Submitted:
01 April 2024
Posted:
02 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
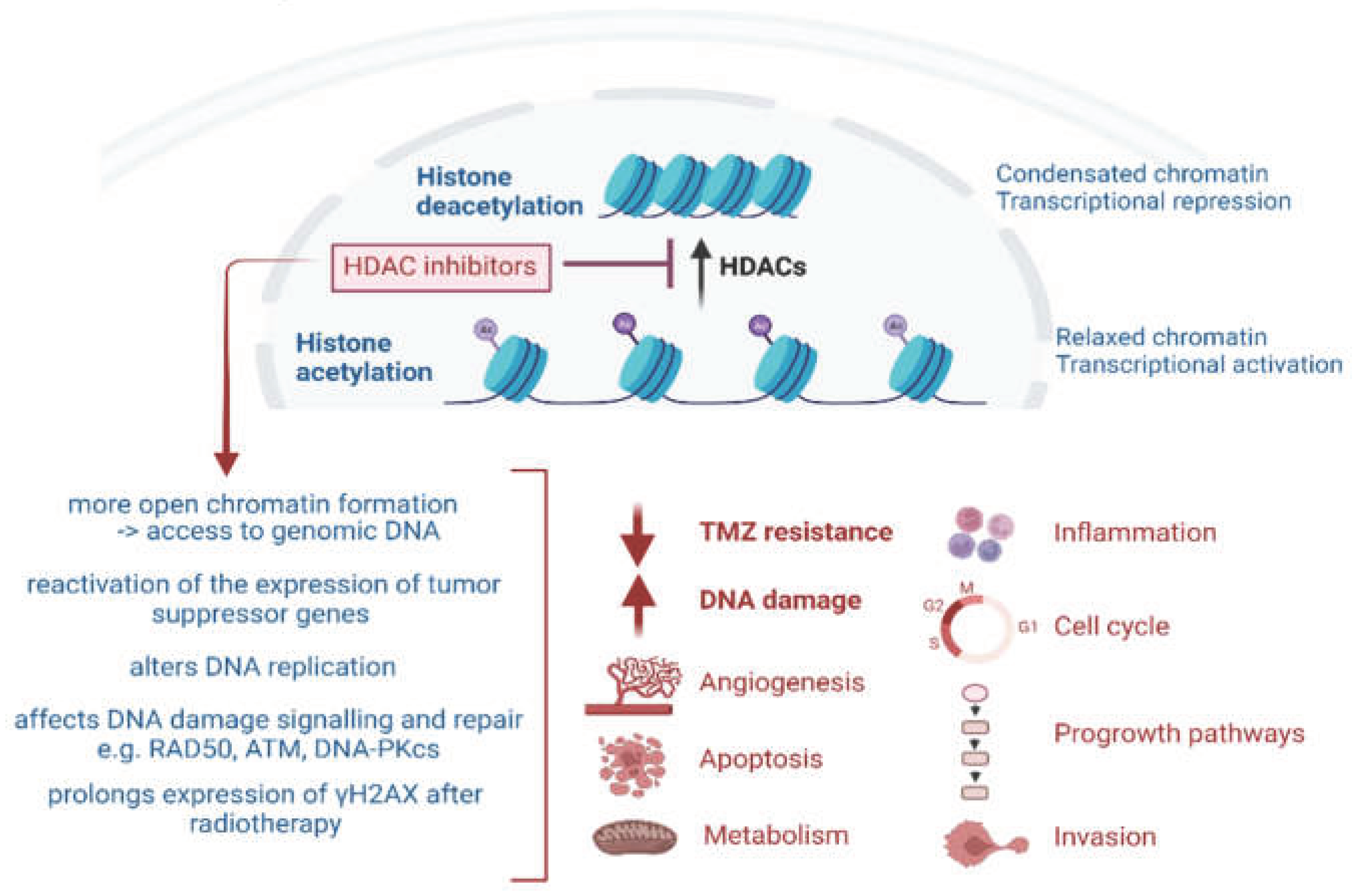
2. Epigenetic Modulation by HDACs and HDAC Inhibitors
2.1. HDACi Induced Apoptosis and Autophagy
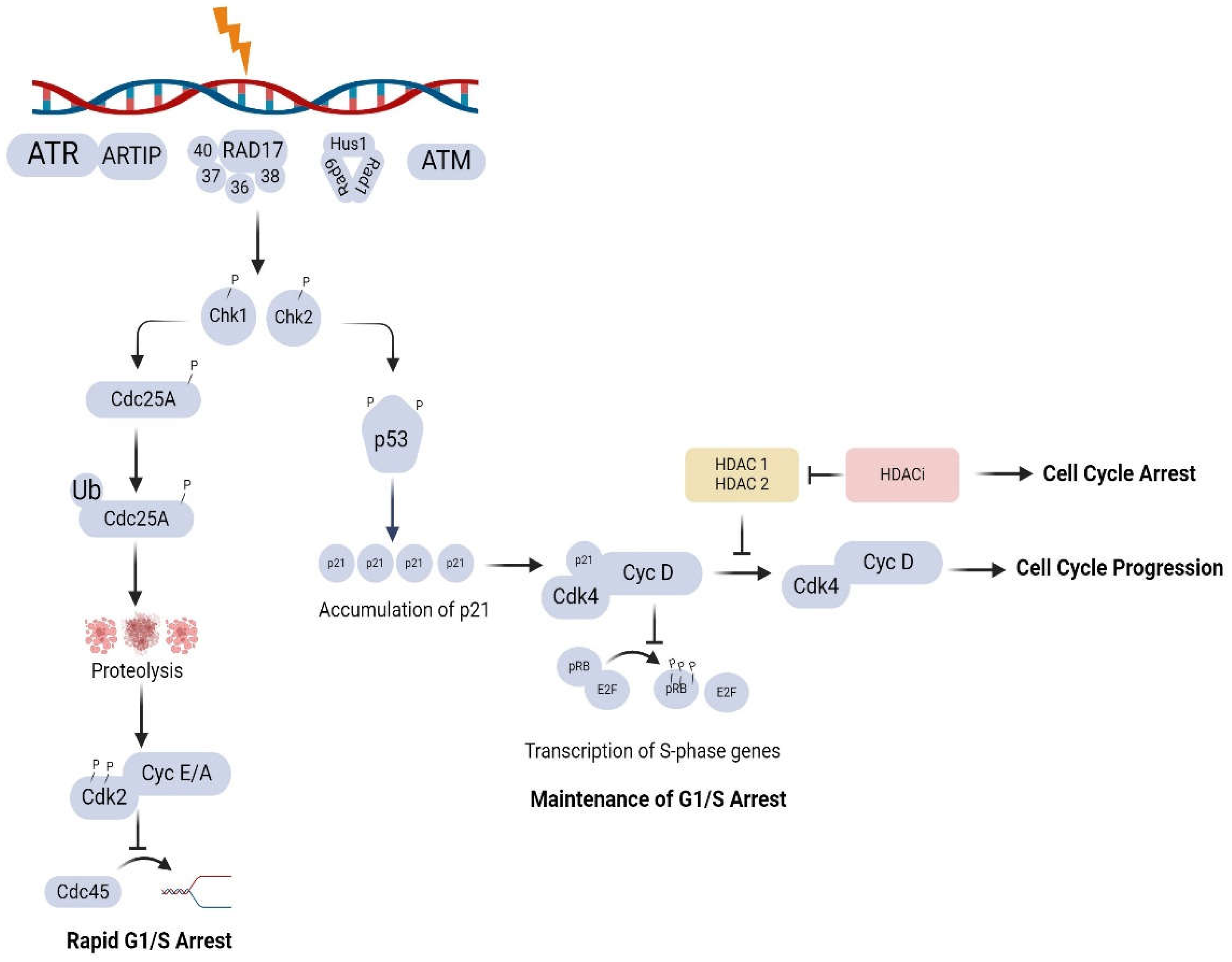
2.2. HDACi-Induced Upregulation of p21 and Cell Cycle Arrest
2.3. HDACi-Induced Inhibition of Angiogenesis
3. Radiosensitisation by HDAC Inhibitors
3.1. DNA DSB Induction and DNA Damage Repair (DDR)
3.2. Role of HDACs and HDACi in the DNA Damage Response (DDR)
4. Impact of Chromatin Structure on Radiation Response
4.1. Influence of Chromatin Structure on DNA Damage Induction , Detection and Repair
4.2. DNA Damage Induced Chromatin Modifications
4.3. Chromatin Modification and Type of Radiation
5. Sequencing of HDACi Treatment and Radiation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, J. H.; Choy, M. L.; Ngo, L.; Foster, S. S.; Marks, P. A. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc Natl Acad Sci U S A 2010, 107, 14639–14644. [Google Scholar] [CrossRef] [PubMed]
- Mrakovcic, M.; Bohner, L.; Hanisch, M.; Fröhlich, L. F. Epigenetic Targeting of Autophagy via HDAC Inhibition in Tumor Cells: Role of p53. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Camphausen, K.; Scott, T.; Sproull, M.; Tofilon, P. J. Enhancement of xenograft tumor radiosensitivity by the histone deacetylase inhibitor MS-275 and correlation with histone hyperacetylation. Clin Cancer Res 2004, 10 Pt 1, 6066–6071. [Google Scholar] [CrossRef] [PubMed]
- Camphausen, K.; Tofilon, P. J. Inhibition of histone deacetylation: a strategy for tumor radiosensitization. J Clin Oncol 2007, 25, 4051–4056. [Google Scholar] [CrossRef] [PubMed]
- Groselj, B.; Sharma, N. L.; Hamdy, F. C.; Kerr, M.; Kiltie, A. E. Histone deacetylase inhibitors as radiosensitisers: effects on DNA damage signalling and repair. Br J Cancer 2013, 108, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Damaskos, C.; Garmpis, N.; Valsami, S.; Kontos, M.; Spartalis, E.; Kalampokas, T.; Kalampokas, E.; Athanasiou, A.; Moris, D.; Daskalopoulou, A.; et al. Histone Deacetylase Inhibitors: An Attractive Therapeutic Strategy Against Breast Cancer. Anticancer Res 2017, 37, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Chinnaiyan, P.; Cerna, D.; Burgan, W. E.; Beam, K.; Williams, E. S.; Camphausen, K.; Tofilon, P. J. Postradiation sensitization of the histone deacetylase inhibitor valproic acid. Clin Cancer Res 2008, 14, 5410–5415. [Google Scholar] [CrossRef] [PubMed]
- Schlaff, C. D.; Arscott, W. T.; Gordon, I.; Tandle, A.; Tofilon, P.; Camphausen, K. Radiosensitization Effects of Novel Triple-Inhibitor CUDC-101 in Glioblastoma Multiforme and Breast Cancer Cells In Vitro. International Journal of Radiation Oncology*Biology*Physics 2013, 87. [Google Scholar] [CrossRef]
- Chiu, H. W.; Yeh, Y. L.; Wang, Y. C.; Huang, W. J.; Chen, Y. A.; Chiou, Y. S.; Ho, S. Y.; Lin, P.; Wang, Y. J. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, enhances radiosensitivity and suppresses lung metastasis in breast cancer in vitro and in vivo. PLoS One 2013, 8. [Google Scholar] [CrossRef]
- Baschnagel, A.; Russo, A.; Burgan, W. E.; Carter, D.; Beam, K.; Palmieri, D.; Steeg, P. S.; Tofilon, P.; Camphausen, K. Vorinostat enhances the radiosensitivity of a breast cancer brain metastatic cell line grown in vitro and as intracranial xenografts. Mol Cancer Ther 2009, 8, 1589–1595. [Google Scholar] [CrossRef]
- Chen, X.; Wong, P.; Radany, E.; Wong, J. Y. HDAC inhibitor, valproic acid, induces p53-dependent radiosensitization of colon cancer cells. Cancer Biother Radiopharm 2009, 24, 689–699. [Google Scholar] [CrossRef]
- Munshi, A.; Kurland, J. F.; Nishikawa, T.; Tanaka, T.; Hobbs, M. L.; Tucker, S. L.; Ismail, S.; Stevens, C.; Meyn, R. E. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res 2005, 11, 4912–4922. [Google Scholar] [CrossRef]
- Antrobus, J.; Parsons, J. L. Histone Deacetylases and Their Potential as Targets to Enhance Tumour Radiosensitisation. Radiation 2022, 2, 149–167. [Google Scholar] [CrossRef]
- Gerelchuluun, A.; Maeda, J.; Manabe, E.; Brents, C. A.; Sakae, T.; Fujimori, A.; Chen, D. J.; Tsuboi, K.; Kato, T. A. Histone Deacetylase Inhibitor Induced Radiation Sensitization Effects on Human Cancer Cells after Photon and Hadron Radiation Exposure. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Barazzuol, L.; Jeynes, J. C.; Merchant, M. J.; Wéra, A. C.; Barry, M. A.; Kirkby, K. J.; Suzuki, M. Radiosensitization of glioblastoma cells using a histone deacetylase inhibitor (SAHA) comparing carbon ions with X-rays. Int J Radiat Biol 2015, 91, 90–98. [Google Scholar] [CrossRef]
- Johnson, A. M.; Bennett, P. V.; Sanidad, K. Z.; Hoang, A.; Jardine, J. H.; Keszenman, D. J.; Wilson, P. F. Evaluation of Histone Deacetylase Inhibitors as Radiosensitizers for Proton and Light Ion Radiotherapy. Front Oncol 2021, 11, 735940. [Google Scholar] [CrossRef]
- Yu, J. I.; Choi, C.; Shin, S. W.; Son, A.; Lee, G. H.; Kim, S. Y.; Park, H. C. Valproic Acid Sensitizes Hepatocellular Carcinoma Cells to Proton Therapy by Suppressing NRF2 Activation. Sci Rep 2017, 7. [Google Scholar] [CrossRef]
- Choi, C.; Lee, G. H.; Son, A.; Yoo, G. S.; Yu, J. I.; Park, H. C. Downregulation of Mcl-1 by Panobinostat Potentiates Proton Beam Therapy in Hepatocellular Carcinoma Cells. Cells 2021, 10. [Google Scholar] [CrossRef]
- Groselj, B.; Kerr, M.; Kiltie, A. E. Radiosensitisation of bladder cancer cells by panobinostat is modulated by Ku80 expression. Radiother Oncol 2013, 108, 429–433. [Google Scholar] [CrossRef]
- Jenke, R.; Ressing, N.; Hansen, F. K.; Aigner, A.; Buch, T. Anticancer Therapy with HDAC Inhibitors: Mechanism-Based Combination Strategies and Future Perspectives. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Maier, P.; Hartmann, L.; Wenz, F.; Herskind, C. Cellular Pathways in Response to Ionizing Radiation and Their Targetability for Tumor Radiosensitization. Int J Mol Sci 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Shabason, J. E.; Tofilon, P. J.; Camphausen, K. Grand rounds at the National Institutes of Health: HDAC inhibitors as radiation modifiers, from bench to clinic. J Cell Mol Med 2011, 15, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Takata, H.; Hanafusa, T.; Mori, T.; Shimura, M.; Iida, Y.; Ishikawa, K.; Yoshikawa, K.; Yoshikawa, Y.; Maeshima, K. Chromatin Compaction Protects Genomic DNA from Radiation Damage. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, P.; Panyutin, I. V.; Remeeva, E.; Neumann, R. D.; Panyutin, I. G. Effect of Chromatin Structure on the Extent and Distribution of DNA Double Strand Breaks Produced by Ionizing Radiation; Comparative Study of hESC and Differentiated Cells Lines. International Journal of Molecular Sciences 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.; Arlinghaus, L. R.; Cardin, D. B.; Goff, L.; Berlin, J. D.; Parikh, A.; Abramson, R. G.; Yankeelov, T. E.; Hiebert, S.; Merchant, N.; et al. Phase I trial of vorinostat added to chemoradiation with capecitabine in pancreatic cancer. Radiotherapy and Oncology 2016, 119, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Galanis, E.; Anderson, S. K.; Miller, C. R.; Sarkaria, J. N.; Jaeckle, K.; Buckner, J. C.; Ligon, K. L.; Ballman, K. V.; Moore, D. F., Jr; Nebozhyn, M.; et al. Phase I/II trial of vorinostat combined with temozolomide and radiation therapy for newly diagnosed glioblastoma: results of Alliance N0874/ABTC 02. Neuro-Oncology 2017, 20, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Gurbani, S. S.; Yoon, Y.; Weinberg, B. D.; Salgado, E.; Press, R. H.; Cordova, J. S.; Ramesh, K. K.; Liang, Z.; Vega, J. V.; Voloschin, A.; et al. Assessing Treatment Response of Glioblastoma to an HDAC Inhibitor Using Whole-Brain Spectroscopic MRI. Tomography 2019, 5, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Teknos, T. N.; Grecula, J.; Agrawal, A.; Old, M. O.; Ozer, E.; Carrau, R.; Kang, S.; Rocco, J.; Blakaj, D.; Diavolitsis, V.; et al. A phase 1 trial of Vorinostat in combination with concurrent chemoradiation therapy in the treatment of advanced staged head and neck squamous cell carcinoma. Investigational New Drugs 2019, 37, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Richon, V.; Ni, X.; Talpur, R.; Duvic, M. Selective induction of apoptosis by histone deacetylase inhibitor SAHA in cutaneous T-cell lymphoma cells: relevance to mechanism of therapeutic action. J Invest Dermatol 2005, 125, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Seto, E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med 2016, 6. [Google Scholar] [CrossRef]
- Mrakovcic, M.; Kleinheinz, J.; Frohlich, L. F. Histone Deacetylase Inhibitor-Induced Autophagy in Tumor Cells: Implications for p53. Int J Mol Sci 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Bolden, J. E.; Peart, M. J.; Johnstone, R. W. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 2006, 5, 769–784. [Google Scholar] [CrossRef]
- Fotheringham, S.; Epping, M. T.; Stimson, L.; Khan, O.; Wood, V.; Pezzella, F.; Bernards, R.; La Thangue, N. B. Genome-wide loss-of-function screen reveals an important role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer Cell 2009, 15, 57–66. [Google Scholar] [CrossRef]
- Everix, L.; Seane, E. N.; Ebenhan, T.; Goethals, I.; Bolcaen, J. Introducing HDAC-Targeting Radiopharmaceuticals for Glioblastoma Imaging and Therapy. Pharmaceuticals 2023, 16. [Google Scholar] [CrossRef] [PubMed]
- Dokmanovic, M.; Clarke, C.; Marks, P. A. Histone Deacetylase Inhibitors: Overview and Perspectives. Molecular Cancer Research 2007, 5, 981–989. [Google Scholar] [CrossRef]
- Smalley, J. P.; Cowley, S. M.; Hodgkinson, J. T. Bifunctional HDAC Therapeutics: One Drug to Rule Them All? Molecules 2020, 25. [Google Scholar] [CrossRef]
- Rajak, H.; Singh, A.; Raghuwanshi, K.; Kumar, R.; Dewangan, P. K.; Veerasamy, R.; Sharma, P. C.; Dixit, A.; Mishra, P. A structural insight into hydroxamic acid based histone deacetylase inhibitors for the presence of anticancer activity. Curr Med Chem 2014, 21, 2642–2664. [Google Scholar] [CrossRef]
- Passaro, E.; Papulino, C.; Chianese, U.; Toraldo, A.; Congi, R.; Del Gaudio, N.; Nicoletti, M. M.; Benedetti, R.; Altucci, L. HDAC6 Inhibition Extinguishes Autophagy in Cancer: Recent Insights. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Zhang, J.; Zhong, Q. Histone deacetylase inhibitors and cell death. Cell Mol Life Sci 2014, 71, 3885–3901. [Google Scholar] [CrossRef]
- Frew, A. J.; Johnstone, R. W.; Bolden, J. E. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett 2009, 280, 125–133. [Google Scholar] [CrossRef]
- Mrakovcic, M.; Kleinheinz, J.; Fröhlich, L. F. p53 at the Crossroads between Different Types of HDAC Inhibitor-Mediated Cancer Cell Death. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Insinga, A.; Minucci, S.; Pelicci, P. G. Mechanisms of selective anticancer action of histone deacetylase inhibitors. Cell Cycle 2005, 4, 741–743. [Google Scholar] [CrossRef] [PubMed]
- Peart, M. J. Novel mechanisms of apoptosis induced by histone deacetylase inhibitors.full. CANCER RESEARCH 2003, 63, 4460–4471. [Google Scholar]
- Gong, P.; Wang, Y.; Jing, Y. Apoptosis Induction byHistone Deacetylase Inhibitors in Cancer Cells: Role of Ku70. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Diao, H.; Dong, N.; Su, X.; Wang, B.; Mo, Q.; Yu, H.; Wang, X.; Chen, C. Histone deacetylase inhibitor induces cell apoptosis and cycle arrest in lung cancer cells via mitochondrial injury and p53 up-acetylation. Cell Biol Toxicol 2016, 32, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. J.; Chee, C. E.; Huang, S.; Sinicrope, F. A. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011, 10, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Gao, Z.; Marks, P. A.; Jiang, X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A 2004, 101, 18030–18035. [Google Scholar] [CrossRef] [PubMed]
- Rebecca, V. W.; Amaravadi, R. K. Emerging strategies to effectively target autophagy in cancer. Oncogene 2016, 35, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kimmelman, A. C. The dynamic nature of autophagy in cancer. Genes Dev 2011, 25, 1999–2010. [Google Scholar] [CrossRef]
- Pagotto, A.; Pilotto, G.; Mazzoldi, E. L.; Nicoletto, M. O.; Frezzini, S.; Pastò, A.; Amadori, A. Autophagy inhibition reduces chemoresistance and tumorigenic potential of human ovarian cancer stem cells. Cell Death & Disease 2017, 8, e2943–e2943. [Google Scholar] [CrossRef]
- El-Khoury, V.; Pierson, S.; Szwarcbart, E.; Brons, N. H.; Roland, O.; Cherrier-De Wilde, S.; Plawny, L.; Van Dyck, E.; Berchem, G. Disruption of autophagy by the histone deacetylase inhibitor MGCD0103 and its therapeutic implication in B-cell chronic lymphocytic leukemia. Leukemia 2014, 28, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Hrzenjak, A.; Kremser, M. L.; Strohmeier, B.; Moinfar, F.; Zatloukal, K.; Denk, H. SAHA induces caspase-independent, autophagic cell death of endometrial stromal sarcoma cells by influencing the mTOR pathway. J Pathol 2008, 216, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, L. F.; Mrakovcic, M.; Smole, C.; Zatloukal, K. Molecular mechanism leading to SAHA-induced autophagy in tumor cells: evidence for a p53-dependent pathway. Cancer Cell Int 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. L.; Yang, P. M.; Shun, C. T.; Wu, M. S.; Weng, J. R.; Chen, C. C. Autophagy potentiates the anti-cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy 2010, 6, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Gilardini Montani, M. S.; Granato, M.; Santoni, C.; Del Porto, P.; Merendino, N.; D'Orazi, G.; Faggioni, A.; Cirone, M. Histone deacetylase inhibitors VPA and TSA induce apoptosis and autophagy in pancreatic cancer cells. Cell Oncol (Dordr) 2017, 40, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Richon, V. M.; Sandhoff, T. W.; Rifkind, R. A.; Marks, P. A. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A 2000, 97, 10014–10019. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Yang, Y.; Liu, S.; Lu, J.; Huang, B.; Zhang, Y. HDAC inhibitor PAC-320 induces G2/M cell cycle arrest and apoptosis in human prostate cancer. Oncotarget 2018, 9, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Lee, H. A.; Chu, K. B.; Moon, E. K.; Kim, S. S.; Quan, F. S. Sensitization to oxidative stress and G2/M cell cycle arrest by histone deacetylase inhibition in hepatocellular carcinoma cells. Free Radic Biol Med 2020, 147, 129–138. [Google Scholar] [CrossRef]
- Hrgovic, I.; Doll, M.; Kleemann, J.; Wang, X.-F.; Zoeller, N.; Pinter, A.; Kippenberger, S.; Kaufmann, R.; Meissner, M. The histone deacetylase inhibitor trichostatin a decreases lymphangiogenesis by inducing apoptosis and cell cycle arrest via p21-dependent pathways. BMC Cancer 2016, 16. [Google Scholar] [CrossRef]
- Schoepflin, Z. R.; Shapiro, I. M.; Risbud, M. V. Class I and IIa HDACs Mediate HIF-1α Stability Through PHD2-Dependent Mechanism, While HDAC6, a Class IIb Member, Promotes HIF-1α Transcriptional Activity in Nucleus Pulposus Cells of the Intervertebral Disc. J Bone Miner Res 2016, 31, 1287–1299. [Google Scholar] [CrossRef]
- Jeong, J. W.; Bae, M. K.; Ahn, M. Y.; Kim, S. H.; Sohn, T. K.; Bae, M. H.; Yoo, M. A.; Song, E. J.; Lee, K. J.; Kim, K. W. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell 2002, 111, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Deroanne, C. F.; Bonjean, K.; Servotte, S.; Devy, L.; Colige, A.; Clausse, N.; Blacher, S.; Verdin, E.; Foidart, J. M.; Nusgens, B. V.; et al. Histone deacetylases inhibitors as anti-angiogenic agents altering vascular endothelial growth factor signaling. Oncogene 2002, 21, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Kuljaca, S.; Tee, A.; Marshall, G. M. Histone deacetylase inhibitors: multifunctional anticancer agents. Cancer Treat Rev 2006, 32, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Iordache, F.; Buzila, C.; Constantinescu, A.; Andrei, E.; Maniu, H. Histone deacetylase (HDAC) inhibitors down-regulate endothelial lineage commitment of umbilical cord blood derived endothelial progenitor cells. Int J Mol Sci 2012, 13, 15074–15085. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.; Pili, R. Histone Deacetylase Inhibitors: Advancing Therapeutic Strategies in Hematological and Solid Malignancies. Pharmaceuticals (Basel) 2010, 3, 2411–2469. [Google Scholar] [CrossRef]
- Munshi, A.; Tanaka, T.; Hobbs, M. L.; Tucker, S. L.; Richon, V. M.; Meyn, R. E. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of γ-H2AX foci. Molecular Cancer Therapeutics 2006, 5, 1967–1974. [Google Scholar] [CrossRef]
- Ediriweera, M. K.; Tennekoon, K. H.; Samarakoon, S. R. Emerging role of histone deacetylase inhibitors as anti-breast-cancer agents. Drug Discov Today 2019, 24, 685–702. [Google Scholar] [CrossRef] [PubMed]
- Luu, T. H.; Morgan, R. J.; Leong, L.; Lim, D.; McNamara, M.; Portnow, J.; Frankel, P.; Smith, D. D.; Doroshow, J. H.; Wong, C.; et al. A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin Cancer Res 2008, 14, 7138–7142. [Google Scholar] [CrossRef]
- Vitti, E. T.; Parsons, J. L. The Radiobiological Effects of Proton Beam Therapy: Impact on DNA Damage and Repair. Cancers (Basel) 2019, 11. [Google Scholar] [CrossRef]
- Bian, L.; Meng, Y.; Zhang, M.; Li, D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: implications for cancer treatment. Molecular Cancer 2019, 18. [Google Scholar] [CrossRef]
- Hall Eric J, A. J. G. Radiobiology for the Radiologist; LIPPINCOT WILLIAMS AND WILKINS, 2012.
- Fontana, A. O.; Augsburger, M. A.; Grosse, N.; Guckenberger, M.; Lomax, A. J.; Sartori, A. A.; Pruschy, M. N. Differential DNA repair pathway choice in cancer cells after proton- and photon-irradiation. Radiother Oncol 2015, 116, 374–380. [Google Scholar] [CrossRef]
- Kachhap, S. K.; Rosmus, N.; Collis, S. J.; Kortenhorst, M. S.; Wissing, M. D.; Hedayati, M.; Shabbeer, S.; Mendonca, J.; Deangelis, J.; Marchionni, L.; et al. Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS One 2010, 5. [Google Scholar] [CrossRef]
- Miller, K. M.; Tjeertes, J. V.; Coates, J.; Legube, G.; Polo, S. E.; Britton, S.; Jackson, S. P. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 2010, 17, 1144–1151. [Google Scholar] [CrossRef]
- Thurn, K. T.; Thomas, S.; Raha, P.; Qureshi, I.; Munster, P. N. Histone deacetylase regulation of ATM-mediated DNA damage signaling. Mol Cancer Ther 2013, 12, 2078–2087. [Google Scholar] [CrossRef]
- Kao, G. D.; McKenna, W. G.; Guenther, M. G.; Muschel, R. J.; Lazar, M. A.; Yen, T. J. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol 2003, 160, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Cubizolles, F.; Zhang, Y.; Reichert, N.; Kohler, H.; Seiser, C.; Matthias, P. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev 2010, 24, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Hai, R.; He, L.; Shu, G.; Yin, G. Characterization of Histone Deacetylase Mechanisms in Cancer Development. Front Oncol 2021, 11, 700947. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Fernández, A.; Cabrero, J. R.; Serrador, J. M.; Sánchez-Madrid, F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol 2008, 18, 291–297. [Google Scholar] [CrossRef]
- Bhaskara, S.; Knutson, S. K.; Jiang, G.; Chandrasekharan, M. B.; Wilson, A. J.; Zheng, S.; Yenamandra, A.; Locke, K.; Yuan, J. L.; Bonine-Summers, A. R.; et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010, 18, 436–447. [Google Scholar] [CrossRef]
- Nishimoto, K.; Niida, H.; Uchida, C.; Ohhata, T.; Kitagawa, K.; Motegi, A.; Suda, T.; Kitagawa, M. HDAC3 Is Required for XPC Recruitment and Nucleotide Excision Repair of DNA Damage Induced by UV Irradiation. Mol Cancer Res 2020, 18, 1367–1378. [Google Scholar] [CrossRef]
- M, Z. HDAC6 Deacetylates and Ubiquitinates MSH2 to Maintain Proper Levels of MutSα. Molecular Cell 2014, 55, 31–46. [Google Scholar]
- Wang, R.-H. Impaired DNA Damage Response, Genome Instability, and Tumorigenesis in SIRT1 Mutant Mice.
- Polo, S. E.; Almouzni, G. Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model. DNA Repair (Amst) 2015, 36, 114–121. [Google Scholar] [CrossRef]
- Fortuny, A.; Polo, S. E. The response to DNA damage in heterochromatin domains. Chromosoma 2018, 127, 291–300. [Google Scholar] [CrossRef]
- Etier, A.; Dumetz, F.; Chéreau, S.; Ponts, N. Post-Translational Modifications of Histones Are Versatile Regulators of Fungal Development and Secondary Metabolism. Toxins (Basel) 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Warters, R. L.; Lyons, B. W. Variation in radiation-induced formation of DNA double-strand breaks as a function of chromatin structure. Radiat Res 1992, 130, 309–318. [Google Scholar] [CrossRef]
- Cowell, I. G.; Sunter, N. J.; Singh, P. B.; Austin, C. A.; Durkacz, B. W.; Tilby, M. J. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One 2007, 2. [Google Scholar] [CrossRef]
- Kim, J. A.; Kruhlak, M.; Dotiwala, F.; Nussenzweig, A.; Haber, J. E. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol 2007, 178, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Nygren, J.; Ljungman, M.; Ahnström, M. Chromatin Structure and Radiation-induced DNA Strand Breaks in Human Cells: Soluble Scavengers and DNA-bound Proteins Offer a Better Protection Against Single- than Double-strand Breaks. International Journal of Radiation Biology 1995, 68, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Kruhlak, M. J.; Celeste, A.; Dellaire, G.; Fernandez-Capetillo, O.; Müller, W. G.; McNally, J. G.; Bazett-Jones, D. P.; Nussenzweig, A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol 2006, 172, 823–834. [Google Scholar] [CrossRef]
- Lisby, M.; Mortensen, U. H.; Rothstein, R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol 2003, 5, 572–577. [Google Scholar] [CrossRef]
- Aten, J. A.; Stap, J.; Krawczyk, P. M.; van Oven, C. H.; Hoebe, R. A.; Essers, J.; Kanaar, R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 2004, 303, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Amaral, N.; Ryu, T.; Li, X.; Chiolo, I. Nuclear Dynamics of Heterochromatin Repair. Trends in Genetics 2017, 33, 86–100. [Google Scholar] [CrossRef] [PubMed]
- Murr, R.; Loizou, J. I.; Yang, Y. G.; Cuenin, C.; Li, H.; Wang, Z. Q.; Herceg, Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 2006, 8, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, Y.; Jiang, X.; Ayrapetov, M. K.; Moskwa, P.; Yang, S.; Weinstock, D. M.; Price, B. D. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol 2010, 191, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, P. M.; Borovski, T.; Stap, J.; Cijsouw, T.; ten Cate, R.; Medema, J. P.; Kanaar, R.; Franken, N. A.; Aten, J. A. Chromatin mobility is increased at sites of DNA double-strand breaks. J Cell Sci 2012, 125 Pt 9, 2127–2133. [Google Scholar] [CrossRef] [PubMed]
- Kim, J. H.; Kim, I. H.; Shin, J. H.; Kim, H. J.; Kim, I. A. Sequence-Dependent Radiosensitization of Histone Deacetylase Inhibitors Trichostatin A and SK-7041. Cancer Res Treat 2013, 45, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Van Nifterik, K. A.; Van den Berg, J.; Slotman, B. J.; Lafleur, M. V.; Sminia, P.; Stalpers, L. J. Valproic acid sensitizes human glioma cells for temozolomide and gamma-radiation. J Neurooncol 2012, 107, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Moertl, S.; Payer, S.; Kell, R.; Winkler, K.; Anastasov, N.; Atkinson, M. J. Comparison of Radiosensitization by HDAC Inhibitors CUDC-101 and SAHA in Pancreatic Cancer Cells. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Munshi, A.; Tanaka, T.; Hobbs, M. L.; Tucker, S. L.; Richon, V. M.; Meyn, R. E. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of gamma-H2AX foci. Mol Cancer Ther 2006, 5, 1967–1974. [Google Scholar] [CrossRef]
- Shanthi Adimoolam*, M. S. , Jun Chen*, Patti Thiemann*, James M. Ford†, and Joseph J. Buggy. HDAC-inhibitor-PCI24781-decreases-RAD51-expression-and-inhibits-homologous-recombination. PNAS 104, 9482. [Google Scholar] [CrossRef]
- Mueller, S.; Yang, X.; Sottero, T. L.; Gragg, A.; Prasad, G.; Polley, M. Y.; Weiss, W. A.; Matthay, K. K.; Davidoff, A. M.; DuBois, S. G.; et al. Cooperation of the HDAC inhibitor vorinostat and radiation in metastatic neuroblastoma: efficacy and underlying mechanisms. Cancer Lett 2011, 306, 223–229. [Google Scholar] [CrossRef]
- Chen X, W. P. , Radany E, Wong JY. HDAC inhibitor, valproic acid, induces p53-dependent radiosensitization of colon cancer cells. Cancer Biother Radiopharm. [CrossRef]
- Flatmark, K.; Nome, R. V.; Folkvord, S.; Bratland, A.; Rasmussen, H.; Ellefsen, M. S.; Fodstad, O.; Ree, A. H. Radiosensitization of colorectal carcinoma cell lines by histone deacetylase inhibition. Radiat Oncol 2006, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Saelen, M. G.; Ree, A. H.; Kristian, A.; Fleten, K. G.; Furre, T.; Hektoen, H. H.; Flatmark, K. Radiosensitization by the histone deacetylase inhibitor vorinostat under hypoxia and with capecitabine in experimental colorectal carcinoma. Radiation Oncology 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Shoji, M.; Ninomiya, I.; Makino, I.; Kinoshita, J.; Nakamura, K.; Oyama, K.; Nakagawara, H.; Fujita, H.; Tajima, H.; Takamura, H.; et al. Valproic acid, a histone deacetylase inhibitor, enhances radiosensitivity in esophageal squamous cell carcinoma. Int J Oncol 2012, 40, 2140–2146. [Google Scholar] [CrossRef] [PubMed]
- Perona, M.; Thomasz, L.; Rossich, L.; Rodriguez, C.; Pisarev, M. A.; Rosemblit, C.; Cremaschi, G. A.; Dagrosa, M. A.; Juvenal, G. J. Radiosensitivity enhancement of human thyroid carcinoma cells by the inhibitors of histone deacetylase sodium butyrate and valproic acid. Mol Cell Endocrinol 2018, 478, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Oike, T.; Hirota, Y.; Dewi Maulany Darwis, N.; Shibata, A.; Ohno, T. Comparison of Clonogenic Survival Data Obtained by Pre- and Post-Irradiation Methods. J Pers Med 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Frankenberg-Schwager, M.; Frankenberg, D.; Harbich, R. Potentially lethal damage repair is due to the difference of DNA double-strand break repair under immediate and delayed plating conditions. Radiat Res 1987, 111, 192–200. [Google Scholar] [CrossRef]
- Reddy, N. M.; Kapiszewska, M.; Lange, C. S. Detection of X-ray damage repair by the immediate versus delayed plating technique is dependent on cell shape and cell concentration. Scanning Microsc 1992, 6, 543–555; [Google Scholar]
| HDAC | Role | Reference | |
|---|---|---|---|
| HDAC 1 and 2 | DNA-damage signalling | data | |
| Stabilise broken ends during NHEJ | [74] | ||
| Influence persistence of Ku70 and Artemis at DNA damage sites- promoting NHEJ | [13] | ||
| Deactivates the function of p21 and p27 | [77,78] | ||
| Hypo-phosphorylation of RB gene | [79] | ||
| HDAC 3 | Maintenance of chromatin structure and genomic stability | [80] | |
| Essential for DNA DSB repair | [13] | ||
| Recruits Xeroderma Pigmentosum C (XPC) during Nucleotide excision repair (NER) | [81] | ||
| HDAC 4 | Silencing of chromatin near broken ends. | [76] | |
| Co-localises with 53BP foci, and contributes to stability of 53BP | [13] | ||
| HDAC 6 | Reduces cellular sensitivity to damaging agents | [82] | |
| Repair of DNA mismatch | [13] | ||
| HDAC 9 and 10 | DSB repair via the HR pathway | [13] | |
| G2/M transition -regulates transcription of cyclin A2 | [78] | ||
| SIRT1 | Reduces activity of p53Modulation of γ-H2AX foci, BBRCA1, Rad51, NBS foci formation | [83] | |
| SIRT6 | Facilitates DSB repair by activating PARP1 | [78] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





