Submitted:
26 March 2024
Posted:
27 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. Morphological Features
2.3. Molecular Methods
2.4. Sequence Alignments and Phylogenetic Analysis
3. Results
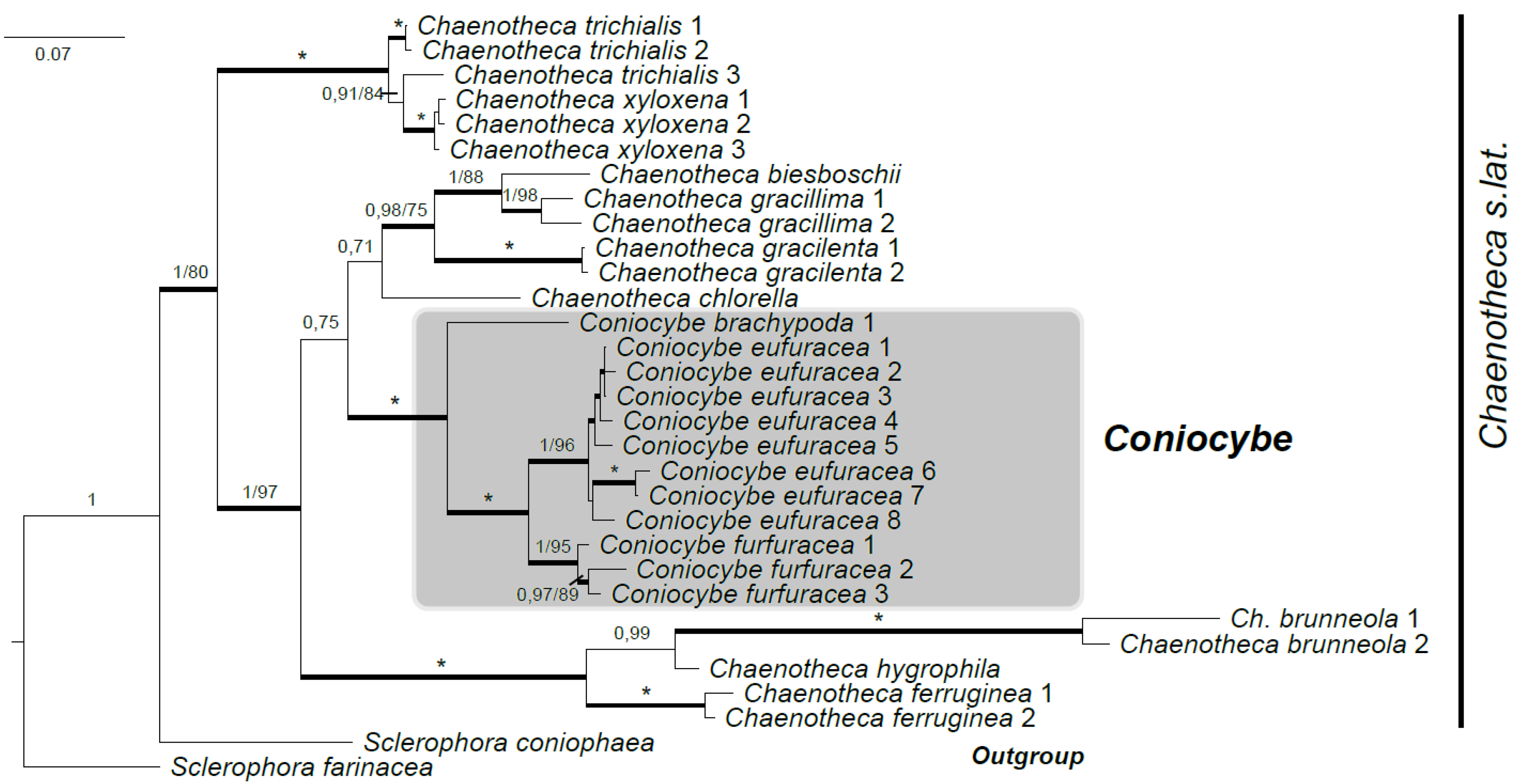
3.1. Phylogeny of Chaenotheca s. lat.
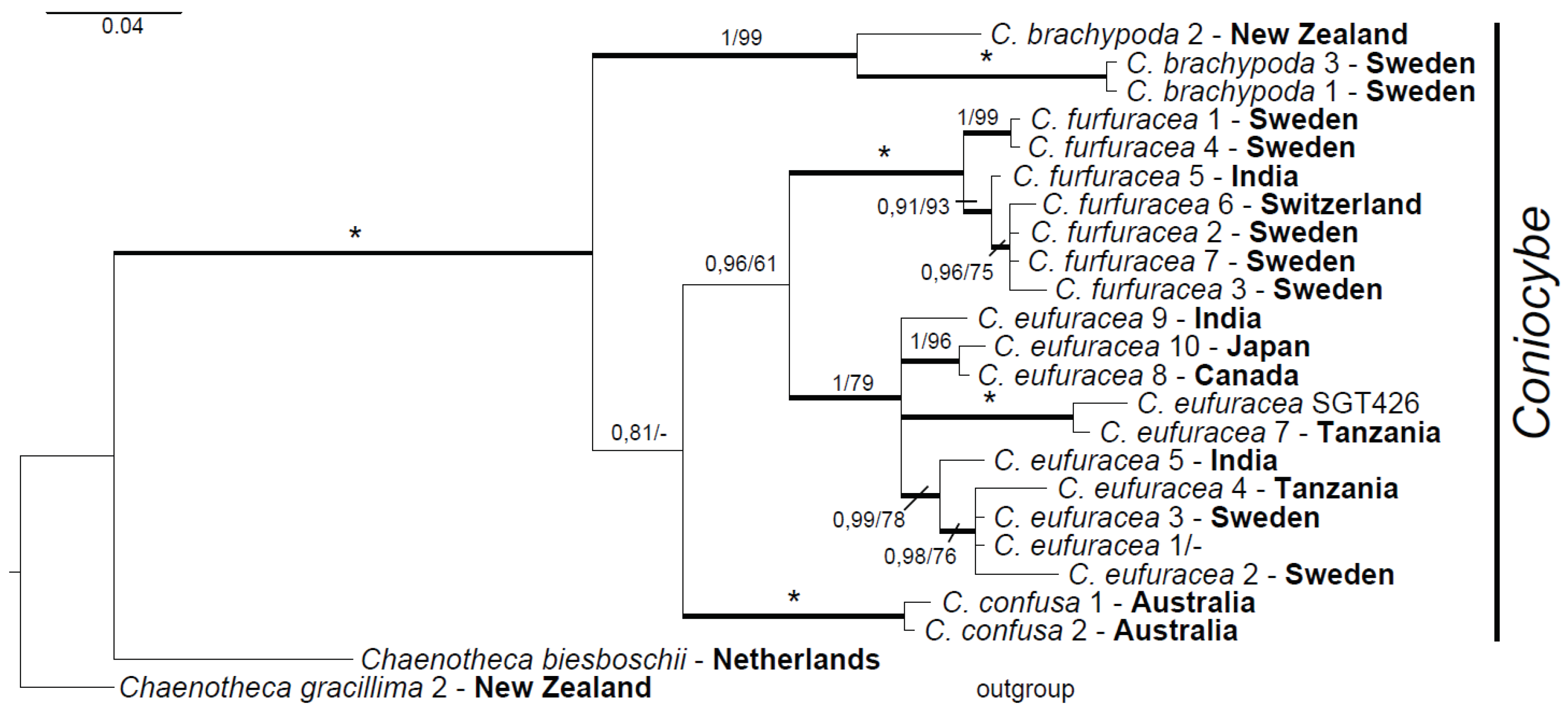
3.2. Phylogeny of Coniocybe
3.3. Taxonomy

- 1.1. Apothecia 0.4 – 1.4 mm high, mazaedium medium brown..............................................................................C. brachypoda
- 1.2. Apothecia 0.6 – 3.0 mm high high, mazedium pale brown..................................................................................................2
- 2.1. Spore surface with short, irregular ridges and cracks visible under the light microscope…………….……...C. confusa
- 2.2. Spores with reticulate ridges, but without cracks ……………………………...……………………………..…….………3
- 3.1. ITS1 diagnostic sequence: CTTCT; ITS2 diagnostic sequence: TGCAGC ……………………….….….……..C. eufuracea
- 3.2. ITS1 diagnostic sequence: TCGTGC; ITS2 diagnostic sequence: TGTAGT…………………...………….......C. furfuracea
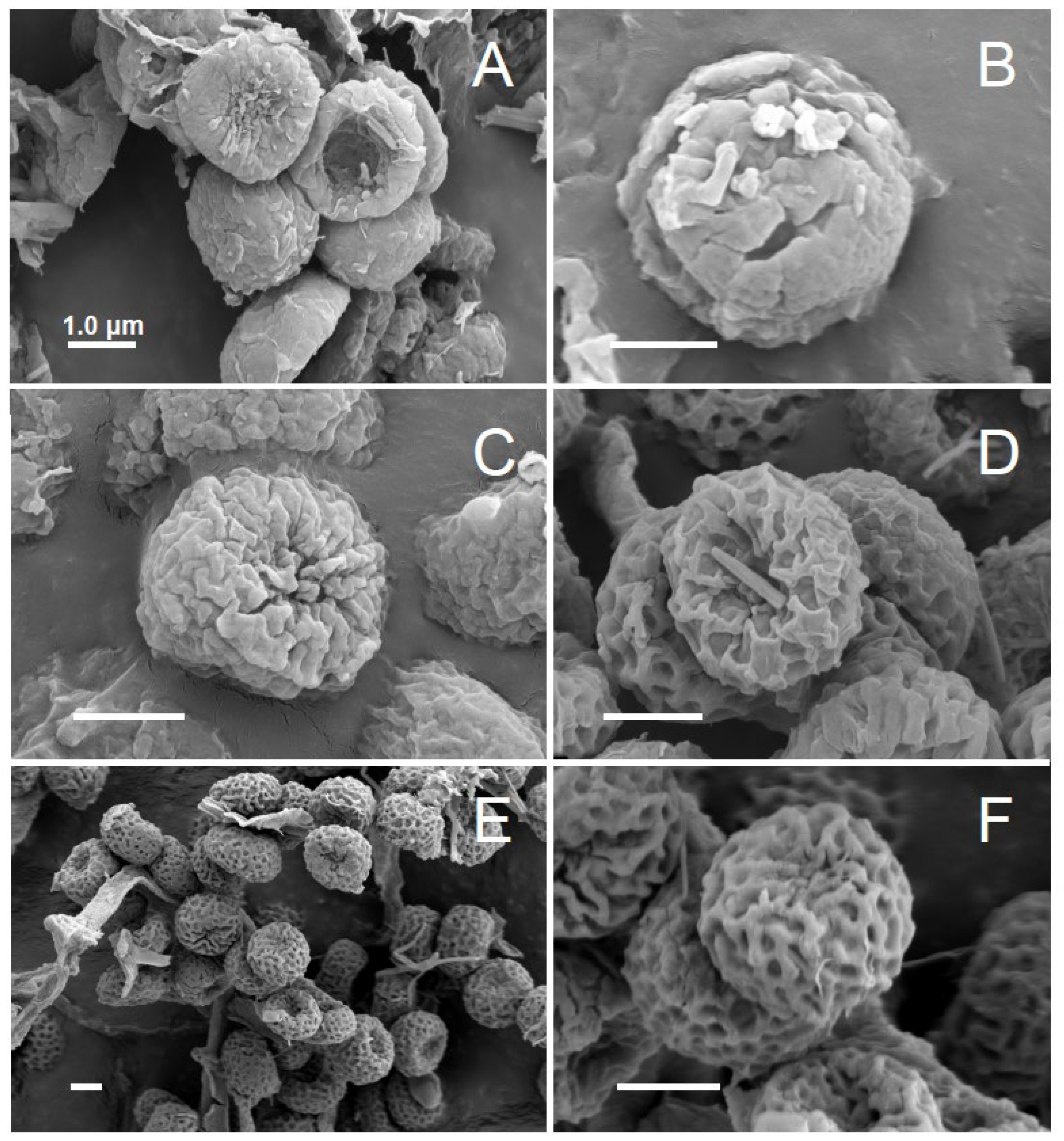
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Acharius, E. Afhandling om de cryptogamiske vexter, som komma under namn af Calicioidea. –1. K. Vetensk. -Akad. Handl. 1815, 246–271. 2. K. Vetensk. -Akad. Handl. 1816, 260–291. 3. K. Vetensk. -Akad. Handl. 1817, 220–244.
- Vainio, E. Lichenographia fennica. III. Acta Soc. Fauna Fl. Fenn. 1927, 57: 1: 1–138.
- Keissler, K. von. Pyrenulaceae bis Mycoporaceae. Coniocarpineae. In Rabenhorst’s Kryptogamen-Flora von Deutschland, Österreich und der Schweiz. 9. 1:2: 1-846 Leipzig. 1936–1938.
- Nádvorník, J. Übersicht der mitteleuropäischen Arten der Flechtenfamilie Caliciaceae. Stud. Bot. Čech. 1942a, 5: 6–46.
- Nádvorník, J. Kurze Übersicht der Flechtenfamilie Caliciaceae. Stud. Bot. Čech. 1942a, 5: 121–128.
- Schmidt, A. Anatomisch-taxonomische Untersuchungen an europäischen Arten der Flechtenfamilie Caliciaceae. Mitt. Staatsinst. Allg. Bot. Hamburg 1970, 13: 111–166.
- Tibell, L. A reappraisal of the taxonomy of Caliciales. Nova Hedwig. 1984, Beiheft 79: 597–713.
- Tibell, L.; Wedin, M. Mycocaliciales, a new order for nonlichenized calicioid fungi. Mycologia 2000, 92: 577–581. [CrossRef]
- Prieto, M.; Baloch, E.; Tehler, A.; Wedin, M. Mazaedium evolution in the Ascomycota (Fungi) and the classification of mazaediate groups of unclear relationship. Cladistics 2013, 29: 1–13. [CrossRef]
- Lücking, R.; Hodkinson, B.P.; Leavitt, S.D. The 2016 classification of lichenized fungi in the Ascomycota and Basidiomycota – Approaching one thousand genera. Bryologist 2016, 119: 361−416. [CrossRef]
- Lumbsch, H.T.; Huhndorf, S.M. Myconet Volume 14. Part One. Outline of Ascomycota—2009. Part Two. Notes on Ascomycete Systematics. Nos. 4751– 5113. Fieldiana Life and Earth Sciences 2010; 1−64.
- Tibell, L. The lichen genus Chaenotheca in the Northern Hemisphere. Symb. Bot. Upsal. 1980, 23(1): 1−65.
- Tibell, L. Typification of names of infrageneric taxa described by Acharius and placed by him in Caliciales. Ann. Bot. Fenn. 1987, 24: 257−280.
- Tibell, L. Australasian Caliciales., Symb. Bot. Upsal. 1987, 27(1): 1–279.
- Fries, E. Lichenographia Europaea reformata. Praemittuntur lichenologiae fundamenta... Conscripsit Elias Fries... Typis Berlingianis. 1831.
- Fries, T.M. Lichenes arctoi Europae Groenlandiaeque hactenus cogniti (Vol. 3). 1860, Leffler.
- Zahlbruckner, A.V. (1926). Lichenes (Flechten). B. Spezieller Teil. Die naturlichen Pflanzenfamilien. Engelmann, Leipzig 61−263.
- Tibell, L. Crustose mazaediate lichens and the Mycocaliciaceae in temperate-South America. Bibl. Lichenol. 1998, 71: 1−107. [CrossRef]
- Tibell, L.; Tibell, S.; Van Der Pluijm, A. Chaenotheca biesboschii a new calicioid lichen from willow forests in the Netherlands. Lichenologist 2019, 51: 123–135. [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2: 113–118. [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: New York, NY, USA, 1990; Volume 18, pp. 315–322.
- Vilgalys, R.; Hester, M. Rapid Genetic Identification and Mapping of Enzymatically Amplified Ribosomal DNA from Several Cryptococcus Species. J. Bacteriol. 1990, 172: 4238–4246. [CrossRef]
- Matheny, P.B.; Liu, Y.J.; Ammirati, J.F.; Hall, B.D. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am. J. Bot. 2002, 89: 688–698. [CrossRef]
- Larsson, A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30: 3276–3278. [CrossRef]
- Posada, D.; Crandall, K.A. Modeltest: testing the model of DNA substitution. Bioinformatics 1998, 14: 817–818. [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27: 171−180. [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 2012, 61: 539– 542. [CrossRef]
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post- analysis of large phylogenies. Bioinformatics 2014, 30: 1312–1313. [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42: 182–192. [CrossRef]
- Rambaut, A.; Drummond, A.J. FigTree v1. 3.1. Institute of Evolutionary Biology; University of Edinburgh: Edinburgh, UK, 2010.
- Jørgensen, P.M.; James, P.W.; Jarvis, C.E. Linnaean lichen names and their typification. Biol. J. Linn. Soc. 1994, 115: 261−405. [CrossRef]
| Species | Isolation | Country | Voucher | GB Acc. No | ||
| ITS | LSU | rpb1 | ||||
|
Chaenotheca biesboschii |
L380 | Netherlands | A.v.d.Pluijm3244 (UPS) | MK514539 | XXX | XXX |
| Ch. brunneola 1 | T076 | Sweden | Tibell22202 (UPS) | AF297964 | XXX | ⸺ |
| Ch. brunneola 2 | T193 | Estonia | TU<EST>: 76415 | KX348127 | XXX | ⸺ |
| Ch. chlorella | T061 | Sweden | Tibell22186 | AF297965 | XXX | ⸺ |
| Ch. ferruginea 1 | T099 | Sweden | Tibell22276 (UPS) | MK514541 | XXX | ⸺ |
| Ch. ferruginea 2 | DF82/ T835 | Switzerland | WSL: DF82 | KX098349 | XXX | ⸺ |
| Ch. gracilenta 1 | T055 | Sweden | Tibell22197 (UPS) | AF410675 | XXX | XXX |
| Ch. gracilenta 2 | T135/ T310 | Sweden | Thor (hb. Thor) | AF410676 | XXX | ⸺ |
| Ch. gracillima 1 | T037 | Sweden | Tibell17052 (UPS) | AF298127 | XXX | XXX |
| Ch. gracillima 2 | T107 | New Zealand | Tibell16725 (UPS) | AF408682 | XXX | XXX |
| Ch. hygrophila | T024 | Japan | Thor 15612 (UPS) | AF298129 | XXX | ⸺ |
| Ch. trichialis 2 | UPSC: 2297/ T038 |
Sweden | Tibell16878 (UPS) | AF298139 | KF157985 | XXX |
| Ch. trichialis 1 | ⸺ | ⸺ | Prieto3028 (S) | JX000102 | JX000085 | JX000136 |
| Ch. trichialis 3 | T129 | Sweden | Tibell22300 (UPS) | AF421203 | XXX | XXX |
| Ch. xyloxena 1 | T066 | Sweden | Tibell22188 (UPS) | AF298140 | XXX | XXX |
| Ch. xyloxena 3 | T181/T131 | Sweden | Tibell22329 (UPS | AF421212 | XXX | XXX |
| Ch. xyloxena 2 | T103 | Sweden | Tibell22171 (UPS) | AF421210 | XXX | XXX |
|
Coniocybe brachypoda 3 |
T030 | Sweden | Tibell17062 (UPS) /UPSC2446 |
AF297962 | ⸺ | ⸺ |
| C. brachypoda 1 | T060/ Prieto3023 |
Sweden | Tibell22193(UPS) /Prieto3023 (S) |
AF297963 | JX000086.1 | JX000135 |
| C. brachypoda 2 | T027 | New Zealand | UPSC2070; Tibell16627 | XXX | ⸺ | ⸺ |
| C. confusa 1 | C1 | Australia | Kantvilas280/19(Ho) | XXX | ⸺ | ⸺ |
| C. confusa 2 | C2 | Australia | Kantvilas280 /19 (Ho) |
XXX | ⸺ | ⸺ |
| C. eufuracea 10 | T036 | Japan | Thor15698 | AF298124 | ⸺ | ⸺ |
| C. eufuracea 3 | T081/T062 | Sweden | Tibell22190 (UPS) | AF298125 | XXX | XXX |
| C. eufuracea 1 | ⸺ | ⸺ | Wedin6366 (UPS) | NR120128_1 | JX000087 | JX000137 |
| C. eufuracea 8 | T439 | Canada | Coffman387 | XXX | XXX | ⸺ |
| C. eufuracea 4 | SGT422 | Tanzania | Temu422 | XXX | XXX | XXX |
| C. eufuracea 2 | SGT443 | Sweden | Temu443 | XXX | XXX | XXX |
| C. eufuracea 5 | T901 | India | Tibell25106 | XXX | XXX | XXX |
| C. eufuracea 9 | T355 | India | Tibell23224 | XXX | ⸺ | ⸺ |
| C. eufuracea 6 | SGT426 | Tanzania | Temu426 | XXX | XXX | ⸺ |
| C. eufuracea 7 | SGT431 | Tanzania | Temu431 | XXX | XXX | XXX |
| C. furfuracea 1 | SGT442 | Sweden | Temu442 | XXX | XXX | ⸺ |
| C. furfuracea 2 | T046 | Sweden | Tibell21829 | XXX | XXX | ⸺ |
| C. furfuracea 3 | T198 | Sweden | Tibell22299 | XXX | XXX | ⸺ |
| C. furfuracea 4 | T155 | Sweden | Tibell22364 (UPS) | AF445357 | ⸺ | ⸺ |
| C. furfuracea 5 | T092 | India | Tibell21874 | XXX | ⸺ | ⸺ |
| C. furfuracea 6 | WSL:DF252 | Switzerland | WSL:DF252 | KX098351_1 | ⸺ | ⸺ |
| C. furfuracea 7 | T199 | Sweden | Tibell22307b | XXX | ⸺ | ⸺ |
| Sclerophora coniophaea | ⸺ | ⸺ | Wedin6367 (UPS) | ⸺ | JX000094 | JX000145 |
| S. farinacea | ⸺ | Estonia | Wedin6414 (UPS) | JX000113 | JX000095 | JX000144 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





