Submitted:
01 April 2024
Posted:
02 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Methods and Software
2.2. Data Analytics
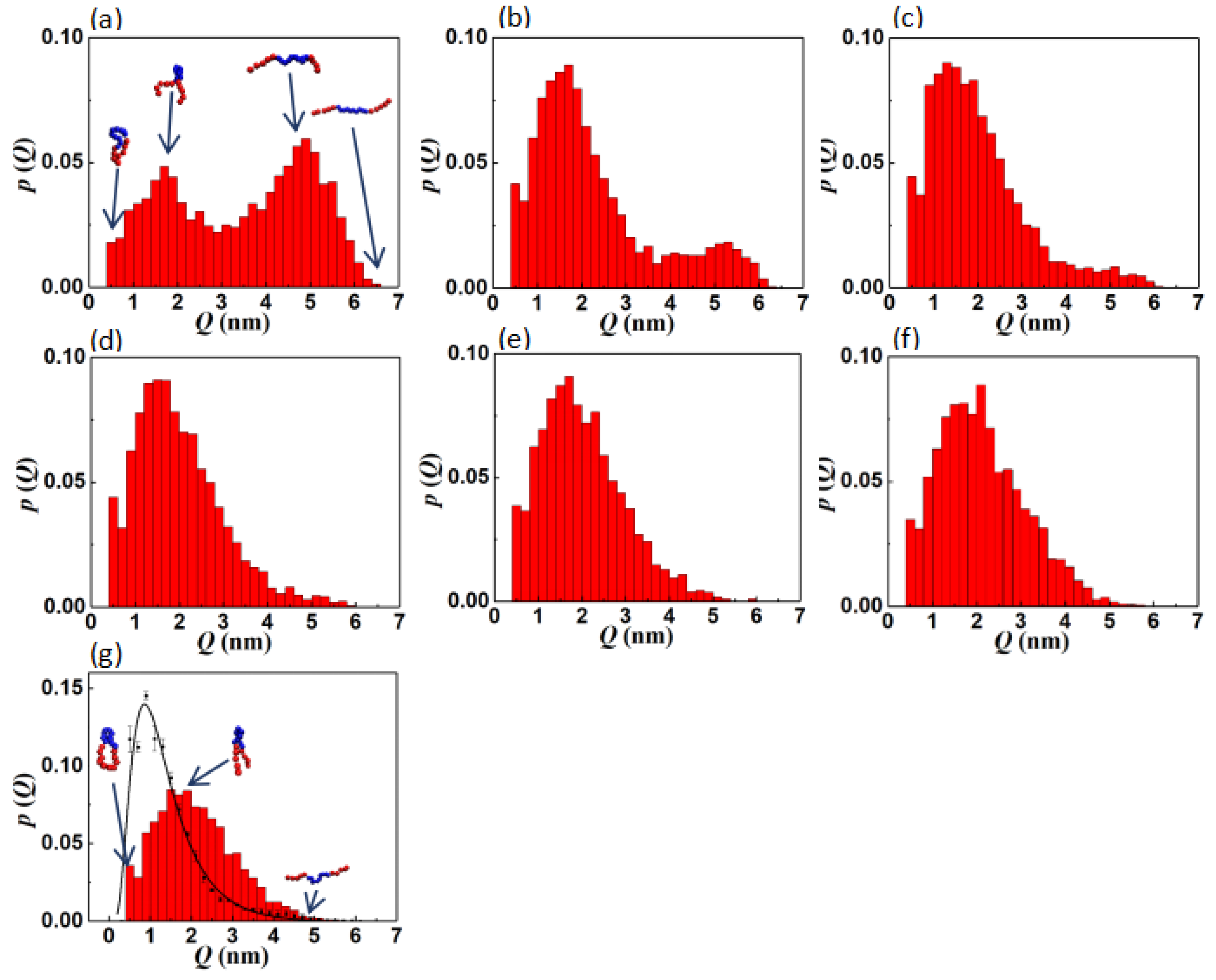
2.2.1. Information Entropy (H)
2.2.2. Pair Correlation Function (
3. Results and Discussion
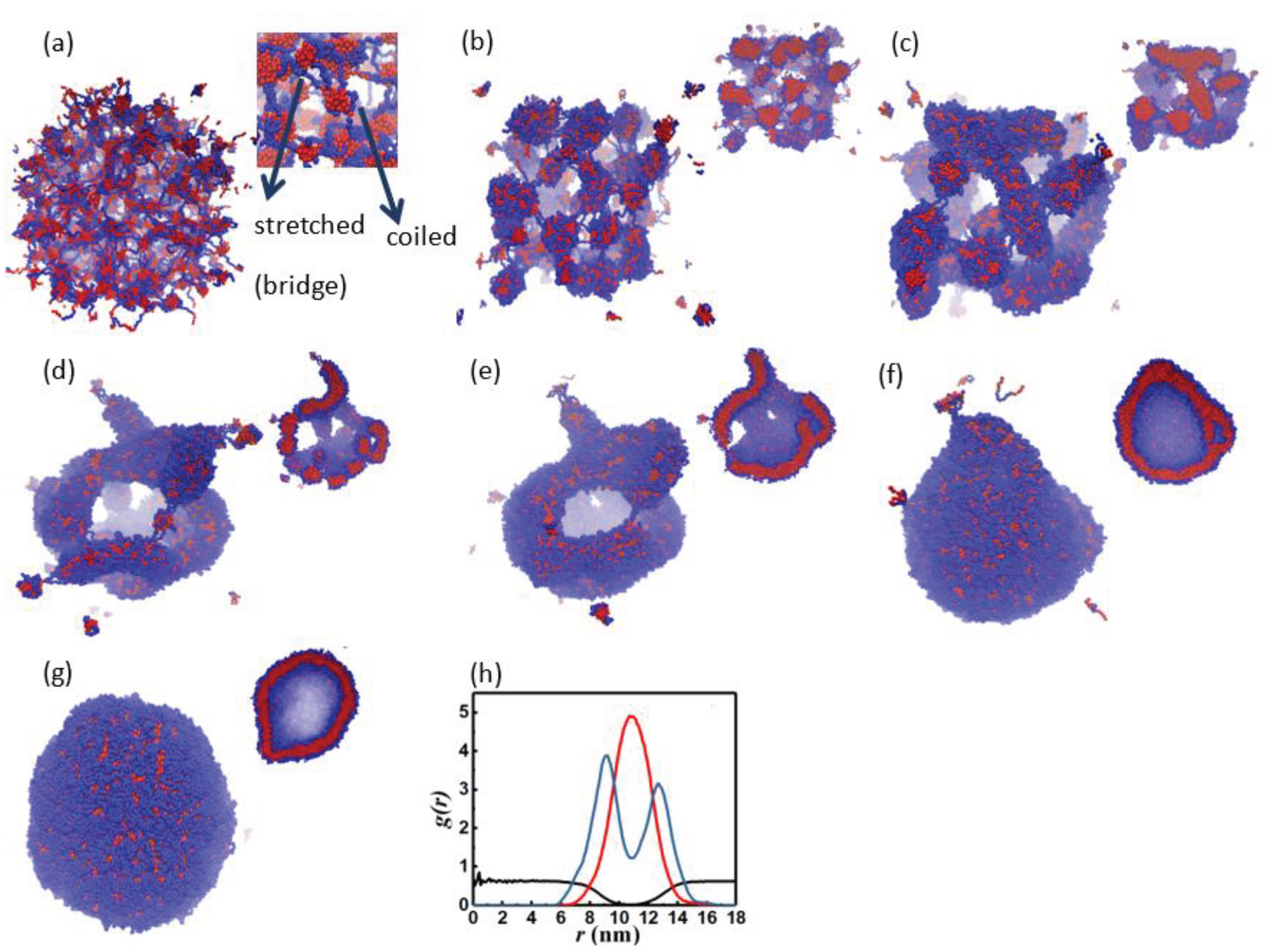
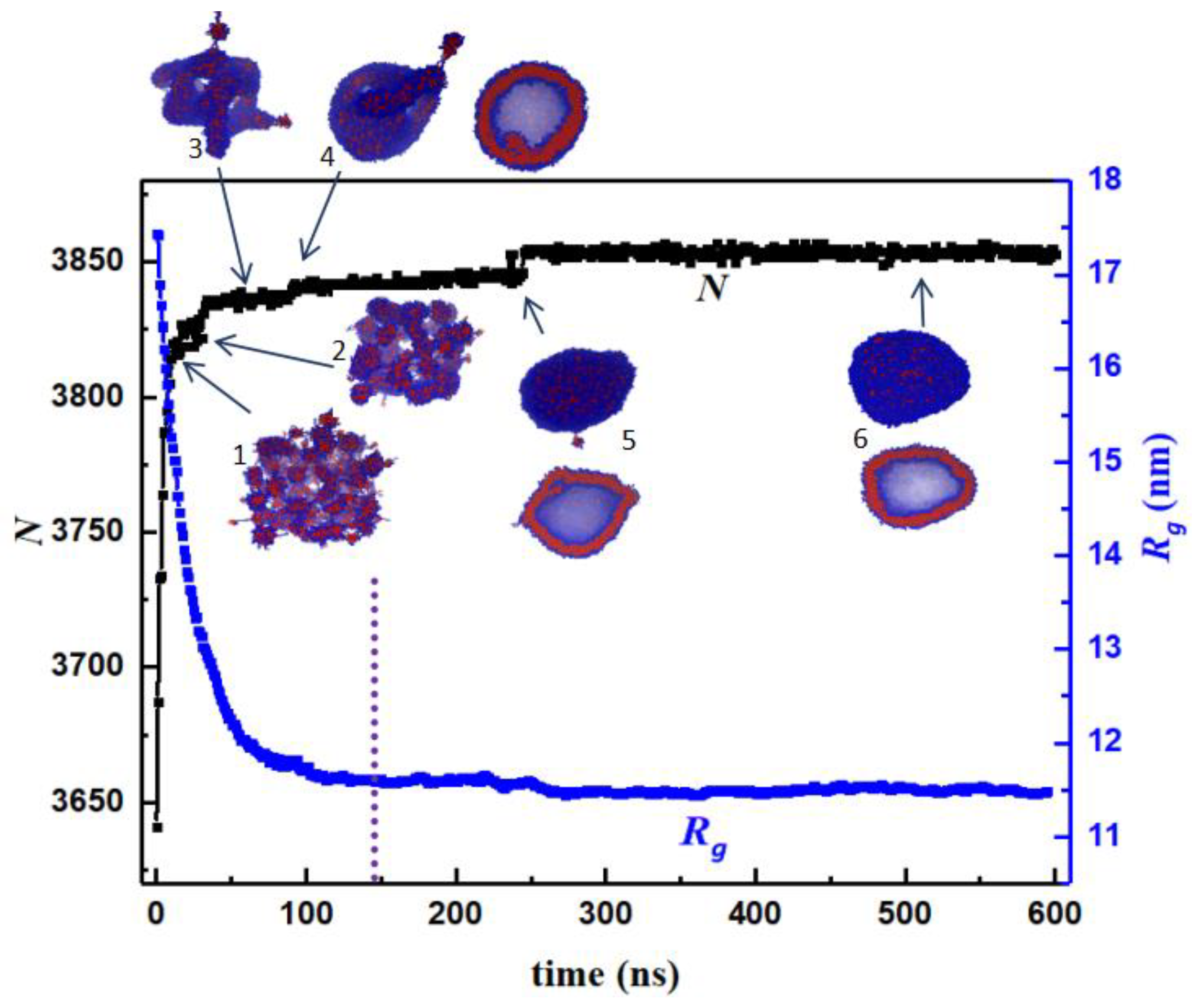
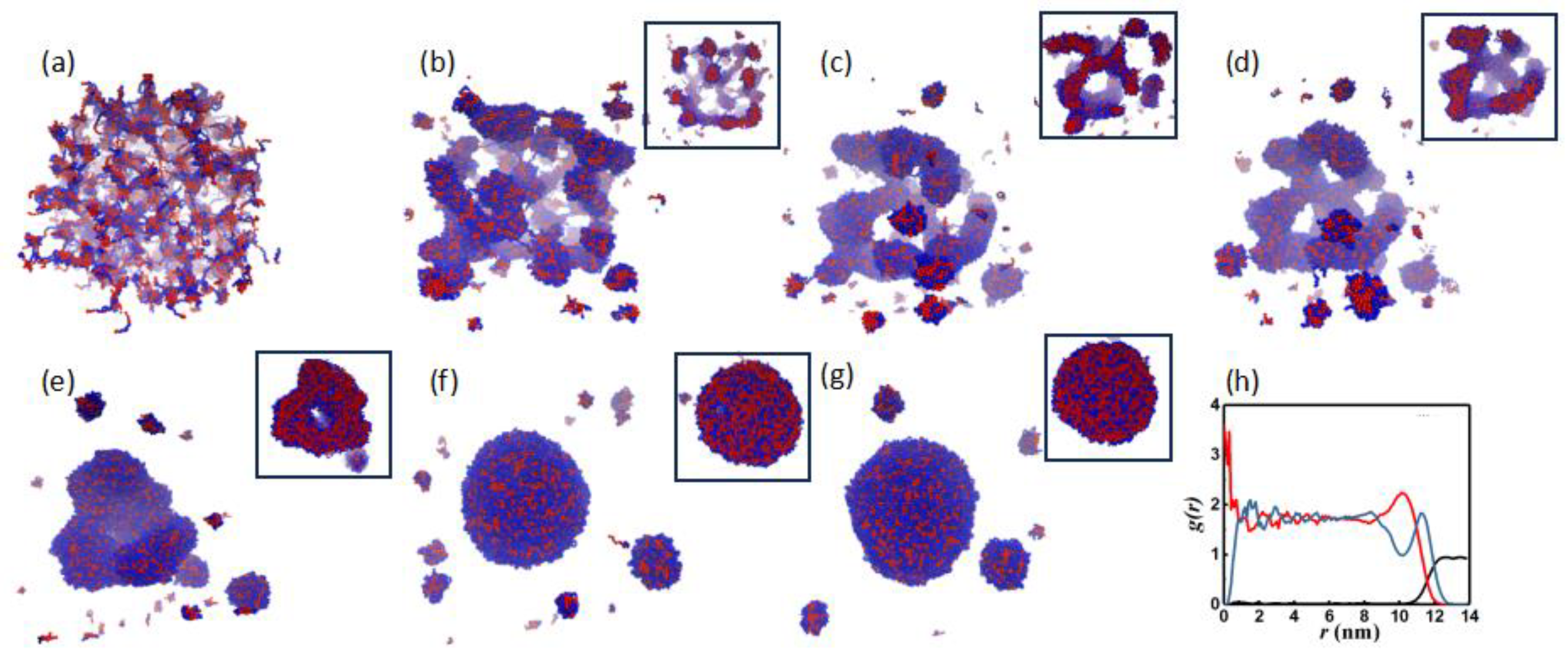
3.1. Morphology Evolution
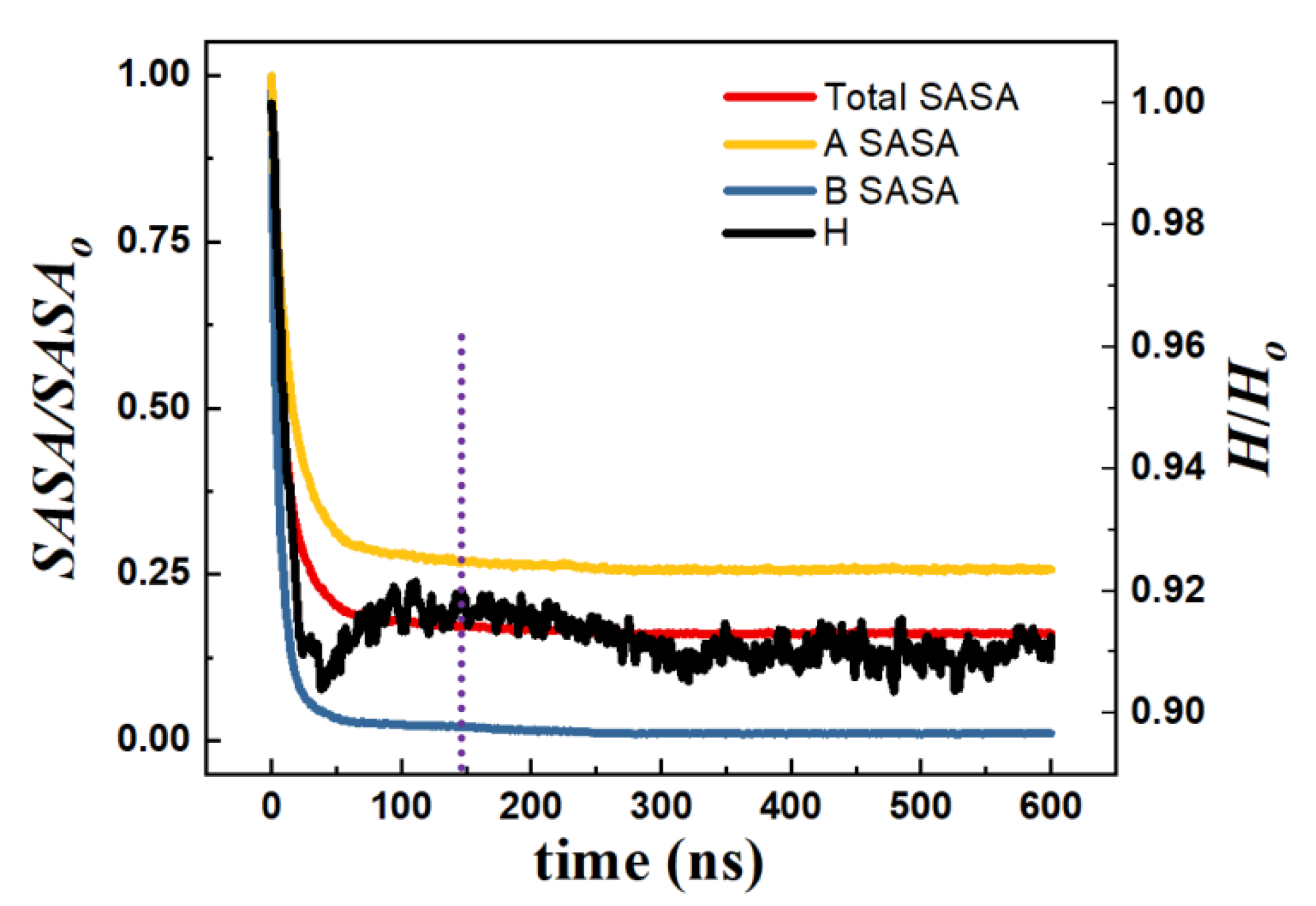
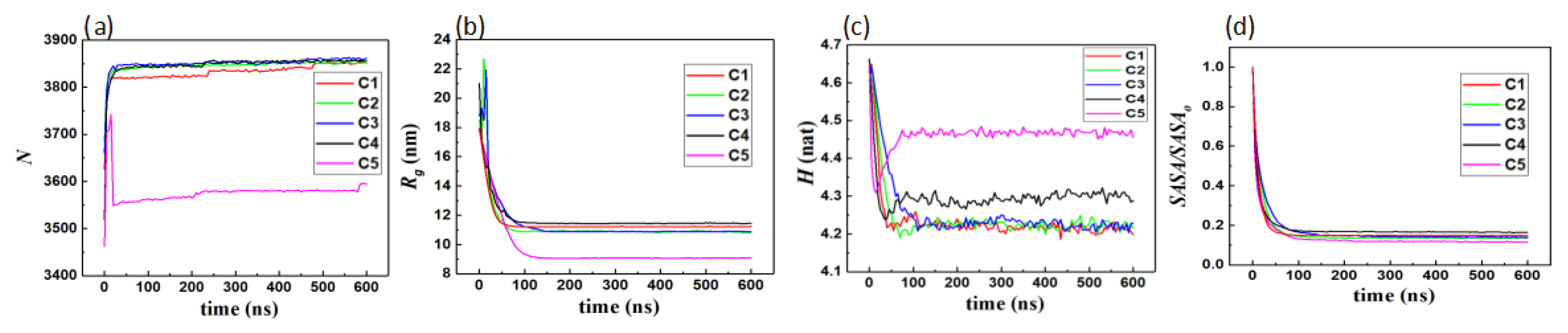
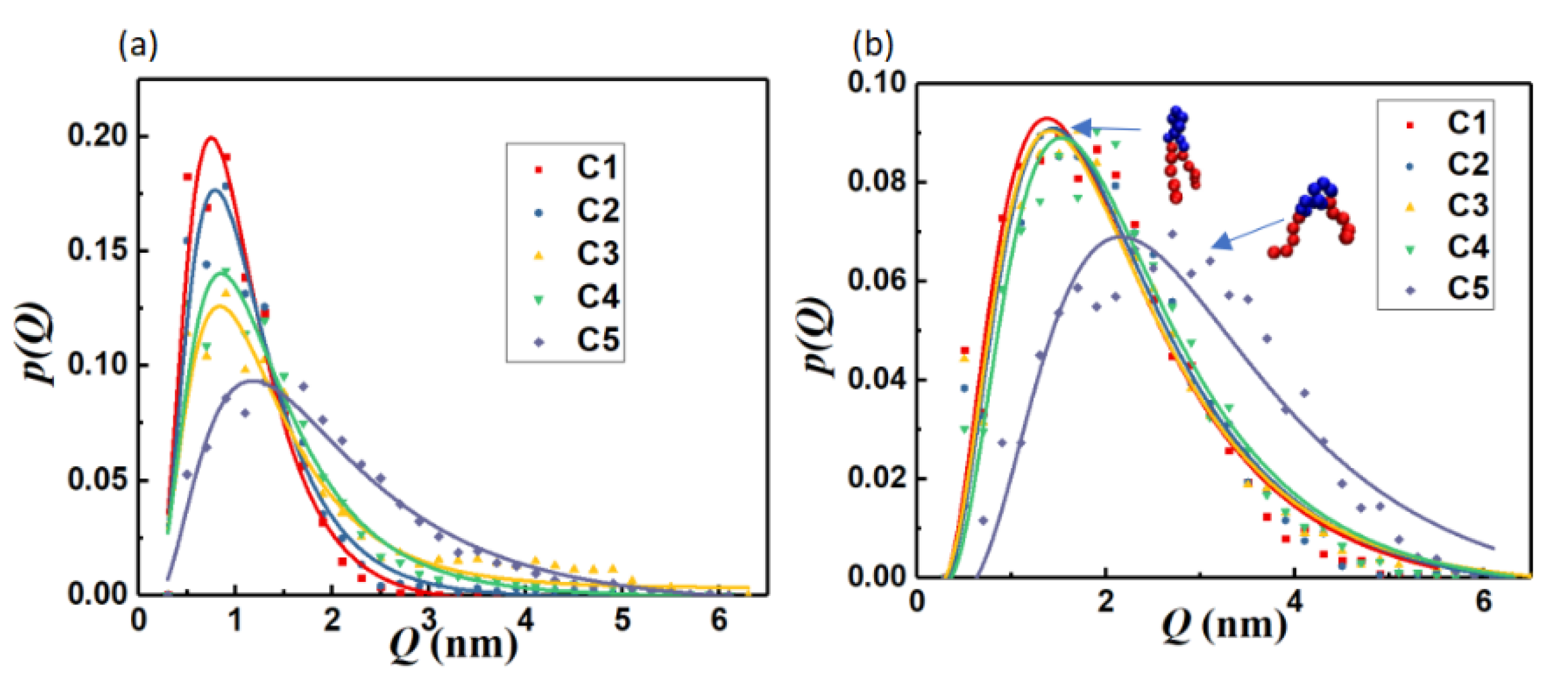
3.1.1. Interfacial Area and Information Entropy
3.2 Effect of Solvent Affinity of the Hydrophobic Segment on Self-Assembly
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bleul, R.; Thiermann, R.; Maskos, M. Techniques to Control Polymersome Size. Macromolecules 2015, 48, 7396–7409. [Google Scholar] [CrossRef]
- Karayianni, M.; Pispas, S. Block Copolymer Solution Self-Assembly: Recent Advances, Emerging Trends, and Applications. J. Polym. Sci. 2021, 59, 1874–1898. [Google Scholar] [CrossRef]
- Li, S.; Byrne, B.; Welsh, J.E.; Palmer, A.F. Self-Assembled Poly(Butadiene)-b-Poly(Ethylene Oxide) Polymersomes as Paclitaxel Carriers. Biotechnol. Prog. 2007, 23, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.; Eisenberg, A.; Mrsic, J.; Maysinger, D.; Eisenberg, A. PCL-b-PEO Micelles as a Delivery Vehicle for Fk506: Assessment of a Functional Recovery of Crushed Peripheral Nerve. Drug Deliv. 2000, 7, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.; Yu, Y.; Maysinger, D.; Eisenberg, A. Polycaprolactone-b-Poly(Ethylene Oxide) Block Copolymer Micelles as a Novel Drug Delivery Vehicle for Neurotrophic Agents Fk506 and l-685,818. Bioconjugate Chem. 1998, 9, 564–572. [Google Scholar] [CrossRef]
- Boucher-Jacobs, C.; Rabnawaz, M.; Katz, J.S.; Even, R.; Guironnet, D. Encapsulation of Catalyst in Block Copolymer Micelles for the Polymerization of Ethylene in Aqueous Medium. Nat. Commun. 2018, 9, 841. [Google Scholar] [CrossRef]
- Cuomo, F.; Ceglie, A.; De Leonardis, A.; Lopez, F. Polymer Capsules for Enzymatic Catalysis in Confined Environments. Catalysts 2018, 9, 1. [Google Scholar] [CrossRef]
- Peters, R.J.; Marguet, M.; Marais, S.; Fraaije, M.W.; van Hest, J.C.; Lecommandoux, S. Cascade Reactions in Multicompartmentalized Polymersomes. Angew. Chem. Int. Ed. 2013, 53, 146–150. [Google Scholar] [CrossRef]
- Wilson, D.A.; Nolte, R.J.; van Hest, J.C. Autonomous Movement of Platinum-Loaded Stomatocytes. Nat. Chem. 2012, 4, 268–274. [Google Scholar] [CrossRef]
- Marguet, M.; Bonduelle, C.; Lecommandoux, S. Multicompartmentalized Polymeric Systems: Towards Biomimetic Cellular Structure and Function. Chem. Soc. Rev. 2013, 42, 512–529. [Google Scholar] [CrossRef]
- Che, H.; van Hest, J.C. Stimuli-Responsive Polymersomes and Nanoreactors. J. Mater. Chem. B 2016, 4, 4632–4647. [Google Scholar] [CrossRef]
- Jacobs, M.L.; Boyd, M.A.; Kamat, N.P. Diblock Copolymers Enhance Folding of a Mechanosensitive Membrane Protein during Cell-free Expression. Proc. Natl. Acad. Sci. USA 2019, 116, 4031–4036. [Google Scholar] [CrossRef] [PubMed]
- Israelachvili, J.N.; Mitchell, D.J.; Ninham, B.W. Theory of Self-Assembly of Lipid Bilayers and Vesicles. Biochim. Biophys. Acta (BBA)-Biomembr. 1977, 470, 185–201. [Google Scholar] [CrossRef]
- Huang, C.; Quinn, D.; Sadovsky, Y.; Suresh, S.; Hsia, K.J. Formation and Size Distribution of Self-Assembled Vesicles. Proc. Natl. Acad. Sci. USA 2017, 114, 2910–2915. [Google Scholar] [CrossRef] [PubMed]
- Thiermann, R.; Bleul, R.; Maskos, M. Kinetic Control of Block Copolymer Self-Assembly in a Micromixing Device – Mechanistical Insight into Vesicle Formation Process. Macromolecular Chemistry and Physics 2016, 218. [Google Scholar] [CrossRef]
- Mai, Y.; Eisenberg, A. Self-Assembly of Block Copolymers. Chem. Soc. Rev. 2012, 41, 5969–5985. [Google Scholar] [CrossRef] [PubMed]
- Antonietti, M.; Förster, S. Vesicles and Liposomes: A Self-Assembly Principle Beyond Lipids. Adv. Mater. 2003, 15, 1323–1333. [Google Scholar] [CrossRef]
- Li, X.; Cooksey, T.J.; Kidd, B.E.; Robertson, M.L.; Madsen, L.A. Mapping Coexistence Phase Diagrams of Block Copolymer Micelles and Free Unimer Chains. Macromolecules 2018, 51, 8127–8135. [Google Scholar] [CrossRef]
- Holder, S.W.; Grant, S.C.; Mohammadigoushki, H. Nuclear Magnetic Resonance Diffusometry of Linear and Branched Wormlike Micelles. Langmuir 2021, 37, 3585–3596. [Google Scholar] [CrossRef]
- Van Hest, J.C.; Delnoye, D.A.; Baars, M.W.; van Genderen, M.H.; Meijer, E.W. Polystyrene-Dendrimer Amphiphilic Block Copolymers with a Generation-Dependent Aggregation. Science 1995, 268, 1592–1595. [Google Scholar] [CrossRef]
- Zhang, L.; Eisenberg, A. Multiple Morphologies of “Crew-Cut” Aggregates of Polystyrene-b-Poly(Acrylic Acid) Block Copolymers. Science 1995, 268, 1728–1731. [Google Scholar] [CrossRef]
- Discher, B.M.; Won, Y.-Y.; Ege, D.S.; Lee, J.C.-M.; Bates, F.S.; Discher, D.E.; Hammer, D.A. Polymersomes: Tough Vesicles Made from Diblock Copolymers. Science 1999, 284, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Bates, F.S. On the Origins of Morphological Complexity in Block Copolymer Surfactants. Science 2003, 300, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Won, Y.-Y.; Brannan, A.K.; Davis, H.T.; Bates, F.S. Cryogenic Transmission Electron Microscopy (Cryo-Tem) of Micelles and Vesicles Formed in Water by Poly(Ethylene Oxide)-Based Block Copolymers. J. Phys. Chem. B 2002, 106, 3354–3364. [Google Scholar] [CrossRef]
- Chen, L.; Shen, H.; Eisenberg, A. Kinetics and Mechanism of the Rod-to-Vesicle Transition of Block Copolymer Aggregates in Dilute Solution. J. Phys. Chem. B 1999, 103, 9488–9497. [Google Scholar] [CrossRef]
- Liu, S.; Sureshkumar, R. Morphological Diversity in Diblock Copolymer Solutions: A Molecular Dynamics Study. Colloids Interfaces 2023, 7, 40. [Google Scholar] [CrossRef]
- Liu, S.; Sureshkumar, R. Energetic and Entropic Motifs in Vesicle Morphogenesis in Amphiphilic Diblock Copolymer Solutions. Colloids and Interfaces 2024, 8, 12. [Google Scholar] [CrossRef]
- Srinivas, G.; Discher, D.E.; Klein, M. L. Self-Assembly and Properties of Diblock Copolymers by Coarse-Grain Molecular Dynamics. Nature Materials 2004, 3, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, G.; Shelley, J.C.; Nielsen, S. O.; Discher, D. E.; Klein, M. L. Simulation of Diblock Copolymer Self-Assembly, Using a Coarse-Grain Model. The Journal of Physical Chemistry B 2004, 108, 8153–8160. [Google Scholar] [CrossRef]
- He, X.; Schmid, F. Spontaneous Formation of Complex Micelles from a Homogeneous Solution. Physical Review Letters 2008, 100, 13. [Google Scholar] [CrossRef]
- He, X.; Schmid, F. Dynamics of Spontaneous Vesicle Formation in Dilute Solutions of Amphiphilic Diblock Copolymers. Macromolecules 2006, 39, 2654–2662. [Google Scholar] [CrossRef]
- Sevink, G.J.; Zvelindovsky, A. V. Self-Assembly of Complex Vesicles. Macromolecules 2005, 38, 7502–7513. [Google Scholar] [CrossRef]
- Ye, X.; Khomami, B. Self-Assembly of Linear Diblock Copolymers in Selective Solvents: From Single Micelles to Particles with Tri-Continuous Inner Structures. Soft Matter 2020, 16, 6056–6062. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Dormidontova, E. E. Equilibrium Chain Exchange Kinetics in Block Copolymer Micelle Solutions by Dissipative Particle Dynamics Simulations. Soft Matter 2011, 7, 4179. [Google Scholar] [CrossRef]
- Javan Nikkhah, S.; Turunen, E.; Lepo, A.; Ala-Nissila, T.; Sammalkorpi, M. Multicore Assemblies from Three-Component Linear Homo-Copolymer Systems: A Coarse-Grained Modeling Study. Polymers 2021, 13, 2193. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, V.; Nielsen, S.O.; Discher, D. E.; Klein, M. L.; Lipowsky, R.; Shillcock, J. Dissipative Particle Dynamics Simulations of Polymersomes. The Journal of Physical Chemistry B 2005, 109, 17708–17714. [Google Scholar] [CrossRef] [PubMed]
- Shillcock, J. C. Spontaneous Vesicle Self-Assembly: A Mesoscopic View of Membrane Dynamics. Langmuir 2012, 28, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Liu, J.; Yang, J.; Wang, R.; Xie, D. Biomimetic Membrane Control of Block Copolymer Vesicles with Tunable Wall Thickness. Soft Matter 2013, 9, 2434. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Wang, Z.; Bao, M. Micelle-Vesicle Transitions in Catanionic Mixtures Of SDS/DTAB Induced by Salt, Temperature, and Selective Solvents: A Dissipative Particle Dynamics Simulation Study. Colloid and Polymer Science 2014, 292, 2349–2360. [Google Scholar] [CrossRef]
- Luo, Z.; Li, Y.; Wang, B.; Jiang, J. Ph-Sensitive Vesicles Formed by Amphiphilic Grafted Copolymers with Tunable Membrane Permeability for Drug Loading/Release: A Multiscale Simulation Study. Macromolecules 2016, 49, 6084–6094. [Google Scholar] [CrossRef]
- Wu, S.; Lu, T.; Guo, H. Dissipative Particle Dynamic Simulation Study of Lipid Membrane. Frontiers of Chemistry in China 2010, 5, 288–298. [Google Scholar] [CrossRef]
- Feng, X.; Yan, N.; Jin, J.; Jiang, W. Disassembly of Amphiphilic AB Block Copolymer Vesicles in Selective Solvents: A Molecular Dynamics Simulation Study. Macromolecules 2023, 56, 2560–2567. [Google Scholar] [CrossRef]
- Kotaka, T.; Tanaka, T.; Hattori, M.; Inagaki, H. Block Copolymer Micelles in Dilute Solution. Macromolecules 1978, 11, 138–145. [Google Scholar] [CrossRef]
- Balsara, N.P.; Tirrell, M.; Lodge, T. P. Micelle Formation of BAB Triblock Copolymers in Solvents that Preferentially Dissolve the A Block. Macromolecules 1991, 24, 1975–1986. [Google Scholar] [CrossRef]
- Yang, Y.-W.; Yang, Z.; Zhou, Z.-K.; Attwood, D.; Booth, C. Association of Triblock Copolymers of Ethylene Oxide and Butylene Oxide in Aqueous Solution. A Study of BnEmBn Copolymers. Macromolecules 1996, 29, 670–680. [Google Scholar] [CrossRef]
- Raspaud, E.; Lairez, D.; Adam, M.; Carton, J.-P. Triblock Copolymers in A Selective Solvent. 1. Aggregation Process in Dilute Solution. Macromolecules 1994, 27, 2956–2964. [Google Scholar] [CrossRef]
- Mortensen, K.; Brown, W.; Joergensen, E. Phase Behavior of Poly(Propylene Oxide)-Poly(Ethylene Oxide)-Poly(Propylene Oxide) Triblock Copolymer Melt and Aqueous Solutions. Macromolecules 1994, 27, 5654–5666. [Google Scholar] [CrossRef]
- Pleštil, J.; Hlavatá, D.; Hrouz, J.; Tuzar, Z. Dilute and Semidilute Solutions of ABA Block Copolymer in Solvents Selective for A or B Blocks: 1. Small-Angle X-Ray Scattering Study. Polymer 1990, 31, 2112–2117. [Google Scholar] [CrossRef]
- Taribagil, R.R.; Hillmyer, M. A.; Lodge, T. P. Hydrogels from ABA and ABC Triblock Polymers. Macromolecules 2010, 43, 5396–5404. [Google Scholar] [CrossRef]
- Giacomelli, F.C.; Riegel, I. C.; Petzhold, C. L.; da Silveira, N. P.; Štěpánek, P. Aggregation Behavior of a New Series of ABA Triblock Copolymers Bearing Short Outer A Blocks in B-Selective Solvent: From Free Chains to Bridged Micelles. Langmuir 2008, 25, 731–738. [Google Scholar] [CrossRef]
- Zhou, Z.; Chu, B.; Nace, V. M. Association Behavior of a Triblock Copolymer of Oxyethylene (E) And Oxybutylene (B). A Study of B5E91B5 in Aqueous Solution. Langmuir 1996, 12, 5016–5021. [Google Scholar] [CrossRef]
- Almehmady, A.; Cavanagh, R.; Mantovani, G.; Stolnik, S. ABA Block Copolymers Comprising a Water Soluble Poly(N-Hydroxyethyl Acrylamide) B Block form Self-Assemblies of Varied Morphologies in an Aqueous Environment. Polymer Chemistry 2023, 14, 4089–4100. [Google Scholar] [CrossRef]
- Singh, V.; Eljaaly, K.; Md, S.; Alhakamy, N.A.; Kesharwani, P. Triblock Copolymeric Drug Delivery as an Emerging Nanocarrier for Treatment of Infectious Diseases. Journal of Drug Delivery Science and Technology 2022, 75, 103691. [Google Scholar] [CrossRef]
- Hoang, N.H.; Lim, C.; Sim, T.; Oh, K. T. Triblock Copolymers for Nano-Sized Drug Delivery Systems. Journal of Pharmaceutical Investigation 2016, 47, 27–35. [Google Scholar] [CrossRef]
- Wanka, G.; Hoffmann, H.; Ulbricht, W. Phase Diagrams and Aggregation Behavior of Poly(Oxyethylene)-Poly(Oxypropylene)-Poly(Oxyethylene) Triblock Copolymers in Aqueous Solutions. Macromolecules 1994, 27, 4145–4159. [Google Scholar] [CrossRef]
- Yu, G.; Eisenberg, A. Multiple Morphologies Formed from an Amphiphilic ABC Triblock Copolymer in Solution. Macromolecules 1998, 31, 5546–5549. [Google Scholar] [CrossRef]
- Nardin, C.; Hirt, T.; Leukel, J.; Meier, W. Polymerized ABA Triblock copolymer vesicles. Langmuir 1999, 16, 1035–1041. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Yang, Y.; Liao, Y.; Wei, Y.; Xie, X. Synthesis of Electroactive Tetraaniline−PEO−Tetraaniline Triblock Copolymer and Its Self-Assembled Vesicle with Acidity Response. Langmuir 2010, 26, 9386–9392. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wu, J.; Zhou, H.; Qu, Y.; Li, B.; Zhang, W. Self-Assembled Blends of AB/BAB Block Copolymers Prepared Through Dispersion Raft Polymerization. Macromolecules 2016, 49, 4490–4500. [Google Scholar] [CrossRef]
- Yuan, J.; Li, Y.; Li, X.; Cheng, S.; Jiang, L.; Feng, L.; Fan, Z. The “Crew-Cut” Aggregates of Polystyrene-B-Poly(Ethylene Oxide)-B-Polystyrene Triblock Copolymers in Aqueous Media. European Polymer Journal 2003, 39, 767–776. [Google Scholar] [CrossRef]
- Wang, J.; Leung, L. M. Self-Assembly and Aggregation of ATRP Prepared Amphiphilic BAB Tri-Block Copolymers Contained Nonionic Ethylene Glycol and Fluorescent 9,10-di(1-Naphthalenyl)-2-Vinyl-Anthracene/1-Vinyl-Pyrene Segments. European Polymer Journal 2013, 49, 3722–3733. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, W.; Chen, D.; Lin, X.; Lu, W. W. Effect of Block Order of ABA- And BAB-Type Nipaam/Hema Triblock Copolymers on Thermoresponsive Behavior of Solutions. Macromolecular Chemistry and Physics 2007, 208, 1773–1781. [Google Scholar] [CrossRef]
- HE, G.; MA, L.; PAN, J.; VENKATRAMAN, S. ABA and BAB Type Triblock Copolymers of PEG and PLA: A Comparative Study of Drug Release Properties and “Stealth” Particle Characteristics. International Journal of Pharmaceutics 2007, 334, (1–2). [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Ramezani, M.; Abnous, K.; Alibolandi, M. Biocompatible Polymersomes-Based Cancer Theranostics: Towards Multifunctional Nanomedicine. Int. J. Pharm. 2017, 519, 287–303. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.J.; Suh, J. M.; Sohn, Y. S.; Bae, Y. H.; Kim, S. W.; Jeong, B. Thermogelling Poly(Caprolactone-b-Ethylene Glycol-b-Caprolactone) Aqueous Solutions. Macromolecules 2005, 38, 5260–5265. [Google Scholar] [CrossRef]
- Naharros-Molinero, A.; Caballo-González, M.Á.; de la Mata, F. J.; García-Gallego, S. Direct and Reverse Pluronic Micelles: Design and Characterization of Promising Drug Delivery Nanosystems. Pharmaceutics 2022, 14, 2628. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, L.; Jiang, S. Effects of a PPO−PEO−PPO Triblock Copolymer on Micellization and Gelation of a PEO−PPO−PEO Triblock Copolymer in Aqueous Solution. Langmuir 2005, 21, 9068–9075. [Google Scholar] [CrossRef] [PubMed]
- Lyubimov, I.; Wessels, M.G.; Jayaraman, A. Molecular Dynamics Simulation and Prism Theory Study of Assembly in Solutions of Amphiphilic Bottlebrush Block Copolymers. Macromolecules 2018, 51, 7586–7599. [Google Scholar] [CrossRef]
- Chakraborty, K.; Shinoda, W.; Loverde, S. M. Molecular Simulation of the Shape Deformation of a Polymersome. Soft Matter 2020, 16, 3234–3244. [Google Scholar] [CrossRef]
- Rodrigues, K.; Mattice, W. L. Micelles and Networks Formed by Symmetric Triblock Copolymers in Dilute Solutions that are Poor Solvents for the Terminal Blocks. Polymer Bulletin 1991, 25, 239–243. [Google Scholar] [CrossRef]
- Rodrigues, K.; Mattice, W. L. Intraparticle Distribution Functions for a Micelle Formed by a Small Symmetric Triblock Copolymer in a Poor Solvent for the Terminal Blocks. Langmuir 1992, 8, 456–459. [Google Scholar] [CrossRef]
- Nguyen-Misra, M.; Mattice, W. L. Micellization and Gelation of Symmetric Triblock Copolymers with Insoluble End Blocks. Macromolecules 1995, 28, 1444–1457. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, X.; Zhang, L.; He, L. Vesicles from the Self-Assembly of Coil–Rod–Coil Triblock Copolymers in Selective Solvents. Polymer 2014, 55, 2921–2927. [Google Scholar] [CrossRef]
- Han, Y.; Yu, H.; Du, H.; Jiang, W. Effect of Selective Solvent Addition Rate on the Pathways for Spontaneous Vesicle Formation of ABA Amphiphilic Triblock Copolymers. Journal of the American Chemical Society 2009, 132, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Kangarlou, B.; Dahanayake, R.; Martin, I J. ; Ndaya, D.; Wu, C.-M.; Kasi, R. M.; Dormidontova, E. E.; Nieh, M.-P. Flower-Like Micelles of Polyethylene Oxide End-Capped with Cholesterol. Macromolecules 2021, 54, 8960–8970. [Google Scholar] [CrossRef]
- Song, Y.; Xie, T.; Jiang, R.; Wang, Z.; Yin, Y.; Li, B.; Shi, A.-C. Effect of Chain Architecture on Self-Assembled Aggregates from Cyclic AB Diblock and Linear ABA Triblock Copolymers in Solution. Langmuir 2018, 34, 4013–4023. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Sureshkumar, R. Anomalous Diffusion and Stress Relaxation in Surfactant Micelles. Phys. Rev. E 2017, 96, 2605. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Sureshkumar, R. Topology, Length Scales, and Energetics of Surfactant Micelles. J. Chem. Phys. 2015, 143, 024905. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Sureshkumar, R. Uniaxial Extension of Surfactant Micelles: Counterion Mediated Chain Stiffening and a Mechanism of Rupture by Flow-Induced Energy Redistribution. ACS Macro Lett. 2015, 5, 108–111. [Google Scholar] [CrossRef]
- Sangwai, A.V.; Sureshkumar, R. Binary Interactions and Salt-Induced Coalescence of Spherical Micelles of Cationic Surfactants from Molecular Dynamics Simulations. Langmuir 2011, 28, 1127–1135. [Google Scholar] [CrossRef]
- Sangwai, A.V.; Sureshkumar, R. Coarse-Grained Molecular Dynamics Simulations of the Sphere to Rod Transition in Surfactant Micelles. Langmuir 2011, 27, 6628–6638. [Google Scholar] [CrossRef] [PubMed]
- Sambasivam, A.; Sangwai, A.V.; Sureshkumar, R. Dynamics and Scission of Rodlike Cationic Surfactant Micelles in Shear Flow. Phys. Rev. Lett. 2015, 114, 8302. [Google Scholar] [CrossRef] [PubMed]
- Sambasivam, A.; Sangwai, A.V.; Sureshkumar, R. Self-Assembly of Nanoparticle–Surfactant Complexes with Rodlike Micelles: A Molecular Dynamics Study. Langmuir 2016, 32, 1214–1219. [Google Scholar] [CrossRef] [PubMed]
- Sambasivam, A.; Dhakal, S.; Sureshkumar, R. Structure and Rheology of Self-Assembled Aqueous Suspensions of Nanoparticles and Wormlike Micelles. Mol. Simul. 2017, 44, 485–493. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [PubMed]
- Darré, L.; Machado, M.R.; Pantano, S. Coarse-Grained Models of Water. WIREs Comput. Mol. Sci. 2012, 2, 921–930. [Google Scholar] [CrossRef]
- Martínez, L.; Andrade, R.; Birgin, E.G.; Martínez, J.M. Packmol: A Package for Building Initial Configurations for Molecular Dynamics Simulations. J. Comput. Chem. 2009, 30, 2157–2164. [Google Scholar] [CrossRef]
- Eisenhaber, F.; Lijnzaad, P.; Argos, P.; Sander, C.; Scharf, M. The Double Cubic Lattice Method: Efficient Approaches to Numerical Integration of Surface Area and Volume and to Dot Surface Contouring of Molecular Assemblies. J. Comput. Chem. 1995, 16, 273–284. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling Through Velocity Rescaling. The Journal of Chemical Physics 2007, 126, 014101. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J. P.; van Gunsteren, W. F.; DiNola, A.; Haak, J. R. Molecular Dynamics with Coupling to an External Bath. The Journal of Chemical Physics 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
| Type | C1 | C2 | C3 | C4 | C5 |
|---|---|---|---|---|---|
| 2.0 | 2.3 | 2.7 | 2.7 | 3.1 | |
| 2.7 | 2.7 | 2.7 | 3.1 | 3.5 | |
| Vesiculation Time (ns) | 84 ± 8 | 105 ± 9 | 142 ± 13 | 130 ± 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




