1. Introduction
Bladder cancer (BC) is among the most prevalent cancers globally and is associated with substantial morbidity, mortality and costs. Currently, cystoscopy is the primary tool for diagnosing and monitoring BC-patients. As cystoscopy is an invasive procedure associated with urinary tract infections [
1], there is a significant interest in identifying biomarkers from non-invasive techniques that could aid in BC diagnosis and treatment.
Small non-coding RNA (sncRNa), such as microRNA (miRNA) and t-RNA-derived fragments (tRFs) have over the last decade received significant attention as potential cancer biomarkers [
2]. While transfer RNA (tRNA) was discovered in the late 1950s [
3], and fractions of tRNAs were identified in cancer patient urine and serum in the 1970s [
4,
5], it was not until the deep sequencing era that tRFs were recognized as a distinct sncRNA subclass [
6].
The maturation of transfer RNA (tRNA) is a complex process, entailing extensive modifications and processing steps [
7]. Initially, RNase Z (ELAC 1/2) and RNase P facilitate the removal of leader sequences at both the 3′ and 5′ ends. Subsequent modifications include the excision of introns by the TSEN complex and the addition of a non-templated CCA sequence, which signifies the completion of tRNA maturation [
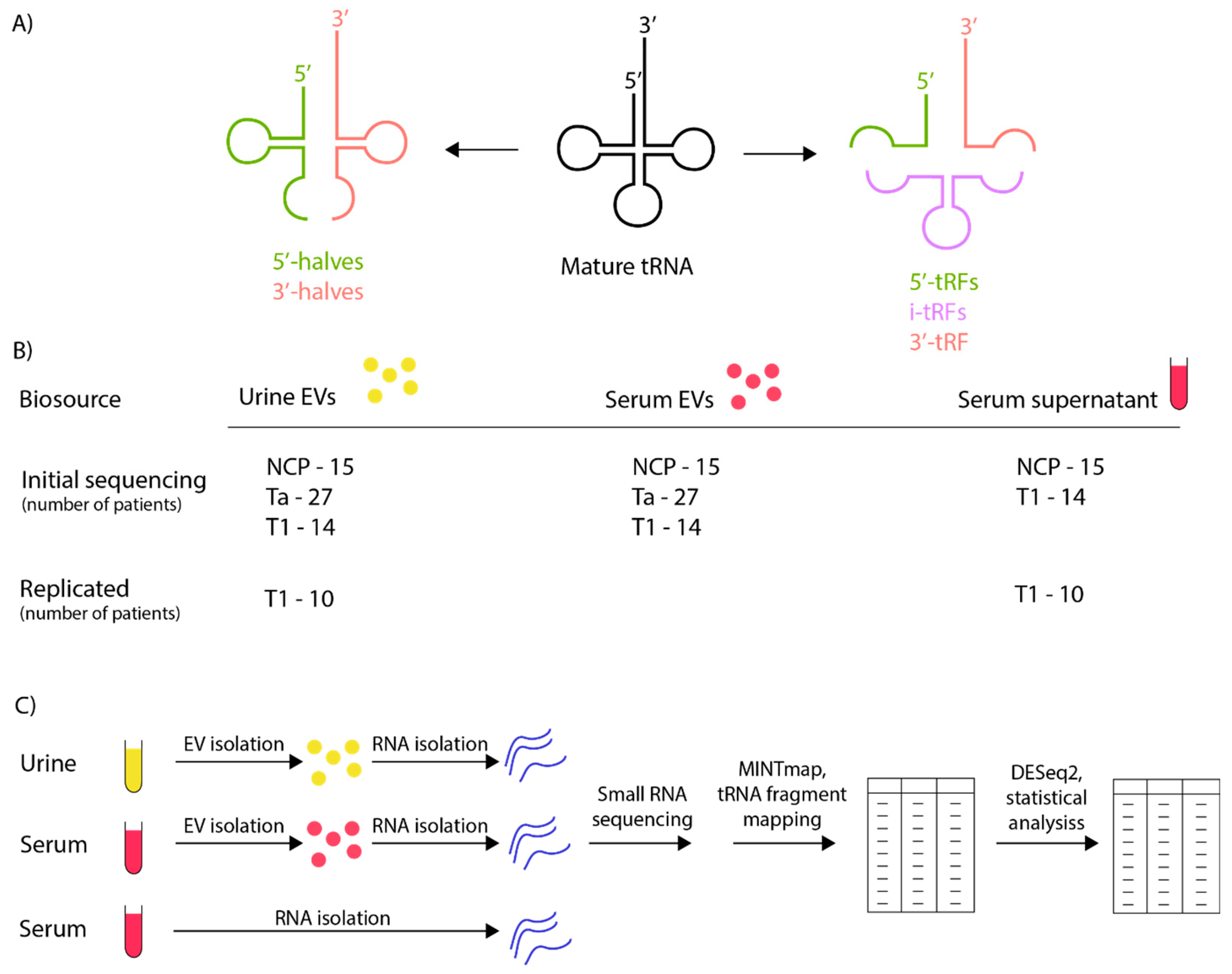
8]. Transfer RNA fragments (tRFs) are derived from specific cleavage events in both pre-tRNA and mature tRNA molecules. The classification of tRFs into two major categories is determined by their location on the mature tRNA and their origin, whether from the 5′ or 3′ end of the parent tRNA molecule. Additionally, tRNA halves, which are 31-40 nucleotides in length, are produced via cleavage by angiogenin within the anticodon loops of mature tRNAs. Concurrently, tRFs, typically ranging from 14-30 nucleotides, originate from cleavages at various sites on either precursor or mature tRNAs. This results in distinct tRF subtypes, including tRNA halves, i-tRF, 3-tRF, and 5-tRF (
Figure 1A) [
7,
9].
One of the main functions of tRFs seems to be regulation of gene expression, both at the transcriptional level through the binding of transcription factors [
10] and at the post-transcriptional level through RNA interference. Recent data has shown that tRFs are abnormally expressed in multiple malignant tumors and related to tumorigenesis [
11]. For instance, tRFs may repress protein translation by binding a complementary mRNA target, resembling miRNAs [
12]. Thus, tRFs may have the potential to function as both oncogenes and tumor suppressors, depending on the individual tRF’s nucleotide sequence and complementary target [
13,
14]. Relatively little is known about the role of tRF in BC pathogenesis, though Su et al. identified a specific tRF methylation pattern in BC cells that attenuated gene silencing of potential proto-oncogenes. tRFs with specific methylation patterns may thus be drivers of oncogenesis in BC [
15,
16,
17].
tRFs may reside in extracellular vesicles (EVs), nanosized vesicles secreted by most cell types and found in bodily fluids such as blood and urine [
7,
18]. The term EV encompasses two distinct subgroups of vesicles; exosomes, which are small EVs (30-100 nm in diameter) and stem from intraluminal vesicles of the endocytic pathway [
19], and microvesicles (MV), which are 100-1000 nm in diameter and stem from budding of the plasma membrane [
20]. Both types of EVs contain molecular cargo, such as mRNA, sncRNA and proteins, which may be transferred from one cell to another. Through the exchange of intravesicular cargo, EVs may play a part in the pathogenesis of a wide range of diseases, including cancer [
21,
22]. As the rate of EV-secretion is higher in cancer cells than healthy cells [
23], and the EV-cargo is shielded from degradation in bodily fluids such as blood, urine and saliva, there has been significant interest in EVs as reservoirs of cancer biomarkers [
24].
Only recently has EV-contained tRFs (EVtRFs) been explored as potential cancer biomarkers. Zhu et al. found that the level of tRFs in plasma exosomes was significantly higher in liver cancer patients than in healthy controls, and they also identified four exosomal tRFs that could have potential as diagnostic biomarkers in liver cancer patients [
25]. Lin et al. identified three exosomal tRFs in plasma of gastric cancer patient plasma, tRF-25, tRF-38 and tRF-18, that were significantly upregulated compared to healthy controls [
26]. Studies have also identified EV-contained tRFs that have potential as diagnostic biomarkers in breast cancer [
27,
28]. While tRFs have been identified in BC patient urinary EVs [
29], a role for EV-contained tRFs in BC diagnosis is largely unexplored.
We previously reported a study identifying potential BC miRNA biomarkers through next generation sequencing (NGS) of urine EVs, serum EVs and serum supernatant for 41 patients with non-muscle invasive BC (NMIBC) and 15 non-cancer patients (NCPs). In this study, the same patient material and methodology were applied to explore tRFs as potential biomarkers in BC.
2. Materials and Methods
2.1. Clinical Samples
Urine and serum supernatant samples were chosen from 41 NMIBC-patients and 15 NCPs recruited to the VESCAN biobank project. The biobank comprises urine and serum samples collected for patients who undergo 1) cystoscopy examinations, 2) BC surgery and 3) BC postsurgery check-ups. For this study we included samples from NMIBC patients with the following characteristics; 1) Urine and serum were biobanked prior to BC surgery 2) Urine and serum were biobanked at a minimum of one postsurgery check-up where no evidence of BC recurrence was detected and 3) There was no evidence of concurrent or prior malignancy other than BC. For most patients, samples from an early (usually 3 months) and late postsurgery checkup (usually 12 months) were included. Samples from NCPs were included if patients had no history of malignancy and presented with benign findings on cystoscopy examination. Clinical details for all patients and corresponding samples are described in our previous study [
28].
2.2. Workflow and Study Design
A summary of workflow and study design is outlined in
Figure 1 B and C. Urine and serum was centrifuged at 500 x g for 15 mins, and 12 000 x g for 30 mins and the supernatant stored at -80 degrees before EV/RNA isolation and subsequent small RNA sequencing. 28 BC patients (18 stage Ta and 10 stage T1) and 15 NCPs were sequenced in the first sequencing run and 13 additional BC patients (9 stage Ta and 4 stage T1) as well as replicate samples for the 10 stage T1 patients from the first sequencing run were sequenced in a second sequencing run. MINT map was used for tRF identification which maps all sequencing reads to the complete tRF space. DEseq2 was employed for the statistical analysis.
2.3. RNA isolation
EV-contained RNA was isolated from urine and serum supernatant using the exoRNeasy kit (Qiagen) according to the manufacturer’s instructions. 5 ml of urine and 0,5 ml of serum supernatant was used. The isolation of total RNA from serum supernatant samples was carried out with the miRNeasy kit (Qiagen).
2.4. Small RNA Sequencing
Small RNA sequencing was performed as previously described [
30].
2.5. Statistical Analysis
Atropos was used to trim adapters with the adapter sequence “TGGAATTCTCGGGTGCCAAGG”. Counts for tRFs were assessed using MINTmap [
31]. Differential expression analysis was performed with DESeq2 using the exclusive counts from MINTmap. The parameter biosource_cancer-stage [
32] (for example: SERUM_TA) was used for the differential expression which was performed separately for initial sequencing samples and for replicated samples. tRFs were defined as being differentially expressed (DEtRFs) between sample groups when an adjusted p-value of <0.05 was observed. The R package eulerr was used to create venndiagrams and pheatmap was used to create heatmaps. Data from MINTbase was used to assign information about which tRNA each tRF originated from and what type each tRF was. PCA was performed using the PCA function from sklearn.
2.6. BLAST Search
To examine what regions in the genome tRFs could interact with we performed a BLAST search for each tRF sequence for tRFs of interest. The BLAST search was performed using the nucleotide collection for homo sapiens and the program blastn. Hits longer than 1 million and whole chromosome hits were filtered out as well as hits with “match_diff” < 2 and “hsp_gaps” < 2. A list of phrases of interest was created based upon the filtered blast results and the number of hits with these phrases was summarized.
3. Results
3.1. Sample Characterisation
Patient cohort attributes are outlined in detail as previously described [
30]. Sample source and processing are outlined in
Figure 1B and C. Principal component analysis (PCA) of identified tRFs was carried out on urinary EVs, serum EVs and serum supernatant sequenced for both the initial and replica sequencing runs (
Figure S1A). Nanoparticle tracking analysis had previously confirmed the isolation of particles consistent with the size of EVs [
30]. We noted clustering of the three biosources, though there was some overlap for serum EV and serum supernatant samples. An explanation for the larger separation by urine EV samples may be the higher amount of tRF reads in urine EV samples (
Figure S1B). A larger fraction of tRFs in urine compared to other biofluids has previously been reported [
33].
3.2. Differential Expression of tRFs in Urine EVs, Serum EVs and Serum Supernatant in the Initial Sequencing Run
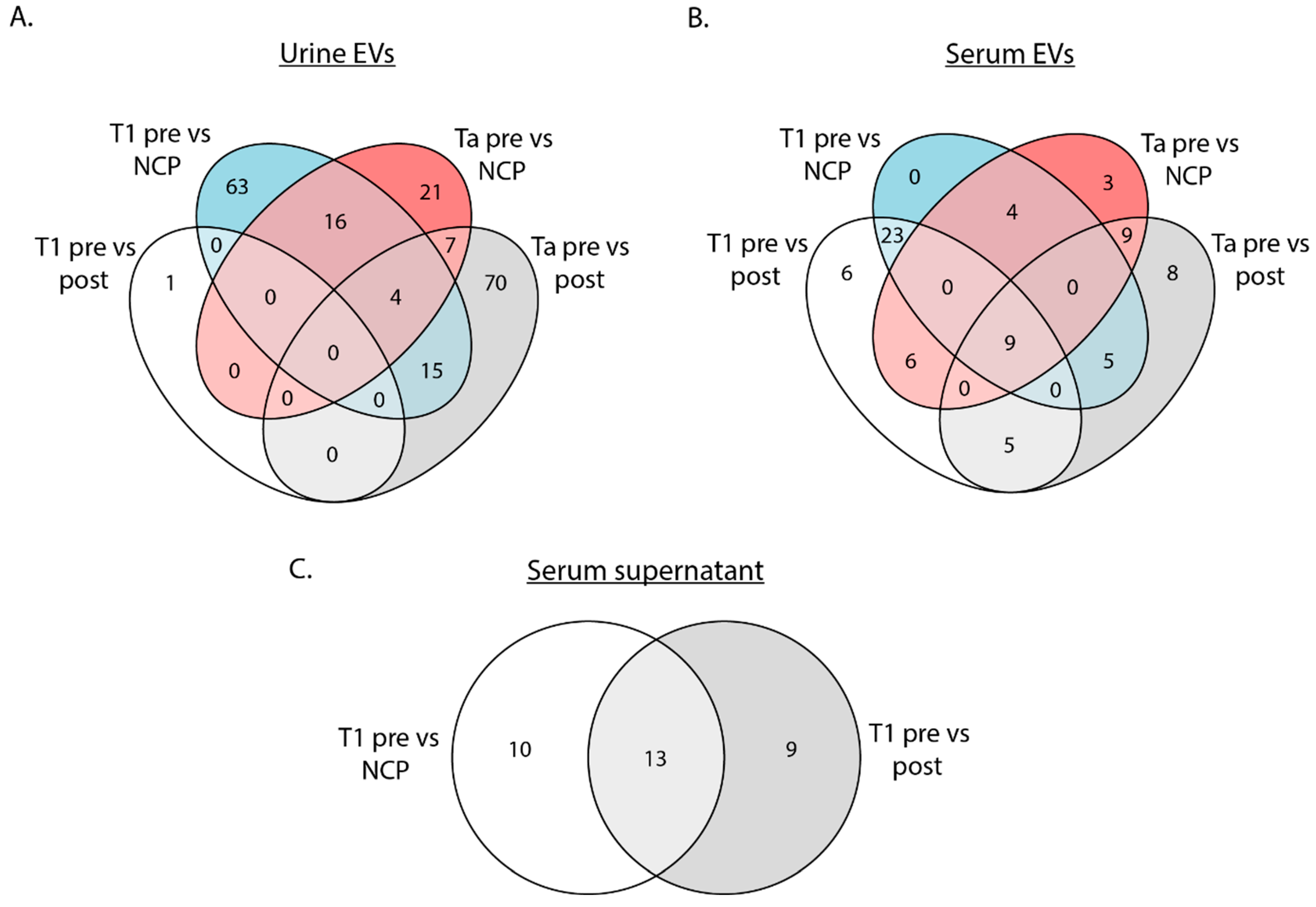
Differentially expressed tRFs (DEtRFs) were identified in both urine EVs, serum EVs and serum supernatant (
Figure 2). For urine EVs from Ta patients, 96 DEtRFs were mutually expressed when comparing presurgery samples to postsurgery samples, while 48 DEtRFs were found when comparing presurgery samples to NCPs. For T1 patients, the opposite trend was observed as 98 DEtRFs were identified when comparing presurgery samples to NCPs and only a single DEtRF was found when comparing presurgery to postsurgery samples (
Figure 2A).
In serum EVs from Ta patients, 36 DEtRFs were identified when comparing presurgery samples to postsurgery samples and 31 when comparing presurgery to NCP samples. There was a significant overlap of DEtRFs between the two comparisons, as 18 tRFs being mutually differentially expressed. A similar result was observed for T1 patients, where 49 DEtRFs were identified when comparing presurgery to postsurgery samples and 41 when comparing presurgery to NCPs, while 32 of these DEtRFs were identified in both comparisons (
Figure 2B). In serum supernatant, a total of 32 DEtRFs were identified, of which 13 individual tRFs were mutually differentially expressed in both the presurgery vs postsurgery and presurgery vs NCP comparison (
Figure 2C).
To summarize, the number of DEtRFs depends both on the sample source and disease.
3.3. tRFs Are Confirmed as Differentially Expressed among Patients with Stage T1 Disease by Replica Sequencing
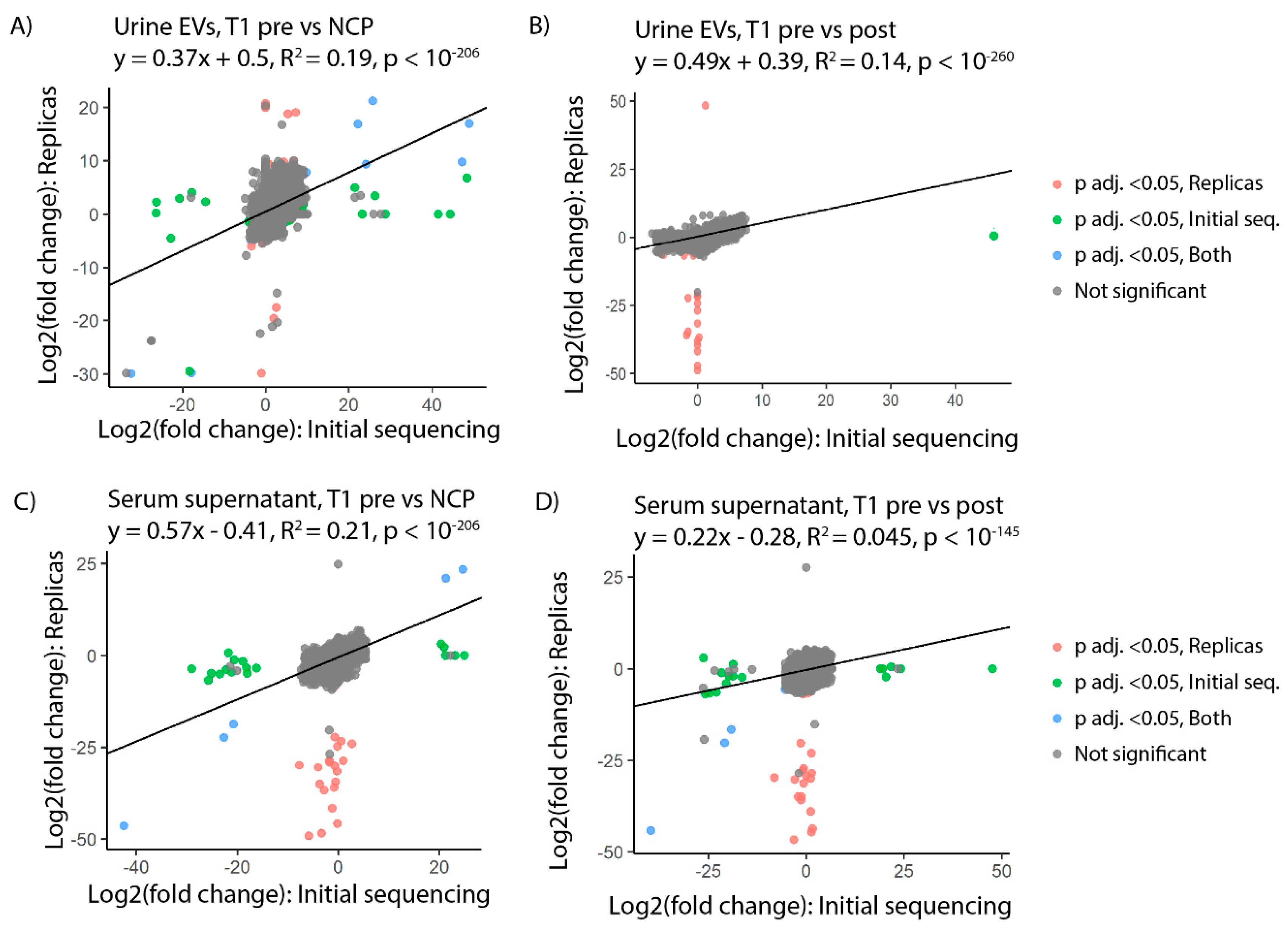
A replica EV/sncRNA isolation and sequencing run was carried out on new aliquots of urine and serum supernatant from 10 patients with stage T1 disease from the original cohort. Log2 fold change was compared between the initial and replica sequencing for all identified tRFs. A notable correlation was seen in urinary EVs between the two runs with a calculated R-squared value of 0.19 for T1 presurgery vs NCP (
Figure 3A) and 0.14 for T1 presurgery vs postsurgery (
Figure 3B). In serum supernatant, R-squared values of 0.21 for T1 presurgery vs NCP (
Figure 3C) and 0.045 for the T1 presurgery vs postsurgery comparisons were obtained. (
Figure 3D).
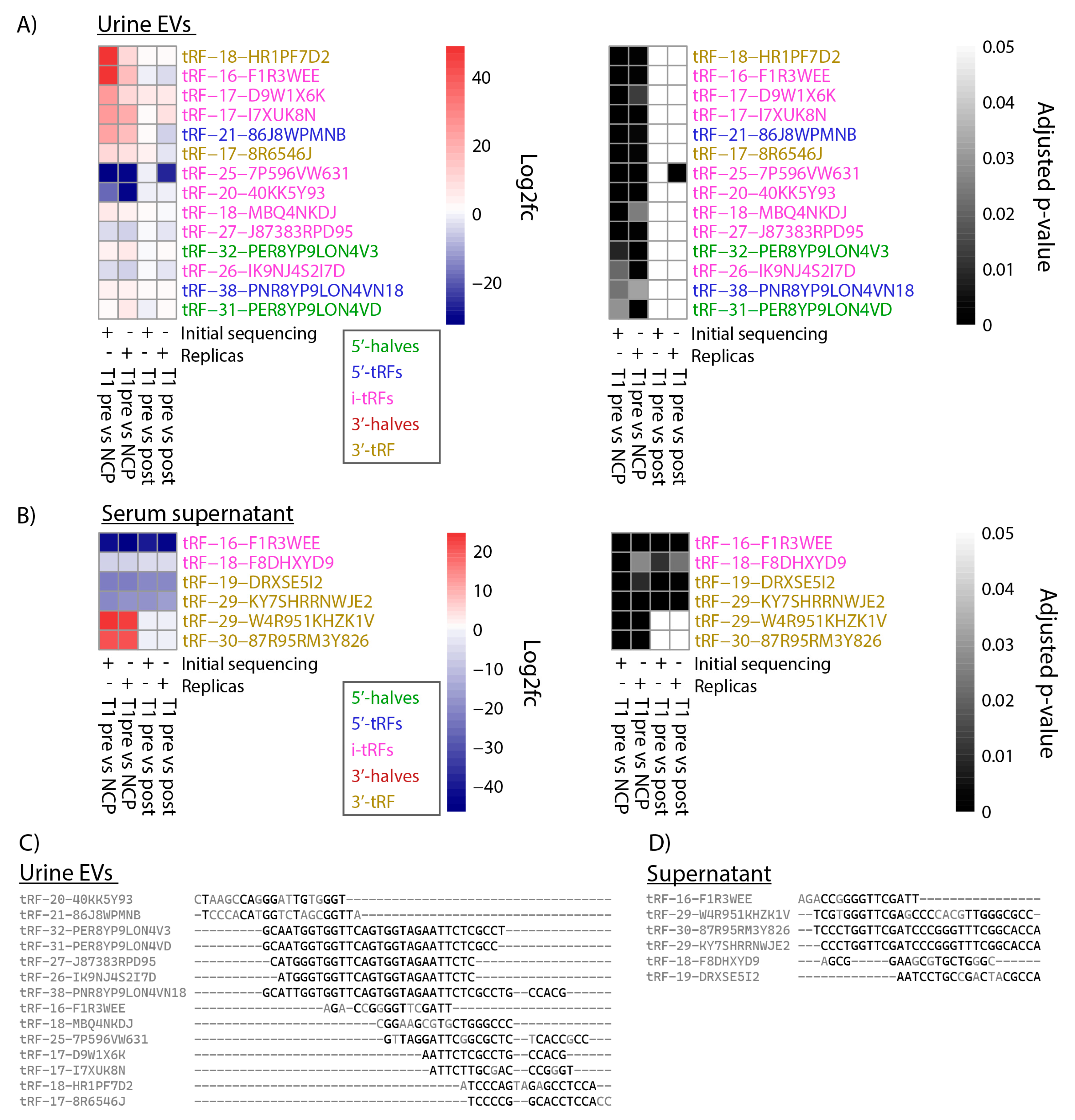
tRFs that were differentially expressed in both sequencing runs were plotted on heatmaps based on log2 fold change and adjusted p-value (
Figure 4A and 4B). In urine EVs, 14 tRFs were identified as differentially expressed in both the initial and replica sequencing, all of which were found when comparing T1 presurgery samples to NCPs (
Figure 4A). In serum supernatant, 6 DEtRFs were identified in both sequencing runs. Interestingly, four of these were found to be differentially expressed when comparing presurgery samples to both postsurgery and NCP samples. Meanwhile the remaining two samples were differentially expressed solely when comparing presurgery samples to NCPs (
Figure 4B).
The 14 tRF sequences for urine EVs and 6 tRF sequences for serum supernatant were aligned using clustal omega. The alignment of the tRFs showed that there are overlapping sequences between the DEtRFs fragments in both biological sources (
Figure 4C and 4D). As MINTmap requires that reads match a tRF exactly in order to be counted, even a single nucleotide difference will change the count associated with a tRF. This would also explain why some tRFs, although similar in sequence, have different log foldchange. However, the sequence similarity between tRFs should be considered when interpreting these results.
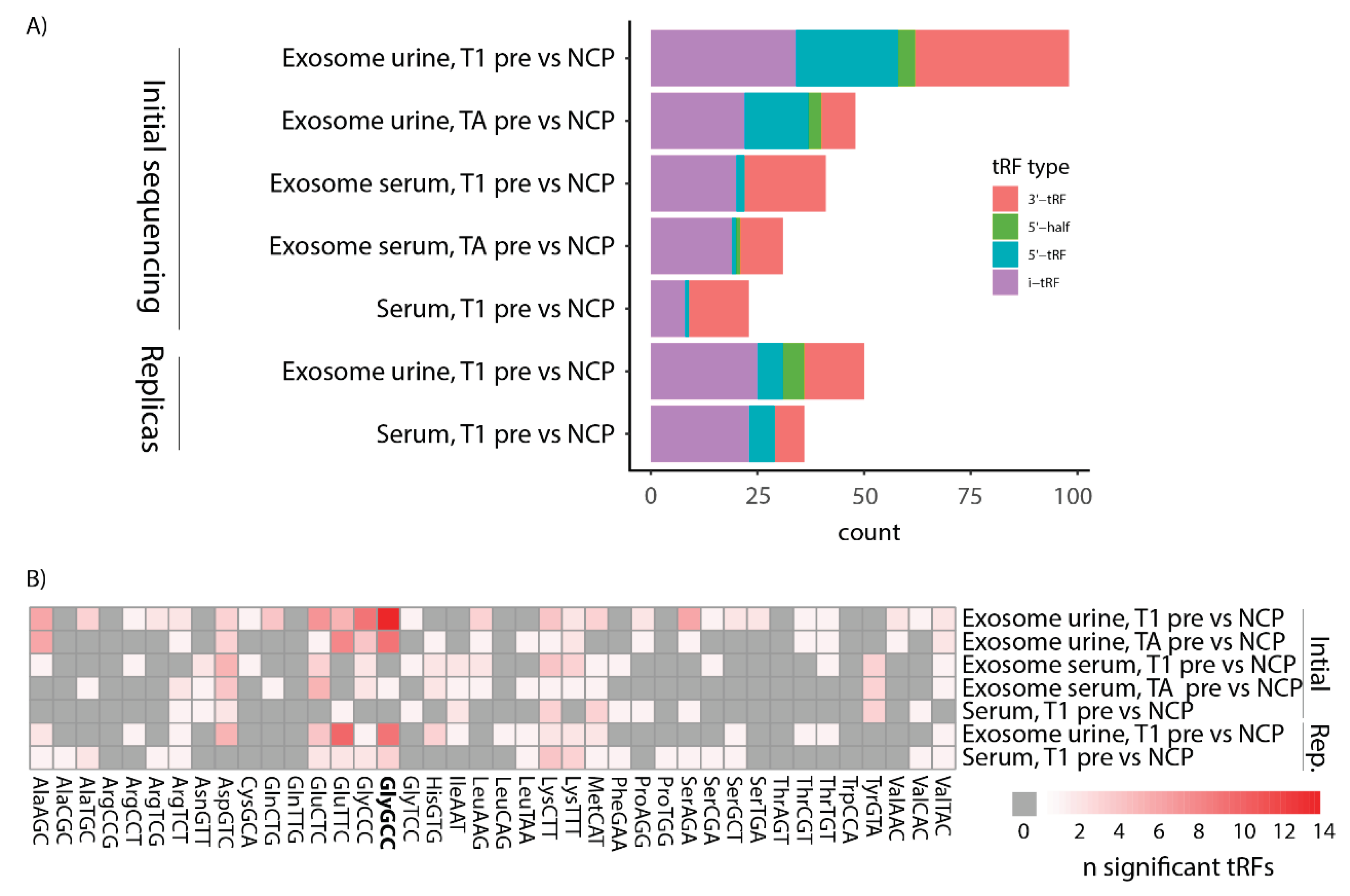
3.4. tRFs Differentially Expressed between Bladder Cancer and Non-Cancer Patients Are of Diverse Types and Originate from a Multitude of tRNAs
Each tRF can be categorized depending on where in the tRNA it originates from and which tRNA it is derived from. Here we used the definitions of tRF type as defined by MINTmap (
Figure 1A). Regarding the comparison between presurgery and NCP samples, we found DEtRFs across all tRF types except 3′-halves (
Figure 5A). However, the distribution is not even, and 5’-halves are underrepresented. To investigate whether there was a bias towards a certain origin of the tRFs, we plotted a heatmap showing which parent tRNA each DEtRF originated from (
Figure 5B). This heatmap shows that DEtRFs originate from most tRNAs. However, there is an enrichment of DEtRFs originating from certain parental tRNAs, such as tRNAsGlyGCC, which are particularly prevalent. Together, these results show that DEtRFs are of diverse types and originate from a multitude of tRNAs.
3.5. Assessment of Potential Role for tRFs in Biological Processes
tRFs can interact with a range of cellular processes, often mediated by binding through their nucleotide sequence [
34]. To generate hypothesis about which biological processes DEtRFs may be involved in, we performed a blast search using the tRF nucleotide sequences (Supplementary
Table 1). We chose to focus on DEtRFs identified in patients with stage T1, as these were confirmed as differentially expressed by replica sequencing. A list of interesting phrases obtained by the blast search was manually created for urine EVs (
Table 1) and serum supernatant (
Table 2). We found the phrase “tRNA” for 12 out of 14 DEtRFs in urine EVs and five out of six DEtRFs in supernatant, confirming the identification of tRFs.
However, we also obtained several other hits. Notably, 3 DEtRFs match with piRNA which can influence cellular function through epigenetic or post-transcriptional silencing. Furthermore, some of the tRFs have matching sequences to enhancer sequences, regulatory elements or CpG islands which is another indication that they could influence gene regulation. Finally, a number of gene names also turn up in the search such as HES7 which is an important part of the Notch signalling pathway. Further evidence is needed to see if these tRFs can influence such biological processes and to confirm that the sequences we find in our data originate from tRFs and not these non-tRF sequences themselves.
4. Discussion
tRF is a recently acknowledged class of sncRNA that have been shown to play a role in carcinogenesis and may have potential as cancer biomarkers. In the present study, the main findings can be summarized in two main points; 1) in both urine EVs and serum supernatant we identified DEtRFs that were confirmed through replica sequencing, 2) sequences of DEtRFs were found to be aligned to genomic sequences linked to processes associated with carcinogenesis, such as enhancers, regulatory elements and CpG islands.
Taking replica sequencing results into account, we identified 14 DEtRFs in urine EVs. Of these,12 were upregulated and two downregulated, when presurgery samples were compared to NCP samples. Three of the tRFs that were upregulated, tRF-18-MBQ4NKDJ, tRF-20-40KK5Y93 and tRF-31-PER8YP9LON4VD have previously been identified in cancer biomarker studies. In a study of gastric cancer patients, the expression of both tRF-18-MBQ4NKDJ and tRF-31-PER8YP9LON4VD was significantly upregulated in gastric cancer patient plasma compared to healthy controls [
34]. In a study on tRFs in breast cancer however, the expression of tRF-20-40KK5Y93 and tRF-31-PER8YP9LON4VD was downregulated in breast cancer tissue, as compared to healthy adjacent tissue [
35]. Regarding results from serum supernatant, 6 DEtRFs were identified when comparing presurgery samples to NCPs, and 4 DEtRFs when comparing presurgery to postsurgery samples.
In similar ways to other sncRNAs, tRFs can regulate cellular processes through complementary sequence binding in processes such as RNA silencing In this way, tRFs can potentially influence cancer progression depending on their sequence. In support of these DEtRFs influencing cancer progression, the BLAST search performed here revealed overlap with cellular processes regulating gene regulation and genes involved in the development of cancer. These hits include enhancer regions, CpG islands and regulatory elements which are non-mutational epigenetic processes and, in some cases, can be an enabling characteristic in oncogenesis The association with enhancer regions may be of particular importance. Interestingly, mammalian-wide interspersed tandem repeats (MIR), which are conserved parts of the human genome that are derived from tRNAs, have been found to act as enhancers of gene expression. It would thus be of particular interest if DEtRFs found in our study could directly interact with MIRs, potentially affecting the regulation of a wide set of genes Other hits are found in genes such as FCGR2A, GPD2 and HES7. Some care should be taken when interpreting the number of hits in the BLAST search results as some of the DEtRFs have sequence overlap as shown in
Figure 4C, D. Unlike MINTmap, the BLAST algorithm allows for some mismatches in the nucleotide sequence and therefore two similar tRFs could overlap with the same hits in the BLAST search results.
As cancer normally develops after changes in more than one gene, it can be difficult to establish the exact role of changes in these genes to development of cancer. However, the gene HES7 is part of the notch signalling pathway which can influence bladder cancer progression [
37]. It is interesting that 6 out of 20 DEtRFs overlap with sequences annotated as piRNAs. piRNAs are the same length as tRFs and some of the piRNAs are complete matches to the tRFs. piRNAs can guide proteins to cleave target RNA, methylate DNA and promote heterochromatin assembly and it would therefore be interesting to investigate how tRFs might influence their biology [
38]. However, a hit in the BLAST search does not necessarily mean that there is a functional connection between the DEtRF and the annotated biological process. It is possible that the tRF counting algorithm match reads in the sequencing data to tRFs that do not originate from tRFs or that the annotation in the database used for the blast search is wrong. The results from the BLAST search is therefore best used to generate hypothesis and future functional studies should therefore investigate the possible connection between the DEtRFs discussed here and cancer progression.
While previous studies have investigated tRF expression in BC-tissue among muscle-invasive bladder cancer (MIBC) patients [40] and tRFs in early disease progression vs poor treatment outcome in BC [41], the present work is to the best of our knowledge the first to identify EV-contained tRFs as potential biomarkers in BC. Some of the study’s limitations need to be addressed, however. The patient sample size is small, with the stage T1 group consisting of 14 patients, and the NCP group consisting of 15 patients. Furthermore, only samples from T1 patients were subjected to a replica EV/RNA isolation and sequencing run, limiting the basis for identifying biomarkers in the Ta group.
In the present study, 14 DEtRFs in urine EVs and six DEtRFs in serum supernatant were identified as potential diagnostic biomarkers for BC patients with stage T1 disease. Four of the DEtRFs identified in serum supernatant, tRF-16-F1R3WEE, tRF-18-F8DHXYD9, tRF-19-DRXSE5I2 and tRF-29-KY7SHRRNWJE2, were downregulated in presurgery samples, both compared to postsurgery and NCP samples, suggesting this downregulation may be specific to BC-patients with stage T1 disease. As postsurgery samples were collected from patients with no clinically detected recurrence at the time of sampling, these four tRFs may hold potential as biomarkers of both primary diagnosis and recurrence-free survival of T1 patients.
5. Conclusions
In the present study, we identified 14 DEtRFs urine EVs and 6 DEtRFs serum supernatants from BC patients with stage T1 disease. A number of these DEtRFs were found to be associated with parts of the genome involved in key carcinogenic processes. We therefore argue that the DEtRFs identified in this work may warrant further investigation into their potential as BC-biomarkers and as players in BC pathogenesis.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
C.-J.A., G.B. were leading study designers with contributions from O.S., K.A.H., H.T.L and M.O.. O.S. and K.A.H. wrote the article. O.S. and K.A.H. carried out nanosized tracking analysis. H.T.L. conducted data analysis and contributed to manuscript input. All authors have read and agreed to the published version of the manuscript.
Funding
.The study was funded by grants from: Liaison Committee between the central Norway Regional Health Authority (RHA) and the Norwegian University of Science and technology (NTNU) (SO) Reference Number: 2017/22110. The Joint Research Committee between St. Olav’s Hospital and the Faculty of Medicine and Health Sciences, NTNU (FFU) Reference Number: 2019/38882. Kreftfondet (Cancer Research Fund), St. Olav’s Hospital. Reference Number: 2016/43/16. Olav Raagholt og Gerd Meidel Raagholts stiftelse.
Institutional Review Board Statement
.The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of NTNU/REK midt (protocol code: 2367 and date of approval: 31 May 2018).
Informed Consent Statement
All use of patient material is approved by Regional Ethic Committee (REK). REK 2017/2367. All patients gave informed consent in accordance with the Declaration of Helsinki.
Data Availability Statement
.The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The authors would like to thank the nurses at the urological department, St. Olav’s for their dedicated effort in organizing sample collection and the recruiting of participants. The authors would also like to thank the research nurses at the department of surgery for their work with sample collection. We would additionally like to thank the staff in Biobank 1 for establishing the VESCAN biobank and cooperating in obtaining and isolating samples for the study. We thank all staff at the Genomic Core Facility for carrying out the sequencing analysis. We would also like to thank Siri Backe and Kirsti Margrethe Olsen at Institute for Clinical and Molecular Medicine for practical assistance in the lab.
Conflicts of Interest
The authors claim no conflict of interest.
References
- Trail M, Cullen J, Fulton E, Clayton F, McGregor E, McWilliam F, et al. Evaluating the Safety of Performing Flexible Cystoscopy When Urinalysis Suggests Presence of “Infection”: Results of a Prospective Clinical Study in 2350 patients. Eur Urol Open Sci. 2021, 31, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Toden S, Goel A. Non-coding RNAs as liquid biopsy biomarkers in cancer. Br J Cancer. 2022, 126, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958, 231, 241–257. [Google Scholar] [CrossRef]
- Speer J, Gehrke CW, Kuo KC, Waalkes TP, Borek E. tRNA breakdown products as markers for cancer. Cancer. 1979, 44, 2120–2123. [Google Scholar] [CrossRef]
- Waalkes TP, Gehrke CW, Zumwalt RW, Chang SY, Lakings DB, Tormey DC, et al. The urinary excretion of nucleosides of ribonucleic acid by patients with advanced cancer. Cancer. 1975, 36, 390–398. [Google Scholar] [CrossRef]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Weng Q, Wang Y, Xie Y, Yu X, Zhang S, Ge J, et al. Extracellular vesicles-associated tRNA-derived fragments (tRFs): biogenesis, biological functions, and their role as potential biomarkers in human diseases. J Mol Med (Berl). 2022, 100, 679–695. [Google Scholar] [CrossRef] [PubMed]
- Wilson B, Dutta A. Function and Therapeutic Implications of tRNA Derived Small RNAs. Front Mol Biosci. 2022, 9, 888424. [Google Scholar]
- Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef]
- Zhang M, Li F, Wang J, He W, Li Y, Li H, et al. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. Onco Targets Ther. 2019, 12, 6371–6383. [Google Scholar] [CrossRef]
- Mao M, Chen W, Huang X, Ye D. Role of tRNA-derived small RNAs(tsRNAs) in the diagnosis and treatment of malignant tumours. Cell Commun Signal. 2023, 21, 178. [Google Scholar] [CrossRef] [PubMed]
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
- Fu M, Gu J, Wang M, Zhang J, Chen Y, Jiang P, et al. Emerging roles of tRNA-derived fragments in cancer. Mol Cancer. 2023, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Endres, L. tRNA modification and cancer: potential for therapeutic prevention and intervention. Futire Medicinal Cemistry. 2019, 8. [Google Scholar] [CrossRef]
- Keam SP, Hutvagner G. tRNA-Derived Fragments (tRFs): Emerging New Roles for an Ancient RNA in the Regulation of Gene Expression. Life (Basel). 2015, 5, 1638–1651. [Google Scholar]
- Xie Y, Yao L, Yu X, Ruan Y, Li Z, Guo J. Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduct Target Ther. 2020, 5, 109. [Google Scholar] [CrossRef] [PubMed]
- Su Z, Monshaugen I, Klungland A, Ougland R, Dutta A. Characterization of novel small non-coding RNAs and their modifications in bladder cancer using an updated small RNA-seq workflow. Front Mol Biosci. 2022, 9, 887686. [Google Scholar]
- van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef]
- Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef]
- Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest. 2016, 126, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Bebelman MP, Janssen E, Pegtel DM, Crudden C. The forces driving cancer extracellular vesicle secretion. Neoplasia. 2021, 23, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lane RE, Korbie D, Hill MM, Trau M. Extracellular vesicles as circulating cancer biomarkers: opportunities and challenges. Clin Transl Med. 2018, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019, 18, 74. [Google Scholar] [CrossRef]
- Lin, C. tRFs as Potential Exosome tRNA-Derived Fragment Biomarkers for Gastric Carcinoma. Clin Lab. 2020, 1. [Google Scholar] [CrossRef]
- Guzman N, Agarwal K, Asthagiri D, Yu L, Saji M, Ringel MD, Paulaitis ME. Breast Cancer-Specific miR Signature Unique to Extracellular Vesicles Includes “microRNA-like” tRNA Fragments. Mol Cancer Res. 2015, 13, 891–901. [Google Scholar] [CrossRef]
- Koi Y, Tsutani Y, Nishiyama Y, Ueda D, Ibuki Y, Sasada S, et al. Predicting the presence of breast cancer using circulating small RNAs, including those in the extracellular vesicles. Cancer Sci. 2020, 111, 2104–2115. [Google Scholar] [CrossRef]
- Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer. 2015, 14, 194. [Google Scholar] [CrossRef]
- Stromme O, Heck KA, Brede G, Lindholm HT, Otterlei M, Arum CJ. Differentially Expressed Extracellular Vesicle-Contained microRNAs before and after Transurethral Resection of Bladder Tumors. Curr Issues Mol Biol. 2021, 43, 286–300. [Google Scholar] [CrossRef]
- Loher P, Telonis AG, Rigoutsos I. MINTmap: fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci Rep. 2017, 7, 41184. [Google Scholar] [CrossRef] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550.
- Srinivasan S, Yeri A, Cheah PS, Chung A, Danielson K, De Hoff P, et al. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell. 2019, 177, 446–462. [Google Scholar] [CrossRef] [PubMed]
- Chan C, Pham P, Dedon PC, Begley TJ. Lifestyle modifications: coordinating the tRNA epitranscriptome with codon bias to adapt translation during stress responses. Genome Biol. 2018, 19, 228. [Google Scholar]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Djingo, D. Mammalian-wide interspersed repeat (MIR)-derived enhancers and the regulation of human gene expression. Mobile DNA. 2014, 5. [Google Scholar]
- Maraver A, Fernandez-Marcos PJ, Cash TP, Mendez-Pertuz M, Duenas M, Maietta P, et al. NOTCH pathway inactivation promotes bladder cancer progression. J Clin Invest. 2015, 125, 824–830. [Google Scholar] [CrossRef]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).