Submitted:
27 March 2024
Posted:
28 March 2024
You are already at the latest version
Abstract
Keywords:
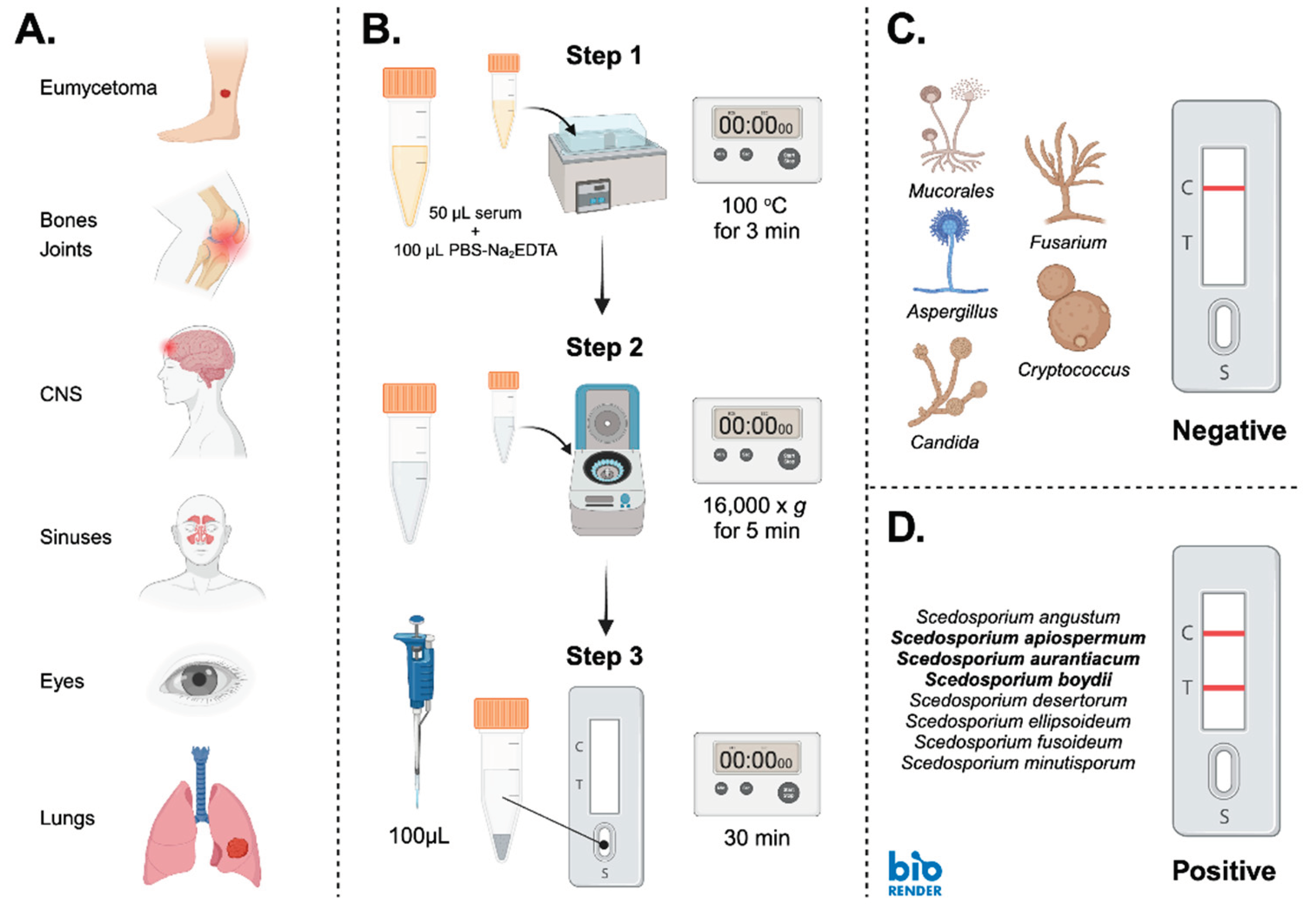
1. Introduction
2. Materials and Methods
2.1. Monoclonal Antibody
2.2. Fungal Culture
2.3. Antibody Purification and Enzyme Conjugation
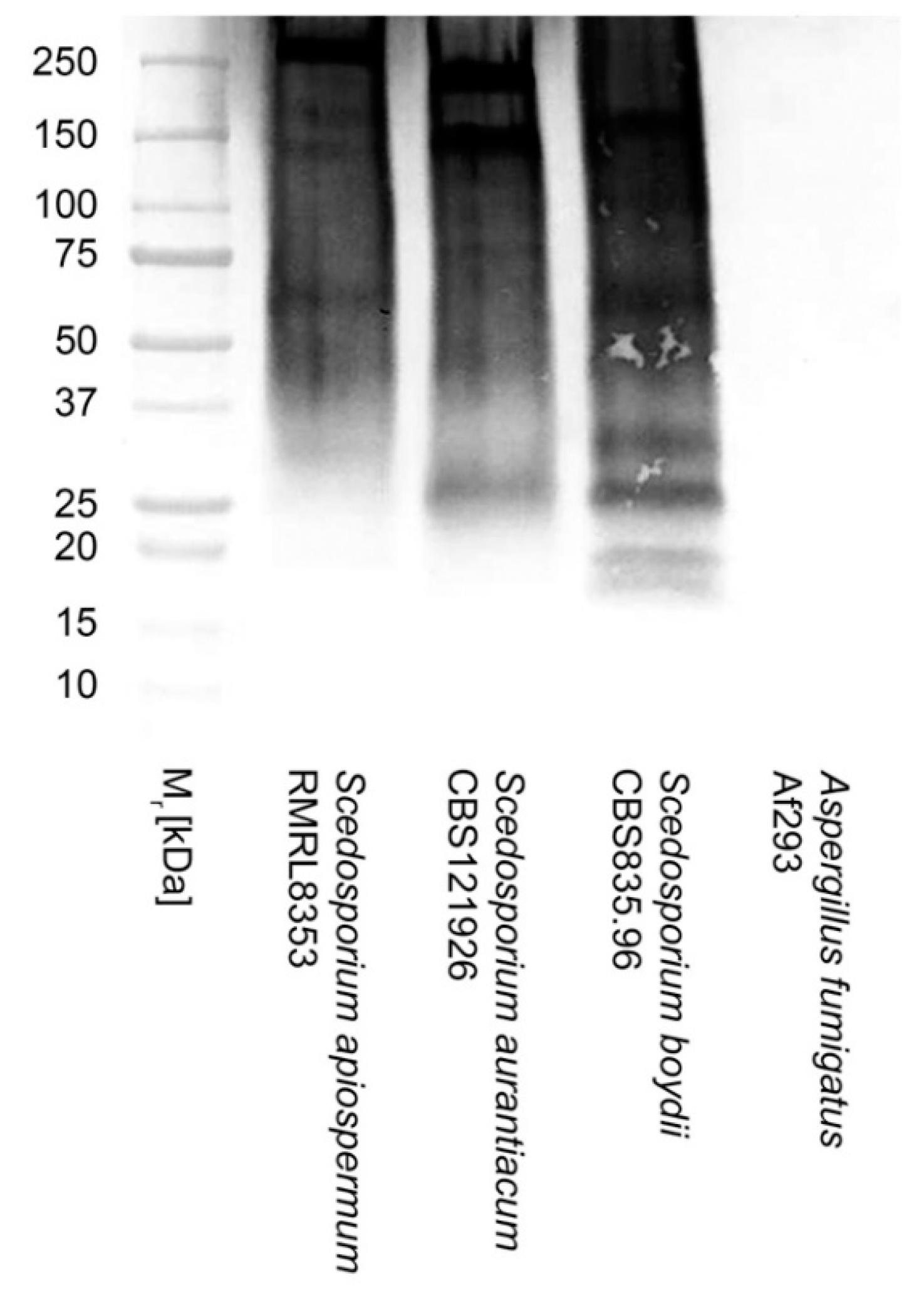
2.4. Polyacrylamide Gel Electrophoresis and Western Blotting
2.5. LFD and ELISA Specificities
2.5.1. Lateral-Flow Device
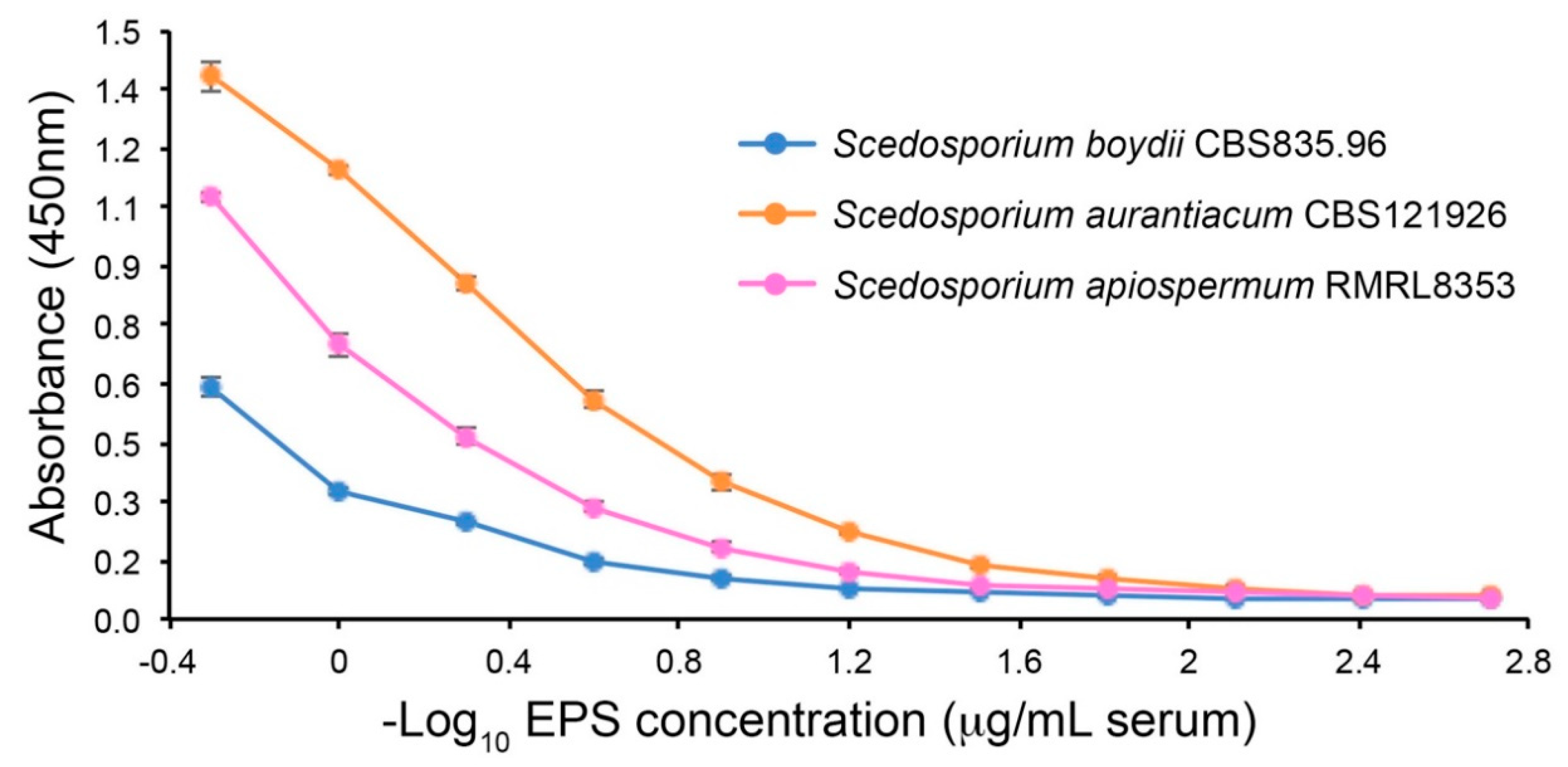
2.5.2. Sandwich Enzyme-Linked Immunosorbent Assay
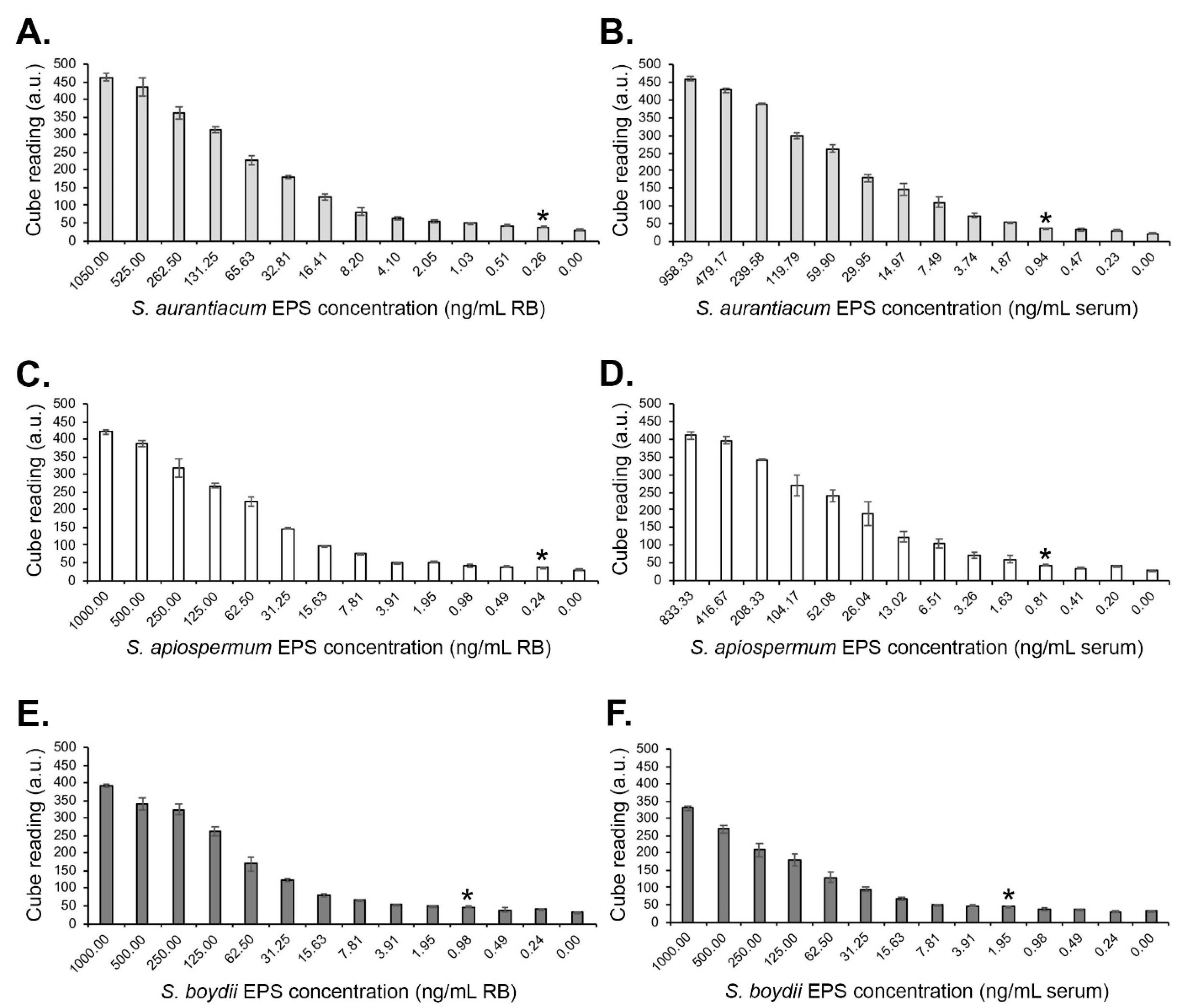
2.6. Serological Detection and Limits of Detection
2.7. Statistical Analysis
3. Results
3.1. Lateral-Flow Device and ELISA
3.1.1. Specificities
3.1.2. Limits of Detection with Human Serum
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gulati, V. , Bakare, S, Tibrewal, S., Ismail, N., Sayani, J., Baghla, D.P.S. A rare presentation of concurrent Scedosporium apiospermum and Madurella grisea eumycetoma in an immunocompetent host. Case Rep. Pathol. 2012, 2012, 154201. [Google Scholar] [PubMed]
- Gupta, M.K., Banerjee, T, Kumar, D., Rastogi, A., Tilak, R. White grain mycetoma caused by Scedosporium apiospermum in north India: as case report. Int. J. Low. Extrem. Wounds. 2013, 12, 286–288.
- Oliveira, F.d.M.; Unis, G.; Hochhegger, B.; Severo, L.C. Scedosporium apiospermum eumycetoma successfully treated with oral voriconazole: report of a case and review of the Brazilian reports on scedosporiosis. Rev. Inst. Med. Trop. Sao Paulo 2013, 55, 121–123. [CrossRef]
- Tóth, E.J.; Nagy, G.R.; Homa, M.; Ábrók, M.; Kiss, I. .; Nagy, G.; Bata-Csörgő, Z.; Kemény, L.; Urbán, E.; Vágvölgyi, C.; et al. Recurrent Scedosporium apiospermum mycetoma successfully treated by surgical excision and terbinafine treatment: a case report and review of the literature. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 31. [Google Scholar] [CrossRef] [PubMed]
- Emery, D.; Denning, D.W. The global distribution of actinomycetoma and eumycetoma. PLOS Neglected Trop. Dis. 2020, 14, e0008397. [Google Scholar] [CrossRef] [PubMed]
- Cortez, K.J., Roilides, E, Quiroz-Telles, F., Meletiadis, J., Antachopoulos, C., Knudsen, T., Buchanan, W., Milanovich, J., Sutton, D.A., Fothergill, A., et al. Infections caused by Scedosporium spp. Clin. Microbiol. Rev. 2008, 21, 157–197.
- Ramirez-Garcia, A.; Pellon, A.; Rementeria, A.; Buldain, I.; Barreto-Bergter, E.; Rollin-Pinheiro, R.; de Meirelles, J.V.; Xisto, M.I.D.S.; Ranque, S.; Havlicek, V.; et al. Scedosporium and Lomentospora: an updated overview of underrated opportunists. Med Mycol. 2018, 56, S102–S125. [Google Scholar] [CrossRef] [PubMed]
- Bouchara, J.-P. , Papon, N. Scedosporium apiospermum. Trends Microbiol. 2019, 27, 12. [Google Scholar]
- Seidel, D. , Meißner, A. , Lackner, M., Piepenbrock, E., Salmanton-García, J., Stecher, M., Mellinghoff, S., Hamprecht, A., Graeff, L.D., Köhler, P., et al. Prognostic factors in 264 adults with invasive Scedosporium spp. and Lomentospora prolificans infection reported in the literature and FungiScope. Crit. Rev. Microbiol. 2019, 45, 1–21. [Google Scholar]
- Bronnimann, D.; Garcia-Hermoso, D.; Dromer, F.; Lanternier, F.; Maulin, L.; Leprince, Y.; Brieu, N.; Gruson, B.; El-Samad, Y.; Chouaki, T.; et al. Scedosporiosis/lomentosporiosis observational study (SOS): Clinical significance of Scedosporium species identification. Med Mycol. 2020, 59, 486–497. [Google Scholar] [CrossRef]
- Koehler, P. , Tacke, D. , Cornely, O.A. Bone and joint infections by Mucorales, Scedosporium, Fusarium and even rarer fungi. Crit. Rev. Microbiol. 2016, 42, 158–171. [Google Scholar]
- Mello, T.P.; Bittencourt, V.C.B.; Liporagi-Lopes, L.C.; Aor, A.C.; Branquinha, M.H.; Santos, A.L. Insights into the social life and obscure side of Scedosporium/Lomentospora species: ubiquitous, emerging and multidrug-resistant opportunistic pathogens. Fungal Biol. Rev. 2018, 33, 16–46. [Google Scholar] [CrossRef]
- Shi, X.-W.; Li, S.-T.; Lou, J.-P.; Xu, B.; Wang, J.; Wang, X.; Liu, H.; Li, S.-K.; Zhen, P.; Zhang, T. Scedosporium apiospermum infection of the lumbar vertebrae: A case report. World J. Clin. Cases 2022, 10, 3251–3260. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Utsumi, Y.; Suzuki, N.; Nakajima, Y.; Murata, O.; Sasaki, N.; Nitanai, H.; Nagashima, H.; Miyamoto, S.; Yaegashi, J.; et al. Multiple Scedosporium apiospermum abscesses in a woman survivor of a tsunami in northeastern Japan: a case report. J. Med Case Rep. 2011, 5, 526–526. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y. , Suzuki, N. , Nakajima, Y., Utsumi, Y., Murata, O., Nagashima, H., Saito, H., Sasaki, N., Fujimura, I., Ogino, Y., et al. Scedosporium auarantiacum brain abscess after near-drowning in a survivor of a tsunami in Japan. Respir. Investig. 2013, 51, 207–211. [Google Scholar]
- Miceli, M.H. Central Nervous System Infections Due to Aspergillus and Other Hyaline Molds. J. Fungi 2019, 5, 79. [Google Scholar] [CrossRef] [PubMed]
- Lauerer, R.J.; Rosenow, E.; Beschorner, R.; Hempel, J.-M.; Naros, G.; Hofmann, A.; Berger, K.; Sartor-Pfeiffer, J.; Mengel, A.; Ziemann, U.; et al. Rapid Diagnosis of Central Nervous System Scedosporiosis by Specific Quantitative Polymerase Chain Reaction Applied to Formalin-Fixed, Paraffin-Embedded Tissue. J. Fungi 2021, 8, 19. [Google Scholar] [CrossRef]
- zkan, A. , Susever, S. , Erturan, Z., Uzun, M., Alparslan, N., Öz, Y., Yeenoglu, Y. A case of keratitis caused by Scedosporium apiospermum. J. Microbiol. Infect. Dis. 2013, 3, 45–48. [Google Scholar]
- Hayashi, Y.; Eguchi, H.; Toibana, T.; Mitamura, Y.; Yaguchi, T. Polymicrobial Sclerokeratitis Caused by Scedosporium apiospermum and Aspergillus cibarius. Cornea 2014, 33, 875–877. [Google Scholar] [CrossRef]
- Palanisamy, M. , Venkatapathy, N. , Rajendram, V., Shobana, C.S. Keratomycosis caused by Graphium eumorphum (Graphium state of Scedosporium apiospermum). J. Clin. Diagn. Res. 2015, 9, DD03–DD04. [Google Scholar]
- Baskaran, P.; Ramakrishnan, S.; Mandlik, K.; Sathe, T.S.; Gubert, J.; Krishnan, T. Ocular infections caused by Scedosporium apiospermum: A case series. Indian J. Ophthalmol. 2018, 66, 137. [Google Scholar] [CrossRef]
- Carnovale, S.; Epelbaum, C.; Abrantes, R.; Córdoba, S.; Cabrera, C.; Caracciolo, B. Scedosporium aurantiacum: First isolation in Argentina from a previously healthy patient after traumatic inoculation. 2022, 54, 318–321. [CrossRef]
- Karaca, U. Scedosporium apiospermum keratitis: a case report. J. Med. Case Rep. 2022, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Ali, D.; Vijayan, A.; Shenoy, K.; Antony, A.T.; Ramachandran, R. Challenges in the diagnosis and management of atypical fungal keratitis during the COVID-19 pandemic: a case series. Access Microbiol. 2023, 5, 000570–v3. [Google Scholar] [CrossRef] [PubMed]
- Motokawa, N.; Miyazaki, T.; Hara, A.; Fukuda, Y.; Morino, S.; Nakamura, H.; Iwasaki, K.; Soda, H.; Izumikawa, K.; Yanagihara, K.; et al. Pulmonary Scedosporium apiospermum Infection with Pulmonary Tumorlet in an Immunocompetent Patient. Intern. Med. 2018, 57, 3485–3490. [Google Scholar] [CrossRef] [PubMed]
- Liu, W. , Feng, R. , Jiang, H. Scedosporium spp. lung infection in immunocompetent patients. A systematic review and MOOSE-compliant meta-analysis. Medicine 2019, 98, 41. [Google Scholar]
- Liu, W.; Feng, R.; Jiang, H. Management of pulmonary Scedosporium apiospermum infection by thoracoscopic surgery in an immunocompetent woman. J. Int. Med Res. 2020, 48. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.; Nachbaur, E.; Jaksch, P.; Dehlink, E. Update on Respiratory Fungal Infections in Cystic Fibrosis Lung Disease and after Lung Transplantation. J. Fungi 2020, 6, 381. [Google Scholar] [CrossRef]
- Sav, H.; Altinbas, R.; Dursun, Z.B. A fatal invasive Scedosporium apiospermum pulmonary infection in an adult patient with malignant lung adenocarcinoma. Curr. Med Mycol. 2020, 6, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Ghasemian, R.; Bandegani, A.; Kermani, F.; Faeli, L.; Roohi, B.; YousefiAbdolmaleki, E.; Hedayati, M.T.; Roilides, E.; Shokohi, T. Fatal pulmonary Scedosporium aurantiacum infection in a patient after near-drowning: A case report. Curr. Med Mycol. 2022, 7, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Ibáñez-Martínez, E.; Solé, A.; Cañada-Martínez, A.; Muñoz-Núñez, C.F.; Pastor, A.; Montull, B.; Falomir-Salcedo, P.; Valentín, A.; López-Hontangas, J.L.; Pemán, J. Invasive scedosporiosis in lung transplant recipients: A nine-year retrospective study in a tertiary care hospital. 2021, 38, 184–187. [CrossRef]
- Mizusawa, M.; Totten, M.; Zhang, S.X. A Case of Scedosporium aurantiacum Infection in the United States. Mycopathologia 2021, 186, 127–130. [Google Scholar] [CrossRef]
- Puerta-Alcalde, P. , Garcia-Vidal, C. Non-Aspergillus mould lung infections. Eur. Respir. Rev. 2022, 31, 220104. [Google Scholar]
- Kishimoto, I.; Shinohara, S.; Ueda, T.; Tani, S.; Yoshimura, H.; Imai, Y. Orbital apex syndrome secondary to a fungal nasal septal abscess caused by Scedosporium apiospermum in a patient with uncontrolled diabetes: a case report. BMC Infect. Dis. 2017, 17, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Loh, U.L.; Tai, P.Y.; Hussein, A.; A Qamarruddin, F. Scedosporium apiospermum: A Rare Cause of Aggressive Orbital Apex Syndrome. Cureus 2018, 10, e3743. [Google Scholar] [CrossRef] [PubMed]
- Singla, S.; Singh, V.; Bansal, A.; Sharma, J.P.; Wadhwa, T.; Sarma, S. Post COVID-19 acute invasive fungal rhinosinusitis caused by Scedosporium apiospermum: a covert pathogen. Int. J. Otorhinolaryngol. Head Neck Surg. 2021, 7, 1187–1192. [Google Scholar] [CrossRef]
- K, S.; B, P.; M, P.; M, A.; V, L. A case of bilateral injection abscesses caused by Graphium type of Scedosporium apiospermum. Med Mycol. Case Rep. 2022, 37, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.-F.; Huang, S.-M.; Xie, L.; Zhang, Y.-Y.; Tang, Y.-R.; Wang, X.-Z.; Pan, &. S.-F.; Ast; Huang, &.S.-M.; Lian Xie, Yuan-Yuan Zhang, Yu-Rong Tang, Xiao-Zhen Wang Department of Clinical Laboratory, Shengli Oilfield Central Hospital, Dongying, People’s Republic of China& et al. A Case of Invasive Fungal Infection Due to Scedosporium apiospermum in a Patient with Psoriasis. Infect. Drug Resist. 2023, ume 16, 5085–5090. [Google Scholar] [CrossRef]
- Pieta, A.; Venetsanopoulou, A.I.; Kittas, C.; Christaki, E.; Voulgari, P.V. Recurrent Scedosporium apiospermum Cutaneous Infection in a Patient with Rheumatoid Arthritis: The Potent Role of IL-6 Signaling Pathway Blockade: A Case-Based Review. J. Fungi 2023, 9, 683. [Google Scholar] [CrossRef]
- Hirschi, S.; Letscher-Bru, V.; Pottecher, J.; Lannes, B.; Jeung, M.Y.; Degot, T.; Santelmo, N.; Sabou, A.M.; Herbrecht, R.; Kessler, R. Disseminated Trichosporon mycotoxinivorans, Aspergillus fumigatus, and Scedosporium apiospermum Coinfection after Lung and Liver Transplantation in a Cystic Fibrosis Patient. J. Clin. Microbiol. 2012, 50, 4168–4170. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, C.; Brandt, C.; Melichar, V.; Runge, C.; Heuer, E.; Sahly, H.; Schebek, M.; Köster, H.; Bouchara, J.-P.; Biedermann, T.; et al. Combined antifungal therapy is superior to monotherapy in pulmonary scedosporiosis in cystic fibrosis. J. Cyst. Fibros. 2019, 18, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Viñado, C. , Girón, R. M., Ilbáñez, E., García-Ortega, A., Pérez, I., Polanco, D., Pemán, J., Solé, A. Filamentous fungi in the airway of patients with cystic fibrosis: just spectators? Rev. Iberoam. Micol. 2012, 38, 168–174. [Google Scholar]
- Marr, K.A.; Carter, R.A.; Crippa, F.; Wald, A.; Corey, L. Epidemiology and Outcome of Mould Infections in Hematopoietic Stem Cell Transplant Recipients. Clin. Infect. Dis. 2002, 34, 909–917. [Google Scholar] [CrossRef]
- Kubak, B.M.; Huprikar, S.S. ; the AST Infectious Diseases Community of Practice Emerging & Rare Fungal Infections in Solid Organ Transplant Recipients. Am. J. Transplant. 2009, 9, S208–S226. [Google Scholar] [CrossRef]
- Rahi, M.S.; Jindal, V.; Pednekar, P.; Parekh, J.; Gunasekaran, K.; Sharma, S.; Stender, M.; Jaiyesimi, I.A. Fungal infections in hematopoietic stem-cell transplant patients: a review of epidemiology, diagnosis, and management. Ther. Adv. Infect. Dis. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Bourlond, B.; Cipriano, A.; Regamey, J.; Papadimitriou-Olivgeris, M.; Kamani, C.; Seidel, D.; Lamoth, F.; Muller, O.; Yerly, P. Case report: Disseminated Scedosporium apiospermum infection with invasive right atrial mass in a heart transplant patient. Front. Cardiovasc. Med. 2022, 9, 1045353. [Google Scholar] [CrossRef]
- Zeng, C.; Ma, Y.-S.; Zhou, J.-Y.; Xue, C.-B.; Xiong, Y.; Zhou, W.; Zhou, L.-H.; Li, J.-G.; Ye, S.-J.; Ye, Q.-F. Donor-Derived Transmission of Scedosporiosis in Kidney Transplant Recipients from a Systemic lupus erythematosus donor. Curr. Med Sci. 2023, 43, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, L. , Buonomo, A. R., Iacovazzo, C., Giaccone, A., Scotto, R., Viceconte, G., Mercinelli, S., Vargas, M., Roscetto, E., Cacciatore, F., et al. Invasive fungal infections in hospitalised patients with COVID-19: a non-intensive care single-centre experience during the first pandemic. J. Fungi 2023, 9, 86. [Google Scholar]
- Lee, M.-G.; Choi, J.-G.; Son, B.-C. Scedosporium apiospermum: An emerging fatal cause of fungal abscess and ventriculitis after near-drowning. Asian J. Neurosurg. 2018, 13, 792–796. [Google Scholar] [CrossRef]
- Mavrouli, M.; Mavroulis, S.; Lekkas, E.; Tsakris, A. Respiratory Infections Following Earthquake-Induced Tsunamis: Transmission Risk Factors and Lessons Learned for Disaster Risk Management. Int. J. Environ. Res. Public Heal. 2021, 18, 4952. [Google Scholar] [CrossRef]
- Warkentien, T.; Rodriguez, C.; Lloyd, B.; Wells, J.; Weintrob, A.; Dunne, J.R.; Ganesan, A.; Li, P.; Bradley, W.; Gaskins, L.J.; et al. Invasive Mold Infections Following Combat-related Injuries. Clin. Infect. Dis. 2012, 55, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Angelini, A.; Drago, G.; Ruggieri, P. Post-tsunami primary Scedosporium apiospermum osteomyelitis of the knee in an immunocompetent patient. Int. J. Infect. Dis. 2013, 17, e646–e649. [Google Scholar] [CrossRef]
- Tribble, D.R. , Rodriguez, C. J. Combat-related fungal invasive wound infections. Curr. Fungal Infect. Rep. 2014, 8, 277–286. [Google Scholar]
- Kronen, R.; Liang, S.Y.; Bochicchio, G.; Bochicchio, K.; Powderly, W.G.; Spec, A. Invasive Fungal Infections Secondary to Traumatic Injury. Int. J. Infect. Dis. 2017, 62, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, A.; Shaikh, F.; Bradley, W.; Blyth, D.M.; Bennett, D.; Petfield, J.L.; Carson, M.L.; Wells, J.M.; Tribble, D.R. ; Infectious Disease Clinical Research Program Trauma Infectious Disease Outcomes Study Group Classification of Trauma-Associated Invasive Fungal Infections to Support Wound Treatment Decisions. Emerg. Infect. Dis. 2019, 25, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- WHO (2022). WHO fungal priority pathogens list to guide research, development and public health action (Geneva: World Health Organization). License: CC BY-NC-SA 3.0 IGO.
- Delhaes, L. , Harun, A. , Chen, S.C.A., Nguyen, Q., Slavin, M., Heath, C.H., Maszewska, K., Halliday, C., Robert, V., Sorrell, T.C., et al. Molecular typing of Australian Scedosporium isolates showing genetic variability and numerous S. aurantiacum. Emerg. Infect. Dis. 2008, 14, 282–290. [Google Scholar]
- Giraud, S.; Bouchara, J.-P. Scedosporium apiospermum Complex: Diagnosis and Species Identification. Curr. Fungal Infect. Rep. 2014, 8, 211–219. [Google Scholar] [CrossRef]
- Subedi, S.; Chen, S.C.-A. Epidemiology of Scedosporiosis. Curr. Fungal Infect. Rep. 2015, 9, 275–284. [Google Scholar] [CrossRef]
- Abrantes, R.A. , Refojo, N. , Hevia, A.I., Fernández, J., Isla, G., Córdoba, S., Dávalos, M.F., Lubovich, S., Maldonado, I., Davel, G.O., et al. Scedosporium spp. from clinical setting in Argentina, with the proposal of the new pathogenic species Scedosporium americanum. J. Fungi 2021, 7, 160. [Google Scholar]
- Neoh, C.F. , Chen, S. C-A., Crowe, A., Hamilton K., Nguyen, Q.A., Marriot, D., Trubiano, J.A., Spelman, T., Kong, D.C.M., Slavin, M.A. Invasive Scedosporium and Lomentospora prolificans infections in Australia: a multicentre retrospective cohort study. Open Forum Infect. Dis. 2023, 10, ofad059. [Google Scholar]
- Hayden, R.T. , Isotalo, P. A., Parrett, T., Wolk, D.M., Qian, X., Roberts, G.D., Lloyd, R.V. In situ hybridisation for the differentiation of Aspergillus, Fusarium, and Pseudallescheria species in tissue section. Diagn. Mol. Pathol. 2003, 12, 21–26. [Google Scholar] [PubMed]
- Chen, S.C.-A.; Halliday, C.L.; Hoenigl, M.; Cornely, O.A.; Meyer, W. Scedosporium and Lomentospora Infections: Contemporary Microbiological Tools for the Diagnosis of Invasive Disease. J. Fungi 2021, 7, 23. [Google Scholar] [CrossRef]
- Rai, P.; Singh, A.K.; Anand, K.B.; Singh, S.P.; Tomar, K. Time versus tissue: Timely identification of Scedosporium Rhinosinusitis in a post-COVID-19 case by MALDI-TOF MS leading to successful management. Med J. Armed Forces India 2022, 78, 360–364. [Google Scholar] [CrossRef]
- Mina, S. , Marot-Leblond, A. , Cimon, B., Fleury, M.J.J., Larcher, G., Bouchara, J.-P. Purification and characterisation of a mycelial catalase from Scedosporium boydii, a useful tool for specific antibody detection patients with cystic fibrosis. Clin. Vacc. Immunol. 2015, 22, 37–45. [Google Scholar]
- Martin-Souto, L. , Antoran, A. , Areitio, M., Aparicio-Fernandez, L., Martín-Gómez, M.T., Fernandez, R., Astigarraga, E., Barreda- Gómez, G., Schwarz, C., Rickerts, V., et al. Dot immunoblotting assay for the rapid serodetection of Scedosporium/Lomentospora in cystic fibrosis patients. J. Fungi 2023, 9, 158. [Google Scholar]
- Thornton, C.R. Tracking the Emerging Human Pathogen Pseudallescheria boydii by Using Highly Specific Monoclonal Antibodies. Clin. Vaccine Immunol. 2009, 16, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.E. , Thornton, C. R. Development of a monoclonal antibody and a serodiagnostic lateral-flow device specific to Rhizopus arrhizus (Syn. R. oryzae), the principal global agent of mucormycosis in humans. J. Fungi 2022, 8, 756. [Google Scholar]
- Thornton, C.R.; Davies, G.E.; Dougherty, L. Development of a monoclonal antibody and a lateral-flow device for the rapid detection of a Mucorales-specific biomarker. Front. Cell. Infect. Microbiol. 2023, 13, 1305662. [Google Scholar] [CrossRef]
- Cerne, C.; Seyedmousavi, S.; Bennett, J.E. Cutaneous hyalohyphomycosis due to Petriella setifera following traumatic inoculation in an immunocompetent host. Med Mycol. Case Rep. 2021, 32, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Marques, D.S. , Vaz, C. P., Branca, R., Campilho, F., Lamelas, C., Afonso, L.P., et al. Rhizomucor and Scedosporium infection post hematopoietic stem-cell transplant. Case Rep. Med. 2011, 2011, 830769. [Google Scholar]
- Shand, J.M.; Albrecht, R.M.; Burnett, H.F.; Miyake, A. Invasive fungal infection of the midfacial and orbital complex due to Scedosporium apiospermum and mucormycosis. J. Oral Maxillofac. Surg. 2004, 62, 231–234. [Google Scholar] [CrossRef]
- Song, Y.; Zhou, M.; Gong, Q.; Guo, J. Scedosporium apiospermum and Lichtheimia corymbifera Co-Infection Due to Inhalation of Biogas in Immunocompetent Patients: A Case Series. Infect. Drug Resist. 2022, ume 15, 6423–6430. [Google Scholar] [CrossRef]
- Kanaujia, R.; Muthu, V.; Singh, S.; Rudramurthy, S.M.; Kaur, H. Rapidly progressive lung coinfection due to Rhizopus and Scedosporium in a diabetic marijuana smoker. Clin. Microbiol. Infect. 2022, 29, 51–53. [Google Scholar] [CrossRef]
- Pinto, M.R.; Mulloy, B.; Haido, R.M.T.; Travassos, L.R.; Bergter, E.B. A peptidorhamnomannan from the mycelium of Pseudallescheria boydii is a potential diagnostic antigen of this emerging human pathogen. Microbiology 2001, 147, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Lopes, L.C.L.; Rollin-Pinheiro, R.; Guimarães, A.J.; Bittencourt, V.C.B.; Martinez, L.R.; Koba, W.; Farias, S.E.; Nosanchuk, J.D.; Barreto-Bergter, E. Monoclonal Antibodies Against Peptidorhamnomannans of Scedosporium apiospermum Enhance the Pathogenicity of the Fungus. PLOS Neglected Trop. Dis. 2010, 4, e853. [Google Scholar] [CrossRef] [PubMed]
- Thornton, C.R. Detection of the ‘big five’ mold killers of humans: Aspergillus, Fusarium, Lomentospora, Scedosporium and Mucormycetes. Adv. Appl. Microbiol. 2020, 110, 1–61. [Google Scholar] [PubMed]
- Thornton, C.R. Development of an Immunochromatographic Lateral-Flow Device for Rapid Serodiagnosis of Invasive Aspergillosis. Clin. Vaccine Immunol. 2008, 15, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Di Nardo, F.; Chiarello, M.; Cavalera, S.; Baggiani, C.; Anfossi, L. Ten Years of Lateral Flow Immunoassay Technique Applications: Trends, Challenges and Future Perspectives. Sensors 2021, 21, 5185. [Google Scholar] [CrossRef] [PubMed]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2018, 4, 46–54. [Google Scholar] [CrossRef]
- Thornton, C.R. The potential for rapid antigen testing for mucormycosis in the context of COVID-19. Expert Rev. Mol. Diagn. 2023, 1–7. [Google Scholar] [CrossRef]
| Species | Isolate Number | Source1 |
ScedLFD a.u.2 |
ScedELISA (Abs 450nm)3 |
|
|---|---|---|---|---|---|
| Apophysomyces variabilis | 658.93 | CBS | 28.8 | 0.052 | |
| Aspergillus fumigatus | Af293 | FGSC | 35.7 | 0.046 | |
| Aspergillus flavus | 144B | CRT | 27.9 | 0.049 | |
| Aspergillus nidulans | A4 | FGSC | 35.1 | 0.044 | |
| Aspergillus niger | 102.4 | CBS | 32.7 | 0.052 | |
| Aspergillus terreus var. terreus | 601.65 | CBS | 30.3 | 0.049 | |
| Candida albicans | SC5314 | SB | 37.1 | 0.060 | |
| Cryptococcus neoformans | 8710 | CBS | 46.5 | 0.055 | |
| Cunninghamella bertholletiae | 151.80 | CBS | 34.5 | 0.053 | |
| Fusarium oxysporum | 167.3 | CBS | 34.0 | 0.052 | |
| Fusarium solani | 224.34 | CBS | 31.7 | 0.047 | |
| Lichtheimia corymbifera | 109940 | CBS | 30.5 | 0.053 | |
| Lichtheimia hyalospora | 146576 | CBS | 33.0 | 0.045 | |
| Lichtheimia ornata | 142195 | CBS | 33.7 | 0.050 | |
| Lichtheimia ramosa | 124197 | CBS | 32.4 | 0.049 | |
| Lomentospora prolificans | 3.1 | CRT | 33.0 | 0.090 | |
| Lomentospora prolificans | 742.96 | CBS | 41.3 | 0.048 | |
| Lomentospora prolificans | 100390 | CBS | 43.0 | 0.046 | |
| Mucor circinelloides | 123973 | CBS | 33.8 | 0.056 | |
| Mucor circinelloides | 124429 | CBS | 38.2 | 0.056 | |
| Mucor circinelloides f. circinelloides | 120582 | CBS | 36.7 | 0.051 | |
| Mucor indicus | 120.08 | CBS | 34.4 | 0.044 | |
| Petriella setifera | 109039 | CBS | 622.3 | 1.431 | |
| Rhizomucor pusillus | 120586 | CBS | 31.7 | 0.051 | |
| Rhizopus arrhizus var. arrhizus | 112.07 | CBS | 37.5 | 0.056 | |
| Rhizopus arrhizus var. delemar | 607.68 | CBS | 38.8 | 0.051 | |
| Rhizopus rhizopodiformis | 102277 | CBS | 32.3 | 0.050 | |
| Scedosporium angustum | 254.72 | CBS | 489.4 | 1.042 | |
| Scedosporium apiospermum | 8353 | RMRL | 426.2 | 0.815 | |
| Scedosporium apiospermum | 117407 | CBS | 469.5 | 1.337 | |
| Scedosporium aurantiacum | 118934 | CBS | 691.1 | 1.403 | |
| Scedosporium aurantiacum | 121926 | CBS | 737.1 | 1.427 | |
| Scedosporium boydii | 835.96 | CBS | 373.4 | 1.190 | |
| Scedosporium boydii | 100393 | CBS | 593.5 | 1.394 | |
| Scedosporium boydii | 100395 | CBS | 435.4 | 0.786 | |
| Scedosporium boydii | 100870 | CBS | 474.3 | 1.126 | |
| Scedosporium boydii | Exton 22A | CRT | 449.7 | 1.121 | |
| Scedosporium dehoogii | 117406 | CBS | 28.1 | 0.049 | |
| Scedosporium desertorum | 489.72 | CBS | 500.2 | 1.310 | |
| Scedosporium ellipsoideum | 438.75 | CBS | 525.1 | 1.315 | |
| Scedosporium fusoideum | 106.53 | CBS | 598.0 | 1.290 | |
| Scedosporium minutisporum | 116911 | CBS | 383.3 | 0.592 | |
| YNB+G only | - | - | 29.8 | 0.048 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





