writing. Moreover, the biological assays and data collection were performed by [W.S.]. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Department of Genetics and Microbiology and the Doctoral School of Quantitative and Natural Sciences, University of Maria Curie-Skłodowska in Lublin.
Data Availability Statement
The gene sequences data presented in this study are available in GenBank repository at
https://www.ncbi.nlm.nih.gov/genbank, reference numbers of sequences are cited in the main text.
Acknowledgments
The authors thank Joanna Puławska (Institute of Horticulture - National Research Institute, Skierniewice, Poland) and Marek Kopacki (University of Life Sciences, Lublin, Poland) for sending the fungal phytopathogens used in this study.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Trujillo, M.E.; Fernández-Molinero, C.; Velázquez, E.; Kroppensted, R.M.; Schumann, P.; Mateos, P.F.; Martínez-Molina, E. Micromonospora mirobrigensis sp. nov. Int. J. Syst. Evol. 2005, 55, 877–880. [Google Scholar] [CrossRef] [PubMed]
- Kuncharoen, N.; Kudo, T.; Ohkuma, M.; Tanasupawat, S. Micromonospora azadirachtae sp. nov., isolated from roots of Azadirachta indica A. Juss. var. siamensis Valeton. Antonie van Leeuwenhoek 2019, 112, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Kara Ali, M.; Ait Kaki, A.; Wahiba, K.; Bramki, A.; Benchabbi, A.; Kacem Chaouche, N. First study of Micromonospora echinospora isolation from a rocky site of Eastern Algeria and first report of its potential use in cementitious materials biohealing. SAJEB 2022, 12, 800–810. [Google Scholar] [CrossRef]
- Zhao, H.; Kassama, Y.; Young, M.; Kell, D.; Goodacre, R. Differentiation of Micromonospora isolates from a coastal sediment in Wales on the basis of fourier transform infrared spectroscopy, 16S rRNA sequence analysis, and the amplified fragment length polymorphism technique. AEM 2004, 70, 6619–6627. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.M.; Valdés, M. Micromonospora: an important microbe for biomedicine and potentially for biocontrol and biofuels. Soil Biol. Biochem. 2010, 42, 536–542. [Google Scholar] [CrossRef]
- Ran, Y.; Zhang, Y.; Wang, X.; Li, G. Nematicidal metabolites from the actinomycete Micromonospora sp. WH06. Microorganisms 2022, 10, 2274. [Google Scholar] [CrossRef] [PubMed]
- Kumas, A.; Ertekin, S.G.; Gurbanov, R.; Şimşek, Y.E.; Kocak, F.O.; Değirmenci, L. Effect of Micromonospora sp. KSC08 on nitrogen conservation throughout composting. Biomass Convers. 2023, 13, 2375–2390. [Google Scholar] [CrossRef]
- Chen, S.J.; Lam, M.Q.; Thevarajoo, S.; Abd Manan, F.; Yahya, A.; Chong, S.C. Genome analysis of cellulose and hemicellulose degrading Micromonospora sp. CP22. 3 Biotech 2020, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Pavitra, R.; Raja, A. Optimization of conditions (influence of shaking, static and pH) for biodecolourization of reactive azo-based textile dye by Micromonospora sp. Appl. Ecol. Environ. Sci. 2020, 8, 282–286. [Google Scholar]
- Schneider, B.; Pfeiffer, F.; Dyall-Smith, M.; Kunte, H. Genome sequence of Micromonospora aurantiaca strain G9, a member of a bacterial consortium capable of polyethylene degradation. Microbiol. resour. announc. 2022, 11, e01148–21. [Google Scholar] [CrossRef]
- Carro, L.; Pujic, P.; Trujillo, M.E.; Normand, P. Micromonospora is a normal occupant of actinorhizal nodules. J. Biosci. 2013, 38, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Prashith Kekuda, T.R.; Shobha, K.S.; Onkarappa, R. Fascinating diversity and potent biological activities of actinomycete metabolites. J. Pharm. Res. 2010, 3, 250–256. [Google Scholar]
- Ay, H.; Nouioui, N.; Klenk, H.P.; Cetin, D.; Igual, J.M.; Sahin, N.; Isik, K. Genome-based classification of Micromonospora craterilacus sp. nov., a novel actinobacterium isolated from Nemrut Lake. Antonie van Leeuwenhoek 2020, 113, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Riesco, R.; Ortúzar, M.; Fernández-Ábalos, J.M.; Trujillo, M.E. Deciphering genomes: genetic signatures of plant-associated Micromonospora. Front. Plant Sci. 2022, 13, 872356. [Google Scholar] [CrossRef]
- Vincent, J.M. A manual for the practical study of root-nodule bacteria; Blackwell Scientific Publishers, Oxford, 1970.
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species1. Int. J. Syst. Evol. 1966, 16, 313–340. [Google Scholar]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Parker, M.A. Relationships of bradyrhizobia from the legumes Apios americana and Desmodium glutinosum. AEM 1999, 65, 4914–4920. [Google Scholar] [CrossRef]
- Carro, L. Avances en la sistemática del género Micromonospora: estudio de cepas aisladas de la rizosfera y nódulos de Pisum sativum. Doctoral Thesis, University of Salamanca, Salamanca, 2011. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda Koraichi, S. Methods for in vitro evaluating antimicrobial activity: a review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Hopwood, D.A. Genetic analysis and genome structure in Streptomyces coelicolor. Bacteriol. Rev. 1967, 31, 373–403. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics, Innes, Norwich, 2000.
- Beever, R.E.; Bollard, E.G. The nature of the stimulation of fungal growth by potato extract. Microbiology 1970, 60, 273–279. [Google Scholar] [CrossRef]
- Mehnert, M.; Retamal-Morales, G.; Schwabe, R.; Vater, S.; Heine, T.; Levicán, G.J.; Schlömann, M.; Tischler, D. Revisiting the Chrome Azurol S assay for various metal ions. Solid State Phenom. 2017, 262, 509–512. [Google Scholar] [CrossRef]
- Rungin, S.; Indananda, C.; Suttiviriya, P.; Kruasuwan, W.; Jaemsaeng, R.; Thamchaipenet, A. Plant growth enhancing effects by a siderophore–producing endophytic streptomycete isolated from a Thai jasmine rice plant (Oryza sativa L. Cv. KDML105). Antonie van Leeuwenhoek 2012, 102, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Gangwar, M.; Rani, S.; Sharma, N. Diversity of endophytic actinomycetes from wheat and its potential as plant growth promoting and biocontrol agents. J. adv. lab. 2012, 3, 13–19. [Google Scholar]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rosselló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, S.P.; Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Ferraz Helene, C.L.; Klepa, M.S.; Hungria, M. New insights into the taxonomy of bacteria in the genomic era and a case study with rhizobia. Int. J. Microbiol. 2022, 2022, 4623713. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular evolution and phylogenetics. Oxford University Press, New York, 2000.
- Carro, L.; Golinska, P.; Nouioui, I.; Bull, A.T.; Igual, J.M.; Andrews, B.A.; Klenk, H.P.; Goodfellow, M. Micromonospora acroterricola sp. nov., a novel actinobacterium isolated from a high altitude Atacama Desert soil. Int. J. Syst. Evol. 2019, 69, 3426–3436. [Google Scholar] [CrossRef] [PubMed]
- El-Tarabily, K.A.; Nassar, A.H.; Hardy, G.E.; Sivasithamparam, K. Plant growth promotion and biological control of Pythium aphanidermatum, a pathogen of cucumber, by endophytic actinomycetes. J. Appl. Microbiol. 2009, 106, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, M.; Duarah, A.; Talukdar, S.; Gohain, M.B.; Debnath, R.; Yadav, A.; Jha, D.K.; Bora, T.C. Bioprospecting Micromonospora from Kaziranga National Park of India and their anti–infective potential. World J. Microbiol. Biotechnol. 2012, 28, 2703–2712. [Google Scholar] [CrossRef]
- Igarashi, Y.; Matsuyuki, Y.; Yamada, M.; Fujihara, N.; Harunari, E.; Oku, N. Structure determination, biosynthetic origin, and total synthesis of akazaoxime, an enteromycin–class metabolite from a marine–derived actinomycete of the genus Micromonospora. J. Org. Chem. 2021, 86, 6528–6537. [Google Scholar] [CrossRef] [PubMed]
- Amin, D.H.; Abolmaaty, A.; Tolba, S.; Abdallah, N.A.; Wellington, E.M.H. Phylogenic characteristics of a unique antagonistic Micromonospora sp. Rc5 to S. aureus isolated from Sinai Desert of Egypt. Annu. Res. Rev. Biol. 2018, 22, 1–15. [Google Scholar] [CrossRef]
- Sineva, O.N.; Bychkova, O.P.; Terekhova, L.P. Acidotolerant actinomycetes of the genus Micromonospora are producers of antibiotic compounds. In Proceedings of the first International Electronic Conference on Antibiotics, on-line. 8–17 May 2021. [Google Scholar]
- Martínez–Hidalgo, P.; García, J.M.; Pozo, M.J. Induced systemic resistance against Botrytis cinerea by Micromonospora strains isolated from root nodules. Front. Microbiol. 2015, 6, 922. [Google Scholar] [CrossRef]
- Mark, D.; DeWald, J.; Dorador, C.; Tucker, N.; Herron, P. Characterisation of anti–pseudomonad activity of hyper–arid Micromonospora species. Access Microbiol. 2023, 2, 7A. [Google Scholar] [CrossRef]
- Ortúzar, M.; Trujillo, M.E.; Román-Ponce, B.; Carro, L. Micromonospora metallophores: a plant growth promotion trait useful for bacterial–assisted phytoremediation? Sci. Total Environ. 2020, 739, 139850. [Google Scholar] [CrossRef]
- Yang, X.; Zhu, Z.-H.; Ji, X.; Liu, Z.-M.; Zhang, H.; Wei, B. Complete genome sequence of Micromonospora craniellae LHW63014T, a potential metal ion–chelating agent producer. Mar. Genom. 2021, 57, 100830. [Google Scholar] [CrossRef]
- Glick, B.R. Introduction to plant growth–promoting bacteria. In Beneficial plant–bacterial interactions, Glick, B.R., Ed.; Springer International Publishing, Cham, 2020; pp. 1–37.
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Simultaneous detection and quantification of indole–3–acetic acid (IAA) and indole–3–butyric acid (IBA) produced by rhizobacteria from L–tryptophan (Trp) using HPTLC. J. Microbiol. Methods. 2015, 110, 7–14. [Google Scholar] [CrossRef]
- Shutsrirung, A.; Chromkaew, Y.; Pathom-Aree, W.; Choonluchanon, S.; Boonkerd, N. Diversity of endophytic actinomycetes in mandarin grown in northern Thailand, their phytohormone production potential and plant growth promoting activity. Soil Sci. Plant Nutr. 2013, 59, 322–330. [Google Scholar] [CrossRef]
- Martínez–Hidalgo, P.; Galindo-Villardón, P.; Trujillo, M.E.; Igual, J.M.; Martínez-Molina, E. Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.). a new promising plant probiotic bacteria. Sci. Rep. 2014, 4, 6389. [Google Scholar] [CrossRef] [PubMed]
- Della Mónica, I.F.; Novas, M.V.; Iannone, L.J.; Querejeta, G.; Scervino, J.M.; Pitta-Alvarez, S.I.; Regalado, J.J. Infection with Micromonospora strain SB3 promotes in vitro growth of Lolium multiflorum plantlets. PCTOC 2018, 134, 445–455. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).