Submitted:
27 March 2024
Posted:
29 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
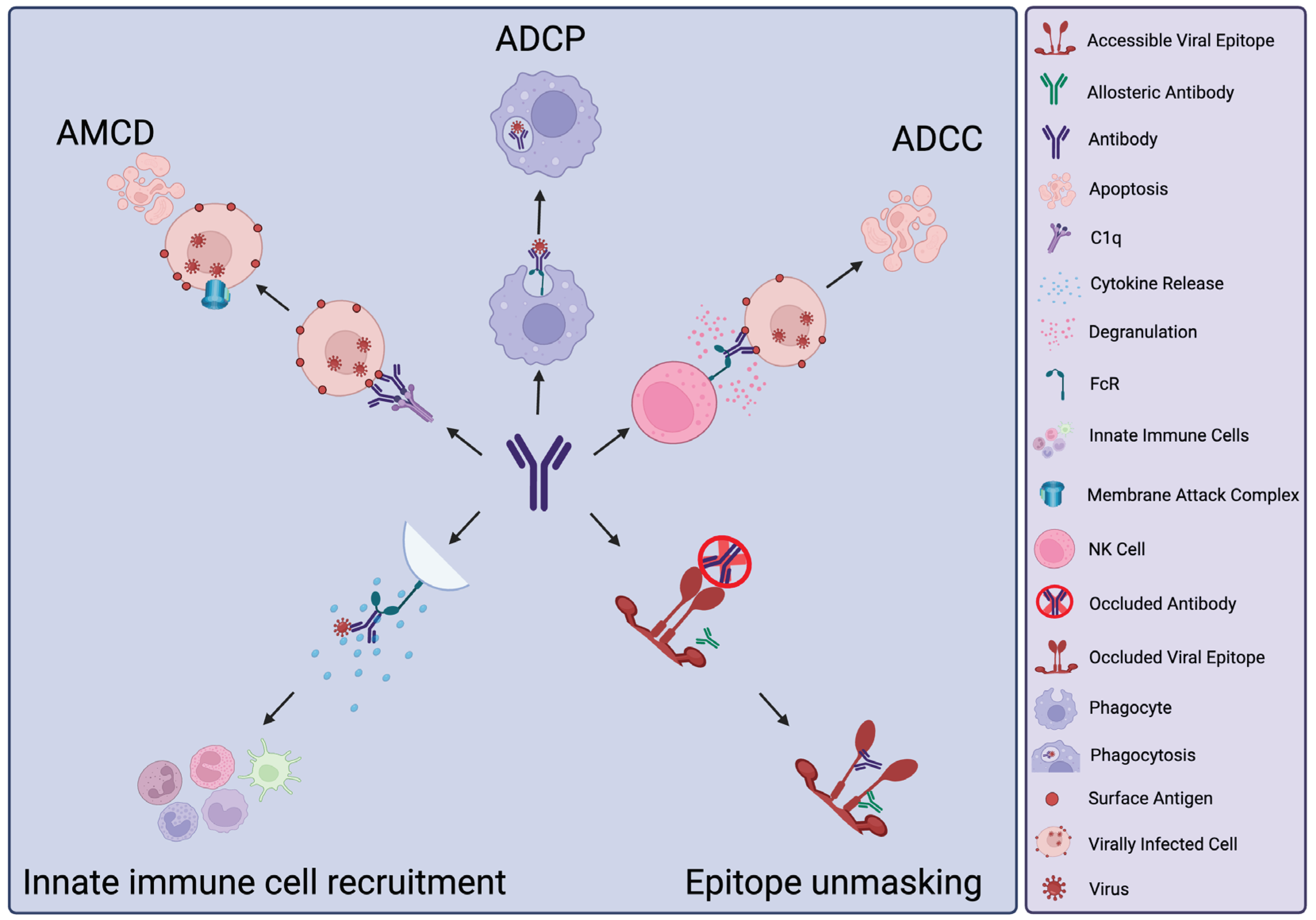
2. Antibody Activities beyond Neutralisation
3. Protective Functions of nNAbs
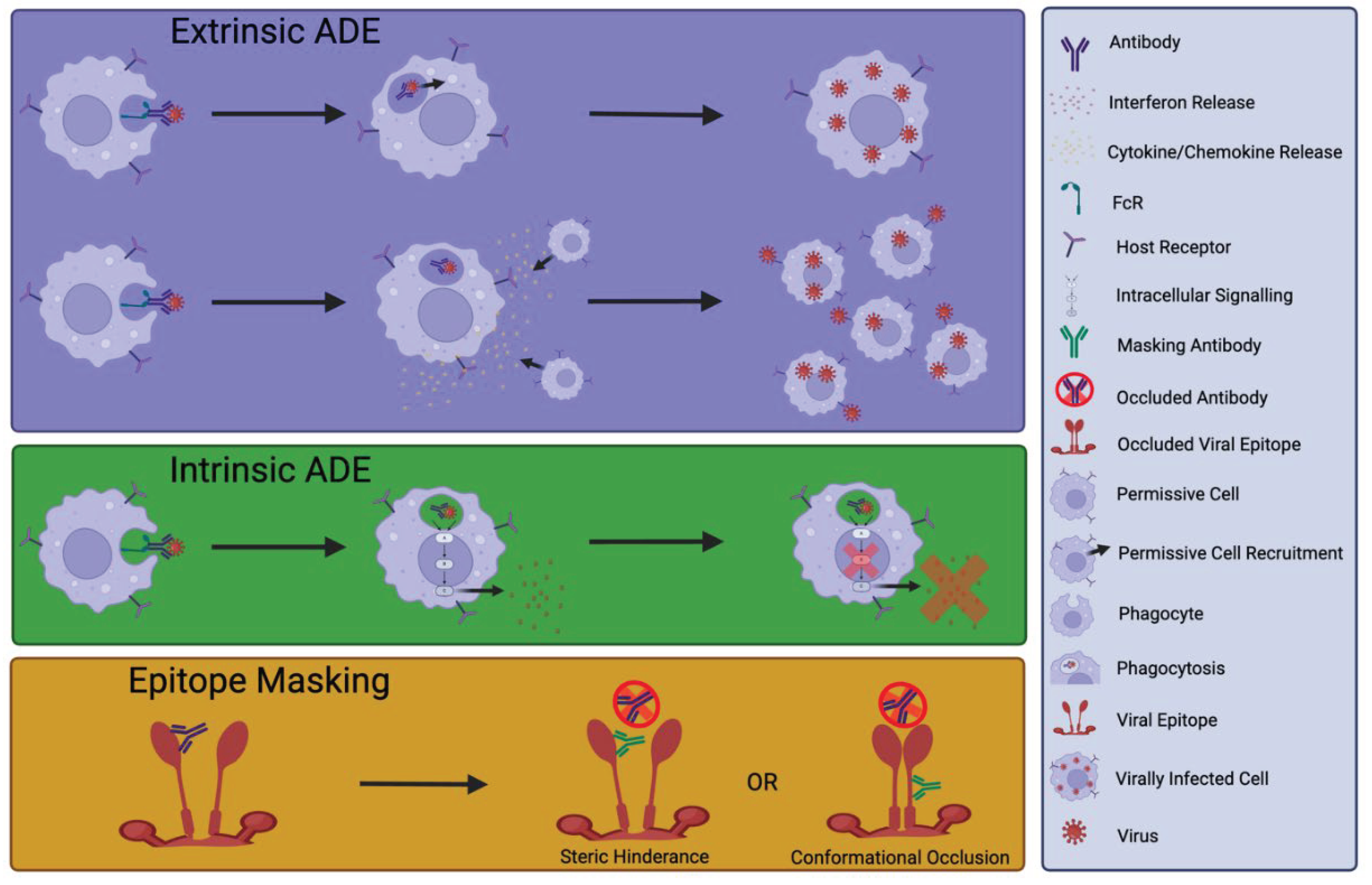
4. Detrimental Effects of nNAbs or NAb
5. Measuring Fc-Dependent Effector Functions
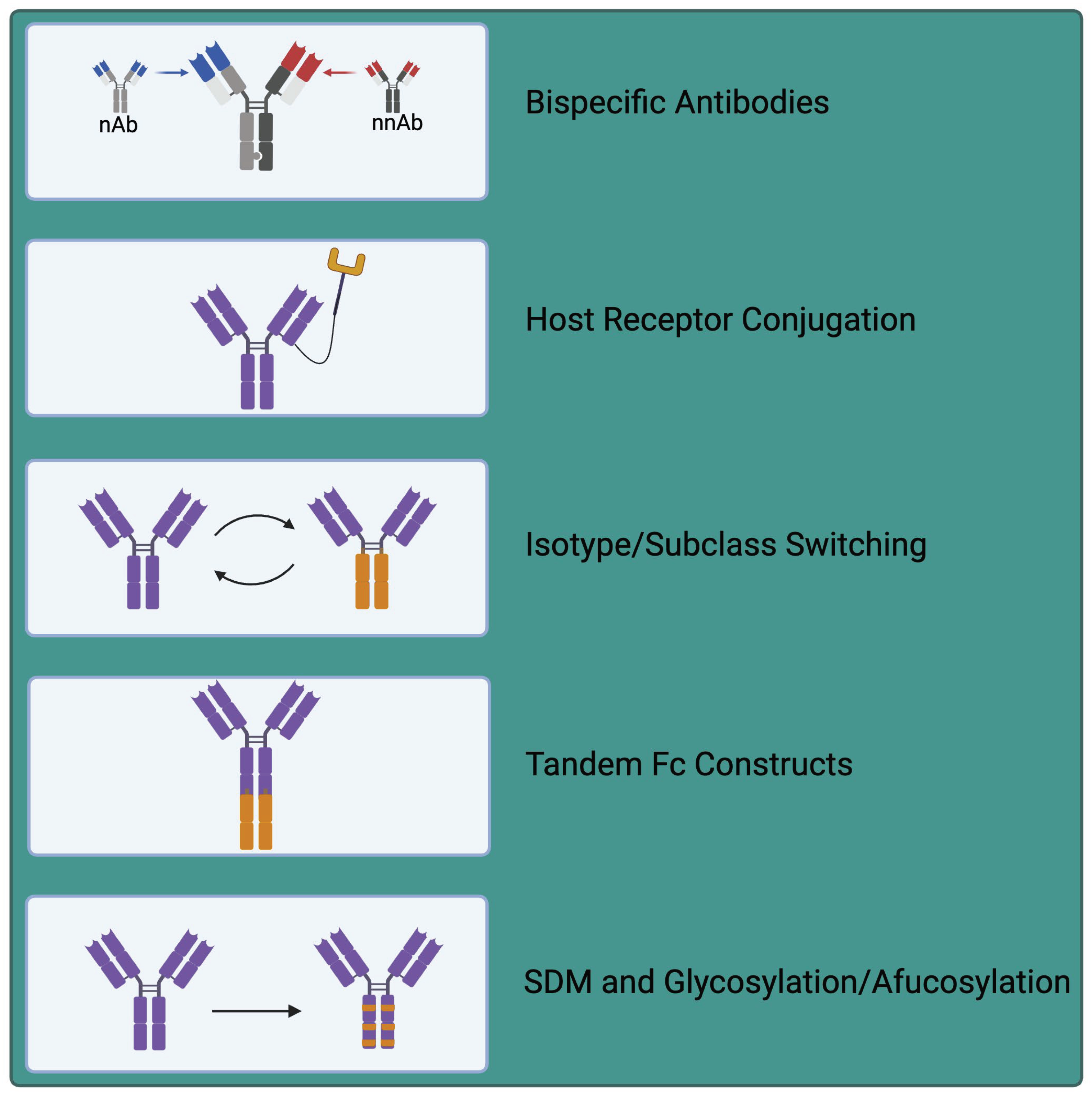
6. Exploiting nNAbs for Therapeutic Applications
7. Future Outlooks and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations List
| ADE | Antibody Dependent Enhancement |
| ADCP | Antibody Dependent Cellular Phagocytosis |
| ADCC | Antibody Dependent Cellular Cytotoxicity |
| AMCD | Antibody Mediated Complement Deposition |
| BNAbs | Broadly Neutralising Antibodies |
| CDC | Complement Dependent Cytotoxicity |
| CMV | Cytomegalovirus |
| DENV | Dengue Virus |
| ELISA | Enzyme Linked Immunosorbent Assay |
| Fab | Fragment Antigen Binding |
| Fc | Fragment Crystallisable |
| FcR | Fragment Crystallisable Receptor |
| HCV | Hepatitis C Virus |
| HA | Hemagglutinin |
| HIV | Human Immunodeficiency Virus |
| HPV | Human Papilloma Virus |
| IAV | Influenza A Virus |
| ITAM | Immunoreceptor Tyrosine-Based Activation Motif |
| LCMV | Lymphocytic Choriomeningitis Virus |
| MAC | Membrane Attack Complex |
| MARV | Marburg virus |
| mAb | Monoclonal Antibody |
| MHC | Major Histocompatibility Complex |
| NAb | Neutralising Antibody |
| NK cell | Natural Killer Cell |
| nNAb | Non-neutralising Antibody |
| pIgR | Poly Ig Receptor |
| RBD | Receptor Binding Domain |
| ScFv | Single Chain Variable Fragment |
References
- Klasse, P.J. Neutralization of Virus Infectivity by Antibodies: Old Problems in New Perspectives. Adv Biol 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.R. Antiviral neutralizing antibodies: from in vitro to in vivo activity. Nat Rev Immunol 2023, 23, 720–734. [Google Scholar] [CrossRef]
- Bottermann, M.; Caddy, S.L. Virus neutralisation by intracellular antibodies. Semin Cell Dev Biol 2022, 126, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Suryadevara, N.; Shrihari, S.; Gilchuk, P.; VanBlargan, L.A.; Binshtein, E.; Zost, S.J.; Nargi, R.S.; Sutton, R.E.; Winkler, E.S.; Chen, E.C.; et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 2021, 184, 2316–2331 e2315. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347e2316. [Google Scholar] [CrossRef] [PubMed]
- Chandler, T.L.; Yang, A.; Otero, C.E.; Permar, S.R.; Caddy, S.L. Protective mechanisms of nonneutralizing antiviral antibodies. PLoS Pathog 2023, 19, e1011670. [Google Scholar] [CrossRef] [PubMed]
- Gunn, B.M.; Bai, S. Building a better antibody through the Fc: advances and challenges in harnessing antibody Fc effector functions for antiviral protection. Hum Vaccin Immunother 2021, 17, 4328–4344. [Google Scholar] [CrossRef] [PubMed]
- Prager, I.; Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J Leukoc Biol 2019, 105, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Erbe, A.K.; Hank, J.A.; Morris, Z.S.; Sondel, P.M. NK Cell-Mediated Antibody-Dependent Cellular Cytotoxicity in Cancer Immunotherapy. Front Immunol 2015, 6, 368. [Google Scholar] [CrossRef]
- Tay, M.Z.; Wiehe, K.; Pollara, J. Antibody-Dependent Cellular Phagocytosis in Antiviral Immune Responses. Front Immunol 2019, 10, 332. [Google Scholar] [CrossRef]
- Schonrich, G.; Raftery, M.J. Neutrophil Extracellular Traps Go Viral. Front Immunol 2016, 7, 366. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, B.S.; Ackerman, M.E. Antibody-mediated complement activation in pathology and protection. Immunol Cell Biol 2020, 98, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Keyt, B.A.; Baliga, R.; Sinclair, A.M.; Carroll, S.F.; Peterson, M.S. Structure, Function, and Therapeutic Use of IgM Antibodies. Antibodies (Basel) 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- de Taeye, S.W.; Bentlage, A.E.H.; Mebius, M.M.; Meesters, J.I.; Lissenberg-Thunnissen, S.; Falck, D.; Senard, T.; Salehi, N.; Wuhrer, M.; Schuurman, J.; et al. FcgammaR Binding and ADCC Activity of Human IgG Allotypes. Front Immunol 2020, 11, 740. [Google Scholar] [CrossRef] [PubMed]
- Muthana, S.M.; Xia, L.; Campbell, C.T.; Zhang, Y.; Gildersleeve, J.C. Competition between serum IgG, IgM, and IgA anti-glycan antibodies. PLoS One 2015, 10, e0119298. [Google Scholar] [CrossRef] [PubMed]
- de Sousa-Pereira, P.; Woof, J.M. IgA: Structure, Function, and Developability. Antibodies (Basel) 2019, 8. [Google Scholar] [CrossRef]
- Biswas, S.; Mandal, G.; Anadon, C.M.; Chaurio, R.A.; Lopez-Bailon, L.U.; Nagy, M.Z.; Mine, J.A.; Hanggi, K.; Sprenger, K.B.; Innamarato, P.; et al. Targeting intracellular oncoproteins with dimeric IgA promotes expulsion from the cytoplasm and immune-mediated control of epithelial cancers. Immunity 2023, 56, 2570–2583 e2576. [Google Scholar] [CrossRef]
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claer, L.; Quentric, P.; Fadlallah, J.; Devilliers, H.; Ghillani, P.; et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med 2021, 13. [Google Scholar] [CrossRef]
- Mestecky, J.; Fultz, P.N. Mucosal immune system of the human genital tract. J Infect Dis 1999, 179 Suppl 3, S470–474. [Google Scholar] [CrossRef]
- Yan, H.; Lamm, M.E.; Bjorling, E.; Huang, Y.T. Multiple functions of immunoglobulin A in mucosal defense against viruses: an in vitro measles virus model. J Virol 2002, 76, 10972–10979. [Google Scholar] [CrossRef]
- Izadi, A.; Godzwon, M.; Ohlin, M.; Nordenfelt, P. Protective non-neutralizing mAbs Ab94 and Ab81 retain high-affinity and potent Fc-mediated function against SARS-CoV-2 variants from Omicron to XBB1.5. https://doi.org/10.1101/2023.09.29.560084. bioRxiv. [CrossRef]
- Murin, C.D.; Wilson, I.A.; Ward, A.B. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat Microbiol 2019, 4, 734–747. [Google Scholar] [CrossRef] [PubMed]
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Chapter 3 (27-37) Virion Structure and Composition. In Fenner and White's Medical Virology (Fifth Edition); Academic Press: London, 2017. [Google Scholar] [CrossRef]
- Muriaux, D.; Darlix, J.L. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol 2010, 7, 744–753. [Google Scholar] [CrossRef]
- Dacon, C.; Tucker, C.; Peng, L.; Lee, C.D.; Lin, T.H.; Yuan, M.; Cong, Y.; Wang, L.; Purser, L.; Williams, J.K.; et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science 2022, 377, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Landais, E.; Moore, P.L. Development of broadly neutralizing antibodies in HIV-1 infected elite neutralizers. Retrovirology 2018, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.C.; Wilson, I.A. Influenza Hemagglutinin Structures and Antibody Recognition. Cold Spring Harb Perspect Med 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, P.; Gruell, H.; Nogueira, L.; Pai, J.A.; Butler, A.L.; Millard, K.; Lehmann, C.; Suarez, I.; Oliveira, T.Y.; Lorenzi, J.C.C.; et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 2018, 561, 479–484. [Google Scholar] [CrossRef]
- Sun, X.; Ma, H.; Wang, X.; Bao, Z.; Tang, S.; Yi, C.; Sun, B. Broadly neutralizing antibodies to combat influenza virus infection. Antiviral Res 2024, 221, 105785. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Song, G.; Liu, H.; Yuan, M.; He, W.T.; Beutler, N.; Zhu, X.; Tse, L.V.; Martinez, D.R.; Schafer, A.; et al. Broadly neutralizing anti-S2 antibodies protect against all three human betacoronaviruses that cause deadly disease. Immunity 2023, 56, 669–686 e667. [Google Scholar] [CrossRef] [PubMed]
- Kinchen, V.J.; Zahid, M.N.; Flyak, A.I.; Soliman, M.G.; Learn, G.H.; Wang, S.; Davidson, E.; Doranz, B.J.; Ray, S.C.; Cox, A.L.; et al. Broadly Neutralizing Antibody Mediated Clearance of Human Hepatitis C Virus Infection. Cell Host Microbe 2018, 24, 717–730 e715. [Google Scholar] [CrossRef]
- Haynes, B.F.; Wiehe, K.; Borrow, P.; Saunders, K.O.; Korber, B.; Wagh, K.; McMichael, A.J.; Kelsoe, G.; Hahn, B.H.; Alt, F.; et al. Strategies for HIV-1 vaccines that induce broadly neutralizing antibodies. Nat Rev Immunol 2023, 23, 142–158. [Google Scholar] [CrossRef]
- Forthal, D.N. Functions of Antibodies. Microbiol Spectr 2014, 2, AID-0019-2014. [Google Scholar] [CrossRef] [PubMed]
- Sedova, E.S.; Scherbinin, D.N.; Lysenko, A.A.; Alekseeva, S.V.; Artemova, E.A.; Shmarov, M.M. Non-neutralizing Antibodies Directed at Conservative Influenza Antigens. Acta Naturae 2019, 11, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Weidenbacher, P.A.; Waltari, E.; de Los Rios Kobara, I.; Bell, B.N.; Morris, M.K.; Cheng, Y.C.; Hanson, C.; Pak, J.E.; Kim, P.S. Converting non-neutralizing SARS-CoV-2 antibodies into broad-spectrum inhibitors. Nat Chem Biol 2022, 18, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Guo, Z.; Subiaur, S.; Benegal, A.; Vahey, M.D. Antibody inhibition of influenza A virus assembly and release. J Virol 2024, 98, e0139823. [Google Scholar] [CrossRef] [PubMed]
- Hughey, P.G.; Roberts, P.C.; Holsinger, L.J.; Zebedee, S.L.; Lamb, R.A.; Compans, R.W. Effects of antibody to the influenza A virus M2 protein on M2 surface expression and virus assembly. Virology 1995, 212, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Phanthong, S.; Densumite, J.; Seesuay, W.; Thanongsaksrikul, J.; Teimoori, S.; Sookrung, N.; Poovorawan, Y.; Onvimala, N.; Guntapong, R.; Pattanapanyasat, K.; et al. Human Antibodies to VP4 Inhibit Replication of Enteroviruses Across Subgenotypes and Serotypes, and Enhance Host Innate Immunity. Front Microbiol 2020, 11, 562768. [Google Scholar] [CrossRef]
- Chehadeh, W.; Lobert, P.E.; Sauter, P.; Goffard, A.; Lucas, B.; Weill, J.; Vantyghem, M.C.; Alm, G.; Pigny, P.; Hober, D. Viral protein VP4 is a target of human antibodies enhancing coxsackievirus B4- and B3-induced synthesis of alpha interferon. J Virol 2005, 79, 13882–13891. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.A.; Zhou, D.; Fry, E.E.; Kotecha, A.; Huang, P.N.; Yang, S.L.; Tsao, K.C.; Huang, Y.C.; Lin, T.Y.; Ren, J.; et al. Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71. Nat Commun 2020, 11, 5253. [Google Scholar] [CrossRef] [PubMed]
- Che Nordin, M.A.; Teow, S.Y. Review of Current Cell-Penetrating Antibody Developments for HIV-1 Therapy. Molecules 2018, 23. [Google Scholar] [CrossRef]
- Zhou, D.; Yang, Y.; Zhao, B.; Yu, J.; Cao, Y.; Yan, H.; Zhao, W.; Chen, L.; Chen, F.; Li, X.; et al. IgA targeting on the alpha-molecular recognition element (alpha-MoRE) of viral phosphoprotein inhibits measles virus replication by interrupting formation and function of P-N complex intracellularly. Antiviral Res 2019, 161, 144–153. [Google Scholar] [CrossRef]
- Caddy, S.L.; Vaysburd, M.; Papa, G.; Wing, M.; O'Connell, K.; Stoycheva, D.; Foss, S.; Terje Andersen, J.; Oxenius, A.; James, L.C. Viral nucleoprotein antibodies activate TRIM21 and induce T cell immunity. EMBO J 2021, 40, e106228. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.L.; Laidlaw, S.M.; Dustin, L.B. TRIM21/Ro52 - Roles in Innate Immunity and Autoimmune Disease. Front Immunol 2021, 12, 738473. [Google Scholar] [CrossRef] [PubMed]
- McEwan, W.A.; Tam, J.C.; Watkinson, R.E.; Bidgood, S.R.; Mallery, D.L.; James, L.C. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol 2013, 14, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Rijnink, W.F.; Stadlbauer, D.; Puente-Massaguer, E.; Okba, N.M.A.; Kirkpatrick Roubidoux, E.; Strohmeier, S.; Mudd, P.A.; Schmitz, A.; Ellebedy, A.; McMahon, M.; et al. Characterization of non-neutralizing human monoclonal antibodies that target the M1 and NP of influenza A viruses. J Virol 2023, 97, e0164622. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M.; Tamiya, S.; Kawai, A.; Hirai, T.; Cragg, M.S.; Yoshioka, Y. Synergistic effect of non-neutralizing antibodies and interferon-gamma for cross-protection against influenza. iScience 2021, 24, 103131. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.S.; Leon, P.E.; Albrecht, R.A.; Margine, I.; Hirsh, A.; Bahl, J.; Krammer, F. Broadly-Reactive Neutralizing and Non-neutralizing Antibodies Directed against the H7 Influenza Virus Hemagglutinin Reveal Divergent Mechanisms of Protection. PLoS Pathog 2016, 12, e1005578. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Reber, A.J.; Kumar, A.; Ramos, P.; Sica, G.; Music, N.; Guo, Z.; Mishina, M.; Stevens, J.; York, I.A.; et al. Non-neutralizing antibodies induced by seasonal influenza vaccine prevent, not exacerbate A(H1N1)pdm09 disease. Sci Rep 2016, 6, 37341. [Google Scholar] [CrossRef]
- Ko, Y.A.; Yu, Y.H.; Wu, Y.F.; Tseng, Y.C.; Chen, C.L.; Goh, K.S.; Liao, H.Y.; Chen, T.H.; Cheng, T.R.; Yang, A.S.; et al. A non-neutralizing antibody broadly protects against influenza virus infection by engaging effector cells. PLoS Pathog 2021, 17, e1009724. [Google Scholar] [CrossRef]
- Dai, H.S.; Griffin, N.; Bolyard, C.; Mao, H.C.; Zhang, J.; Cripe, T.P.; Suenaga, T.; Arase, H.; Nakano, I.; Chiocca, E.A.; et al. The Fc Domain of Immunoglobulin Is Sufficient to Bridge NK Cells with Virally Infected Cells. Immunity 2017, 47, 159–170 e110. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Tomioka, Y.; Takakuwa, H.; Uechi, G.I.; Yabuta, T.; Ozaki, K.; Suyama, H.; Yamamoto, S.; Morimatsu, M.; Mai, L.Q.; et al. Cross-protective potential of anti-nucleoprotein human monoclonal antibodies against lethal influenza A virus infection. J Gen Virol 2016, 97, 2104–2116. [Google Scholar] [CrossRef]
- Carragher, D.M.; Kaminski, D.A.; Moquin, A.; Hartson, L.; Randall, T.D. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J Immunol 2008, 181, 4168–4176. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, J.; Cainelli-Gebara, V.; Mercier, G.; Mansour, S.; Talbot, P.J.; Lussier, G.; Oth, D. Protection from mouse hepatitis virus type 3-induced acute disease by an anti-nucleoprotein monoclonal antibody. Brief report. Arch Virol 1987, 97, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Barcena, M.; Oostergetel, G.T.; Bartelink, W.; Faas, F.G.; Verkleij, A.; Rottier, P.J.; Koster, A.J.; Bosch, B.J. Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc Natl Acad Sci U S A 2009, 106, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Ploner, A.; Elfstrom, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundstrom, K.; Dillner, J.; Sparen, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N Engl J Med 2020, 383, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Falcaro, M.; Castanon, A.; Ndlela, B.; Checchi, M.; Soldan, K.; Lopez-Bernal, J.; Elliss-Brookes, L.; Sasieni, P. The effects of the national HPV vaccination programme in England, UK, on cervical cancer and grade 3 cervical intraepithelial neoplasia incidence: a register-based observational study. Lancet 2021, 398, 2084–2092. [Google Scholar] [CrossRef]
- Quang, C.; Chung, A.W.; Frazer, I.H.; Toh, Z.Q.; Licciardi, P.V. Single-dose HPV vaccine immunity: is there a role for non-neutralizing antibodies? Trends Immunol 2022, 43, 815–825. [Google Scholar] [CrossRef]
- Bahnan, W.; Wrighton, S.; Sundwall, M.; Blackberg, A.; Larsson, O.; Hoglund, U.; Khakzad, H.; Godzwon, M.; Walle, M.; Elder, E.; et al. Spike-Dependent Opsonization Indicates Both Dose-Dependent Inhibition of Phagocytosis and That Non-Neutralizing Antibodies Can Confer Protection to SARS-CoV-2. Front Immunol 2021, 12, 808932. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin-Bussieres, G.; Chen, Y.; Ullah, I.; Prevost, J.; Tolbert, W.D.; Symmes, K.; Ding, S.; Benlarbi, M.; Gong, S.Y.; Tauzin, A.; et al. A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep 2022, 38, 110368. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Shorman, M. Cytomegalovirus. In StatPearls; Treasure Island (FL) 2024. https://www.ncbi.nlm.nih.gov/books/NBK459185/.
- Bootz, A.; Karbach, A.; Spindler, J.; Kropff, B.; Reuter, N.; Sticht, H.; Winkler, T.H.; Britt, W.J.; Mach, M. Protective capacity of neutralizing and non-neutralizing antibodies against glycoprotein B of cytomegalovirus. PLoS Pathog 2017, 13, e1006601. [Google Scholar] [CrossRef]
- Semmes, E.C.; Miller, I.G.; Wimberly, C.E.; Phan, C.T.; Jenks, J.A.; Harnois, M.J.; Berendam, S.J.; Webster, H.; Hurst, J.H.; Kurtzberg, J.; et al. Maternal Fc-mediated non-neutralizing antibody responses correlate with protection against congenital human cytomegalovirus infection. J Clin Invest 2022, 132. [Google Scholar] [CrossRef]
- Semmes, E.C.; Miller, I.G.; Rodgers, N.; Phan, C.T.; Hurst, J.H.; Walsh, K.M.; Stanton, R.J.; Pollara, J.; Permar, S.R. ADCC-activating antibodies correlate with decreased risk of congenital human cytomegalovirus transmission. JCI Insight 2023, 8. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, I.; Kropff, B.; Ambrose, L.; McIntosh, M.; McLean, G.R.; Pichon, S.; Atkinson, C.; Milne, R.S.B.; Mach, M.; Griffiths, P.D.; et al. Protection from cytomegalovirus viremia following glycoprotein B vaccination is not dependent on neutralizing antibodies. Proc Natl Acad Sci U S A 2018, 115, 6273–6278. [Google Scholar] [CrossRef]
- Goodwin, M.L.; Webster, H.S.; Wang, H.Y.; Jenks, J.A.; Nelson, C.S.; Tu, J.J.; Mangold, J.F.; Valencia, S.; Pollara, J.; Edwards, W.; et al. Specificity and effector functions of non-neutralizing gB-specific monoclonal antibodies isolated from healthy individuals with human cytomegalovirus infection. Virology 2020, 548, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Mayr, L.M.; Su, B.; Moog, C. Non-Neutralizing Antibodies Directed against HIV and Their Functions. Front Immunol 2017, 8, 1590. [Google Scholar] [CrossRef] [PubMed]
- Hioe, C.E.; Li, G.; Liu, X.; Tsahouridis, O.; He, X.; Funaki, M.; Klingler, J.; Tang, A.F.; Feyznezhad, R.; Heindel, D.W.; et al. Non-neutralizing antibodies targeting the immunogenic regions of HIV-1 envelope reduce mucosal infection and virus burden in humanized mice. PLoS Pathog 2022, 18, e1010183. [Google Scholar] [CrossRef] [PubMed]
- Alter, G.; Yu, W.H.; Chandrashekar, A.; Borducchi, E.N.; Ghneim, K.; Sharma, A.; Nedellec, R.; McKenney, K.R.; Linde, C.; Broge, T.; et al. Passive Transfer of Vaccine-Elicited Antibodies Protects against SIV in Rhesus Macaques. Cell 2020, 183, 185–196 e114. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, J.A.; Bar-On, Y.; Lu, C.L.; Fera, D.; Lockhart, A.A.K.; Lorenzi, J.C.C.; Nogueira, L.; Golijanin, J.; Scheid, J.F.; Seaman, M.S.; et al. Non-neutralizing Antibodies Alter the Course of HIV-1 Infection In Vivo. Cell 2017, 170, 637–648 e610. [Google Scholar] [CrossRef] [PubMed]
- Corey, L.; Gilbert, P.B.; Tomaras, G.D.; Haynes, B.F.; Pantaleo, G.; Fauci, A.S. Immune correlates of vaccine protection against HIV-1 acquisition. Sci Transl Med 2015, 7, 310rv317. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.S.; Coote, C.; Moreau, Y.; Isaac, J.E.; Ewing, A.C.; Kourtis, A.P.; Sagar, M. Antibody-dependent cellular cytotoxicity responses and susceptibility influence HIV-1 mother-to-child transmission. JCI Insight 2022, 7. [Google Scholar] [CrossRef]
- Stieh, D.J.; King, D.F.; Klein, K.; Liu, P.; Shen, X.; Hwang, K.K.; Ferrari, G.; Montefiori, D.C.; Haynes, B.; Pitisuttithum, P.; et al. Aggregate complexes of HIV-1 induced by multimeric antibodies. Retrovirology 2014, 11, 78. [Google Scholar] [CrossRef]
- Ilinykh, P.A.; Huang, K.; Santos, R.I.; Gilchuk, P.; Gunn, B.M.; Karim, M.M.; Liang, J.; Fouch, M.E.; Davidson, E.; Parekh, D.V.; et al. Non-neutralizing Antibodies from a Marburg Infection Survivor Mediate Protection by Fc-Effector Functions and by Enhancing Efficacy of Other Antibodies. Cell Host Microbe 2020, 27, 976–991 e911. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.A.; Brannan, J.M.; Bryan, C.; McNeal, A.; Davidson, E.; Turner, H.L.; Vu, H.; Shulenin, S.; He, S.; Kuehne, A.; et al. Cooperativity Enables Non-neutralizing Antibodies to Neutralize Ebolavirus. Cell Rep 2017, 19, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Gunn, B.M.; Yu, W.H.; Karim, M.M.; Brannan, J.M.; Herbert, A.S.; Wec, A.Z.; Halfmann, P.J.; Fusco, M.L.; Schendel, S.L.; Gangavarapu, K.; et al. A Role for Fc Function in Therapeutic Monoclonal Antibody-Mediated Protection against Ebola Virus. Cell Host Microbe 2018, 24, 221–233 e225. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Oxenius, A. Non-neutralizing antibodies protect from chronic LCMV infection independently of activating FcgammaR or complement. Eur J Immunol 2013, 43, 2349–2360. [Google Scholar] [CrossRef] [PubMed]
- Nicasio, M.; Sautto, G.; Clementi, N.; Diotti, R.A.; Criscuolo, E.; Castelli, M.; Solforosi, L.; Clementi, M.; Burioni, R. Neutralization interfering antibodies: a "novel" example of humoral immune dysfunction facilitating viral escape? Viruses 2012, 4, 1731–1752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhong, L.; Struble, E.B.; Watanabe, H.; Kachko, A.; Mihalik, K.; Virata, M.L.; Alter, H.J.; Feinstone, S.; Major, M. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci U S A 2009, 106, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Verrier, F.; Nadas, A.; Gorny, M.K.; Zolla-Pazner, S. Additive effects characterize the interaction of antibodies involved in neutralization of the primary dualtropic human immunodeficiency virus type 1 isolate 89.6. J Virol 2001, 75, 9177–9186. [Google Scholar] [CrossRef] [PubMed]
- Matveev, A.L.; Pyankov, O.V.; Khlusevich, Y.A.; Tyazhelkova, O.V.; Emelyanova, L.A.; Timofeeva, A.M.; Shipovalov, A.V.; Chechushkov, A.V.; Zaitseva, N.S.; Kudrov, G.A.; et al. Novel B-Cell Epitopes of Non-Neutralizing Antibodies in the Receptor-Binding Domain of the SARS-CoV-2 S-Protein with Different Effects on the Severity of COVID-19. Biochemistry (Mosc) 2023, 88, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Yang, Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol 2020, 20, 339–341. [Google Scholar] [CrossRef]
- Barba-Spaeth, G.; Dejnirattisai, W.; Rouvinski, A.; Vaney, M.C.; Medits, I.; Sharma, A.; Simon-Loriere, E.; Sakuntabhai, A.; Cao-Lormeau, V.M.; Haouz, A.; et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 2016, 536, 48–53. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Supasa, P.; Wongwiwat, W.; Rouvinski, A.; Barba-Spaeth, G.; Duangchinda, T.; Sakuntabhai, A.; Cao-Lormeau, V.M.; Malasit, P.; Rey, F.A.; et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat Immunol 2016, 17, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Sawant, J.; Patil, A.; Kurle, S. A Review: Understanding Molecular Mechanisms of Antibody-Dependent Enhancement in Viral Infections. Vaccines (Basel) 2023, 11. [Google Scholar] [CrossRef]
- Narayan, R.; Tripathi, S. Intrinsic ADE: The Dark Side of Antibody Dependent Enhancement During Dengue Infection. Front Cell Infect Microbiol 2020, 10, 580096. [Google Scholar] [CrossRef]
- Guzman, M.G.; Alvarez, M.; Halstead, S.B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch Virol 2013, 158, 1445–1459. [Google Scholar] [CrossRef] [PubMed]
- Katzelnick, L.C.; Gresh, L.; Halloran, M.E.; Mercado, J.C.; Kuan, G.; Gordon, A.; Balmaseda, A.; Harris, E. Antibody-dependent enhancement of severe dengue disease in humans. Science 2017, 358, 929–932. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Shanmugam, R.K.; Ramasamy, V.; Arora, U.; Batra, G.; Acklin, J.A.; Krammer, F.; Lim, J.K.; Swaminathan, S.; Khanna, N. Zika virus envelope nanoparticle antibodies protect mice without risk of disease enhancement. EBioMedicine 2020, 54, 102738. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Xu, K.; Li, J.; Huang, Q.; Song, J.; Han, Y.; Zheng, T.; Gao, P.; Lu, X.; Yang, H.; et al. Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch. Nat Immunol 2021, 22, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Beesetti, H.; Brown, J.A.; Ahuja, R.; Ramasamy, V.; Shanmugam, R.K.; Poddar, A.; Batra, G.; Krammer, F.; Lim, J.K.; et al. Dengue and Zika virus infections are enhanced by live attenuated dengue vaccine but not by recombinant DSV4 vaccine candidate in mouse models. EBioMedicine 2020, 60, 102991. [Google Scholar] [CrossRef]
- Willey, S.; Aasa-Chapman, M.M.; O'Farrell, S.; Pellegrino, P.; Williams, I.; Weiss, R.A.; Neil, S.J. Extensive complement-dependent enhancement of HIV-1 by autologous non-neutralising antibodies at early stages of infection. Retrovirology 2011, 8, 16. [Google Scholar] [CrossRef]
- Wieczorek, L.; Zemil, M.; Merbah, M.; Dussupt, V.; Kavusak, E.; Molnar, S.; Heller, J.; Beckman, B.; Wollen-Roberts, S.; Peachman, K.K.; et al. Evaluation of Antibody-Dependent Fc-Mediated Viral Entry, as Compared With Neutralization, in SARS-CoV-2 Infection. Front Immunol 2022, 13, 901217. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Yu, X.; Jiang, W.; Chen, S.; Wang, R.; Wang, M.; Jiao, S.; Yang, Y.; Wang, W.; et al. Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires FcgammaRIIB and virus-antibody complex with bivalent interaction. Commun Biol 2022, 5, 262. [Google Scholar] [CrossRef]
- Sejdic, A.; Frische, A.; Jorgensen, C.S.; Rasmussen, L.D.; Trebbien, R.; Dungu, A.; Holler, J.G.; Ostrowski, S.R.; Eriksson, R.; Soborg, C.; et al. High titers of neutralizing SARS-CoV-2 antibodies six months after symptom onset are associated with increased severity in COVID-19 hospitalized patients. Virol J 2023, 20, 14. [Google Scholar] [CrossRef] [PubMed]
- Legros, V.; Denolly, S.; Vogrig, M.; Boson, B.; Siret, E.; Rigaill, J.; Pillet, S.; Grattard, F.; Gonzalo, S.; Verhoeven, P.; et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol 2021, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Zanella, I.; Degli Antoni, M.; Marchese, V.; Castelli, F.; Quiros-Roldan, E. Non-neutralizing antibodies: Deleterious or propitious during SARS-CoV-2 infection? Int Immunopharmacol 2022, 110, 108943. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol 2020, 5, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Chen, Y.; Tan, J.; Wang, X.; Zhang, D. Does potential antibody-dependent enhancement occur during SARS-CoV-2 infection after natural infection or vaccination? A meta-analysis. BMC Infect Dis 2022, 22, 742. [Google Scholar] [CrossRef] [PubMed]
- Dugan, H.L.; Stamper, C.T.; Li, L.; Changrob, S.; Asby, N.W.; Halfmann, P.J.; Zheng, N.Y.; Huang, M.; Shaw, D.G.; Cobb, M.S.; et al. Profiling B cell immunodominance after SARS-CoV-2 infection reveals antibody evolution to non-neutralizing viral targets. Immunity 2021, 54, 1290–1303 e1297. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Gonzalez, J.C.; Sievers, B.L.; Mallajosyula, V.; Chakraborty, S.; Dubey, M.; Ashraf, U.; Cheng, B.Y.; Kathale, N.; Tran, K.Q.T.; et al. Early non-neutralizing, afucosylated antibody responses are associated with COVID-19 severity. Sci Transl Med 2022, 14, eabm7853. [Google Scholar] [CrossRef] [PubMed]
- Gogesch, P.; Dudek, S.; van Zandbergen, G.; Waibler, Z.; Anzaghe, M. The Role of Fc Receptors on the Effectiveness of Therapeutic Monoclonal Antibodies. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Bailey, M.J.; Broecker, F.; Leon, P.E.; Tan, G.S. A Method to Assess Fc-mediated Effector Functions Induced by Influenza Hemagglutinin Specific Antibodies. J Vis Exp 2018. [Google Scholar] [CrossRef]
- Fischinger, S.; Fallon, J.K.; Michell, A.R.; Broge, T.; Suscovich, T.J.; Streeck, H.; Alter, G. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J Immunol Methods 2019, 473, 112630. [Google Scholar] [CrossRef]
- Ackerman, M.E.; Moldt, B.; Wyatt, R.T.; Dugast, A.S.; McAndrew, E.; Tsoukas, S.; Jost, S.; Berger, C.T.; Sciaranghella, G.; Liu, Q.; et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J Immunol Methods 2011, 366, 8–19. [Google Scholar] [CrossRef]
- de Neergaard, T.; Sundwall, M.; Wrighton, S.; Nordenfelt, P. High-Sensitivity Assessment of Phagocytosis by Persistent Association-Based Normalization. J Immunol 2021, 206, 214–224. [Google Scholar] [CrossRef] [PubMed]
- de Neergaard, T.; Nordenfelt, P. Quantification of Phagocytosis Using Flow Cytometry. Methods Mol Biol 2023, 2674, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, H.; Crowley, A.R.; Butler, S.E.; Xu, S.; Weiner, J.A.; Bloch, E.M.; Littlefield, K.; Wieland-Alter, W.; Connor, R.I.; Wright, P.F.; et al. Markers of Polyfunctional SARS-CoV-2 Antibodies in Convalescent Plasma. mBio 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Izadi, A.; Hailu, A.; Godzwon, M.; Wrighton, S.; Olofsson, B.; Schmidt, T.; Soderlund-Strand, A.; Elder, E.; Appelberg, S.; Valsjo, M.; et al. Subclass-switched anti-spike IgG3 oligoclonal cocktails strongly enhance Fc-mediated opsonization. Proc Natl Acad Sci U S A 2023, 120, e2217590120. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, L.B.; Tedla, N.; Bull, R.A. Broadly-Neutralizing Antibodies Against Emerging SARS-CoV-2 Variants. Front Immunol 2021, 12, 752003. [Google Scholar] [CrossRef] [PubMed]
- Wilen, C.B.; Tilton, J.C.; Doms, R.W. HIV: cell binding and entry. Cold Spring Harb Perspect Med 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.; Nguyen, D.N.; Tolbert, W.D.; Gasser, R.; Ding, S.; Vezina, D.; Yu Gong, S.; Prevost, J.; Gendron-Lepage, G.; Medjahed, H.; et al. Across Functional Boundaries: Making Nonneutralizing Antibodies To Neutralize HIV-1 and Mediate Fc-Mediated Effector Killing of Infected Cells. mBio 2021, 12, e0140521. [Google Scholar] [CrossRef]
- Beaudoin-Bussieres, G.; Prevost, J.; Gendron-Lepage, G.; Melillo, B.; Chen, J.; Smith Iii, A.B.; Pazgier, M.; Finzi, A. Elicitation of Cluster A and Co-Receptor Binding Site Antibodies are Required to Eliminate HIV-1 Infected Cells. Microorganisms 2020, 8. [Google Scholar] [CrossRef]
- Nolan, O.; O'Kennedy, R. Bifunctional antibodies: concept, production and applications. Biochim Biophys Acta 1990, 1040, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Longo, D.L. The Expanding Clinical Role of Bifunctional Antibodies. N Engl J Med 2022, 387, 2287–2290. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.A.; Gramespacher, J.A.; Pance, K.; Rettko, N.J.; Solomon, P.; Jin, J.; Lui, I.; Elledge, S.K.; Liu, J.; Bracken, C.J.; et al. Bispecific VH/Fab antibodies targeting neutralizing and non-neutralizing Spike epitopes demonstrate enhanced potency against SARS-CoV-2. MAbs 2021, 13, 1893426. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Ravetch, J.V. Functional diversification of IgGs through Fc glycosylation. J Clin Invest 2019, 129, 3492–3498. [Google Scholar] [CrossRef]
- Delidakis, G.; Kim, J.E.; George, K.; Georgiou, G. Improving Antibody Therapeutics by Manipulating the Fc Domain: Immunological and Structural Considerations. Annu Rev Biomed Eng 2022, 24, 249–274. [Google Scholar] [CrossRef]
- van der Horst, H.J.; Nijhof, I.S.; Mutis, T.; Chamuleau, M.E.D. Fc-Engineered Antibodies with Enhanced Fc-Effector Function for the Treatment of B-Cell Malignancies. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Oldham, R.J.; Teal, E.; Beers, S.A.; Cragg, M.S. Fc-Engineering for Modulated Effector Functions-Improving Antibodies for Cancer Treatment. Antibodies (Basel) 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.L.; Chen, H.; Karki, S.; Lazar, G.A. Engineered Fc variant antibodies with enhanced ability to recruit complement and mediate effector functions. MAbs 2010, 2, 181–189. [Google Scholar] [CrossRef]
- Borrok, M.J.; Luheshi, N.M.; Beyaz, N.; Davies, G.C.; Legg, J.W.; Wu, H.; Dall'Acqua, W.F.; Tsui, P. Enhancement of antibody-dependent cell-mediated cytotoxicity by endowing IgG with FcalphaRI (CD89) binding. MAbs 2015, 7, 743–751. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




