1. Introduction
The
Orthoflavivirus zikaense (class
Flasuviricetes, order
Amarillovirales family
Flaviviridae; genus
Orthoflavivirus; Zika virus or ZIKV) [
1,
2,
3] is a single-stranded positive-sense RNA virus whose genome is arranged in a linear, no segmented configuration with two flanking noncoding regions: 5’ NCR and 3’ NCR [
4]. Viral open reading frame (ORF) codes for a polyprotein that can be cleaved into ten proteins. Three are structural proteins: capsid (C), pre-membrane/membrane (PrM), and envelope (E). The remaining seven are nonstructural proteins: NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 [
4,
5,
6,
7]. Historically, the Zika virus was named based on the locality of the initial isolation in Zika Forest in Uganda, Africa. This African isolated, also known as MR766, was identified in 1947 from rhesus monkey serum during surveillance for YFV (Species:
Orthoflavivirus flavi) [
7]. Phylogenetically, ZIKV can be divided into two lineages: African and Asian. These lineages may be associated with differences in virulence or tropism. African and Asian lineages are phenotypically different due to mutations acquired during viral evolution [
5].
ZIKV has been sporadically detected in in
Aedes aegypti mosquitoes from Africa and Asia [
4,
7]. Over the years, the Asian lineage has caused outbreaks in Southeast Asia, several Pacific Islands, and the Americas. In Brazil, for example, ZIKV has been identified as the etiologic agent behind the outbreak of an illness with symptoms of rash, mild fever, and arthralgia between 2014 and 2015 [
8,
9]. Moreover, during the Brazilian epidemic, it was observed that ZIKV can cross the maternal-fetal barrier during pregnancy and cause severe problems to the infant, such as neuronal death, abnormal vasculature, including leaky blood-brain barrier, cell cycle arrest, and apoptosis of neural progenitor cells resulting in postnatal microcephaly with brain damage, which was later described as Congenital ZIKV Syndrome (CZS) [
9,
10].
The placenta is a transient organ involved with reproduction, connecting mother and fetus in order to confer nutrition, oxygenation and immune protection for the fetus [
11]. While the Asian lineage has been linked to CZS, studies indicated that the African isolate may be associated with stronger virulence, higher viral replication, and lytic infections of placental cells [
12]. These characteristics may lead to an early termination of pregnancy, while the Asian isolated can be less destructive to placental cells, allowing the pregnancy to proceed but interfering with the embryo development [
4,
10,
13].
The placenta is a heterogeneous organ formed by different tissues and can support the expression of Endogenous Retrovirus (ERVs) [
14]. In human ERV represent around 8% of the human genome and are found in various vertebrate species [
15,
16]. These ERVs may have resulted from an ancient retroviral infection that integrated into the host genome, especially in the germline, and became endogenous [
16]. Due mutation accumulation, most ERVs are inactive, but some can be expressed [
17,
18,
19]. Furthermore, they are regulated by DNA methylation [
20,
21] and are active mainly in the testis and placenta [
19]. Some ERV families are involved with the placentation process through the syncytiotrophoblast formation by trophoblastic cell fusion [
14].
In humans, ERVW-1 and ERVFRD-1 are the families that produce functional proteins during placentation [
22,
23]. The
env genes of ERVW-1 and ERVFRD-1 encodes for Syncytin-1 and Syncytin-2 proteins, respectively [
22,
23], which, unlike most retrovirus sequences that are defective, are conserved and perform essential functions in the placentation process, promoting the placental syncytiotrophoblast development. On the one hand, Syncytin-1 is a glycoprotein involved in the fusion of trophoblast cells that occurs by interactions between Syncytin-1 and its RDR-SLC receptor (a neutral amino acid transporter and type D mammalian retrovirus receptor) [
22]. On the other hand, Syncytin-2 also has fusogenic activity, and its receptor is a member of the carbohydrate transporter superfamily MFSD2 (major facilitator superfamily domain containing 2) [
24].
Pregnancy is a complex immunological state in which shifts occur on T helper cells (CD4
+) balance: Th1 and Th2 [
25,
26]. T helper cells form a subset of T cells according to cytokine production in which T helper 1 (Th1) secrete proinflammatory cytokines such as Interferon-gamma (IFN-y), whereas the T helper 2 (Th2) secrete anti-inflammatory cytokines such as interleukin-4 and IL-10 [
27,
28]. In normal conditions, down-regulation of the Th1 response and up-regulation of the Th2 response occurs during pregnancy to induce maternal tolerance and suppression [
25,
26]. ERVs may also have immunomodulatory activity during pregnancy. For example, Syncytin-1 protein may participate in placentation maintenance by inhibition of Th1 cytokines (TNF-α and IFN-γ) and chemokine CXCL10 in human blood cells [
29], and Syncytin-2 has an immunosuppressive characteristic may play a role to maternal-fetal immune tolerance during pregnancy [
30].
Th1 and Th2 cells arise from the same precursor naïve T lymphocytes [
31,
32]. The presence of IFNG determines the differentiation of these cells in Th1 lymphocytes [
31], whereas IL4 induces the differentiation of naïve T cells in Th2 lymphocytes [
32].
Viral infections may activate antiviral and inflammatory pathways [
33]. The host immune response plays a vital role in the clinical course of patients with viral infection, with cellular immunity and components of the innate immune response, such as interferons and other cytokines, playing an essential role in viral control [
33]. ZIKV can induce an intense immune response in the developing fetus, with probable involvement of ERVs [
34,
35]. Several studies observed that ERVW-1 has a role in host defense against exogenous viruses in non-placental cells [
36,
37]. However, in cases of Zika infection, the regulatory mechanisms and expression of the ERVs in the placental environment are still unknown. In the current study, we aimed to observe if the infection of placental cell lineages by two different ZIKV isolates can modulate the expression of host defense genes and ERVs compared to controls without infection. In addition, we tried to observe if the modulation of ERVs could be correlated with the modulation of a group of genes involved with the host defense.
2. Materials and Methods
2.1. Cells, Viruses and Infections
Based on results from viral kinetics previously published [
12], we reproduced the three time-points that yielded the highest signal of viral particle production in the monolayer (24h and 48h). BeWo CCL-98™ (ATCC
®) derived from human choriocarcinoma [
38] and HTR-8/SVneo CRL-3271™ (ATCC
®) derived from human trophoblast [
39] are placental cell lines and were kindly provided by Dr. Estela Bevilacqua from the Institute of Biomedical Sciences, University of São Paulo, Brazil.
For the infection, 90% confluent BeWo and HTR-8 T175 flasks were exposed to MR766 low-passage (African lineage, with a few passages in mice; GenBank: AY632535.2) and IEC-Paraíba (Asian lineage isolated by Institute Evandro Chagas; GenBank: KX2800260) ZIKV lineages. After one hour of incubation, the cell media was supplemented with 2% FBS and the flasks were incubated at 37 °C, 5% CO2 until harvest. Cell supernatants and monolayers were separated and kept at -80 °C preparatory for microarray and Taqman® assays.
2.2. Nucleic acid Extraction
RNA was extracted using the Qiagen Real-Time PCR for RT2 RNA QC PCR Array column extraction kit (Qiagen Inc. Germantown MD 20874 USA, cat.# 330291), following the manufacturer’s protocol for column purification with the AllPrep DNA/RNA/Protein Mini Kit (Qiagen Inc. Germantown MD 20874 USA, cat.# 80004). The integrity of the extracted RNA was assessed on the 2100 Bioanalyzer Instrument (Agilent Tecnologies Inc. Santa Clara CA 95051 USA) by comparing the ratio between the quantities of ribosomal RNA 28S and 18S. Nucleic acids were quantified using a NanoDrop® 2000 (Thermo Fisher Scientific Inc. Wilmington, DE 19810 USA) spectrophotometer in 260 nm and normalized to an average RNA concentration value of 200 ng/µL.
2.3. Molecular Detection
RNA extraction was performed using 250 µL of each time-point supernatant. For viral detection in RT-PCR, the AgPath-ID™ One-Step RT-PCR Reagent kit (Thermo Fisher Scientific Inc. Carlsbad CA USA 92008, cat.# 4387391) was used. A total of 5 μL of RNA, extracted from each condition, was combined with 5.7 μL of RNase-free water, 3.5 μL of Buffer, 0.3 μL of enzyme, and 1.5 μL of each primer/probe. The primers/probes used were ZIKV 835 10 μM and ZIKV 1086 10 μM (Lanciotti
et al., 2008). The readings were performed in 7500 Real-time PCR System (Applied Biosystems Foster City CA 94404 USA) with the following cycling conditions: 10 minutes incubation at 45°C for cDNA synthesis, 10 minutes at 95°C for reverse transcriptase inactivation and 40 cycles of 15 seconds at 95°C and 45 seconds at 60°C for annealing and extension. The TaqMan
® primers with probes selected for experimentation are described in Lanciotti
et al. (2008) [
40] and listed in
Appendix A,
Table A3. The primers target Zika virus envelope gene ZIKV-MR766 835 sense primer (position 835-857), with the sequence TTG GTC ATG ATA CTG CTG CTG ATT GC, and ZIKV 911c antisense primer (position 911-890), with the sequence CCT TCC ACA AAG TCC CTA TTG C.
2.4. Detection of ERV Expression in Placenta Cell Line
Appendix A,
Table A1, lists all the analysed ERVs and the corresponding literature used to justify their inclusion in this experiment. TaqMan
® qPCR Assays. The cDNAs obtained were quantified using NanoDrop
® 2000 and the values in ng/μL for the ratio of absorbances measured at 260 nm and 280 nm were obtained and standardized to the same input of 350 ng/mL. The manufacturer’s protocol TaqMan
® Fast Advanced Master Mix for qPCR (Thermo Fisher Scientific Inc. Carlsbad CA 92008 USA, cat.# 4444556) was used with some adaptations to prepare the reaction. For each reaction, 5.0 μL of TaqMan
® Fast Advanced Master Mix, 0.5 μL of 20x primers/probes, 3.5 μL of DEPC-Treated water (Thermo Fisher Scientific Inc. Carlsbad CA 92008 USA, cat.#AM9920), and 1.0 μL of cDNA were added. The material was distributed in 96-well microplates Axygen
® (Corning Glendale Arizona USA cat.# 12799438), sealed with optical adhesive (Sarstedt Sarstedtstraße 1 51588 Nümbrecht Germany, cat.# 951994), and subjected to the 7500 Real-time PCR System. For annealing and extension steps, the equipment was programmed to reach 50°C for 2 minutes, followed by 95°C for 10 minutes, and then 40 cycles of 15 seconds at 95°C and 60°C for 1 minute.
The relative expression of ERVs was calculated using the ΔCt method and 2^(-ΔΔCt) method [
41,
42]. The ΔCt values were computed as the discrepancy between the target genes and the control gene whenever both Ct values were available. ΔΔCt values were calculated as the difference between the ΔCt of infected and non-infected cells unless missing data precluded this calculation. Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) and RNA, 18S ribosomal 1 (RNA 18S1) were used as reference genes for normalization (
Appendix A Table A2), as they exhibited consistent constitutive expression across different samples.
2.5. Expression of Interleukins and Transcription Factors Related to the Immune Response
The expression of interleukins and transcription factors related to the immune response was detected using custom microarrays Qiagen Human RT2 RNA QC PCR Array (Qiagen Inc. Germantown MD 20874 USA, cat# 330291). This system allows the analysis of the expression profile of 20 genes encoding regulatory enzymes (
Appendix A Table A5). The assay includes 2 reference genes used as controls for the experiment: Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) and RNA, 18S ribosomal 1 (RNA 18S1). The test also contains controls for reverse transcription, positive amplification controls, and genomic DNA contamination control.
For the detection of the targets (controls) mentioned in
Appendix A,
Table A4, concentrations of 800 ng/mL of cDNA per reaction were used. The protocol employed was the RT2 RNA QC PCR Array Handbook provided by the manufacturer, with some adaptations. In summary, each reaction consisted of 12.5 μL of 2x RT2 SyberGreen Master Mix, 11.5 μL of RNase-free water, and 1 μL of cDNA. For the plate quality control (RTC), 12.5 μL of 2x RT2 SyberGreen Master Mix, 11.5 μL of RNase-free water, and 1 μL of cDNA with a 1:100 dilution, were used. For the quality control standards PPC and GDC, 12.5 μL of 2x RT2 SyberGreen Master Mix and 12.5 μL of RNase-free water were used. The reactions were dispensed into the corresponding wells, with a final volume of 25 μL per well. The plate was processed in the 7500 Real-time PCR System with the following program: 2 minutes at 50°C, 95°C for 10 minutes for Hot-Start DNA Taq Polymerase activation; 40 cycles of 15 seconds at 95°C followed by 1 minute at 60°C for fluorescence data acquisition.
Gene expression analyses and statistical calculations were performed according to the manufacturer’s instructions (
http://sabiosciences.com/pcrarraydataanalysis.php). Relative quantification was carried out by comparing Relative Fluorescence Units and Ct values of the amplification of each target gene to the control genes (DCt), following a quantification method established by the manufacturer [
43].
2.6. Statistical Analysis
The results were analysed using the GraphPad Prism 5.01 software (GraphPad Software San Diego California USA). Comparisons were conducted by comparing the mRNA expression values of ERVs and components of the immune response in cells with the control groups (non-infected) and those infected with ZIKV isolates.
3. Results
3.1. Gene Expression
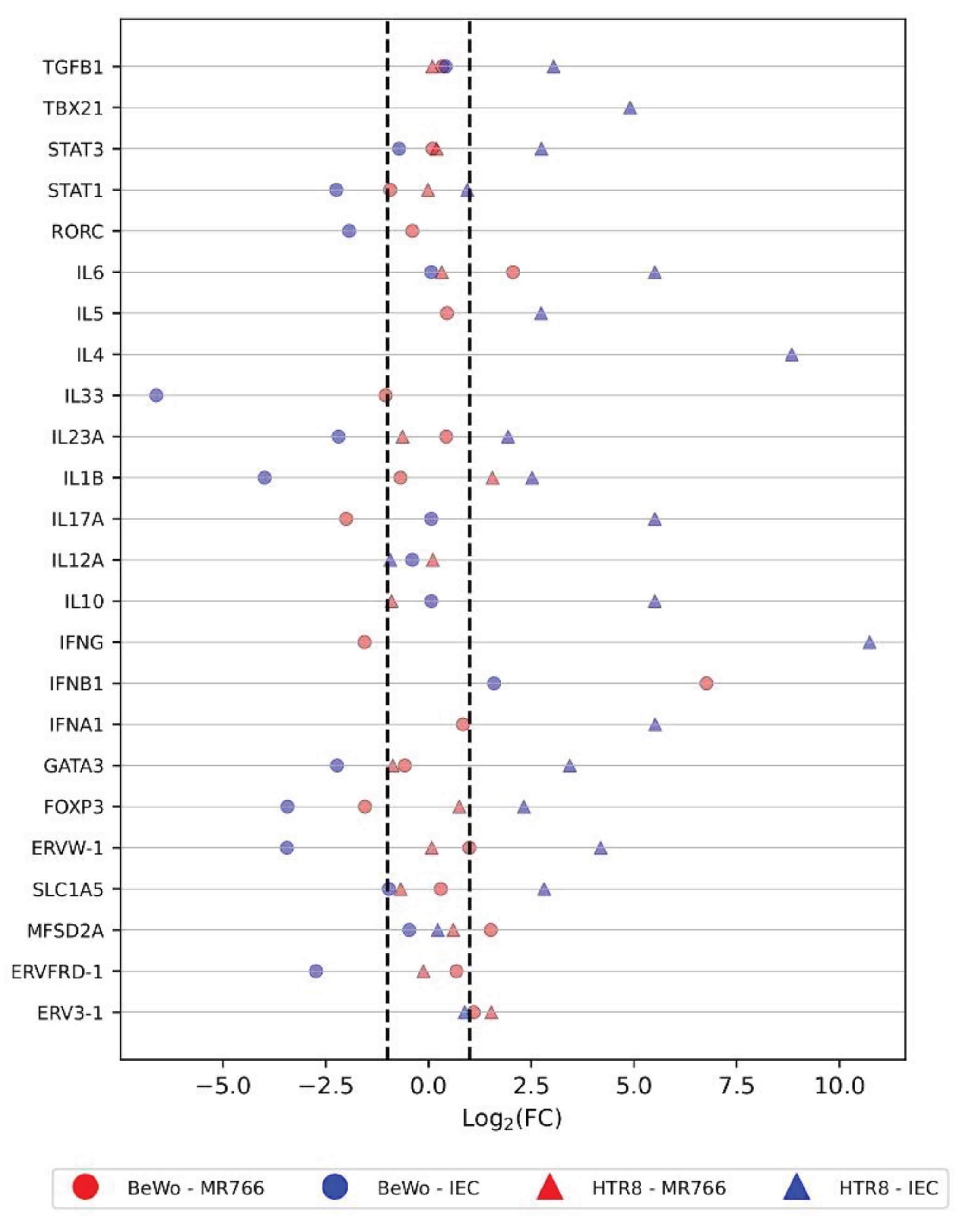
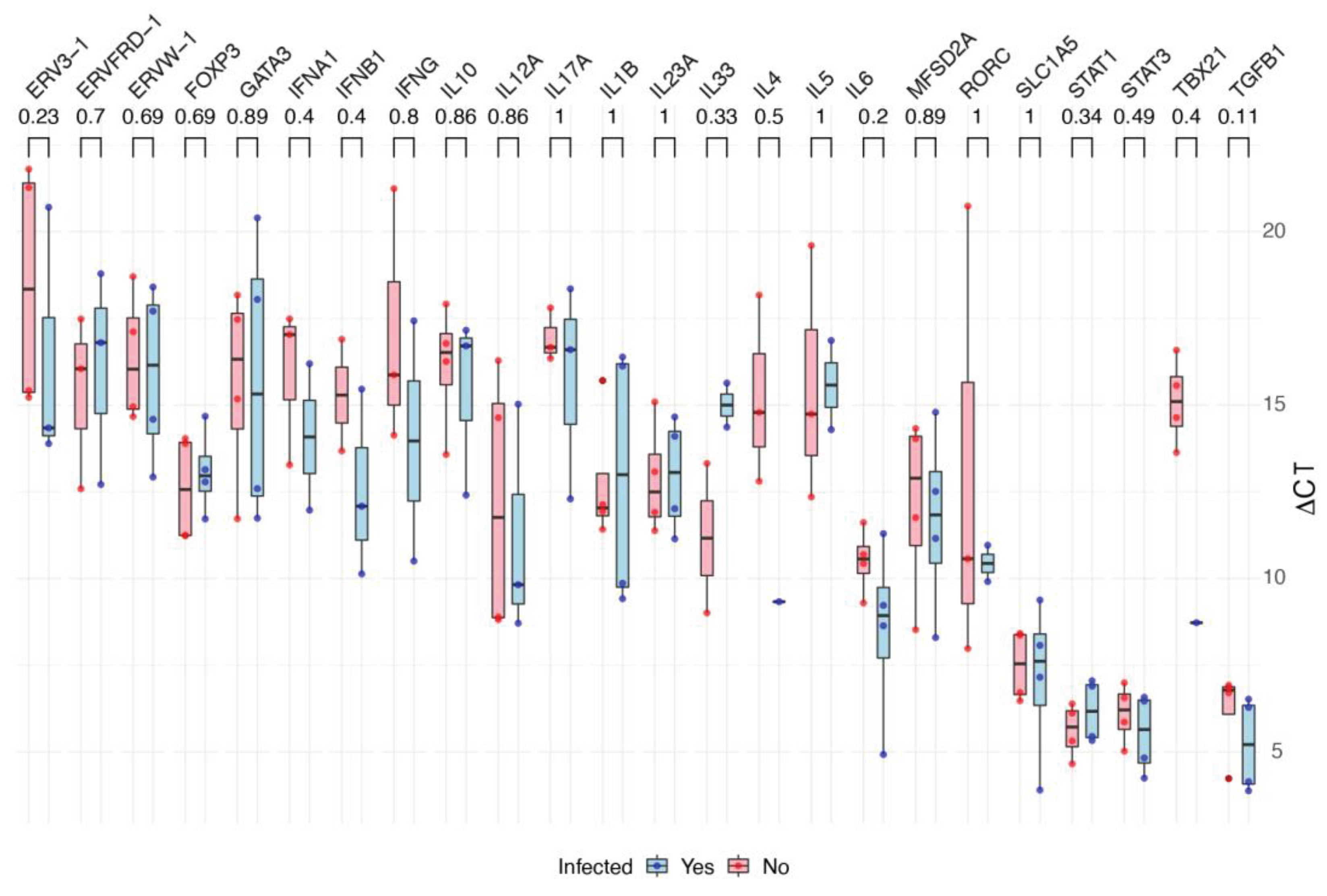
The scatterplot (
Figure 1) shows the fold change of each gene in BeWo and HTR8 cell lineages under two conditions: infected or not infected by ZIKV. There was no statistical difference between each gene’s ∆CT values from infected and not infected cells although there were Log2(FC) values smaller than -1 or greater than +1 (
Figure 2).
While most genes showed no significant change in expression upon infection, a few exceptions emerged: TBX21 and IL-4 were slightly under expressed in infected cells, while IL-33 was slightly over expressed in infected cells. In contrast, genes in IEC-infected cells showed a tendency towards upregulation, with only STAT1, IL12A, MFSD2A, and ERV3-1 showing minimal change. This suggests a complex interplay between infection and gene expression, with some genes exhibiting subtle alterations and others responding more robustly.
3.2. Pairwise Correlation Analyses of HERVs and Interleukins Expression Profiles
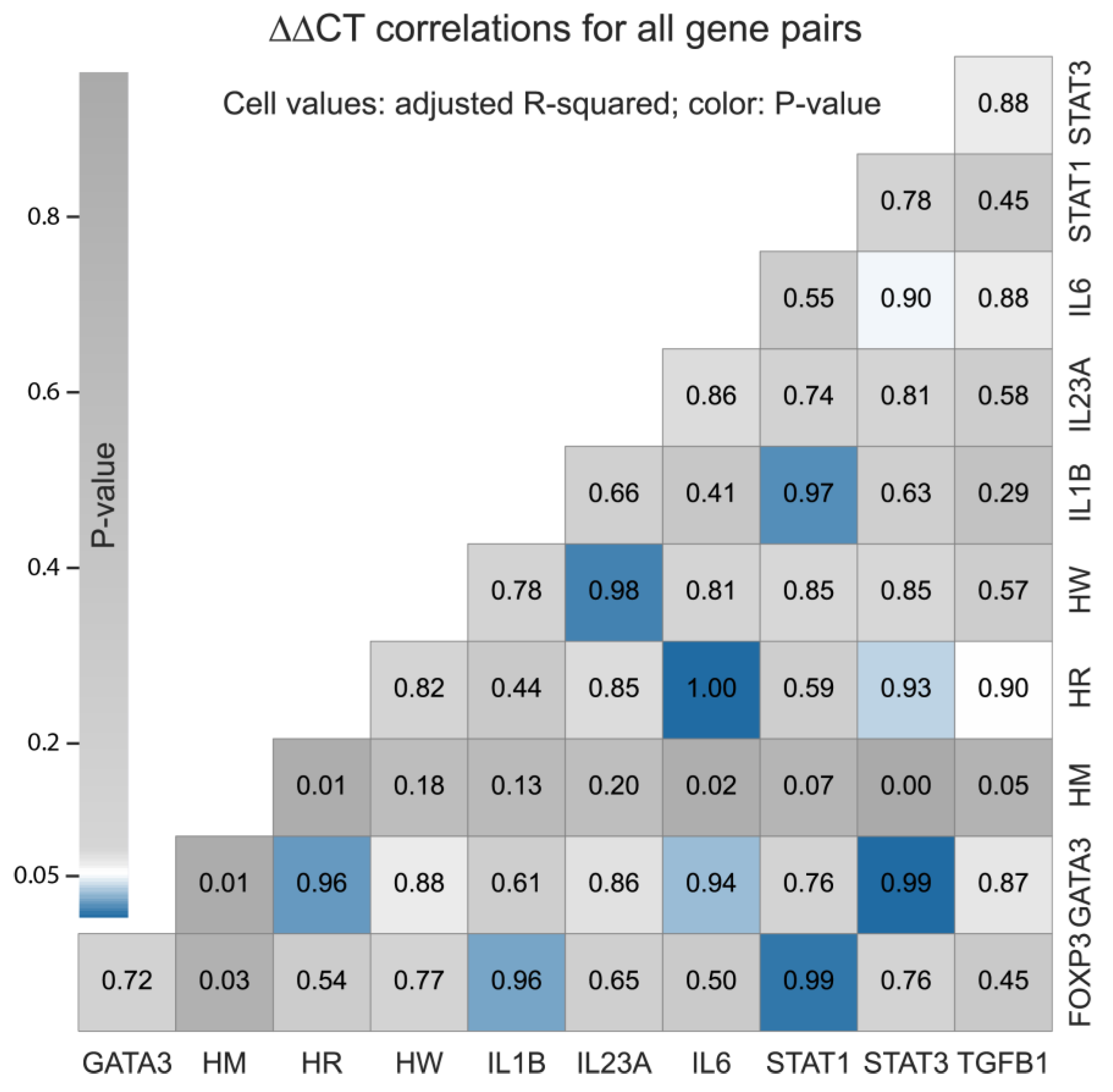
We calculated the ∆∆CT values for one HERVs (ERVW-1), two syncytin receptors (MFSD2A and SLC1A5) and eight interleukins (FOXP3, GATA3, IL1B, IL23A, IL6, STAT1, STAT3, and TGFB1), as illustrated in
Figure 3. ∆∆CT values were not computed for 13 HERVs and defensive genes, for which we did not have all the required expression data points. Four correlations had P-value < 0.01 and adjusted-R
2 greater than 98%: SLC1A5 vs. IL-6, GATA3 vs. STAT3, FOXP3 vs. STAT1, and ERVW-1 vs. IL-23A. Another five had P-value < 0.05 and adjusted-R
2 < 97%: IL1B vs. STAT1, GATA3 vs. SLC1A5, FOXP3 vs. IL1B, GATA3 vs. IL-6, and SLC1A5 vs. STAT3.
4. Discussion
Barbosa
et al. (2023) [
12] reported data from in vitro experiments demonstrating that BeWo and HTR-8 trophoblastic cells are susceptible and permissive to two Zika virus (ZIKV) strains, ZIKV-MR766 (African or MR766) and ZIKV-IEC-Paraíba (Asian-Brazilian or IEC). Barbosa
et al. (2023) [
12] also demonstrated the existence of a different viral dynamic between African and Asian-Brazilian lineages in vitro. Geddes
et al. (2021) [
44] presented a comprehensive genome-wide transcriptome analysis of human primary astrocytes infected with Chikungunya, Mayaro, Oropouche, or Zika viruses. These authors also showed a co-evolution in the mechanisms involved in the escape of arboviruses to antiviral immune response mediated by the interferon (IFN) pathway. Castro
et al. (2022) [
45] quantified the modulation of HERV expression by four different encephalitic arboviruses during infection of human primary astrocytes. Their data show common HERVs expression modulation by both alphaviruses, suggesting conserved evolutionary routes of transcription regulation. Furthermore, Castro
et al.’s (2022) [
46] results support the role of HERVs induction in the transcription regulation process of genes during arboviral infections. Put together, this literature shows that trophoblastic cells are susceptive to ZIKV infection and that arbovirus infection potentially affects the expression of HERVs and interleukins. Here, we reproduced Barbosa
et al.’s (2023) [
12] methodology to test if these trophoblastic cell lineages would show differential expression of selected HERVs and interleukins during the early stage of ZIKV infection as suggested by previous studies such as Geddes
et al. (2021) [
44] and Castro
et al. (2022) [
45].
Our study targeted two lineages of trophoblasts derived from placental cells, human placental choriocarcinoma (BeWo) cells and HTR-8. Specifically, we measured the expression of the selected ERVs, syncytin receptors, and interleukins in both cell types. We compared control cells to cells infected with two ZIKV variants, MR766 and IEC. Gene expression levels under both (infected and not-infected) conditions were measured using custom SABiosciences RT2 RNA QC PCR arrays for interleukins and Thermo Fisher’s Taqman assays for HERVs and syncytin receptors.
The Fold-Change (FC), calculated as Log2(2-∆∆CT), indicates that some selected genes could be up- or down-regulated following ZIKV infection. Moreover, we found a strong and positive correlation between the expression of nine gene pairs: SLC1A5 and IL6; GATA3 and STAT3; FOXP3 and STAT1; ERVW-1 and IL23A; IL1B and STAT1; GATA3 and SLC1A5; FOXP3 and IL1B; GATA3 and IL6; and SLC1A5 and STAT3. However, we determined that none of the selected genes were differently expressed under a confidence interval of 95%. This can be explained if the target placental cells do not express the selected genes differently in response to ZIKV infection under the conditions of this experimental study.
The lack of differentially expressed genes in our analyses occurred in the early stage of ZIKV infection in the tested trophoblast lineages. It is necessary to contextualize our observations considering previous studies such as Geddes et al. (2021) [
44], Castro et al. (2022) [
45], and others that focused on different sets of ERV and interleukin genes at later time points. For example, in contrast to our study, Rabelo
et al. [
46] showed different expression profiles of immune response in the later stage of ZIKV infection during pregnancy in the placenta. Rabelo
et al. [
46] observed a release of the proinflammatory cytokines TNF and IFNG, which are involved in Th1 response, in human placental tissues infected by ZIKV.
Da Silva
et al. [
47] showed an upregulation of IFNG, IFNA1 and IFNB1in blood samples of patients infected with ZIKV during acute infection (until five days after the beginning of symptoms) in comparison with healthy subjects. The expressions of IL6 and IL-12 were similar for both groups. However, placenta presents immunosuppressive properties [
29,
30,
48,
49,
50]. Chang
et al. [
48] demonstrated that pretreatment with IFNG increased the immunosuppression performed by placenta-derived multipotent cells due to a substantial augmentation of TGFB1 expression, an anti-inflammatory cytokine. Furthermore, syncytin-1 [
29] and syncytin-2 [
30] are immunosuppressive proteins expressed in the placenta and inhibit the release of Th1 cytokines, especially TNF and IFNG. Such suppression may contribute to maternal immune tolerance.
Noteworthy, syncytin-1 has been shown to weaken the antiviral response against Influenza, leading to reduced production of IFNA1, IFNL1, and IFNG, and inducing IL10 release by peripheral monocytes (PBMCs). However, it was associated with an increase in the levels of some proinflammatory cytokines (IL6 and IL1B) [
51]. Additionally, upregulation of Th2 cytokines (IL4, IL10, and IL13) and suppression of Th1 cytokines (IL2, TNF, and IFNG) in the feto-maternal interface contribute to a successful pregnancy and immune tolerance to the fetus [
25,
26,
52,
53]. In contrast, increased expression of Th1 and lower levels of Th2 cytokines are associated with fetal resorption and abortion [
54,
55].
Equally important, IL10 [
56,
57] is the main immunosuppressive cytokine and negatively regulates IL-12 production, which induces Th1 response and cell-mediated cytotoxicity against intracellular pathogens, such as viruses [
58,
59,
60]. As a final point, Lu
et al. [
61] observed that glutamine, mediated by SLC1A5 (also called ASCT2), induced STAT3 phosphorylation and activation, causing an increase in the CCL5 expression and infiltration of T cells in oral lichen planus.
We combine our results with the cited literature to shed light on ZIKV infections in the placenta. The ZIKV epidemic that occurred in Brazil between 2015-2016 made clear the potential of these infections to affect the embryo/fetus, resulting in many cases of permanent neurological sequelae, including microcephaly. Despite our original expectations, we did not detect differentially expressed HERVs, syncytin receptors, and interleukin genes after 24 to 48 hours of ZIKV infection in two lineages of trophoblast cells, BeWo and HTR-8. Therefore, ZIKV infection did not cause correlations between pairs of gene expression profiles that we observed under our experimental conditions. We hypothesize that the suppression of immune response by the placenta may explain these findings. Additional investigations are warranted to validate this hypothesis.
While our results can be a window towards early ZIKV infection in some trophoblastic cell lineages, future studies that include additional trophoblastic cell lineages and more advanced time points in viral kinetics are needed to reveal the response of these cells to ZIKV over time. We anticipate that longer infection time will trigger differential expression of ERVs, interleukins, and cytokines necessary for the formation and functioning of the placenta.
Author Contributions
A.L.C., P.P.O. and M.D.B, these authors contributed equally and shared the first authorship. D.J.M. and L.M.R.J. are jointly credited as last authors. Conceptualization, L.M.R.J. methodology and validation, A.L.C., D.J.M., M.D.B, P.P.O., and T.P.B; formal analysis, T.P.B; investigation, A.L.C., M.D.B, P.P.O. and T.P.B; resources, L.M.R.J and P.M.A.Z.; data curation, C.M.P, D.J.M, L.M.R.J, M.D.B, R.A.S. and P.P.O.; writing—original draft preparation, A.L.C, J.T.M, L.M.R.J, M.D.B and P.P.O; writing—review, editing and visualization, C.M.P., D.J.M., L.M.R.J, M.D.B, R.A.S. and P.P.O; other relevant contributions, F.A., I.M.V.G.C., M.R.S.B., R.D.C; supervision, D.J.M., L.M.R.Z. and M.D.B.; project administration, L.M.R.J; funding acquisition, L.M.R.J and P.M.A.Z. All authors have read and agreed to the published version of the manuscript.
Funding
Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP): Postdoctoral fellows scholarship number: 2023/07287-5 (M.D.B.), 2023/10230-5 (C.M.P) and 2021/05661-1 (R.A.S.); Young Investigator Program 2019/01255-9 and 2021/03684-4 (R.D.C.); FAPESP theme project number 2020/08943-5 (L.M.R.J.).
Data Availability Statement
Original experimental data in material can be downloaded.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix A
Table A1.
List of analyzed ERVs.
Table A1.
List of analyzed ERVs.
| ERV |
Immune System Funtion |
Reference |
| H-W (syncytin1) |
Immunoregulator |
Blaise et al. (2005) [61] |
| H-FRD (syncytin2) |
Immunoregulator |
Blaise et al. (2005) [61] |
| H-R (HERV-3) |
Immunoregulator |
Mangeney et al. (2007) [62] |
Table A2.
List of transcriptions factors and cytokines analyzed during array assay.
Table A2.
List of transcriptions factors and cytokines analyzed during array assay.
| |
ERV |
Oligonucleotide |
Transcription Factors
Array Catalog Number 330171
CLAH25383 |
Foxp3 forkhead box P3 |
PPH00029C |
| Gata-3 Transcription Factor |
PPH02143A |
| Rorc receptor C |
PPH05877A |
| Stat1 transcription activator 1 |
PPH00811C |
| Stat3 transcription activator 3 |
PPH00708F |
Cytokines
Array Catalog Number 330171
CLAH25383 |
IL-10 interleukin 10 |
PPH00572C |
| IL17a interleukin17a |
PPH00537C |
| IL-6 interleukin 6 |
PPH00560C |
| IL-12a_p35 interleukin 12a |
PPH00544B |
| IL-23a_p19 interleukin 23 α |
PPH01688B |
| IL-33 interleukin 33 |
PPH17375E |
| IL-1B interleukin 1 beta |
PPH00171C |
| TGF-B1 growth factor 1 beta |
PPH00508A |
| IL-4 interleukin 4 |
PPH00565B |
| IL-5 interleukin 5 |
PPH00692B |
| IFN-a interferon alpha |
PPH01321B |
| IFN-b interferon beta |
PPH00384F |
| IFN-g interferon gamma |
PPH00380C |
| GAPDH – endogenous control |
PPH00150F |
| 18SrRNA – endogenous control |
PPH05666E |
Table A3.
Oligonucleotide primer sequences used for ERV detection for Taqman® assays.
Table A3.
Oligonucleotide primer sequences used for ERV detection for Taqman® assays.
| ERV |
Oligonucleotide |
| WE8_F (syncytin-1) |
TaqManAssayThermo |
| WE8_R (syncytin-1) |
Hs01926764_ul |
| FRD (syncytin-2) F |
TaqManAssayThermo |
| FRD (syncytin-2) R |
Hs01942443_sl |
| HR_F |
TaqManAssayThermo |
| HR_R |
Hs04184598_sl |
| RDR F (receptor for syncytin1) |
TaqManAssayThermo |
| RDR R (receptor for syncytin1) |
Hs01056542_ml |
| MFSD-2 F (receptor parasyncytin2) |
TaqManAssayThermo |
| MFSD-2 R (receptor parasyncytin2) |
Hs00293017_m1 |
Table A4.
Reference genes used as controls for the experiment.
Table A4.
Reference genes used as controls for the experiment.
| ERV |
Oligonucleotide |
| GAPDH (Ref.) |
TaqManAssayThermo |
| |
Hs03929097_g1 |
| 18SrRNA (Ref.) |
TaqManAssayThermo |
| |
Hs99999901_s1 |
Figure A1.
Resume of methodology.
Figure A1.
Resume of methodology.
References
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Davison, A.J.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; Harrach, B. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses. Archives of Virology 2021, 166, 2633–2648. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses Executive Committee. The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nature Microbiology 2020, 5, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Postler, T.S.; Beer, M.; Blitvich, B.J.; Bukh, J.; de Lamballerie, X.; Drexler, J.F.; Imrie, A.; Kapoor, A.; Karganova, G.G.; Lemey, P.; Lohmann, V. Renaming of the genus flavivirus to orthoflavivirus and extension of binomial species names within the family Flaviviridae. Archives of virology 2023, 168, 224. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Gubler, D.J. 2016. Zika virus. Clinical Microbiology Reviews 2016, 29, 487–524. [Google Scholar] [CrossRef] [PubMed]
- Faye, O.; Freire, C.C.M.; Iamarino, A.; Faye, O.; de Oliveira, J.V.C.; Diallo, M.; Zanotto, P.M.A.; Sall, A.A. Molecular evolution of Zika virus during its emergence in the 20th century. PLoS neglected tropical diseases 2014, 8, e2636. [Google Scholar] [CrossRef] [PubMed]
- Sirohi, D.; Kuhn, R.J. Zika virus structure, maturation, and receptors. The Journal of infectious diseases 2017, 8, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus (I). Isolations and serological specificity. Transactions of the royal society of tropical medicine and hygiene 1952, 46, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Olson, J.; Ksiazek, T.G. Zika virus, a cause of fever in Central Java, Indonesia. Transactions of the Royal Society of Tropical Medicine and Hygiene 1981, 75, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Campos, G.S.; Bandeira, A.C.; Sardi, S.I. Zika virus outbreak, Bahia, Brazil. Emerging infectious diseases 2015, 21, 1885. [Google Scholar] [CrossRef]
- Zanluca, C.; Melo, V.C.; Mosimann, A.L.; Santos, G.I.; Santos, C.N.; Luz, K. First report of autochthonous transmission of Zika virus in Brazil. Memórias do Instituto Oswaldo Cruz 2015, 110, 569–572. [Google Scholar] [CrossRef]
- Burton, G.J.; Fowden, A.L. The placenta: a multifaceted, transient organ. Philos Trans R Soc Lond B Biol Sci. 2015, 370, 20140066. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.D.; Costa, A.; Prieto-Oliveira, P.; Andreata-Santos, R.; Peter, C.M.; Zanotto, P.M.; Janini, L.M.R. Proposal of Model for Evaluation of Viral Kinetics of African/Asian/Brazilian—Zika virus Strains (Step Growth Curve) in Trophoblastic Cell Lines. Viruses 2023, 15, 1446. [Google Scholar] [CrossRef]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.; Guimarães, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Blaise, S.; de Parseval, N.; Heidmann, T. Functional characterization of two newly identified Human Endogenous Retrovirus coding envelope genes. Retrovirology 2005, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.; Cornwallis, C.K.; Jern, P. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proceedings of Nationwl Academics of Sciences of United States of America 2015, 112, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Coffin, J.M. Constructing primate phylogenies from ancient retrovirus sequences. Proceedings of Nationwl Academics of Sciences of United States of America 1999, 96, 10254–10260. [Google Scholar] [CrossRef] [PubMed]
- Tristem, M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. Journal of Virology 2000, 74, 3715–3730. [Google Scholar] [CrossRef] [PubMed]
- de Parseval, N.; Lazar, V.; Casella, J.F.; Benit, L.; Heidmann, T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. Journal of Virology 2003, 77, 10414–10422. [Google Scholar] [CrossRef]
- Szpakowski, S.; Sun, X.; Lage, J.M.; Dyer, A.; Rubinstein, J.; Kowalski, D.; Sasaki, C.; Costa, J.; Lizardi, P.M. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene 2009, 448, 151–167. [Google Scholar] [CrossRef]
- Lavie, L.; Kitova, M.; Maldener, E.; Meese, E.; Mayer, J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). Journal of Virology 2005, 79, 876–883. [Google Scholar] [CrossRef]
- Blond, J.L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. Journal of Virology 2000, 74, 3321–3329. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.P.; Chen, L.F.; Yang, S.R.; Chen, C.Y.; Ko, C.C.; Chang, G.D.; Chen, H. Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biology of reproduction 2008, 79, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Esnault, C.; Priet, S.; Ribet, D.; Vernochet, C.; Bruls, T.; Lavialle, C.; Weissenbach, J.; Heidmann, T. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proceedings of the National Academy of Sciences 2008, 105, 17532–17537. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Mosmann, T.R.; Guilbert, L.; Tuntipopipat, S.; Wegmann, T.G. Synthesis of T helper 2-type cytokines at the maternal-fetal interface. Journal of Immunology 1993, 151, 4562–4573. [Google Scholar] [CrossRef]
- Marzi, M.; Vigano, A.; Trabattoni, D.; Villa, M.L.; Salvaggio, A.; Clerici, E.; et al. Characterization of type 1 and type 2 cytokine production profile in physiologic and pathologic human pregnancy. Clinical and Experimental Immunology 2003, 106, 127–133. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. Journal of Immunology 1986, 136, 2348–2357. [Google Scholar] [CrossRef]
- Del Prete, G.F.; De Carli, M.; Mastromauro, C.; Biagiotti, R.; Macchia, D.; Falagiani, P.; Ricci, M.; Romagnani, S. Purified protein derivative of Mycobacterium tuberculosis and excretory-secretory antigen(s) of Toxocara canis expand in vitro human T cells with stable and opposite (type 1 T helper or type 2 T helper) profile of cytokine production. Journal of Clinical Investigation 1991, 88, 346–350. [Google Scholar] [CrossRef]
- Tolosa, J.M.; Schjenken, J.E.; Clifton, V.L.; Vargas, A.; Barbeau, B.; Lowry, P.; Maiti, K.; Smith, R. The endogenous retroviral envelope protein syncytin-1 inhibits LPS/PHA-stimulated cytokine responses in human blood and is sorted into placental exosomes. Placenta 2012, 33, 933–941. [Google Scholar] [CrossRef]
- Lokossou, A.G.; Toudic, C.; Nguyen, P.T.; Elisseeff, X.; Vargas, A.; Rassart, É.; Lafond, J.; Leduc, L.; Bourgault, S.; Gilbert, C.; Scorza, T.; Tolosa, J.; Barbeau, B. Endogenous retrovirus-encoded Syncytin-2 contributes to exosome-mediated immunosuppression of T cells†. Biology of Reproduction 2020, 102, 185–198. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Joyce, J.; Fitch, F.W. Antiproliferative effect of IFN-gamma in immune regulation. III. Differential selection of TH1 and TH2 murine helper T lymphocyte clones using recombinant IL-2 and recombinant IFN-gamma. Journal of Immunology 1989, 143, 15–22. [Google Scholar] [CrossRef]
- Seder, R.A.; Paul, W.E.; Davis, M.M.; Fazekas de St Groth, B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. Journal of Experimental Medicine 1992, 176, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, S.; Mandrekar, P. Cellular stress response and innate immune signaling: integrating pathways in host defense and inflammation. Journal of leukocyte biology 2013, 94, 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Schust, D.J.; Bonney, E.A.; Sugimoto, J.; Ezashi, T.; Roberts, R.M.; Choi, S.; Zhou, J. The immunology of syncytialized trophoblast. International journal of molecular sciences 2021, 22, 1767. [Google Scholar] [CrossRef]
- Girsch, J.H.; Mejia Plazas, M.C.; Olivier, A.; Farah, M.; Littlefield, D.; Behl, S.; Punia, S.; Sakemura, R.; Hemsath, J.R.; Norgan, A.; Enninga, E.A. Host-viral interactions at the maternal-fetal interface. What we know and what we need to know. Frontiers 2022, 2, 833106. [Google Scholar] [CrossRef]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nature Reviews Microbiology 2019, 17, 355–370. [Google Scholar] [CrossRef]
- Kassiotis, G. The Immunological Conundrum of Endogenous Retroelements. Annual Review of Immunology 2023, 41, 99–125. [Google Scholar] [CrossRef]
- Pattillo, R.A.; Gey, G.O. The establishment of a cell line of human hormone-synthesizing trophoblastic cells in vitro. Cancer Research 1968, 7, 1231–1236. [Google Scholar]
- Graham, C.H.; Hawley, T.S.; Hawley, R.C.; MacDougall, J.R.; Kerbel, R.S.; Khoo, N.; Lala, P.K. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Experimental cell research 1993, 206, 204–211. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerging Infectious Diseases 2008, 14, 1232–1239. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Arikawa, E.; Quellhorst, G.; Han, Y.; Pan, H.; Yang, J. RT2 Profiler PCR Arrays: Pathway-focused gene expression profiling with qRT-PCR. SA Bioscience 2010. [Google Scholar]
- Geddes, V.E.V.; Brustolini, O.J.B.; Cavalcante, L.T.D.F.; Moreira, F.R.R.; De Castro, F.L.; Guimaraes, A.P.D.C.; Gerber, A.L.; Figueiredo, C.M.; Diniz, L.P.; Neto, E.D.A.; Tanuri, A. Common Dysregulation of Innate Immunity Pathways in Human Primary Astrocytes Infected With Chikungunya, Mayaro, Oropouche, and Zika Viruses. Frontiers in Cellular and Infection Microbiology 2021, 11, 641261. [Google Scholar] [CrossRef]
- Castro, F.L.D.; Brustolini, O.J.B.; Geddes, V.E.V.; Souza, J.P.B.M.D.; Alves-Leon, S.V.; Aguiar, R.S.; Vasconcelos, A.T.R. Modulation of HERV Expression by Four Different Encephalitic Arboviruses during Infection of Human Primary Astrocytes. Viruses 2022, 14, 2505. [Google Scholar] [CrossRef]
- Rabelo, .K; de Souza, L.J.; Salomão, N.G.; Machado, L.N.; Pereira, P.G.; Portari, E.A.; Basílio-de-Oliveira, R.; Dos Santos, F.B.; Neves, L.D.; Morgade, L.F.; Provance, D.W., Jr; Higa, L.M.; Tanuri, A; de Carvalho, J.J.; Paes, M.V. Zika Induces Human Placental Damage and Inflammation. Frontiers in Immunology 2020, 11, 2146. [Google Scholar] [CrossRef]
- Da Silva, M.H.M.; Moises, R.N.C.; Alves, B.E.B.; Pereira, H.W.B; . de Paiva, A.A.P.; Morais, I.C.; Nascimento, Y.M.; Monteiro, J.D; de Souto, J.T.; Nascimento, M.S.L.; de Araújo, J.M.G.; da Guedes, P.M.M.; Fernandes, J.V. Innate immune response in patients with acute Zika virus infection. Medical Microbiology and Immunology 2019, 208, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Yen, M.; Chen, Y.; Chien, C.; Huang, H.; Bai, C.; et al. Placenta-Derived Multipotent Cells Exhibit Immunosuppressive Properties That Are Enhanced in the Presence of Interferon-γ. Stem Cells 2006, 24, 2466–2477. [Google Scholar] [CrossRef] [PubMed]
- Culouscou, J.M.; Remacle-Bonnet, M.M.; Pommier, G.; Rance, R.J.; Depieds, R.C. Immunosuppressive properties of human placenta: study of supernatants from short-term syncytiotrophoblast cultures. Journal of Reproductive Immunology 1986, 9, 33–47. [Google Scholar] [CrossRef]
- Menu, E.; Kaplan, L.; Andreu, G.; Denver, L.; Chaouat, G. Immunoactive products of human placenta. Cellular Immunology 1989, 119, 341–352. [Google Scholar] [CrossRef]
- Tolosa, J.M.; Parsons, K.S.; Hansbro, P.M.; Smith, R.; Wark, P.A.B. The Placental Protein Syncytin-1 Impairs Antiviral Responses and Exaggerates Inflammatory Responses to Influenza. PLoS One 2015, 10, 4–e0118629. [Google Scholar] [CrossRef] [PubMed]
- Piccinni, M.P.; Beloni, L.; Livi, C.; Maggi, E.; Scarselli, G.; Romagnani, S. Defective production of both leukemia inhibitory factor and type 2 T-helper cytokines by decidual T cells in unexplained recurrent abortions. Nature Medicine 1998, 4, 1020–1024. [Google Scholar] [CrossRef] [PubMed]
- Raghupathy, R.; Makhseed, M.; Azizieh, F.; Omu, A.; Gupta, M.; Farhat, R. Cytokine production by maternal lymphocytes during normal human pregnancy and in unexplained recurrent spontaneous abortion. Human Reproduction 2000, 15, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, P.L. Increased CD56+ NK cells and enhanced Th1 responses in human unexplained recurrent spontaneous abortion. Genetics and Molecular Research 2015, 14, 18103–18109. [Google Scholar] [CrossRef]
- Sakakibara, M.; Maeda, Y.; Nakamura, K. Fetal loss due to Th1-skewed Th1/Th2 balance with increase (not decrease) of regulatory T cells in abortion-prone mouse model. Journal of Toxicological Sciences 2022, 47, 327–336. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, A.; Aste-Amezaga, M.; Valiante, N.M.; Ma, X.; Kubin, M.; Trinchieri, G. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. The Journal of experimental medicine 1993, 178, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Aste-Amezaga, M.; Ma, X.; Sartori, A.; Trinchieri, G. Molecular mechanisms of the induction of IL-12 and its inhibition by IL-10. Journal of Immunology 1998, 160, 5936–5944. [Google Scholar] [CrossRef]
- Aste-Amezaga, M.; D’Andrea, A.; Kubin, M.; Trinchieri, G. Cooperation of Natural Killer Cell Stimulatory Factor/Interleukin-12 with Other Stimuli in the Induction of Cytokines and Cytotoxic Cell-Associated Molecules in Human T and NK Cells. Cellular Immunology 1994, 156, 480–492. [Google Scholar] [CrossRef]
- Ozmen, L.; Aguet, M.; Trinchieri, G.; Garotta, G. The in vivo antiviral activity of interleukin-12 is mediated by gamma interferon. Journal of Virology 1995, 69, 8147–8150. [Google Scholar] [CrossRef]
- Monteiro, J.M.; Harvey, C.; Trinchieri, G. Role of Interleukin-12 in Primary Influenza Virus Infection. Journal of Virology 1998, 72, 4825–4831. [Google Scholar] [CrossRef]
- Lu, J.; Su, Z.; Li, W.; Ling, Z.; Cheng, B.; Yang, X.; Tao, X. ASCT2-mediated glutamine uptake of epithelial cells facilitates CCL5-induced T cell infiltration via ROS-STAT3 pathway in oral lichen planus. International Immunopharmacology 2023, 119, 110216. [Google Scholar] [CrossRef] [PubMed]
- Mangeney, M.; Renard, M.; Schlecht-Louf, G.; Bouallaga, I.; Heidmann, O.; Letzelter, C.; Richaud, A.; Ducos, B.; Heidmann, T. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proceedings of the National Academy of Sciences 2007, 104, 20534–20539. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).