Submitted:
29 March 2024
Posted:
01 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Exosome Purification
2.2. Proteomics
2.2.1. Chemicals and Reagents:
2.2.2. S-Trap Digestion Protocol:
2.2.3. LC-MS Parameters:
2.2.4. Proteomics Analysis:
2.3. MiRNA Sequencing
3. Results
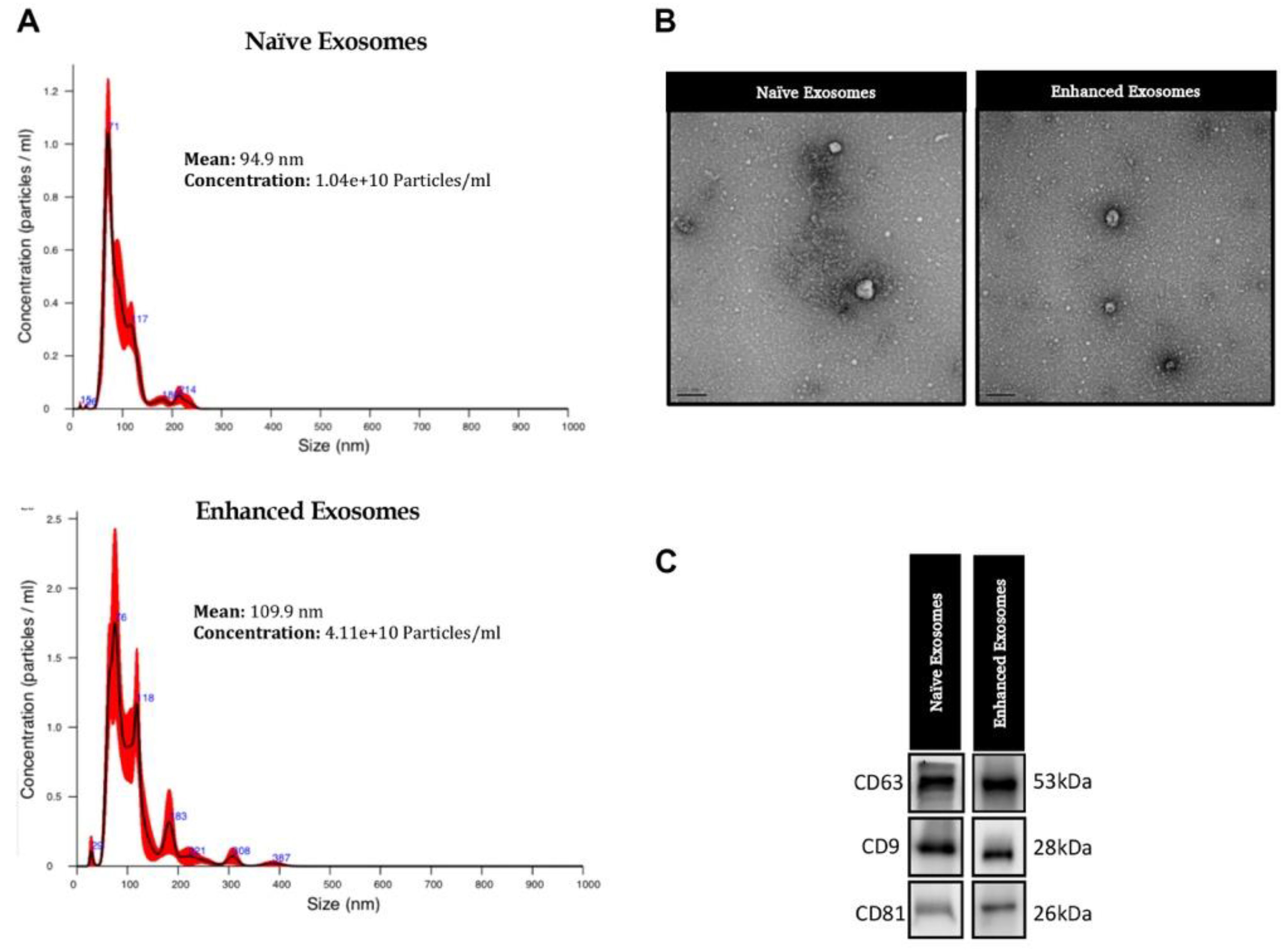
3.1. Exosome Characteristics
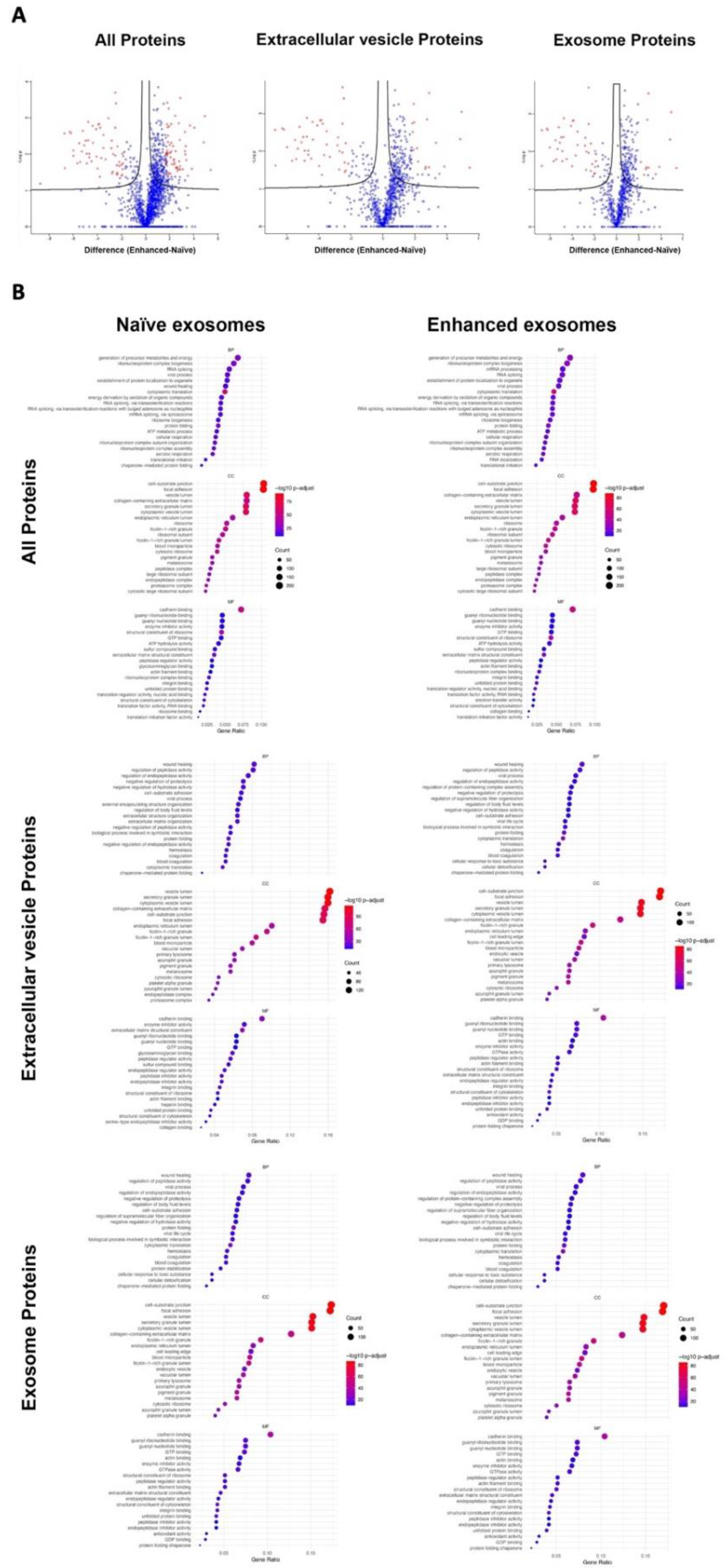
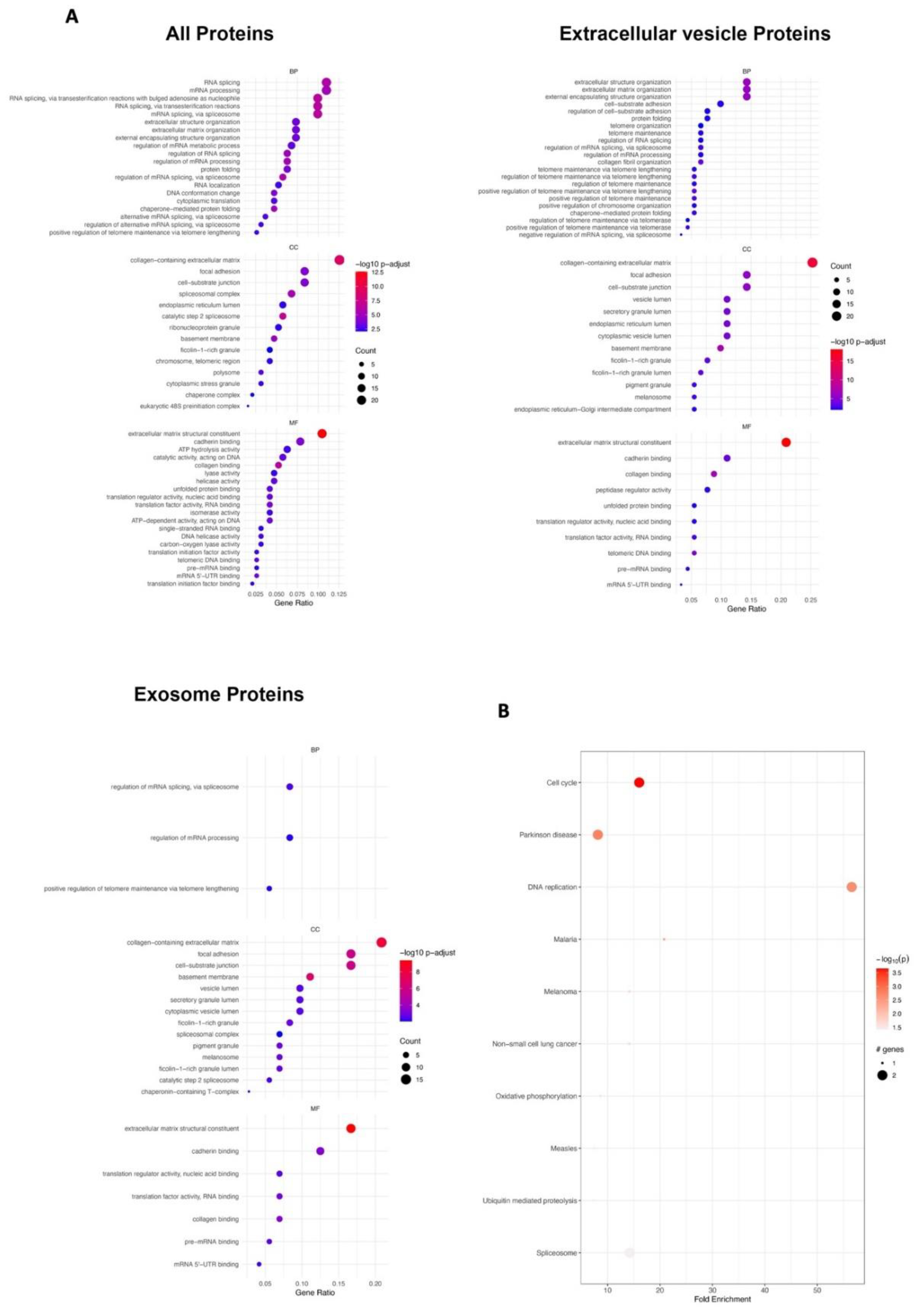
3.2. Bioinformatics Analyses of Proteins Derived from Naïve Exosomes and Enhanced Exosomes
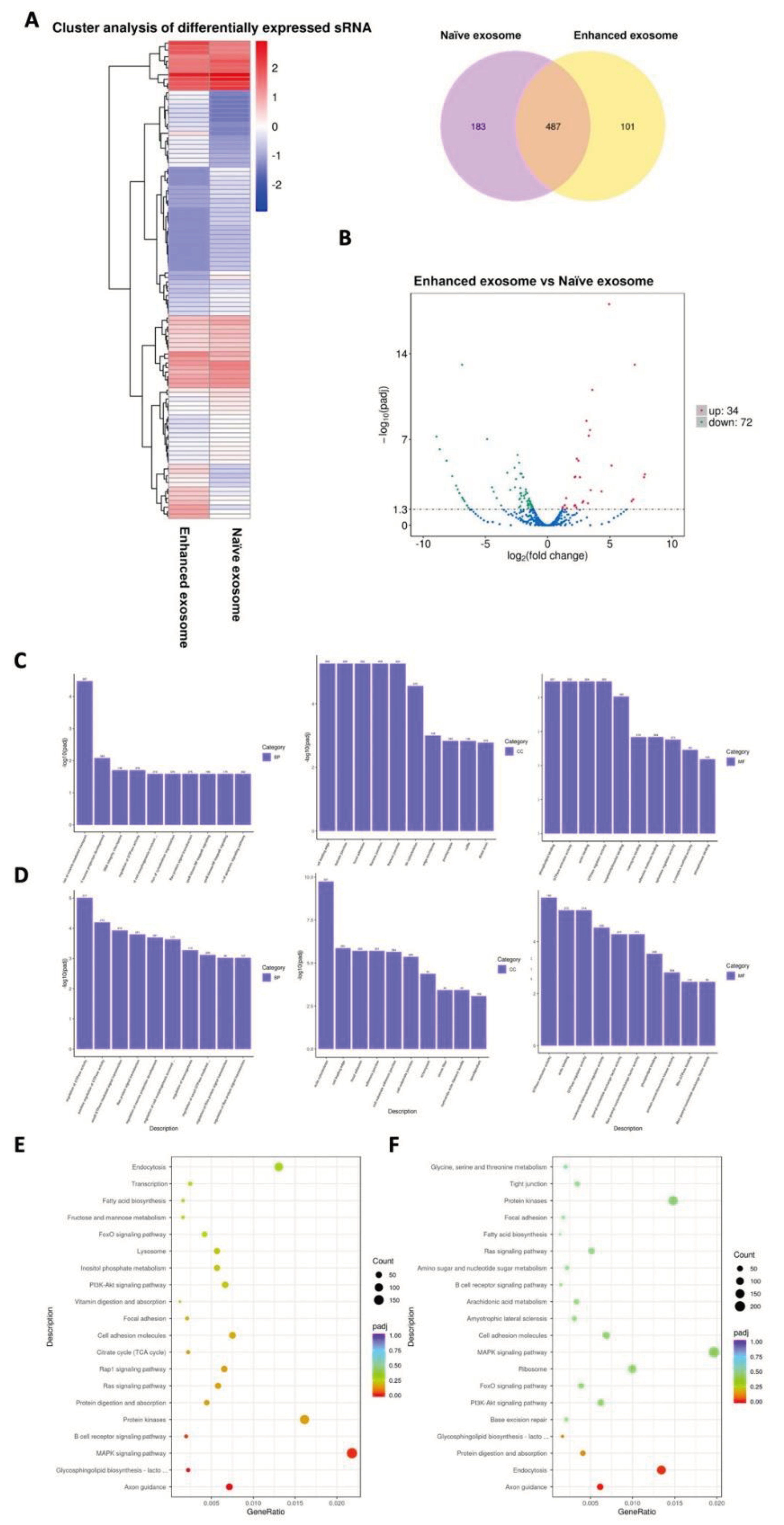
3.3. Bioinformatics Analyses of miRNAs Derived from Naïve Exosomes and Enhanced Exosomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gurung, G.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal 2021, 19, 47. [Google Scholar] [CrossRef] [PubMed]
- McNamara, R.P.; Dittmer, D.P. Modern Techniques for the Isolation of Extracellular Vesicles and Viruses. J Neuroimmune Pharmacol 2020, 15, 459–472. [Google Scholar] [CrossRef]
- Zhao, R.; Zhao, T.; He, Z.; Cai, R.; Pang, W. Composition, isolation, identification and function of adipose tissue-derived exosomes. Adipocyte 2021, 10, 587–604. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Bella, M.A.D. Overview and Update on Extracellular Vesicles: Considerations on Exosomes and Their Application in Modern Medicine. Biology 2022, 11, 804. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.H.; Badierah, R.; Redwan, E.M.; El-Fakharany, E.M. A Comprehensive Insight into the Role of Exosomes in Viral Infection: Dual Faces Bearing Different Functions. Pharmaceutics 2021, 13, 1405. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, A.; Wrzecińska, M.; Czerniawska-Piątkowska, E.; Kupczyński, R. Exosomes–Spectacular role in reproduction. Biomed Pharmacother 2022, 148. [Google Scholar] [CrossRef]
- Baranyai, T.; Herczeh, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.J.; El Andaloussi, S.; Pálinkás, Z.; Kumar, V.; Nagy, P.; Kittel, A.; Buzás, E.I.; Ferdinandy, P.; Giricz, Z. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PloS One 2015, 10. [Google Scholar] [CrossRef]
- Street, J.M.; Koritzinsky, E.H.; Glispie, D.M.; Star, R.A.; Yuen, P.S.T. Urine Exosomes: An Emerging Trove of Biomarkers. Adv Clin Chem 2017, 78, 103–122. [Google Scholar] [CrossRef]
- Cheshmi, B.; Cheshomi, H. Salivary exosomes: properties, medical applications, and isolation methods. Mol Biol Rep 2020, 47, 6295–6307. [Google Scholar] [CrossRef]
- Han, J.S.; Kim, S.E.; Jin, J.Q.; Park, N.R.; Lee, J.Y.; Kim, H.L.; Lee, S.B.; Yang, S.W.; Lim, D.J. Tear-Derived Exosome Proteins Are Increased in Patients with Thyroid Eye Disease. Int J Mol Sci 2021, 22, 1115. [Google Scholar] [CrossRef] [PubMed]
- Galley, J.D.; Besner, G.E. The Therapeutic Potential of Breast Milk-Derived Extracellular Vesicles. Nutrients 2020, 12, 745. [Google Scholar] [CrossRef] [PubMed]
- Zamani, P.; Fereydouni, N.; Butler, A.E.; Navashenaq, J.G.; Sahebkar, A. The therapeutic and diagnostic role of exosomes in cardiovascular diseases. Trends Cardiovasc Med 2019, 6, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tian, L.; Lu, J.; Ng, I.O.L. Exosomes and cancer—Diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis 2022, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.G.; Grizzle, W.E. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. The Am J Pathol 2014, 184, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Kar, K.; Dhar, R.; Mukherjee, S.; Nag, S.; Gorai, S.; Mukerjee, N.; Mukherjee, D.; Vatsa, R.; Jadhav, M.C.; Ghosh, A.; Devi, A.; Krishnan, A.; Thorat, N.D. Exosome-Based Smart Drug Delivery Tool for Cancer Theranostics. ACS Biomater Sci Eng 2023, 9, 577–594. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, L.; Zeng, X.; Schwarz, H.; Nanda, H.S.; Peng, X.; Zhou, Y. Exosomes, a New Star for Targeted Delivery. Front Cell Dev Biol 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Guo, S.; Ren, X.; Wu, Z.; Liu, S.; Yao, X. Current Strategies for Exosome Cargo Loading and Targeting Delivery. Cells 2023, 12, 1416. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Minerva, S.; Shah, R.; Bhat, A.; Verma, S.; Chander, G.; Bhat, G.R.; Thapa, N.; Bhat, A.; Wakhloo, A.; Ganie, M.A. Role of genetic, environmental, and hormonal factors in the progression of PCOS: A review. Journal of Reproductive Healthcare and Medicine 2022, 3. [Google Scholar] [CrossRef]
- Park, H.-S.; Chugh, R.M.; Seok, J.; Cetin, E.; Mohammed, H.; Siblini, H.; Ali, F.L.; Ghasroldasht, M.M.; Alkelani, H.; Elsharoud, A.; Ulin, M.; Esfandyari, S.; Al-Hendy, A. Comparison of the therapeutic effects between stem cells and exosomes in primary ovarian insufficiency: as promising as cells but different persistency and dosage. Stem Cell Res Ther 2023, 14, 165. [Google Scholar] [CrossRef]
- Yin, X.; Fang, S.; Wang, M.; Wang, Q.; Fang, R.; Chen, J. EFEMP1 promotes ovarian cancer cell growth, invasion and metastasis via activated the AKT pathway. Oncotarget 2016, 7, 47938–47953. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.Q.; Zhu, X.T.; Zhang, S.N.; Ma, Y.F.; Han, Y.H.; Jiang, Y.; Zhang, Y.H. Premature ovarian insufficiency: a review on the role of oxidative stress and the application of antioxidants. Front Endocrinol 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Gagné, A.; Têtu, B.; Orain, M.; Turcotte, S.; Plante, M.; Grégoire, J.; Renaud, M.-C.; Bairati, I.; Trudel, D. HtrA1 expression and the prognosis of high-grade serous ovarian carcinoma: a cohort study using digital analysis. Diag Pathol 2018, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Khyrana, A.; Maguire, J.L.; Chien, J.; Shridhar, V. HtrA1 sensitizes ovarian cancer cells to cisplatin-induced cytotoxicity by targeting XIAP for degradation. Int J cancer 2012, 130, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, S.W.; Jung, M.H.; Kim, Y.S.; Kim, K.S.; Suh, D.S.; Kim, K.H.; Choi, E.H.; Kim, J.; Kwon, B.S. Plasma-activated medium inhibits cancer stem cell-like properties and exhibits a synergistic effect in combination with cisplatin in ovarian cancer. Free Radic Biol Med 2022, 182, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Shinozuka, T.; Kanda, M.; Shimizu, D.; Umeda, S.; Takami, H.; Inokawa, Y.; Hattori, N.; Hayashi, M.; Tanaka, C.; Nakayama, G.; Kodera, Y. Identification of stromal cell-derived factor 4 as a liquid biopsy-based diagnostic marker in solid cancers. Sci Rep 2023, 13, 15540. [Google Scholar] [CrossRef]
- Qu, W.; Chen, X.; Wang, J.; Lv, J.; Yan, D. MicroRNA-1 inhibits ovarian cancer cell proliferation and migration through c-Met pathway. Clin Chim Acta 2017, 473, 237–244. [Google Scholar] [CrossRef]
- Zhang, D.; Qu, B.; Hu, B.; Cao, K.; Shen, H. MiR-1-3p enhances the sensitivity of ovarian cancer cells to ferroptosis by targeting FZD7. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2022, 47, 1512–1521. [Google Scholar] [CrossRef]
- Shang, A.; Yang, M.; Shen, F.; Wang, J.; Wei, J.; Wang, W.; Lu, W.; Wang, C.; Wang, C. MiR-1-3p Suppresses the Proliferation, Invasion and Migration of Bladder Cancer Cells by Up-Regulating SFRP1 Expression. Cell Physiol Biochem 2017, 41, 1179–1188. [Google Scholar] [CrossRef]
- Li, S.-M.; Wu, H.-L.; Yu, X.; Tang, K.; Wang, S.-G.; Ye, Z.-Q.; Hu, J. The putative tumour suppressor miR-1-3p modulates prostate cancer cell aggressiveness by repressing E2F5 and PFTK1. J Exp Clin Cancer Res 2018, 37, 219. [Google Scholar] [CrossRef]
- Gao, L.; Yan, P.; Guo, F.F.; Liu, H.J.; Zhao, Z.F. MiR-1-3p inhibits cell proliferation and invasion by regulating BDNF-TrkB signaling pathway in bladder cancer. Neoplasma 2018, 65, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Li, F.; Xu, S.; Hu, J.; Gao, L. The important role of miR-1-3p in cancers. J Transl Med 2023, 21, 769. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, H.; Long, M.; Song, L.; Meng, Z.; Lin, S.; Zhang, Y.; Qin, J. Icariin attenuates the tumor growth by targeting miR-1-3p/TNKS2/Wnt/β-catenin signaling axis in ovarian cancer. Front Oncol 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Song, W.; Xu, D.; Wu, J.; Gao, R. Mechanisms of miR-103a-3p/CHI3L1 in proliferation and vascular mimicry of ovarian cancer cells. J Int Oncol 2020. [Google Scholar] [CrossRef]
- Duan, Y.; Dong, Y.; Dang, R.; Hu, Z.; Yang, Y.; Hu, Y.; Cheng, J. MiR-122 inhibits epithelial mesenchymal transition by regulating P4HA1 in ovarian cancer cells. Cell Biol Int 2018, 42, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, S.; Liu, X. MicroRNA profiling of plasma exosomes from patients with ovarian cancer using high-throughput sequencing. Oncol Lett 2019, 17, 5601–5607. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Luo, Y.; Li, X. Circ_0072995 Promotes Ovarian Cancer Progression Through Regulating miR-122-5p/SLC1A5 Axis. Biochem Genet 2022, 60, 153–172. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; You, T.; Jing, J. MiR-125b inhibits cell biological progression of Ewing's sarcoma by suppressing the PI3K/Akt signalling pathway. Cell Prolif 2014, 47, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Prizant, H.; Light, A.; Biswas, A.; Hayes, E.; Lee, H.-J.; Baead, D.; Gleicher, N.; Hammes, S.R. Androgens regulate ovarian follicular development by increasing follicle stimulating hormone receptor and microRNA-125b expression. Proc Natl Acad Sci USA 2014, 111, 3008–3013. [Google Scholar] [CrossRef]
- Chen, P.; Sun, L.-S.; Shen, H.-M.; Qu, B. LncRNA KCNQ1OT1 accelerates ovarian cancer progression via miR-125b-5p/CD147 axis. Pathol Res Pract 2022, 239. [Google Scholar] [CrossRef]
- Liu, X.; Ma, L.; Rao, Q.; Mao, Y.; Xin, Y.; Xu, H.; Li, C.; Wang, X. MiR-1271 Inhibits Ovarian Cancer Growth by Targeting Cyclin G1. Med Sci Monit 2015, 19, 3152–3158. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Shi, J.; Xu, X. MicroRNA-1271-5p inhibits the tumorigenesis of ovarian cancer through targeting E2F5 and negatively regulates the mTOR signaling pathway. Panminerva Med 2021, 63, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Enolida, H.; Tatarano, S.; Kawahara, K.; Uchida, Y.; Nishiyama, K.; Fujimura, L.; Kikkawa, N.; Seki, N.; Nakagawa, M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer 2010, 1–2, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Chiyomaru, T.; Kawakami, K.; Yoshino, H.; Enokida, H.; Nohata, N.; Fuse, M.; Ichikawa, T.; Naya, Y.; Nakagawa, M.; Seki, N. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer 2012, 106, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Zhang, S.; Shan, C.; Zhou, L.; Zhou, Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J 2013, 280, 3962–3974. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Xia, B.; Meng, F.; Lou, G. miR-133a suppresses ovarian cancer cell proliferation by directly targeting insulin-like growth factor 1 receptor. Tumor Biol 2014, 35, 1557–1564. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Zhang, Y. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol Res Pract 2021, 217. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.-Z.; Lou, X.-Y.; Lv, Q.-L.; Cheng, L.; Wu, N.-Y.; Hu, L.; Zhou, H.-H. MicroRNA-184 acts as a potential diagnostic and prognostic marker in epithelial ovarian cancer and regulates cell proliferation, apoptosis and inflammation. Phaemazie 2015, 70, 668–673. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Zhang, Y.-Y.; Zhu, B.-L.; Feng, F.-Z.; Zhang, H.-T.; Yan, H.; Zhou, B. MiR-203a-3p regulates the biological behaviors of ovarian cancer cells through mediating the Akt/GSK-3β/Snail signaling pathway by targeting ATM. J Ovarian Res 2019, 12, 60. [Google Scholar] [CrossRef]
- Li, T.; Li, Y.; Rehmani, H.; Guo, J.; Padia, R.; Calbay, O.; Ding, Z.; Jiang, Y.; Jin, L.; Huang, S. Attenuated miR-203b-3p is critical for ovarian cancer progression and aptamer/miR-203b-3p chimera can be explored as a therapeutic. Advances in Cancer Biology Metastasis 2022. [Google Scholar] [CrossRef]
- Cheng, Y.; Ma, X.-L.; Wei, Y.-Q.; Wei, X.-W. Potential roles and targeted therapy of the CXCLs/CXCR2 axis in cancer and inflammatory diseases. Biochim Biophys Acta Rev Cancer 2019, 1871, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Xie, Y.; Zhuang, X.; Yuan, Z. MiR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed Pharmacother 2018, 104, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Sheng, N.; Xu, Y.-Z.; Xi, Q.-H.; Jiamg, H.-Y.; Wang, C.-Y.; Zhang, Y.; Ye, Q. Overexpression of KIF2A is Suppressed by miR-206 and Associated with Poor Prognosis in Ovarian Cancer. Cell Physiol Biochem 2018, 50, 810–822. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Guo, R.; Yuan, Z.; Shi, H.; Zhang, D. LncRNA HOTAIR Regulates CCND1 and CCND2 Expression by Sponging miR-206 in Ovarian Cancer. Cell Physiol Biochem 2018, 49, 1289–1303. [Google Scholar] [CrossRef]
- Boscaro, C.; Baggio, C.; Carotti, M.; Sandonà, D.; Trevisi, L.; Cignarella, A.; Bolego, C. Targeting of PFKFB3 with miR-206 but not mir-26b inhibits ovarian cancer cell proliferation and migration involving FAK downregulation. FASEB J 2022, 36. [Google Scholar] [CrossRef] [PubMed]
- Udesen, P.B.; Sørensen, A.E.; Svendsen, R.; Frisk, N.L.S.; Hess, A.L.; Aziz, M.; Wissing, M.L.M.; Englund, A.L.M.; Dalgaard, L.T. Circulating miRNAs in Women with Polycystic Ovary Syndrome: A Longitudinal Cohort Study. Cells 2023, 12, 983. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Li, J.; Zhang, B.; Hu, J.; Ma, J.; Cui, L.; Chen, Z.-J. Differential expression profile of plasma exosomal microRNAs in women with polycystic ovary syndrome. Fertil Steril 2021, 115, 782–792. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, Y.; Zhao, H.; Yng, Z.; Kang, Y. Aberrant miRNA-mRNA regulatory network in polycystic ovary syndrome is associated with markers of insulin sensitivity and inflammation. AnnTransl Med 2021, 9, 1405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xiao, H.; Zhang, X.; Qiukai, E.; Gong, X.; Li, T.; Han, Y.; Ying, X.; Cherrington, B.D.; Xu, B.; Liu, X.; Zhang, X. Decreased microRNA-125b-5p disrupts follicle steroidogenesis through targeting PAK3/ERK1/2 signalling in mouse preantral follicles. Metabolism 2020, 107. [Google Scholar] [CrossRef]
- McAllister, J.M.; Han, A.X.; Modi, B.P.; Teves, M.E.; Mavodza, G.R.; Anderson, Z.L.; Shen, T.; Christenson, L.K.; Archer, K.J.; Strauss, J.F. miRNA Profiling Reveals miRNA-130b-3p Mediates DENND1A Variant 2 Expression and Androgen Biosynthesis. Endocrinology 2019, 160, 1964–1981. [Google Scholar] [CrossRef]
- Kulkarni, R.; Teves, M.E.; Han, A.X.; McAllister, J.M.; Strauss, J.F., 3rd. Colocalization of Polycystic Ovary Syndrome Candidate Gene Products in Theca Cells Suggests Novel Signaling Pathways. J Endocor Soc 2019, 3, 2204–2223. [Google Scholar] [CrossRef] [PubMed]
- Waterbury, J.S.; Teves, M.E.; Gaynor, A.; Han, A.X.; Mavodza, G.; Newell, J.; Strauss, J.F., 3rd; McAllister, J.M. The PCOS GWAS Candidate Gene ZNF217 Influences Theca Cell Expression of DENND1A.V2, CYP17A1, and Androgen Production. J Endocor Soc 2022, 6. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhong, Z.; Chi, H.; Huang, M.; Jiang, R.; Che, J. Genome-Wide Screen of miRNAs and Targeting mRNAs Reveals the Negatively Regulatory Effect of miR-130b-3p on PTEN by PI3K and Integrin β1 Signaling Pathways in Bladder Carcinoma. Int J Mol Sci 2016, 18, 78. [Google Scholar] [CrossRef] [PubMed]
- Gioacchini, G.; Notarstefano, V.; Sereni, E.; Zacà, C.; Coticchio, G.; Giorgini, E.; Vaccari, L.; Carnevali, O.; Borini, A. Does the molecular and metabolic profile of human granulosa cells correlate with oocyte fate? New insights by Fourier transform infrared microspectroscopy analysis. Mol Hum Reprod 2018, 24, 521–532. [Google Scholar] [CrossRef]
- Pangas, S.A. Bone morphogenetic protein signaling transcription factor (SMAD) function in granulosa cells. Mol Cell Endocrinol 2012, 356, 40–47. [Google Scholar] [CrossRef]
| miRNAs | Targets |
| hsa-miR-103a-3p | IL32, P4HA2, DRD4, TMEM101, TEKT2, PPP6R2, PLEK2, CHGA, USP11, PEX11G, NOD1, HSD17B1, PPY, OSCP1, TBX4, FBXO24, POLM, ITPR2, UBE2C, WFDC3, PACSIN1, DDX49, ANGPTL8, MYBPH, MORC4, OS9, SQOR, SLC3A1, VSX2, CCNE1, MOV10L1, SIRPB1, SEMA3A, HPX, ETNK1, ITGAX, NDUFB10, DEF8, ZNF750, SIK1, SLC44A3, REG3G, GPR6, HIKESHI, CC2D1B, OBSCN, GSTT2B, SPON2, PIP, SSU72, OLFML2B, BOLA3, ZDHHC19, FBXW12, SERPINA4, TMED3, PHB, DEFB4A, WDR48, UMOD, ANKS3, DISP3, EXOSC10, PTCRA, REG3A, MTCL1, C17orf62, AVEN, NT5DC2, MYEOV, CEP290, ALAS1, MADD, SLC22A8, HMGA1, MCTP1, RPS6KB2, ST8SIA5, DMAP1, CCNG2, DEFB4B, SLC25A20, TMEM184A, SLC26A10, SPACA4, ZNF491, CD37, PRMT5, CYP4F12, IL32, KDSR, CTAG1B, TRMT12, CLEC4G, SLC52A2, ANKRD11, TMEM89, NAT8L, ACSM5, PGP, CCDC84, VSTM2B, FRMD1, ZFYVE19, ESR1, NAPRT, TBC1D16, OAS2, CD55, SLC35E4, CASZ1, PCNX2, SDSL, ZNF683, GPAA1, KEL, OR8B4, XAB2, KCNN3, LFNG, NIF3L1, MAP4, DISP1, FLNA, TEDC2, SCAMP5, GLMP, TPR, LY9, CD1E, SH2D2A, KHDC4, CALHM6, SELENBP1, ACADSB, RPRD2, LDOC1, ZDHHC16, DIRAS3, SLC29A1, RAPGEF1, CD40, YIPF3, TFAP2E, TMEM35B, TGIF2, MFSD14C, SP5, WDR45, SPATA31A7, BTN3A2, PCDHA4, SEPT8, DCTN3, TTLL10, AVP, SLC22A23, ARHGAP6, MTUS2, SLC13A5, ITSN1, TOM1, ZG16B, CTBP1, ARNTL, KIF19, SIGLEC6, AKT1S1, FCRL6, SORBS2, ANKRD39, SEMA5B, VKORC1, IZUMO4, WDR6, AKAP10, CCDC51, KIR2DL4, DDR1, RAB2A, CHMP1A, TREH, GSTP1, MS4A18, ANO7, NAGA, C2orf73, RHBDD3, RPS14, PIGQ, KCNC4, ADPRHL1, CACNA1B, CHN1, STK11IP, SLC2A6, KIAA0930, A1CF, PGC, FAM229A, IFRD2, SLC2A8, CD6, THNSL2, IFT172, C3orf18, MRPL10, HYAL2, KBTBD2, PSPH, BCS1L, PLA2G6, PHF1, SLCO2B1, PIGV, SENP2, ARHGEF33, SRPK3, ZNF485, RLN3, VAMP8, CLCN1, RNF144A, PSMD2, PFKP, COL6A3, HJURP, SLC12A9, EDEM1, EPM2A, HAUS7, TRIM26, NCDN, SNRK, TST, ZNF382, FBXO34, PDE2A, C16orf91, SEMA3E, FAM90A1, FOLR3, GTF2I, OPN4, OR10C1, CCDC188, BCAS4, GSDME, CCT3, NR5A2, MRPL28, ZNF185, TTC22, ORC5, MBOAT7, TMEM150A, ANAPC1, C2orf92, DOT1L, RBKS, NDUFS8, FAM126B, CLCN4, FOLR2, PDE3B, DCAF6, NELFA, PCSK5, SCMH1, GOLGA2, IRF1, XPNPEP1, KLHDC8A, ATP11C, CFLAR, PPP2R5D, TNNC1, C1orf116, TFPI2, SLC25A48, CBS, PPP1R16B, ALAS2, D2HGDH, ASCC2, MRPS5, PRKD3, PGAP2, IL1RAP, DEGS1, RABL6, PYCR2, GPR157, PLP1, SYP, CYP21A2, MFNG, TMEM8A, PIM3, ARMCX3, CREB3L2, DESI1, INPP4A, TRAIP, SLC35E1, LAS1L, MTA1, OSBPL10, STK36, HKDC1, CKS1B, DPH2, NOM1, RASSF4, PKD1, ACADM, ARF1, NFKBIL1, CNN3, ALPP, DNAL4, TBC1D10B, MFSD4A, UBR4, LAMB1, SPINK5, SACS, CLIC5, AP5Z1, FANCC, ITGB3BP, PQLC1, BCR, SETD5, TM9SF4, KREMEN1, SLC38A5, ZC3H11A, CCM2, PDZD4, KDM4A, XKR8, DTNB, PABPC1L, TP53BP2, EWSR1, RGR, COPS6, PDLIM4, STIMATE, NPDC1, CCZ1B, NEBL, TSPAN32, GSDMB, ANKS6, ALB, LBR, ISM2, AC114490.2, ARIH2, RAB11FIP3, PSD, MMP11, TMCO4, MZT2A, CREM, IARS2, UBAC1, CACUL1, ATRAID, TOP2B, C3orf58, ZNF789, AL080251.1, SLC4A2, EIF3B, JAK1, MFSD2B, SH3BP1, AMT, PCMT1, LY6G6C, KDM4C, LAMC1, NRXN2, STX1A, LZTR1, MAST2, TNIP2, DGKQ, MARCH6, ABCC3, NR1I3, ATP13A2, C5orf34, ZDHHC11, INPP4B, PXYLP1, TLR1, SIL1, THAP9, SLC9B2, CHIT1, AMN1, ARL10, RGS7BP, IRF2, IFT122, SDHA, HK3, TRIP13, SPAG9, ZNF451, NADK2, SH3BP2, CEP72, GYPB, WDR36, PCDH1, ZFAND1, TBC1D9B, ATP6V1C1, PIEZO1, RRNAD1, NSMAF, TRIQK, TSPAN18, STC2, ENTPD4, MROH5, ATP6V1H, MTSS1, PRSS55, ERICH1, PHYHIP, REEP4, AARD, DIS3L, ABCA7, PPP6R3, KCNK7, USP47, NUP214, RPS3, C11orf16, ME3, ST5, PACS1, TK2, PRDM10, RIC8A, ARMS2, SULF1, ANO1, TPP1, CDKL1, CTBP2, INTS11, TP53I11, PCNX3, IGLL5, MOB2, MAJIN, CAMSAP2, RCE1, ACAT1, PPP1R16A, TKFC, POLR2L, NUMA1, PTPRO, PLEKHB1, ARAP1, ACSF3, RELT, PDE1B, NCAPD2, IGHMBP2, SLC22A1, FAAP100, MMP20, M6PR, WDR74, DNAH10, MGP, SLC39A3, SUOX, DDI2, TPH2, HECTD4, HVCN1, KRT78, NTN4, GAS2L3, PFKM, CSAD, DUSP6, PLXNC1, DAZAP2, KRT6A, CA6, DDX23, TSPAN8, FICD, ESPL1, HECTD1, ACTN1, PSEN1, ASB2, PCNX1, MIPOL1, SIPA1L1, PPP1R13B, NIPA1, IDH3A, GATM, MAN2A2, EIF5, RNF31, GABPB1, ADCY4, TLN2, GCNT3, DNAJC17, ALDH1A2, PBX4, GMPR2, SEMA4B, CEP152, FAH, SLC9A5, ATXN2L, PLA2G4F, PRMT7, GALNS, CASKIN1, AC090360.1, EXOC3L1, AMFR, CLCN7, CCDC33, ARMC5, C16orf58, NOL3, CNOT1, TAF1C, ZNF688, VAC14, BAIAP3, DDX19B, NLRP1, NEIL1, ABCC11, GEMIN4, SLC25A11, PELP1, CIC, SPACA6, FN3KRP, TSR1, CLUH, FAM57A, CHRNB1, SGSM2, STRA6, CORO7, AKAP1, HCFC1R1, WNT9B, URI1, NAA60, RASSF5, DUS1L, SMARCD2, TTC39C, MYO15B, ADCYAP1, NARF, TMEM94, ITGB4, RAB31, GDPD1, LRRC45, OGFOD3, CDC6, CCDC57, FLOT2, GHDC, WDR7, TEN1, MAU2, HAUS5, ZNF793, MAP3K14, RBFA, HIGD1B, CXXC1, GPATCH8, FHOD3, ROGDI, TMEM161A, ATP9B, EFTUD2, ACTN4, ZNF532, EIF3G, ALPK2, ZACN, LIG3, EPS8L1, NAGLU, SLC7A10, JSRP1, CTAG1A, ARHGEF1, ADGRE2, ECH1, CD22, ILVBL, MCOLN1, HPN, CCDC97, ZNF17, F2RL3, TMEM38A, PPFIA4, EMG1, KCNN4, LIPE, VAV1, FSD1, HNRNPL, SIGLEC7, ZCCHC18, LCE1C, ARFRP1, GRM4, AL034430.1, MYO19, UNC93B1, PCGF2, MED26, TTLL13P, SPPL2B, AC008162.2, SIK1B |
| hsa-miR-103a-3p | CAMKK1, CNFN, CD69, MXI1, APC, GNB5, TCIRG1, EXPH5, KCTD7, SEC16A, KCNMA1, PKNOX1, TNFSF12, FASTK, HRASLS5, MAP2K1, FAM53A, NOSTRIN, GPR137C, ZAR1, APP, ZFYVE26, MYO9A, SMOC1, SOX13, CFAP46, TTF2, ZNFX1, GLO1, MAP3K8, REL, CYP1A1, TSPAN4, GATD1, NONO, KMT2B, IFT22, MFSD13A, MED20, DGCR6, ZNF879, AL139260.3, DLG2, TMEM72, BRDT, STAT3, PPP1R2, LAMB2, TMEM54, SNX17, MFN1, SLC11A1, ASB18, HECW1, CERCAM, SMARCD3, NDUFV2, DENND2A, TOGARAM2, PLIN2, MMACHC, MRPL38, RNF146, SPTLC1, GTF2IRD2, MPP3, CAPN10, ITGB1BP2, CFP, PTP4A2, TTLL3, SLC2A11, FAM45A, SUB1, MGAT1, ZNF346, CNGA1, FER, ELMOD2, CORIN, WDR17, MZB1, ARL15, UBE2Z, ZNF706, ITK, ACAT1, DDX25, CAPN5, GDPD5, DEAF1, TRMT9B, MRPL16, FKBP4, MLXIP, ARAP1, SLC38A10, LETMD1, ACTR6, ATAD2, DAAM1, TMCO5A, DET1, IVD, RPS27L, KATNBL1, MAN2A2, SQOR, METTL22, STX1B, GINS3, SPG7, CDH3, WDR59, PPP1R27, MPDU1, MYCBPAP, ZZEF1, CCNE1, TMEM105, LASP1, CCDC57, ERBB2, CLPTM1, FOSB, KDSR, GPI, DNAJC7, KEAP1, KDM4B, ZFR2, CACNA1A, DAPK3, SIPA1L3, LIG1, ZNF17, IRF3, SNRPA, SELENOW, USP29, EMC8, ELAC2, MMRN2 |
| hsa-miR-122-5p | PPP2R5B, GATC, VPS53, SLC25A34, XPO6, SWI5, NUDT1, GPRC5C, OR2AK2, BGLAP, TSPAN2, BPIFB3, CFAP74, RAB3IP, CAP2, DENND1A, DOT1L, FAM118A, HECW2, G6PC2, VTCN1, FHIT, SLC11A1, THADA, RAC1, CMTR1, STX7, ZP3, SFXN3, PPP2R2D, INPP5K, ZAP70, DMXL1, ETF1, SLC2A9, FUT10, GEMIN5, PLEKHA7, C11orf49, MROH1, FPGT-TNNI3K, GNS, APPL2, UBAP1L, FANCA, BEAN1, SIRT7, RAB8A, DNMT1, CCDC155, NUMBL, TMEM185A |
| hsa-miR-125b-2-3p | GUCA1A, PTBP3, ISL1, NPC2, SLC52A1, EDAR, HINT2, MUL1, GCKR, SPIB, AWAT2, RPP30, PLEKHG4B, BTG2, SOX8, KCNT2, CAPN13, PF4, CCDC174, B4GALT2, CCPG1, BANF1, TPRX1, WDR45, RPAIN, HCAR2, BRCC3, RTN4RL2, AGAP3, SH2D6, ZKSCAN7, NPAS3, CHRD, THSD4, TBX19, NR1I3, SARDH, MMRN2, KLF17, CLCN5, CYP1A1, CIITA, CHD2, DOK5, NRL, IL25, ARID1B, C19orf24, CD8A, VDAC2, CFAP65, TMEM250, IL1R1, AC037459.1, DPCD, PRSS45, WDCP, AC023509.3, TPR, ZNF674, GLP2R, KCP, ST3GAL5, MICAL3, MRPS31, TRANK1, LAMA5, ABCA2, TFIP11, GTF2I, C2orf42, LETM1, NOL8, PCOLCE, DLG5, ACSS2, ECE1, RNF182, CERCAM, VTI1A, NIT1, ECE2, KRBOX4, SKIL, PPP2R1A, IL11RA, C21orf2, C2orf70, ECHDC2, SPINK5, ROR1, DNER, HLADOA, LRP1B, TMLHE, RGS3, TAB1, IL9R, FAAH, CCT3, ZNF491, LY75, CENPL, PCOLCE2, PER3, MIA3, DGCR8, SSR3, CNIH3, AURKAIP1, GLB1, RPL9, ACAD9, APBB3, TSC22D3, RCHY1, FAM81B, CANX, RNF175, POLR2B, PALLD, CNOT8, CARD8, SPIDR, ZFHX4, ZNF706, ZNF395, MYOZ3, DECR1, CEP57L1, THAP8, IFITM1, PLEKHA7, MTA2, ZNF692, LRP5, HCAR3, C11orf88, SLC37A4, REPS1, SMPD1, C12orf60, NPEPPS, CRACR2B, ATG16L2, ACACB, TM7SF3, C2CD5, MED21, TARSL2, CCDC91, MPHOSPH9, CD163L1, IBA57, CAPS2, ACVRL1, TWF1, MED13L, MAPKAPK5, TNS2, HDAC4, KRT86, LIN52, NAB2, EAPP, PAK6, TIPIN, CDH11, CARHSP1, UBR1, NRG4, AC026464.3, MYO9A, EARS2, IL4R, ATXN2L, FAN1, CPEB1, ANKFY1, P2RX1, TMEM100, RPL23, PPIAL4A, ASPSCR1, PPIAL4F, PPIAL4E, AC243756.1, PPIAL4C, ZNF559, MOB3A, NKIRAS2, TNNI3, HDHD2, GLYR1, EFTUD2, C18orf25, LIN37, KLK2, LTBP4, ZNF749, PEG3, PHLDB3, ZNF772, AC012254.2, DUSP22 |
| hsa-miR-125b-5p | CYP26B1, MLF2, UBOX5, BCKDK, ANAPC15, PXN, DOK1, ARHGAP40, PACSIN1, LRFN3, MAP3K10, USP29, KMT5C, DDB2, TIMM10, AKAP2, HHAT, OSBPL5, MUL1, ANK2, SFRP5, GFOD2, LRRC46, ZNF593, PTGES2, FAM160B2, CRYAA, FAM207A, IKZF3, MAPKAPK2, ANKRA2, ANPEP, TSC22D4, VPS37C, UTF1, MAP3K11, ABLIM3, MATK, SPEG, BTBD9, CDPF1, AMBRA1, ASB2, CSH1, TMEM107, GADD45GIP1, EIF4E1B, TMEM184A, EHD1, ZNF7, LMNB2, IFNLR1, CTAG1B, HDDC3, SNAI3, SIGIRR, ZNF555, TMLHE, SELENOV, GGT7, OPRL1, ZNF385A, CRTC1, SECISBP2, SP100, SLC26A9, HIST1H2AD, VCX, WDR31, COL11A2, COL18A1, NUTM2B, EPB41, LRRC69, PSMD13, NUTM2G, NEIL1, ADAM11, SERPINH1, EPHA2, ZNF707, SSU72, SIRT5, NT5C1B, BAK1, R3HDM4, SMG5, S100A3, CTAG2, STMN3, DNLZ, AIF1L, GDAP1L1, MANBAL, HSPG2, PLA2G5, SKIV2L, PROSER2, NUTM2D, NUTM2A, RPUSD3, WIZ, PTOV1, CSH2, MED20, UBE2D3, ANKRD13B, CENPO, EMP1, CCDC125, TAGLN2, OTOG, HMX1, CCDC134, UBE2I, COMT, HIC2, TTC7A, VWA3B, MAGEA2B, AFF3, RBM44, MTHFR, DENND6B, MUM1, MECP2, RPS28, SLC19A1, ANLN, SERPINE3, ZNF839, RFC5, POR, CTU1, ADORA2A, C20orf173, DBNL, NUTM2E, AKR1D1, ENTPD6, ZNF780B, HLAF, TOP3B, RGSL1, CRP, EXO1, PNCK, RARRES3, ENO2, FZR1, SPAAR, OSBP2, TMEM131L, MED22, NEK6, AC037459.1, SMARCD2, ADAM12, FANCG, ASCC2, OGFR, C8orf58, SEMA4F, ANKRD60, LYRM4, CFAP65, TBC1D2B, HM13, MGST3, CPT1B, UROD, PDGFRA, DUSP18, PRR14L, SGSM3, C1orf61, RNF103, MIB2, SEMA4D, WDR27, CLSTN1, MPZL1, ACAA1, COL11A1, MOV10, GP6, SSR2, COL21A1, ST7L, SFI1, FGFR2, PARVG, EXOC4, TMPRSS2, NOC2L, TTN, SLC4A11, TSC2, PTPRE, TEP1, PARD3B, ARPC1A, P4HTM, TM2D1, DNAJC11, AUP1, STK11IP, DHX30, TLE6, SNTG2, PARP3, CEP250, LCK, SLCO2A1, ARR3, ARHGEF3, MFAP2, SYNGAP1, PLCD1, NEU1, FES, SEPT8, ABLIM1, QARS, EPS8L3, GPRC5C, ZAP70, PCNX2, NDUFA3, ACP1, LRRC29, RHBDF1, PTPN14, TBC1D5, MUSTN1, PHF1, NR1H3, DNAH1, TCTA, HES2, INTS11, C7orf50, PCYT1A, PIGL, ZBTB48, MDC1, CNBD2, AURKAIP1, ASB3, PTPN18, REG3A, CYB561D2, IKBKG, AGRN, PTK7, NCOR2, MAGEA8, PSAP, CLCNKA, TPI1, ACTR8, HDAC4, SLC2A4RG, HHATL, DNAH10, ADAM10, REG3G, HDAC11, ZC3H12D, CEBPA, P2RX4, FLT4, F12, CDK10, WDR41, PRELID2, COL4A5, PIGG, LYRM9, PDK2, FRMD1, IDUA, ALG1L2, SLC52A1, HARS2, CHCHD6, TRIO, RMDN1, STMN4, GLI4, LYPLA1, ADAM18, AP3M2, DECR1, RAB11FIP1, PNMA2, RPS6KA1, SIGLEC10, FHAD1, MCAM, TP53I11, SPAG11A, GSDMD, PTPRU, TMPRSS13, TTC12, PRMT1, ZNF250, CHID1, ZDHHC5, SPAG11B, GATD1, BRMS1, ST14, DPF2, NDUFS8, SIDT2, E2F8, GRK2, ST5, PLEKHA7, GRAMD1B, SART1, DPP3, OLFML1, TMEM258, TMEM134, MPHOSPH9, CD6, SLC6A12, SFSWAP, VWF, VTN, HIP1R, MMAB, PGA4, PGA5, C1RL, MACROD1, ST8SIA1, PAH, PAX8, IFI27L1, SLC25A29, ITGA7, IDH3A, CCDC33, CD79B, ZNF710, DPP8, PHGR1, CMIP, RSRP1, PRSS36, SPIRE2, CASKIN1, MVP, CDH11, KDM8, RBL2, RPAP1, KIF22, EME2, VAC14, CLEC18A, BRICD5, KATNB1, PLEKHG4, ABCC11, IL34, SLC25A10, CNTROB, SPACA6, AC139530.2, ITGAE, DVL2, CHRNB1, SGSM2, TXNDC17, MRM3, NPLOC4, SLC46A1, ARHGAP44, MAP3K3, RBFOX3, MARCH10, ICAM2, RAB11FIP4, CENPV, RPL36, TBC1D29, TNS4, SCN4A, RAC3, VAPA, GHDC, CATSPERG, SPPL2B, STK11, TMEM161A, DOCK6, CARM1, CXXC1, C19orf66, MAP4K1, PRKAR1A, PODNL1, VMP1, CIRBP, ADAMTSL5, POLRMT, FIZ1, ZNF234, ACP5, APOC4, EPS8L1, C19orf12, COQ8B, PET100, C19orf44, AXL, KLK8, GPR108, FCHO1, ZNF414, SPTBN4, ANGPTL4, SNRPA, SIGLEC7, KXD1, NMRK2, KHSRP, CTAG1A, CD37, SLC25A42, ZNF329, NAPA, TP73, AC116565.1, SSTR3, YTHDF3, HTR2A |
| hsa-miR-1271-5p | ADCK2, GNA11, HDAC7, AQP2, CBX6, TRIB3, ZMYND10, IL17C, PDCD2L, SDF2L1, EEFSEC, ESPL1, BCO1, FAF2, GIPR, DBH, BUD23, ERBB3, GPR17, AIG1, EML3, SLC24A2, TYSND1, GOLGA6A, FASTK, C9orf24, PACSIN3, PLK1, NLRP4, C2orf54, ASPHD1, GPR137, CLASP2, TEX44, FUT7, CABP1, ZNF771, RPS15A, NLRC3, OR8B8, MCF2L2, MT1E, ID2, FAM129C, BET1L, MT1B, DOK7, CDK11B, ING4, RPS21, SDSL, ATG16L1, MAGED2, GIT2, ENTPD6, MYH7, CDK11A, ITIH2, GOLGA8A, CDC42SE2, ODF2, SLC29A3, SKIV2L, MGME1, PRDM16, AC026461.4, CPXM1, GREB1, GPRC5C, BBS1, CELF6, MIEF2, CEP250, RGPD2, SAMD12, SPATA13, CRIP1, RGPD1, THRB, CARF, IFI35, MX2, C21orf33, TVP23C, PROC, POLR1C, SHANK2, SPHK2, EFHD1, OXSM, IFRD1, CARD10, MBD3, C11orf68, CBX5, DBNL, HNF4A, FAM160B2, PLA2G6, CABLES2, F8, FAM171A1, GRAMD4, COL21A1, MBOAT2, SLC12A8, GRAMD1C, TSTD1, CHRNA4, MT1HL1, MIB2, FDPS, WDR31, RAPGEF4, ANO10, ADORA2A, PLXNA3, THADA, CRELD1, IL17D, KIAA2012, HPCA, PRKCZ, LAMB1, SACM1L, P2RY11, RIPOR2, DENND2D, ABCF2, RNF220, RAB34, ERGIC3, ASAP3, RUNDC3B, UTP6, PRPSAP2, GUSB, USP19, EPHA2, TIE1, LAMB2, GBP5, STARD3, AGPAT3, GKN2, MGMT, TBRG4, CUL3, MEGF11, EVA1C, CSTL1, KCNIP3, SREBF1, IFRD2, DRG1, ATP13A1, TTC22, CSF3R, DRC3, SHISA5, ANKRD16, TPRG1, SRC, HDAC6, DMAP1, AZIN2, PLP1, LRRC2, LAMA5, C1orf109, MBP, IQGAP2, PCDH1, KIF13A, ANKHD1, PPAT, HSD17B11, SPATA20, NGFR, MXD3, RNF44, QRFPR, FAM200B, METAP1, ARAP3, PAPSS1, ANXA6, OSR2, GRIA1, FAM83A, SLC39A14, TAF2, ENO3, ARFGAP2, GOLGA8G, CTTN, TMEM262, PIDD1, TPP1, GOLGA8F, PPP1R16A, MMP7, IMMP1L, CDKL1, CREB3L1, PEX5, IPO8, CARS2, NLRC5, CCND1, PRH1, LHX6, SPECC1L, MAP1S, ANO2, OLR1, PACS2, TPCN1, GNPTAB, AL928654.3, CNR1, PRKCH, PTGDR, TTLL5, SLC35F4, MTHFD1, ALDH6A1, TVP23CCDRT4, GLDN, SLC12A1, TM2D3, IGDCC3, NIPA1, MTFMT, SQOR, FAM214A, MT3, GOLGA8S, ATXN2L, GOLGA8M, GRAMD2A, AC090527.2, MAP1LC3B, ADCY7, GOLGA8H, GTF3C1, GGA2, GOLGA8J, GOLGA8T, DDX19B, MAN2C1, VPS53, TVP23B, SLC43A2, SIRT7, RNF213, CFAP52, CYB561, TMEM241, TOP2A, ARHGAP44, MKS1, SAP30BP, WIPF2, BAHCC1, LSM7, AES, ARHGAP33, GIPC1, G6PC, USP32, CDC37, MBD1, CIRBP, AC092073.1, GHDC, GTPBP3, ZNF665, TIMM50, HNRNPM, F2RL3, DLL3, KLK2, PNPLA6, DUSP22, CCL15CCL14, NTMT1, MLLT6, SRCIN1, CCL23, CCL15, PTPRQ |
| hsa-miR-130b-3p | PRTN3, BCL2L14, TCEA2, PPA2, MRPL37, CNIH4, STXBP5, IPPK, OR10A4, ATP6V0A1, OR10W1, THOP1, AGAP1, WDR43, TRIM2, ADGRG3, AOX1, ANKS6, CXorf65, DLG5, ACSL6, USP37, EXOSC7, ATG2B, PRKD1, USP34, ABL2, LYRM2, FNTA, MTMR9, MYRF, FKBP4, RAB5C, KCNH3, RPS29, APH1B, CNOT1, AKT2, AC099811.2, NDUFA7, C3, INSR, TIMM50, AC010323.1, MFSD14C |
| hsa-miR-133a-3p | ITGA3, B4GALT7, NGFR, TRO, TRAM2, CDC34, MORC2, NME4, POP4, FBL, SIPA1L3, AEBP1, CDX1, INO80B, DLGAP3, PDZD11, KLRK1, MRPS2, SEC61A1, PYCARD, PIN1, FXR2, CNN1, PRDM12, YIPF2, NAPSA, PPP1R1B, FADS2, TTBK1, CACNG4, GGT5, PPP1R12C, KCNT1, COL5A3, CHFR, SCAMP2, ARHGDIA, SLC47A1, SEC22C, ADAMTSL1, SOGA1, SPARCL1, OSCAR, OBSL1, PDXK, NAGS, FCGR3B, TRIM17, COL6A3, CLEC3B, SENP2, CLEC1B, PRRT2, SLC50A1, KCTD19, DDIT4, SSH3, CLDN15, DNAI2, GABRB3, MMP14, RPL7A, STOML1, RORC, MARCH8, SF3B2, PDZD3, FOXJ1, FOXL2, FAM187A, CMTM1, NELFA, SERPINA11, NCKAP5L, MMP25, MXD4, ZNF385A, FBXL19, BRAT1, TRIP12, TRADD, DHX30, NPHS1, IL27, FGF17, LAT2, PIK3CD, SREBF2, PNMA5, MRPL55, ENAH, FCGR3A, PFDN2, DNASE1L1, EDN3, SLC2A6, PREX1, LRRC73, FAM167B, KIF12, MSANTD3, MGAT5B, STK19, RSG1, MPIG6B, DLG2, ACTN1, CNNM4, PLXDC2, ACSF3, SHROOM1, NUDT2, CDKN2A, FUS, C1QTNF7, LNP1, IL17RE, ABL1, PTTG1, KCNK7, CELF6, FGFR1, GSTP1, COL13A1, MYL5, ARVCF, DPP6, ABLIM2, CCDC103, SLC35G6, ATP5MFPTCD1, PSMB9, C2orf76, TRAF2, DDAH2, GRPEL2, RASAL1, EPCAM, SFXN4, CD22, CERCAM, BRPF1, OGFOD2, CEP250, TTC38, FAM182B, GIGYF2, SDCBP, PRDM1, PICK1, MRPS18A, RPL10, MGAT1, TULP1, LSP1, KCTD15, PIGV, RLN3, MICAL1, PILRA, MLPH, SERPINE2, POLD2, NCOA6, SH3BP2, MMP23B, EMID1, SEC14L2, GCK, PCED1A, FAM20B, TSSC4, LCP1, ELF3, CCT3, ZNF185, PILRB, TTC31, ASCC2, RASA4, ACAP2, SMIM34A, FCAMR, DMKN, ACTR1B, RHBG, GSS, COL9A3, TGM2, PLEKHA3, BACH2, KLC4, ARFGAP3, PLA2G6, TTLL6, CLCN2, PFKL, DXO, ZNF862, STK40, KEL, NADK, MAMDC2, SCAMP4, SHANK1, APP, MRPS5, ABCA2, PEX5L, NDUFB8, TFDP2, CCDC13, LANCL2, RHBDD2, RNF13, PSORS1C1, MSH5, SNORC, CYP2J2, SURF6, B9D1, FAAH, CRAMP1, SH2D3C, WDR54, CXorf56, HSPH1, EFNA3, MYO1A, KLHDC8B, IL1RN, ACAP3, TBC1D2B, MXRA8, AL096711.2, SSBP1, RRAGB, SPAG8, CPA1, FAM129B, HMGCS2, NRP2, DNASE1L3, PDZRN3, NFKBIE, CACNA2D3, MAGED4, BCAR3, WHRN, MAGED4B, EXOSC7, IZUMO4, ZNF783, NT5C3B, CNTN2, AC093668.2, YPEL5, MLF1, ZNF391, PDS5B, EZH2, PKN3, POLR3H, TAF1, COG5, OPHN1, CLASP1, MYCBPAP, ZNHIT1, PPIL2, PRSS53, PRKCB, CDS2, TRIM56, FUBP3, CCDC150, NISCH, NGEF, DDC, PRRC2B, PNPLA7, RREB1, CBS, DOPEY2, ARPC1B, NCOR2, RABL2A, NICN1, DEPDC5, TSC22D1, LRP8, OGDHL, GYG1, UBE2G2, HM13, ARHGAP8, CEBPA, SH3RF1, PGM3, MZB1, TRIML2, PITX1, NFKB1, KLHL3, HK3, ACAD9, SEC24D, FAM193B, IPO11, FGFRL1, SLC10A7, SRPK1, MGAT4D, EXOC3, AXDND1, CFI, CORIN, COL12A1, ANKHD1, ATP8A1, NR1I3, ABHD18, BFSP2, LMAN2, CPLX2, ACOX3, ZNF704, NDRG1, VIRMA, TMPRSS4, AC105052.1, SNX31, GRIA1, MGAT4B, SORBS3, VDAC3, SLC39A14, ST3GAL1, TRIQK, NKX63, TBC1D9B, PITPNM1, CCDC17, C1QTNF5, ZNF250, TESPA1, ZNF195, PPP2R5B, PPP1R16A, NKAPD1, CHMP4A, TYK2, BACE1, ARHGEF4, ECE1, ZNHIT2, LRP5, AMPD2, TMEM86A, SART1, CKLFCMTM1, EFEMP2, MRPL17, SMPD1, NDUFS8, BBOX1, DNHD1, STK33, PCSK7, ATM, SIPA1, P2RX3, TPCN2, GPR162, KIAA1551, PDE2A, RHOF, CARS2, SRGAP1, SLC6A12, TRMT10B, EHD2, VWF, STAT6, IDH2, NDUFA9, RASA4B, NOP2, MEIS1, KLRC4KLRK1, PLEKHB1, WSCD2, ZNF606, MAP1LC3B2, MCRS1, MARCH9, EEA1, SOCS2, FKBP11, GATC, ARHGAP9, TNS2, ITGA5, ITGA7, SDR39U1, SLC7A7, WDR25, CD63, AKT1, RPS6KL1, CRTC3, TPM1, GCNT3, RTF1, SLC22A31, TK2, MYO7B, KCTD13, KLHDC4, CBFA2T3, NEIL1, CDH16, CEMP1, CMIP, ABCA3, DHX38, NTHL1, LMF1, SMPD3, VPS35L, STARD13, SLC38A8, LDHD, TCF4, ADCY7, ZSCAN29, MAPK7, AKAP10, PLD2, ATP2A3, ZP2, RPS15A, SLC26A11, AKAP1, PELP1, ALDH3A1, DPH1, POLR2A, NXN, PER1, RAB11FIP4, ITGB4, LLGL2, PIEZO2, SUPT4H1, PROCA1, SLC25A35, SEPT4, ARL5C, SLC16A3, EIF4A1, PRR29, ERBB2, PRKCA, TSEN54, CD300LF, MTMR4, CDK12, RPL26, TMEM94, MUM1, PCSK4, UBXN6, MATK, TLE2, EDDM13, NAT14, NEDD4L, PALM, BCAM, APLP1, POLRMT, ATP5F1D, AP1M1, RFX2, SPHK2, SH3GL1, SNAPC2, NAPA, PGPEP1, ETHE1, MYDGF, IRF3, KCNN4, MRPL34, SLC17A7, ARHGEF1, VAV1, PPFIA4, PPFIA3, MORN1, TBX18, CCL15-CCL14, DGKK, AC239799.1, CTDP1 |
| hsa-miR-184 | ANKRD54, PEX11G, SF1, MTUS2, AMPD2, TMEM184A, NLGN2, ZBTB46, C20orf196, TCTN2, ATP13A2, CARM1, CACNA1G, TOMM20L, TRIM27, KLF6, TSSC4, PPP1R21, AURKA, MTA1, HMCN2, PTRH1, RLN3, MAP6, CLUH, METTL8, PABPC4, AXIN1, HM13, SLC26A9, PHF21B, TLE1, ABCA2, CDC25B, DPYSL4, FAM83F, S100PBP, SLC2A4RG, ST6GALNAC4, AKT2, TRDMT1, MTMR11, KRTDAP, TARS2, GAL3ST4, ARHGEF10L, CTSA, PHF24, CTSZ, ARHGEF7, DGCR8, TACC2, IFT22, PPP2R3B, LHFPL4, UGT2A1, CDK18, EXOC1L, EFNA5, SPAG9, MGAT4B, STK3, LRRC6, FUT2, STT3A, TMEM80, SIGIRR, UBQLN1, ROBO3, ZSCAN9, C1RL, ARHGEF40, NRDE2, TTC7B, PSMA4, PSME2, CACNA1H, ZKSCAN2, FAM192A, ABAT, RNF167, NXN, ELAC2, CTC1, GPRC5C, PTBP1, AP3D1, PIP5K1C, LTBP4, LGALS4, SIPA1L3, PAFAH1B3, IRGQ, KANTR |
| hsa-miR-203a-3p | SLC26A10, PRKG1, ACVR1, FTSJ3, FOXK2, CCM2, SIM2, SLC35B1, ITGA1, DDX55, GMFB, SUSD6, FUS, LSM7, APOC1 |
| hsa-miR-206 | GTPBP1, LY96, SOCS1, TCF7L2, TCTN3, NOTCH4, TRIM31, BSCL2, ELMOD3, ASCC2, TMEM178A, COL20A1, MRO, ZNF33A, SFTPC, FAM120A, AQP12B, AQP12A, ROBO2, PER3, GSTO2, SLC25A12, OCIAD1, RHOBTB3, ASPH, PDP1, GDPD5, ZC2HC1C, FBRSL1, TRAF3, CD276, FA2H, CTC1, ATP5F1A, SIPA1L3 |
| hsa-miR-335-5p | SGK3 |
| hsa-miR-551a | DVL2, RPL18A, PCDHB6, IFT43, PCDHB8, PCDHB10, NEUROG3, TYMP, TUBGCP2, SLC2A11, COX10, KAT6A, ADAMTSL1, FAM81A, FAM46B, ZFYVE28, ABCF3, GPR182, ANGPTL4, MMP14, TRPM7, C4orf50, TMEM150B, SORCS2, EWSR1, DYNC2H1, SNRPD2, TRPM3, VKORC1, AC008397.2, ANKRD35, ZNF707, C20orf203, CNIH4, RXFP4, CHTOP, CYB5R4, ZNF451, BTBD17, DCTN3, EIF1AX, GMDS, AKAP17A, MPP1, GPT, ATP2A1, SELENOS, ZSWIM8, ITSN1, CENPM, ELF1, PRSS40B, PPT2, POLM, PASK, VILL, RTL10, ANKRD62, CIZ1, CAP1, PBRM1, APOE, NR2E1, MAGEA10, LSG1, ZNF776, GTF2I, LCNL1, TNK2, ADAP1, SULF2, COLEC11, TADA3, GPAT2, SFTPA1, RGPD5, TMEM114, OGT, SNRNP27, MFSD13A, CAV1, AURKA, GNB1L, WDR74, PRODH, SEMA4F, DENND6B, EIF6, ECE1, DDX43, LTC4S, RND3, CLTA, RARRES2, PLCH1, RWDD2B, ZMIZ1, ACAP3, SCAF11, TFR2, MECOM, MATN1, PLCG1, RGS19, HES4, KIF6, HBA2, MX1, AKAP2, MCF2L, PANK4, TSC22D3, RCC1, SLC2A5, LRCH4, HBA1, IFT172, LZTS2, RGPD6, HLAC, KRBA1, RGPD8, SLC22A18, RMND5B, MEF2C, TMEM110MUSTN1, RFC1, DDR1, KIAA1191, BDH2, FAM149A, KCNQ4, CPE, SPON2, SPAG9, SERF1B, RPL26L1, NCOA2, ADAM28, MPP2, KIF13B, SLC39A14, RSF1, DLG2, BCLAF1, CRACR2B, TNKS1BP1, CHID1, AC135050.2, TRIM29, HINFP, EPS8L2, PITPNM2, OGFOD2, C12orf65, DDX11, UBC, VSIG10, P3H3, KDM2B, C12orf57, FAM180B, TIAM2, CRIP2, SPRYD3, KCNH3, NDRG2, KLC1, EAPP, SLC25A29, DGKA, ALDH1A3, ADCY4, PIF1, PHGR1, IGF1R, GLDN, BCKDK, NIP7, SPG7, GPRC5B, APRT, RPUSD1, ITGAL, CTU2, RIPOR1, FAM192A, GSPT1, SGSH, FLYWCH1, CTRL, FUK, CHRNB1, SLC47A1, FN3KRP, NAT9, AMZ2, PSMC5, SLC46A1, PSMD8, ADAMTSL5, CYTH1, SPRED3, CDIP1, ANKRD27, TBXA2R, GIPC1, PRKAR1A, TMEM161A, FAAP24, GADD45B, CPAMD8, ADAMTS10, NFKB2, KLK2, PPFIA4, SYT3, AC003006.1, VAV1, FAM156B, NDOR1, ANKRD20A3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





