1. Introduction
Influenza is a highly contagious respiratory disease caused by influenza A and B viruses. Symptoms are usually cold-like, but some patients develop pneumonia requiring hospitalization. The World Health Organization estimates that influenza epidemics cause 3–5 million cases of severe illness and 290,000–650,000 deaths annually worldwide [
1]. The influenza virus, a single-stranded RNA virus, is prone to genetic mutation and can antigenically “drift,” leading to seasonal epidemics every winter. Seasonal influenza is considered a vaccine-preventable disease, but the current mainstay of needle-and-syringe vaccines has shortcomings, such as relative ineffectiveness in elderly people [
2,
3].
Optimizing administration methods is a key strategy to enhance the effectiveness of influenza vaccines. The influenza virus enters the oral or nasal cavities and establishes infection in the upper respiratory tract mucosae. Therefore, these mucosae serve as the first line of defense against pathogens such as the influenza virus and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Mucosal protection generally operates through antibody-mediated and cytotoxic T-cell responses, which can be triggered to some extent by needle-and-syringe vaccines.
Sublingual vaccines, like nasal vaccines, preferentially induce mucosal immune responses in the upper and lower respiratory tract, in addition to a systemic response [
4]. One advantage of sublingual vaccines is their safety compared to nasal vaccines, which may adversely affect the brain, central nervous system, or lungs [
5,
6]. Additionally, sublingual vaccines are needle-free, providing high patient compliance and the possibility of self-administration without the need for medical personnel.
However, vaccine administration via the sublingual route presents practical challenges, such as a mucin barrier inhibiting vaccine entry into immune cells and a significant volume of saliva that dilutes the vaccine. In a previous study using a non-human primate model, cynomolgus macaques, we addressed these challenges by pre-treating the sublingual surface with
N-acetyl cysteine (NAC), a mild reducing agent that disintegrates the mucin layer [
7]. Additionally, we reduced saliva dilution through anesthesia, using a mixture of medetomidine and ketamine, to minimize saliva secretion during vaccine administration [
7]. Subsequently, we demonstrated the safety and efficacy of a sublingual SARS-CoV-2 vaccine containing the receptor binding domain (RBD) peptide of the spike (S) glycoprotein of SARS-CoV-2 [
8].
Nowadays, adjuvants are integral components of medical and veterinary vaccines, excluding live-attenuated and recombinant adenovirus-vectored vaccines. The primary role of adjuvants is to stimulate innate immune responses and elicit antigen-specific adaptive immune responses [
9]. Adjuvants can be broadly categorized as delivery systems and immunostimulants. Delivery systems consist of carrier materials that load antigens, enhancing their uptake and presentation by antigen-presenting cells (APCs), thereby facilitating antigen presentation. Conversely, immunostimulants are pathogen-associated molecular pattern (PAMP) molecules that induce the maturation and activation of APCs by targeting Toll-like receptors and other pattern recognition receptors [
10].
Examples of delivery system adjuvants include MF59 and AS03, which are both oil-in-water nanoemulsions. These adjuvants have received approval for use in influenza vaccines [
11]. Conversely, double-stranded polyinosinic:polycytidylic acid (Poly(I:C)), a TLR3 agonist, falls under the category of immunostimulant adjuvants. However, Poly(I:C) has not yet received approval due to its side effects, including fever and proinflammatory cytokine production, when administered intramuscularly. In a previous study, we performed a direct comparison between AS03 and Poly(I:C) adjuvants in sublingual vaccination of cynomolgus macaques and found that Poly(I:C) exhibited superior safety profiles compared to AS03 [
8].
In this study, we examined a sublingual influenza vaccine formulated with the influenza hemagglutinin (HA) antigen and the Poly(I:C) adjuvant in nonhuman primates. Additionally, we investigated the molecular mechanisms underlying immune-related responses mediated by the sublingual vaccine using DNA microarray analysis.
2. Materials and Methods
2.1. Reagents and Antibodies
In this study, the following materials were used: phosphate-buffered saline (PBS) (Nissui, Japan), polyester swabs (JCB, Industry Limited, Japan), filter spin columns (Norgen Biotek Corp., Canada), Nunc-immuno module, F8 Maxisorp (Thermo Fisher Scientific K.K., Japan), streptavidin-HRP Conjugate (SA-HRP) (Invitrogen-Thermo Fisher Scientific K.K.), and tetramethyl benzidine (TMB) (Sigma-Aldrich Co. LLC, Japan). NAC, bovine serum albumin, Na-casein, sodium azide (NaN3), and Tween 20 were obtained from FUJIFILM Wako Pure Chemical Corporation (Japan). A quadrivalent FLUBIK HA Syringes™ vaccine (The Research Foundation for Microbial Diseases of Osaka University, Japan), Poly(I:C) HMW vaccine grade (Invitrogen, Waltham, MA, USA), and an ELAST enzyme-linked immunosorbent assay (ELISA) amplification system (PerkinElmer, Inc., USA) were also used. Biotin-labeled (BT) monkey IgA (Mabtech, Inc., USA), biotin-labeled (BT) monkey IgA (alpha-chain) (Merck, Germany), horseradish peroxidase (HRP)-human IgG (EY Laboratories, Inc., USA), and BT IgE antibodies (Bio-Rad Laboratories, Inc., USA) were used.
Furthermore, RNAiso Plus, PrimeScript™ Reverse Transcriptase, Recombinant RNase Inhibitor, TB Green® Premix Ex Taq™ (Tli RNaseH Plus), RR420(Takara Bio Inc., Japan), RNeasy MinElute Cleanup Kit (QIAGEN, USA), dNTP Mix and Oligo (dT)15 Primer (Promega, USA), Low Input Quick Amp Labeling Kit, RNA6000 Nano Kit, Agilent Whole Human Genome DNA Microarray 4 × 44K v2, and Agilent Gene Expression Hybridization Kit (Agilent Technologies, USA) were used in this study.
2.2. Animals
Nine cynomolgus macaques (Macaca fascicularis), comprising both male and female individuals aged 8–19 years old, were used in this study. Adhering to the 3R policy of animal use, the macaque monkeys underwent subsequent washout for 20 months after the termination of certain examinations. Prior to their use in this study, these monkeys tested negative for hepatitis B virus, Simian immunodeficiency virus, tuberculosis, Shigella spp., Salmonella spp., and helminthic parasites.
Animal examinations were conducted according to the guidelines of Institutional Animal Care and Committee Guide of Intelligence and Technology Lab, Inc. (ITL) based on the Guidelines for Proper Conduct of Animal Experiments and approved by the Animal Care Committee of the ITL (approved number: AE2022022, date: 24 November 2022). Other examinations were also approved by the ITL Biosafety Committee (approved number: BS2022022, date: 24 November 2022).
2.3. Vaccination and Sampling
The vaccination antigen contained 120 μg/ml HA, i.e., 30 μg/ml of each HA subtype, including A/Victoria/1/2020 (H1N1), A/Darwin/9/2021 (H3N2), B/Phuket/3073/2013 (Yamagata), and B/Austria/1359417/2021 (Victoria). The HA antigen and Poly(I:C) adjuvant (1 mg/ml) were stored at 4°C and −70°C, respectively, until use. Nine cynomolgus macaques were divided into control/PBS (pP01~03, qP09) and experiment/HA + Poly(I:C) groups (pP05–08, qP10). The animals received either 0.5 ml of PBS for the control group or 0.5 ml containing 30 μg HA antigen and 400 μg Poly(I:C) for the experimental group.
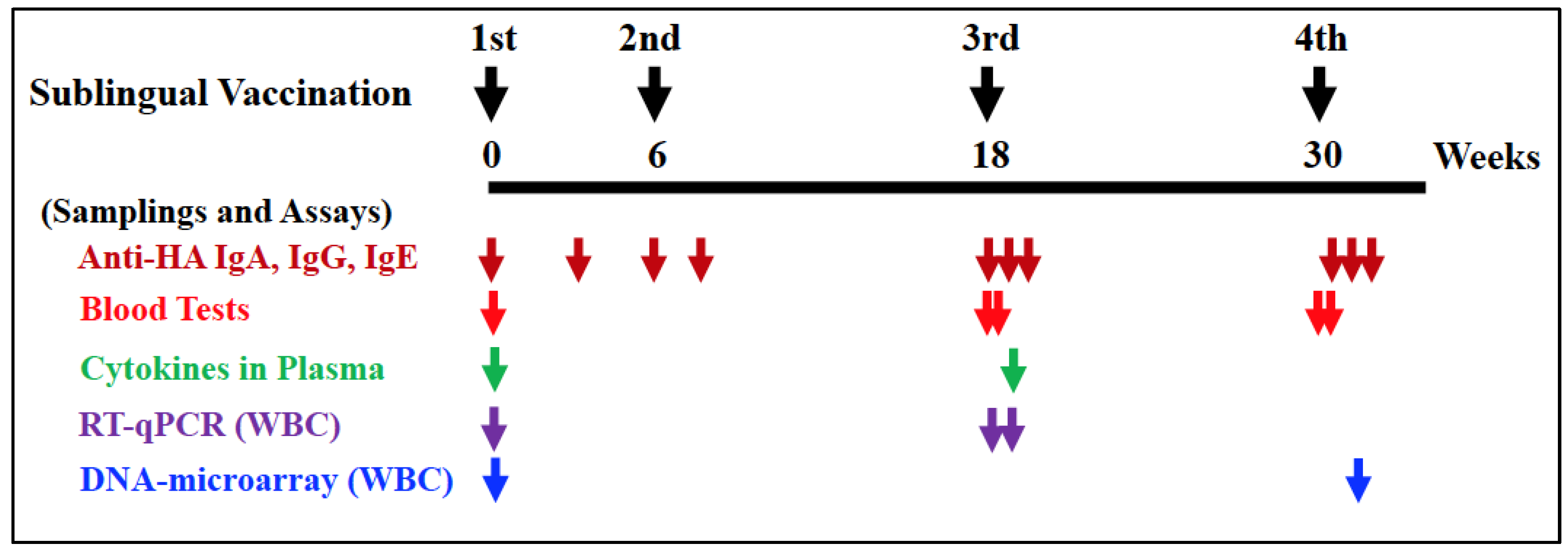
Vaccination procedures were conducted under anesthesia using a mixture of medetomidine and ketamine, with atipamezole administered to awaken the monkeys. Before vaccination, the sublingual surface of the monkeys was pretreated for 5 minutes using wet cotton dipped in 1.5% NAC, followed by saline wash to disintegrate the mucin layer. After wiping the wet mucin surface with dry cotton, 0.5 ml of PBS or HA with Poly(I:C) was administered into the sublingual space using a pipette, followed by a standing time of at least 5 minutes. Sublingual vaccinations were performed twice (at 0W and 6W) with a six-week interval, followed by the third and fourth vaccinations (at 18W and 30W) with twelve-week intervals, as shown in
Figure 1.
Blood, saliva, and nasal washings were collected from each monkey under anesthesia, as mentioned above. Plasma samples were prepared by centrifuging blood to assay HA-specific IgA, IgG, or IgE antibodies. Saliva samples were adsorbed to a swab with polystyrene fiber, and nasal washings were recovered using centrifugation in a spin column. These samples were stored at −40°C until use.
2.4. ELISA for HA-Specific Antibodies
To detect HA-specific IgA, IgG, or IgE antibodies, Nunc-immuno module plates were coated with 100 μl of 0.4 μg/ml HA (300-fold diluted vaccination antigen) in PBS and incubated at 37°C for 1 hour, followed by incubation at 4°C overnight. After washing with PBS containing 0.05% Tween 20, 1% Na-Casein in PBS containing 0.02% NaN3 was added to the plates for blocking, followed by incubation at 37°C for 1 hour and then stored at 4°C until use. Saliva, nasal washings, and plasma samples were diluted 100 to 500-fold with 1% Na-casein-PBS containing 0.02% NaN3. These diluted samples were used as ELISA samples. To perform an ELISA, after removing the blocking reagent, 50 μl of the diluted ELISA samples and 1M NaCl at a final concentration of 0.5 M were added to the plates to eliminate non-specific reactions.
After incubation at 37°C for 1 hour or at 4°C overnight and removing the samples, plates were washed with PBS containing 0.05% Tween 20. Subsequently, for antibody detection, appropriately diluted BT monkey IgA, BT monkey IgA (alpha-chain), HRP-human IgG, or BT IgE antibodies were added, followed by incubation at 37°C for 1 hour. After washing, the plates were amplified using diluted SA-HRP and an ELAST System mixture comprising biotinyl tyramide. The ELISA sensitivity was enhanced 10 to 30-fold by using this amplification.
After amplification, plates were washed with PBS containing 0.05% Tween 20, and diluted SA-HRP was added, followed by incubation at 37°C for 1 hour. Color development was performed with TMB and terminated by adding H2SO4. Absorption was measured at 450 and 600 nm using a plate reader (iMark Microplate reader, Bio-Rad Laboratories, Inc.).
The concentrations of total sIgA in saliva and nasal washings were measured using a Monkey IgA ELISA Basic Kit (Mabtech, Sweden) according to the manufacturer’s procedure.
A relative titer for HA-specific sIgA was estimated from a calculation of the optical density of HA-specific sIgA/concentrations of total sIgA, as the sIgA secreted in saliva and nasal washings varied from sample to sample.
2.5. Blood Tests
Blood samples were collected one day before the first vaccination (W0) and one day before and after the third (W18) and fourth (W30) vaccinations. Fresh whole blood samples were examined for the complete blood count of eight items: red blood cells, white blood cells (WBC), hemoglobin, hematocrit, mean cell volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, and platelets. Plasma samples were assayed for biochemical blood tests of thirteen items: total protein, albumin, albumin/globulin ratio, total bilirubin, aspartate transaminase (glutamic oxaloacetic transaminase), alanine transaminase (glutamic pyruvic transaminase), alkaline phosphatase, gamma-glutamyl transpeptidase, urea nitrogen-BUN, creatinine, total cholesterol, neutral fats, and C-reactive protein (CRP).
2.6. Cytokine Assays
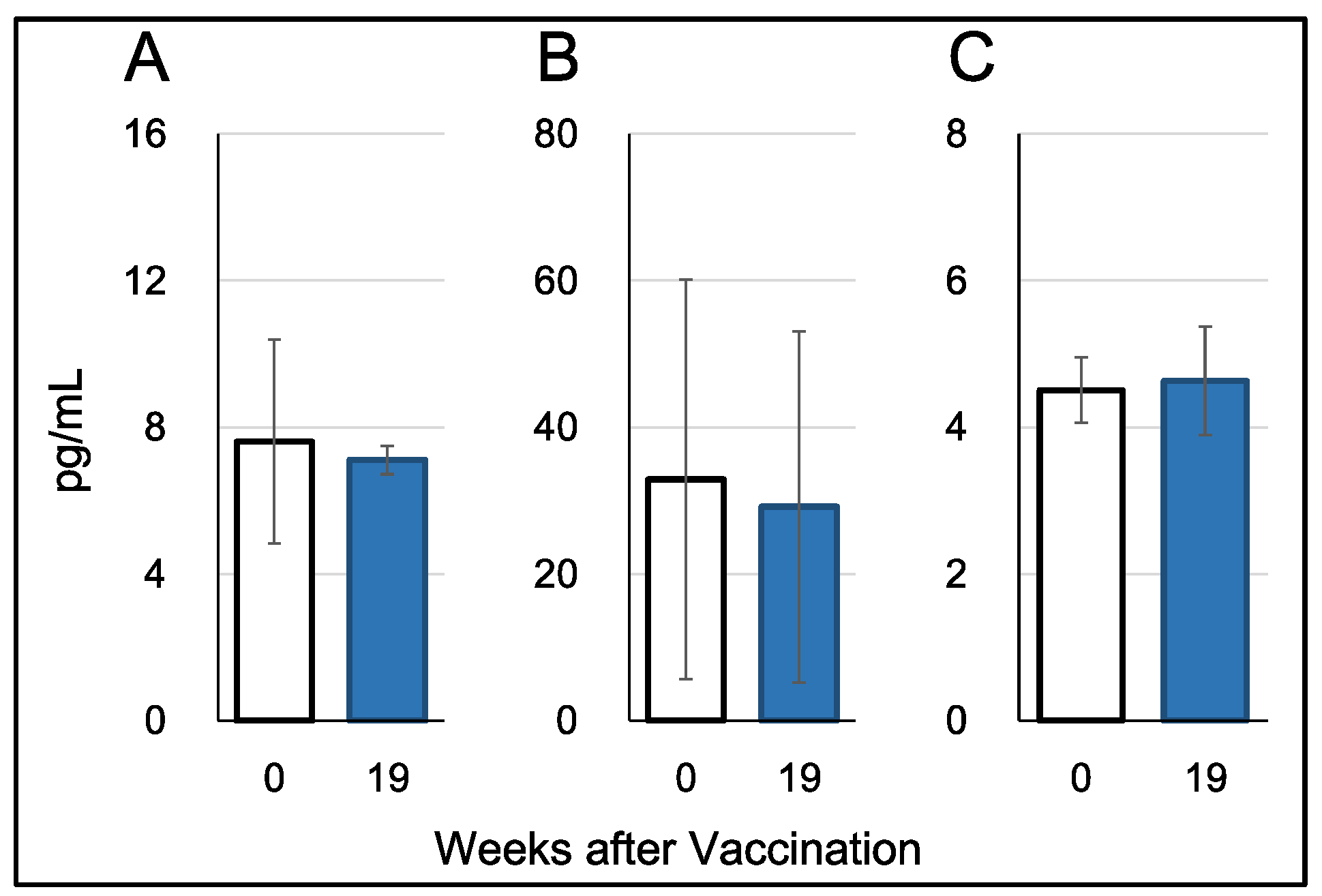
Plasma cytokine levels were measured using plasma samples collected one day before the first vaccination (W0) and one week after the third vaccination (W19). Three cytokines, IFN-alpha, IFN-gamma, and IL-17, were assayed using ELISA with the Monkey IFN-alpha (pan) ELISA Kit (Arigo Biolaboratories Corporation, Taiwan), Monkey IFN-gamma ELISA Kit (U-CyTech Biosciences), and Monkey IL-17 ELISA Kit (U-CyTech Biosciences), respectively.
2.7. WBC Isolation
Blood samples were collected in heparinized syringes, and WBCs were isolated using erythrocyte lysis buffer, following the method described by Hoffman et al. [
12].
2.8. RNA Isolation
Total RNA was extracted from WBCs using the acid guanidine thiocyanate-phenol-chloroform method and silica membrane column-based purification [
13]. Initially, WBCs were homogenized in RNAiso Plus (Takara Bio Inc.), and the total RNA was isolated. The RNA was treated with DNase (QIAGEN, USA) in the aqueous phase and purified using the RNeasy MinElute Cleanup Kit (QIAGEN, USA) following the manufacturer’s instructions. Additionally, total RNA (0.2–0.4 mL) from monkey whole blood was isolated after on-column DNA digestion using NucleoSpin RNA Blood (Macherey-Nagel GmbH & Co., Germany) according to the manufacturer’s instructions. The quantity and purity of RNA were assessed at 230, 260, 280, and 320 nm using an Ultrospec 2000 spectrometer (GE Healthcare Biosciences AB, Uppsala, Sweden).
For DNA microarray analyses, the RNA integrity number (RIN) was determined using an Agilent 2,100 Bioanalyzer (Agilent Technologies Japan Ltd., Tokyo, Japan). Only high-quality RNA samples, defined by an A260/A230 of 1.5, A260/A280 of 1.8, and RIN of 6.0, were used in the microarray analysis.
2.9. Gene Expression Analyses Using Quantitative Reverse Transcription PCR
The mRNA levels of target genes in WBC samples were determined using quantitative reverse transcription PCR (RT-qPCR), as described previously [
14]. Briefly, cDNA was synthesized from the purified RNA using PrimeScript Reverse Transcriptase with RNase Inhibitor (Takara Bio Inc.), dNTP mixture (Promega Corp., Madison, WI, USA), and Oligo dT primers (Invitrogen, Waltham, MA, USA). Real-time PCR was conducted using the Mx3000P QPCR System (Agilent Technologies Inc., Santa Clara, CA, USA) with a SYBR Premix Ex Taq II (Tli RNaseH Plus) Kit (Takara Bio Inc.). Specific primers for monkey
IL12a, IL12b, GZMB, type I IFNs (
IFN-alpha1 and
IFN-beta1),
CD69, and the reference gene low-density lipoprotein receptor-related protein 10 (
Lrp10) were designed using Primer3 and Primer-BLAST [
14]. A standard curve was generated by serial dilution of a known amount of glyceraldehyde 3-phosphate dehydrogenase amplicon to calculate the cDNA copy number of the genes. The PCR conditions included initial denaturation at 95°C for 15 s, followed by 35 cycles of denaturation at 95°C for 10 s and annealing/extension at 63°C for 30 s, with a dissociation curve. The quantity of target gene mRNA was expressed as the ratio against that of a suitable reference gene,
LRP10 [
15].
2.10. DNA Microarray Analyses
DNA microarray analyses were conducted for both the control and experimental groups. Following cDNA synthesis, Cy3-labeled cRNA was synthesized and purified using the Low Input Quick Amp Labeling Kit (Agilent) according to the manufacturer’s instructions. Notably, reverse transcription was conducted using a T7 promoter-oligo(dT) primer. Absorbance was measured at 260, 280, 320, and 550 nm to verify that the labeled cRNA had incorporated >6 pmol/mg of Cy3-CTP. The labeled cRNA was then fragmented using the Gene Expression Hybridization Kit (Agilent) and applied to Whole Human Genome DNA Microarray 4x44K v2 slides (Agilent). After hybridization at 65°C for 17 hours, the slides were washed with Gene Expression Wash Buffers 1 and 2 (Agilent) according to the manufacturer’s instructions. Subsequently, the slides were scanned using a GenePix 4000B scanner (Molecular Devices Japan K.K., Tokyo, Japan). Scanned images were digitized and normalized using GenePix Pro software (Molecular Devices Japan K.K., Tokyo, Japan).
2.11. Bioinformatic Analysis of Microarray Data
By comparing with the control group, genes that exhibited more than a 2-fold upregulation or less than a 0.5-fold downregulation in the experimental group were identified. These genes were annotated, and references were searched using NCBI databases, Google, and related information. The immune-related functions of annotated genes were deduced to understand the possible mechanisms of action of the sublingual vaccine.
4. Discussion
The sublingual vaccine using the influenza HA antigen and the Poly(I:C) adjuvant demonstrated safety in nonhuman primates, as evidenced by the results of blood tests, including chemical tests, in this study. These findings align with our previous research, which indicated the safety of the sublingual vaccine containing the SARS-CoV-2 S RBD peptide antigen and the same adjuvant [
8]. Despite Poly(I:C) being utilized as a vaccine adjuvant in cancer treatment, its clinical use remains unapproved beyond limited applications. Safety concerns regarding Poly(I:C) adjuvants have primarily arisen from studies involving nasal vaccination in mice [
31,
32]. Notably, humans differ anatomically from mice in lacking defined bronchi-associated lymphoid tissue (BALT) but possessing infection-inducible BALT [
33]. Moreover, differences in the immune systems between rodents and primates have been underscored by genome-based evidence [
34]. Hence, adverse events mediated by Poly(I:C) may be overestimated when administered nasally in rodent models. Research into the development of Poly(I:C) as a vaccine adjuvant continues in both preclinical and clinical settings [
35].
As a danger signal, double-stranded Poly(I:C) RNA activates APCs, particularly dendritic cells (DCs) [
36,
37]. Poly(I:C) binds to endosomal TLR3 and cytosolic receptors, such as retinoic acid-inducible gene I and melanoma differentiation-associated gene 5 [
38,
39]. Activation of TLR3 by Poly(I:C) leads to the production of proinflammatory cytokines, IFNs, IL-15, and Natural killer (NK) cell activation [
40]. Previous studies primarily reported the Poly(I:C)-mediated production of proinflammatory cytokines and related factors in studies involving nasal vaccination in mice [
31,
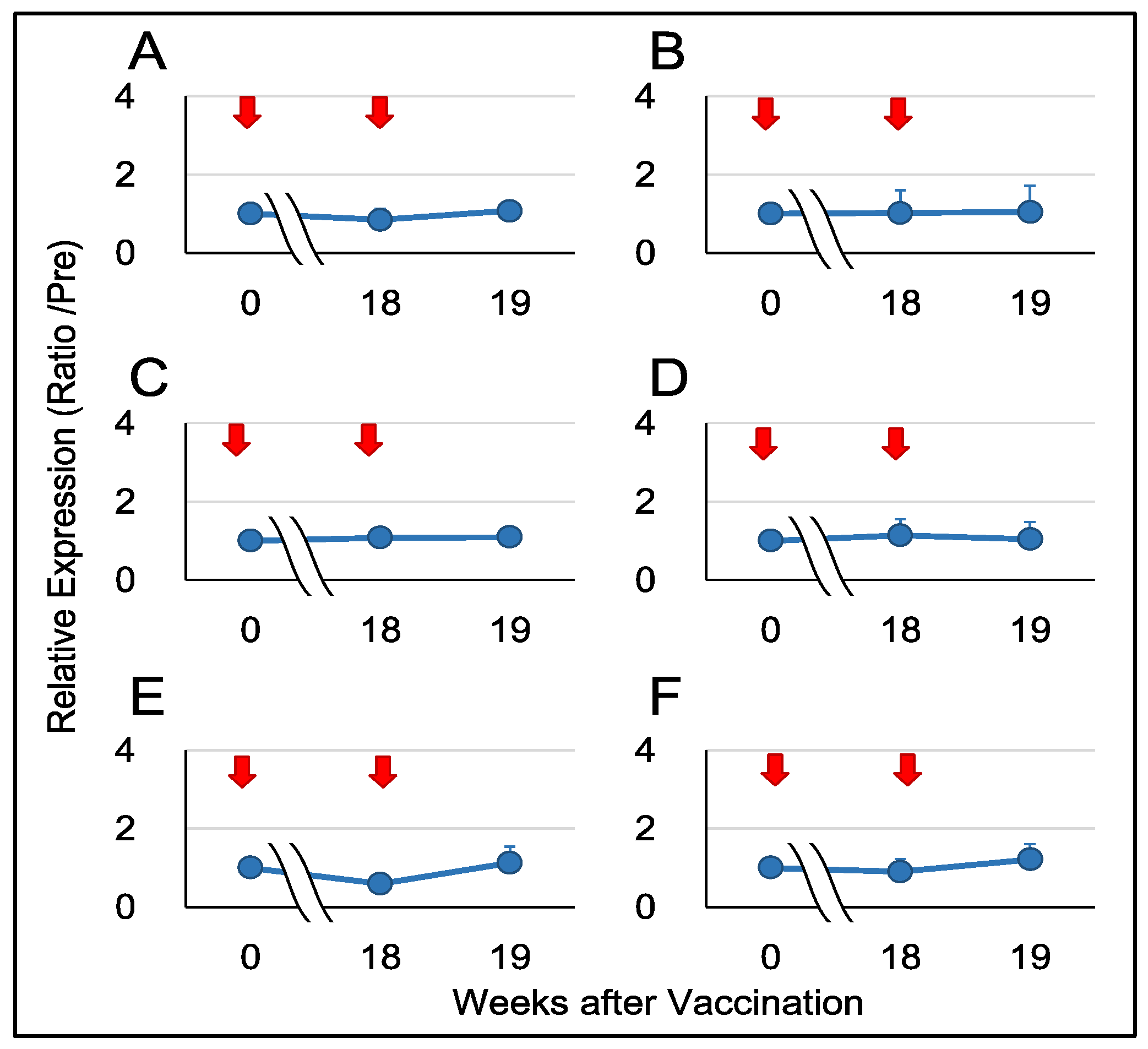
32]. However, when employing sublingual vaccination in nonhuman primates, we observed minimal upregulation in the gene expression of proinflammatory cytokines and related factors, including
IL12A,
IL12B,
IFNA1,
IFNB1,
CD69, and
GZMB (
Figure 4).
Unlike the skin, the sublingual epithelium in humans and monkeys lacks keratinized cell layers, allowing antigens to penetrate the mucosa without specialized devices like microneedles [
41]. Antigens are captured by antigen-presenting DCs, primarily Langerhans cells, dispersed in the mucosa. For example, the ovalbumin antigen was captured by sublingual DCs within 30–60 minutes after sublingual administration in mice [
42]. Studies by Hervouet et al. have demonstrated the presence of antigen-bearing DCs in distant lymph nodes and the spleen [
43]. Typically, humoral immune responses can be observed two weeks after sublingual immunization in mice [
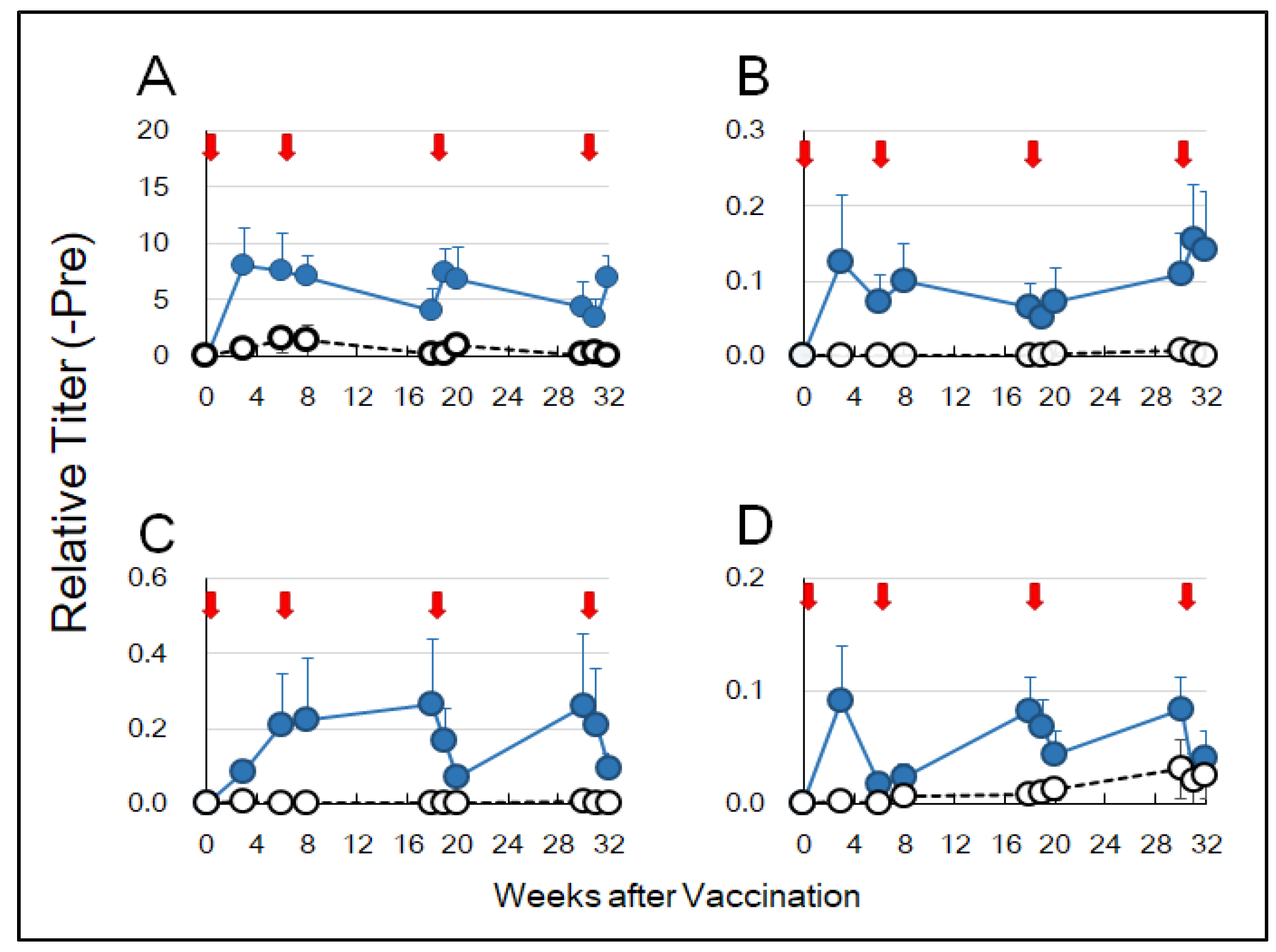
44]. In this study, anti-HA-specific IgA antibodies were detected in saliva and nasal washings three weeks after sublingual vaccination of cynomolgus monkeys (
Figure 2B,C). Anti-HA IgG antibodies were also detected in the blood (
Figure 2D). These results suggest that sublingually administered HA antigen formulated with the Poly(I:C) adjuvant could elicit mucosal immune responses at remote sites, leading to a systemic immune reaction. In our previous study, IgA and IgG antibodies against the SARS-CoV-2 spike RBD were raised in the blood after sublingual vaccination in a non-human primate model [
8].
DNA microarray analyses are a high throughput method used to investigate the expression levels of most genes included in the genome of an organism. In this study, we employed the DNA microarray technique to elucidate molecular events in the immune system and related responses mediated by sublingual vaccination with HA antigens of influenza A and B viruses (two strains each), formulated with the Poly(I:C) adjuvant. The DNA microarray analyses were performed using RNA isolated from WBCs because WBCs are readily available for future clinical studies and can be compared with current preclinical studies conducted on nonhuman primates. The results showed that in the vaccinated group, five genes (
CLEC4G,
PF4V1,
KLF1,
OLFM1, and
GNG11) were significantly upregulated (more than twofold) compared to the control group (
Table 1).
CLEC4G encodes a C-type lectin that binds to complex-type
N-glycans. In humans,
CLEC4G is located on chromosome 19 and is clustered with three genes encoding DC-SIGN, L-SIGN, and CD23, all of which are C-type lectins. CLEC4G serves as an attachment site for the Ebola filovirus and West Nile flavivirus, and it also acts as a receptor for PAMPs of SARS-CoV-2 [
16]. Regarding influenza virus, DC-SIGN serves as an entry site for influenza A virus into DCs [
45]. Recently, Lu et al. demonstrated that myeloid cells, including monocytes, macrophages, and DCs, express
CLEC4G, and its binding to ligands activates inflammatory reactions in these cells [
17]. Notably, we previously sublingually vaccinated cynomolgus macaques with the RBD peptide of SARS-CoV-2 S glycoprotein and found that
CLEC4G was significantly downregulated in WBC [
8]. However, Lu et al. reported that CLEC4G does not bind to the RBD region of the SARS-CoV-2 S protein. This discrepancy between the upregulation of CLEC4G by the influenza HA antigen and its downregulation by the SARS-CoV-2 S RBD peptide antigen warrant further investigation.
Previously,
CLEC4G expression was described as restricted to sinusoidal endothelial cells of the liver and lymph nodes. Tang et al. revealed that CLEC4G is a novel T-cell regulator suppressing the effector functions of activated hepatic T cells [
46]. The liver favors the induction of tolerance rather than immunity, which is critical for maintaining immunologic silence in response to harmless antigenic material present in food. The sublingual epithelium is considered an immunological tolerance-prone site and sublingual application of allergens is a curative therapy for allergic disorders and diseases [
47]. As this study did not collect and examine the sublingual tissue samples, it is uncertain whether
CLEC4G is sublingually expressed and how its expression levels change at the site of administration after sublingual immunization.
After in vitro stimulation with GM-CSF and IL-4,
CLEC4G is upregulated in monocyte-derived macrophages and DCs, and a splice variant encoding a soluble form of CLEC4G, lacking the transmembrane region, is preferentially synthesized [
18]. As mentioned above,
CLEC4G is clustered with the CD23-encoding gene (
FCER2). CD23, a C-type lectin, is also known as a low-affinity Igε Fc receptor on B cells. Splice variants of CD23 are translated into soluble and membrane-bound forms. Soluble CD23 binds to CD21 on B cells and stimulates IgE production, while membrane-bound CD23 binds to the IgE ligand and suppresses IgE production [
48]. Like CD23, soluble and membrane-bound CLEC4G may exert contradictory influences on immune reactions.
PF4V1 is contained in platelets and well-known as a potent inhibitor of angiogenesis. PF4V1 (CXCL4L1) is a ligand of the CXCR3 receptor and chemoattracts activated T cells, NK cells, and immature DCs [
19]. Recently, we found the upregulation of
PF4V1 after sublingual vaccination using the SARS-CoV-2 S RBD peptide antigen formulated with the Poly(I:C) adjuvant [
8]. The upregulation of
PF4V1 observed in the present study using the influenza HA antigen and the same adjuvant suggests that the
PF4V1 induction is not antigen-specific but related to the inflammatory effect of the Poly(I:C) adjuvant. Brandhofer et al. reported that PF4V1 forms a heterocomplex with an atypical chemokine macrophage migration inhibitory factor (MIF) and that the PF4V1-MIF complex does not have chemotactic activity to T cells and monocytes [
49]. The upregulation of PF4V1 may have a suppressive aspect for inflammation caused by adjuvants.
KLF1 is a zinc-finger transcription factor that is indispensable for the transcription of the β-globin gene in hematopoietic cells.
Klf1, the murine ortholog of
KLF1, upregulates
Cd274 which encodes programmed death ligand 1 (PD-L1), in regulatory T cells (Tregs) [
20]. In this study, the upregulation of
KLF1 in WBC may have contributed to Treg induction and immunological tolerance. However, we did not observe the upregulation of either
CD274 or
PDCD1, encoding the PD-1 receptor whose ligand is PD-L1.
In colorectal cancer (CRC) cells, OLFM1 inhibits the non-canonical nuclear factor-kappa B (NF-κB) signaling pathway, which plays a pivotal role in the proliferation and activation of immune cells [
21].
GNG11 is downregulated in splenic marginal zone lymphomas [
22], suggesting that upregulated
GNG11 has an inhibitory effect on the proliferation of B cells. The upregulation of
OLFM1 and
GNG11 in WBC observed in the present study may have suppressed the immune response against the vaccinated antigen.
The results of the DNA microarray analyses using WBC RNA showed that in the vaccinated group, six genes, namely
NEURL1B,
CHST15,
MOB3A,
ANXA6,
DNAJA3, and
HSPH1, were significantly downregulated (over 1/2 fold) compared to the control group (
Table 2).
NEURL1B ubiquitinates Delta, a ligand of Notch in the Notch signaling pathway, and accelerates its proteasomal degradation. NEURL1B plays a pivotal role in the embryonic development of the nervous system, and mRNA levels of
NEURL1B subside postnatally [
23].
NEURL1B is downregulated in CRC tissues, and this downregulation is implicated in the proliferation, invasion, and metastasis of CRC cells [
24]. Recently, we observed the downregulation of
NEURL1B after sublingual vaccination with the SARS-CoV-2 S RBD peptide and the Poly(I:C) adjuvant [
8]. The downregulation of
NEURL1B in this study using the HA antigen and the same adjuvant suggests that the observed downregulation is caused by the Poly(I:C) adjuvant. Similar to CRC cells, the downregulation of
NEURL1B may be involved in the proliferation, secretion of proteases from, and motility of inflammatory cells.
CHST15 is expressed in pre-B cells, pro-B cells, and mature B cells in the B-cell lineage and upregulates recombination activating gene-1 [
25]. The downregulation of
CHST15 observed in this study may have interfered with the humoral immune response against the HA antigen.
MOB3A encodes a protein kinase activator and is involved in intracellular signal transduction. In normal cells, aberrant production/activation of RAS, BRAF, or MEK leads to an irreversible cell-cycle arrest termed oncogene-induced senescence (OIS). MOB3A bypasses OIS by engaging the Hippo pathway, which plays a central role in the regulation of the size of various organs [
50]. Macrophage stimulating 1 (MST1), a member of the Hippo pathway, inhibits the differentiation of follicular helper T cells and the germinal center (GC) reaction [
26]. Moreover, the downregulation of
MOB3A may have stimulated immune response through GC formation. However,
MST1 was not significantly downregulated in this study.
ANXA6 is implicated in the proliferation of and intracellular signaling in T cells. In vivo proliferation of CD4
+ T cells, but not CD8
+ T cells, was impaired in
AnxA6-/- mice [
27]. Our findings suggested that the downregulation of
ANXA6 may disturb helper T-cell function and humoral immune responses.
DNAJA3, an HSP localized to mitochondria, stimulates the ATPase activity of Hsp70 chaperones and facilitates protein folding, degradation, multimeric complex assembly, and so on. Activation of NF-κB, a pivotal transcription factor in immune cells, was suppressed by
DNAJA3 knockdown in vitro [
28]. HSPH1 promotes the replacement of Hsp70-bound ADP with ATP and facilitates the chaperone activity of HSP70. Additionally, HSPH1 plays distinct roles as a holdase that inhibits the aggregation of misfolded proteins and as a disaggregase that resolubilizes aggregates for new folding attempts or proteasomal degradation [
29]. HSPH1 stimulates NF-κB signaling through MyD88 stabilization in activated B-cell diffuse large B-cell lymphoma [
30]. Both DNAJA3 and HSPH1 activate the NF-κB signaling pathway in certain circumstances. The downregulation of
DNAJA3 and
HSPH1 observed in this study may have a negative effect on the immune response after vaccination.
Therefore, the regulation of
CLEC4G and the function of CLEC4G in sublingual vaccination of non-human primates are worth further investigation. The gene expression profile lacked a clear direction as a whole; that is, both potentially immune-enhancing and immune-suppressive changes in mRNA composition were observed in response to the sublingual administration of the HA antigen and the Poly(I:C) adjuvant. Possibly, the inflammatory reaction was enhanced by the upregulation of
CLEC4G and the acquired immune reaction was inhibited by the downregulation of
ANXA6. The NF-κB signaling pathway seems to be inhibited by the upregulation of
OLFM1 and the downregulation of
DNAJA3 and
HSPH1. These observations suggested that the sublingual vaccine yields a balanced state of immune-enhancing and immune-suppressive responses, as seen in a previous study (8) (
Figure 5).