Submitted:
02 April 2024
Posted:
03 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and methods
2.1. Sample preparation
2.2. Strain culture and DNA extraction
2.3. Screening of specific genes of P. aeruginosa and P. fragi
2.4. Primers, probes design for dddPCR and specificity verification
2.5. Establishment of the dddPCR assay
2.6. Establishment of standard curve
2.7 Sensitivity test of dddPCR detection
2.8. Anti-interference ability evaluation
2.9. Evaluation of artificial simulated contamination of actual samples
3. Results and discussions
3.1. Analysis of candidate gene selection
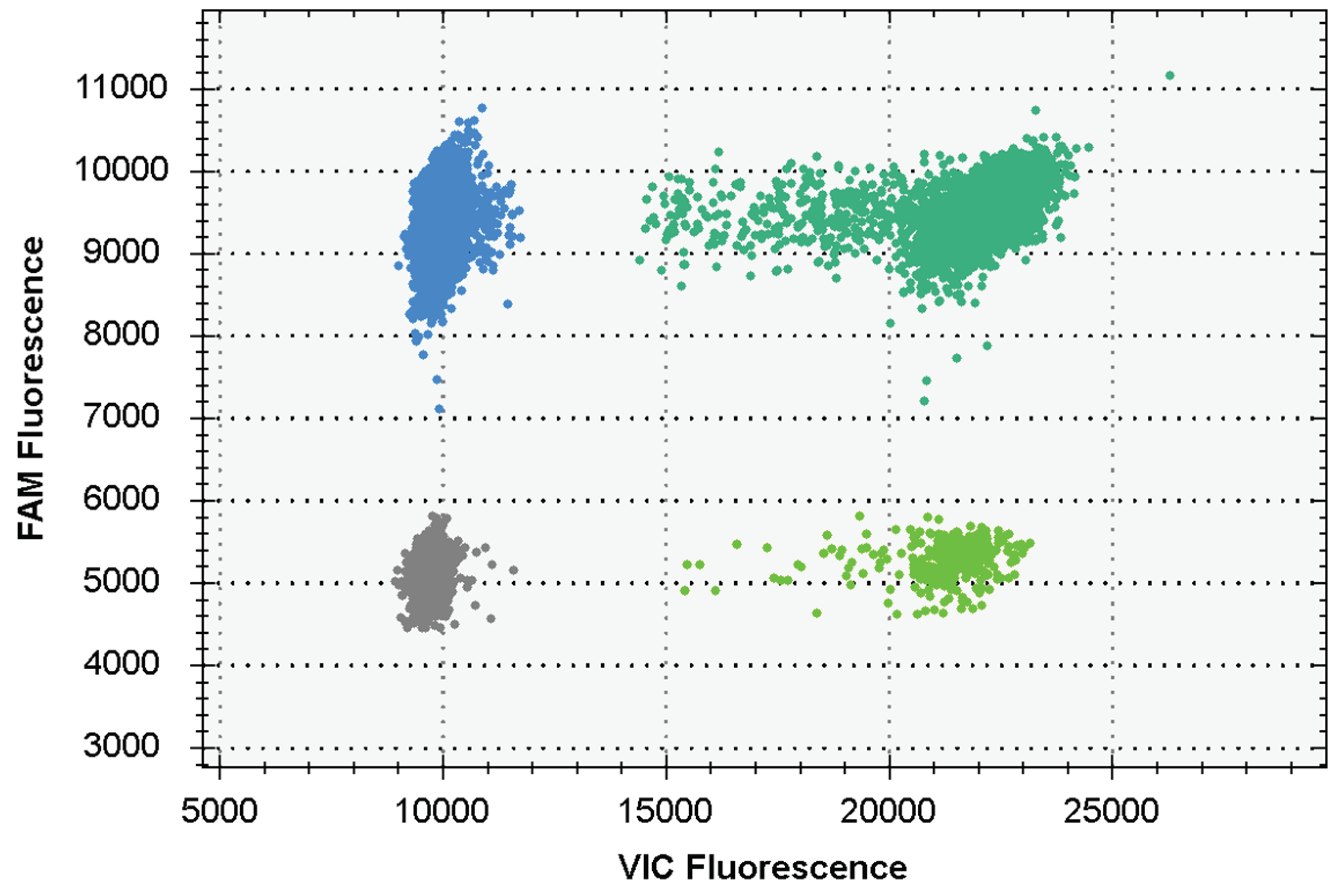
3.2. Evaluation of dddPCR reaction system construction results
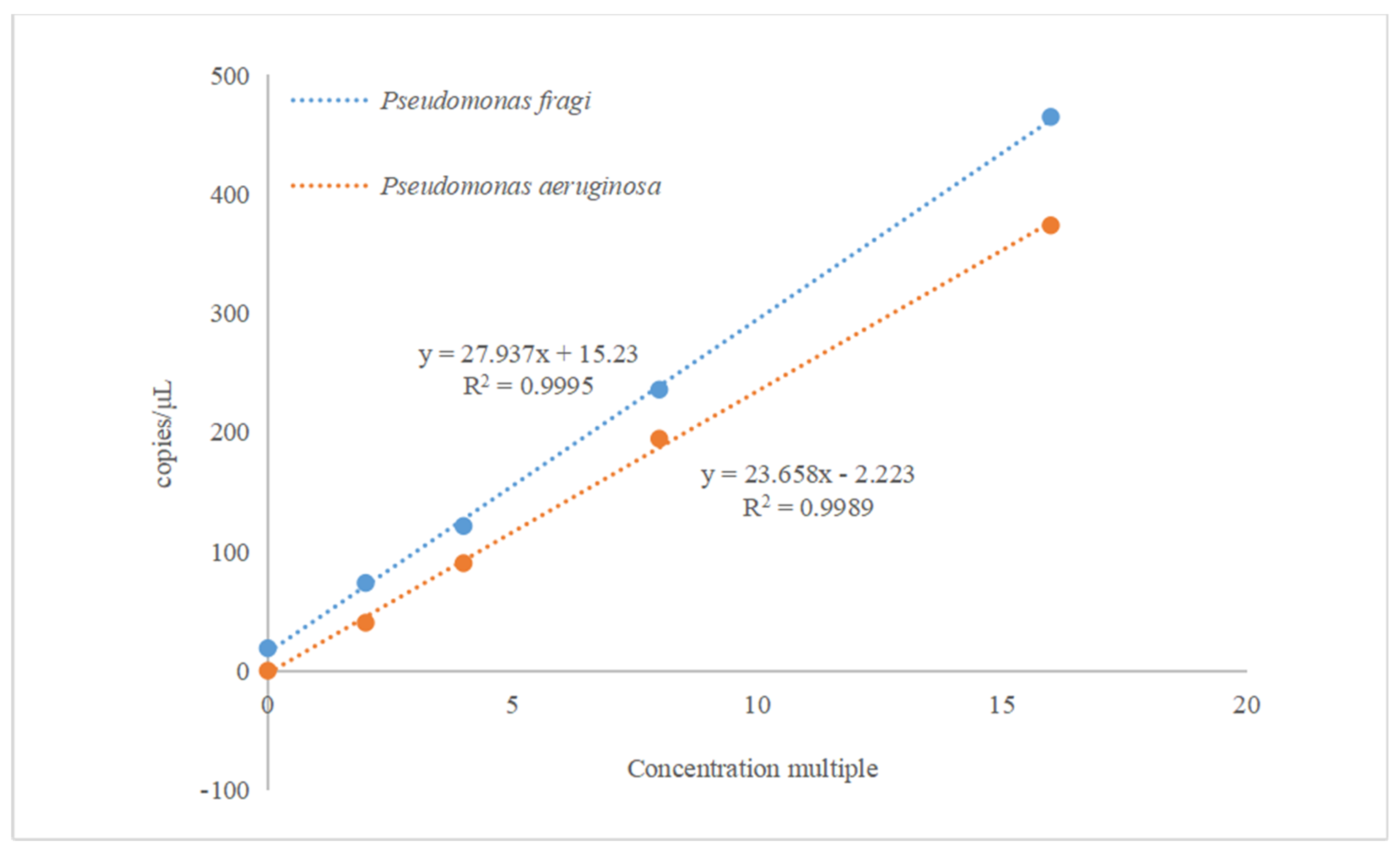
3.3. Linear relationship analysis of the reaction system
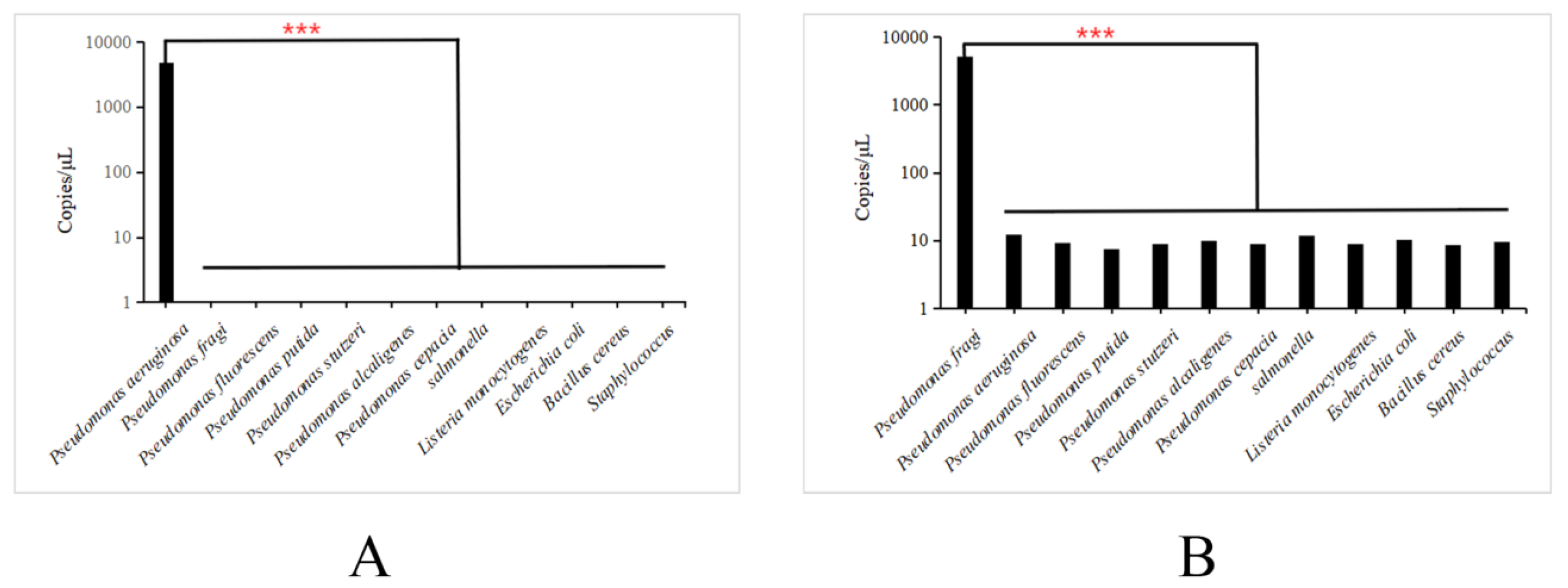
3.4. Analysis of specific primers for dddPCR
3.5. Sensitivity analysis of genome and colony detection by dddPCR
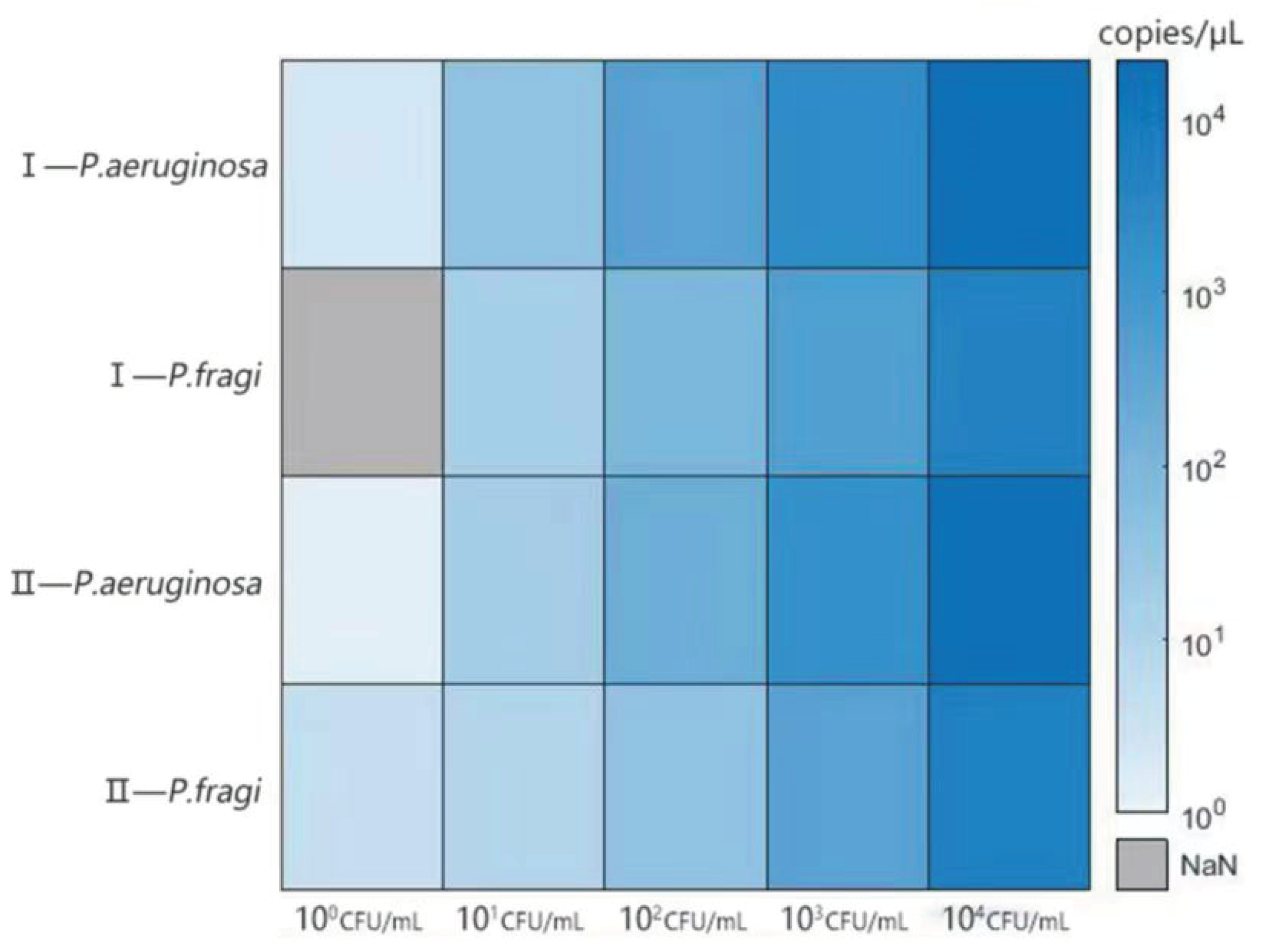
3.6. Analysis of anti-interference ability
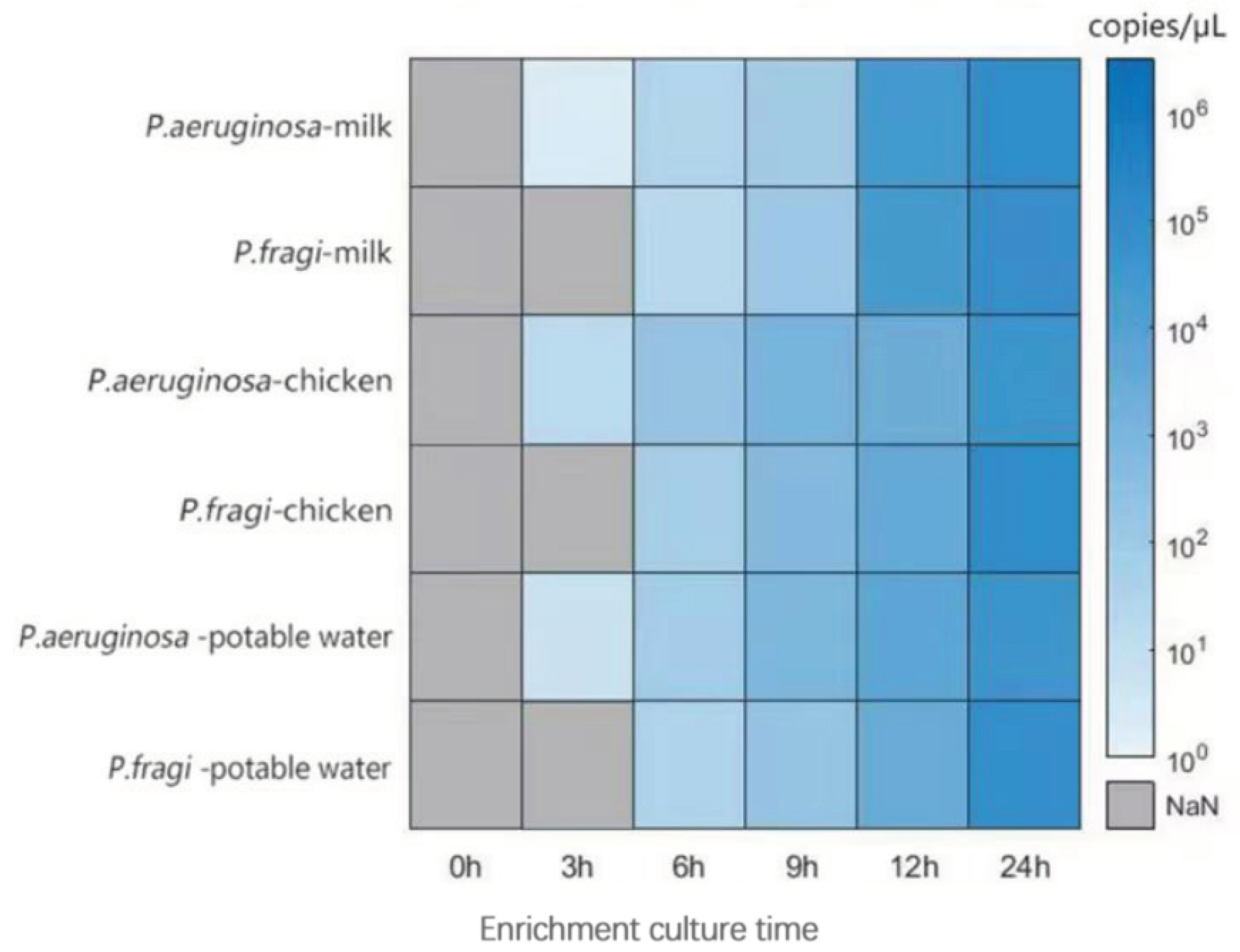
3.7. Analysis of test results of artificially contaminated food
4. Conclusion
Author Contributions
Funding
Data Availability Statement
Acknowledgements
Conflicts of Interest
References
- Tropea, A. Microbial Contamination and Public Health: An Overview. International Journal of Environmental Research and Public Health 2022, 19. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Barranquero, J.A.; Cazorla, F.M.; de Vicente, A. Pseudomonas syringae pv. syringae Associated With Mango Trees, a Particular Pathogen Within the “Hodgepodge” of the Pseudomonas syringae Complex. Frontiers in Plant Science 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman Fiona, S.L. Enhanced annotations and features for comparing thousands ofPseudomonasgenomes in the Pseudomonas genome database. Nucleic Acids Research 2016, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed]
- Bilican, I.; Bahadir, T.; Bilgin, K.; Guler, M.T. Alternative screening method for analyzing the water samples through an electrical microfluidics chip with classical microbiological assay comparison of P. aeruginosa. Talanta 2020, 219. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chen, Y.; Li, M.; Jia, L.; Zhang, L.; Zhu, J. Gelatin-based photonic hydrogels for visual detection of pathogenic Pseudomonas aeruginosa. Sensors and Actuators B: Chemical 2021, 329. [Google Scholar] [CrossRef]
- Huang, S.; Wang, X.; Chen, X.; Liu, X.; Xu, Q.; Zhang, L.; Huang, G.; Wu, J. Rapid and sensitive detection of Pseudomonas aeruginosa by isothermal amplification combined with Cas12a-mediated detection. Scientific Reports 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Garedew, L.; Berhanu, A.; Mengesha, D.; Tsegay, G. Identification of gram-negative bacteria from critical control points of raw and pasteurized cow milk consumed at Gondar town and its suburbs, Ethiopia. BMC Public Health 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Quintieri, Fanelli, Caputo: Antibiotic Resistant Pseudomonas Spp. Spoilers in Fresh Dairy Products: An Underestimated Risk and the Control Strategies. Foods 2019, 8.
- Bloomfield, S.J.; Palau, R.; Holden, E.R.; Webber, M.A.; Mather, A.E. Genomic characterization of Pseudomonas spp. on food: implications for spoilage, antimicrobial resistance and human infection. BMC Microbiology 2024, 24. [Google Scholar] [CrossRef]
- Dong, Q.; Sun, L.; Fang, T.; Wang, Y.; Li, Z.; Wang, X.; Wu, M.; Zhang, H. Biofilm Formation of Listeria monocytogenes and Pseudomonas aeruginosa in a Simulated Chicken Processing Environment. Foods 2022, 11. [Google Scholar] [CrossRef]
- Yang, J.; Liang, R.; Mao, Y.; Dong, P.; Zhu, L.; Luo, X.; Zhang, Y.; Yang, X. Potential inhibitory effect of carbon dioxide on the spoilage behaviors of Pseudomonas fragi in high-oxygen packaged beef during refrigerated storage. Food Microbiol 2023, 112, 104229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, J.; Yuan, Y.; Yue, T. Diversity and characterization of spoilage-associated psychrotrophs in food in cold chain. Int J Food Microbiol 2019, 290, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Quintieri, L.; Caputo, L.; Brasca, M.; Fanelli, F. Recent Advances in the Mechanisms and Regulation of QS in Dairy Spoilage by Pseudomonas spp. Foods 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Chen, S.; Wang, H.; Zhang, J.; Xu, X.; Wang, H. Advances in understanding the predominance, phenotypes, and mechanisms of bacteria related to meat spoilage. Trends in Food Science & Technology 2021, 118, 822–832. [Google Scholar]
- Wang, G.-y.; Wang, H.-h.; Han, Y.-w.; Xing, T.; Ye, K.-p.; Xu, X.-l.; Zhou, G.-h. Evaluation of the spoilage potential of bacteria isolated from chilled chicken in vitro and in situ. Food Microbiology 2017, 63, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Wang, Q.; Liu, J.; Wang, D.; Li, J.; Li, T. Effects of deletion of siderophore biosynthesis gene in Pseudomonas fragi on quorum sensing and spoilage ability. International Journal of Food Microbiology 2023, 396. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, G.; Xue, P.; Dong, X.; Xia, Y.; Regenstein, J.; Du, M.; Sun, L. Spoilage microbes’ effect on freshness and IMP degradation in sturgeon fillets during chilled storage. Food Bioscience 2021, 41. [Google Scholar] [CrossRef]
- Jiang, Y.; Zheng, C.; Jin, M.; Zhou, R.; Wu, Q.; Huang, F.; Lou, Y.; Zheng, L. An Ultrasensitive Colorimetric Foodborne Pathogenic Detection Method Using a CRISPR/Cas12a Mediated Strand Displacement/Hybridization Chain Reaction. J Agric Food Chem 2023, 71, 4193–4200. [Google Scholar] [CrossRef]
- Furet, J.P.; Quenee, P.; Tailliez, P. Molecular quantification of lactic acid bacteria in fermented milk products using real-time quantitative PCR. Int J Food Microbiol 2004, 97, 197–207. [Google Scholar] [CrossRef]
- Li, Z.; Xu, X.; Wang, D.; Jiang, X. Recent advancements in nucleic acid detection with microfluidic chip for molecular diagnostics. TrAC Trends in Analytical Chemistry 2023, 158. [Google Scholar] [CrossRef]
- Hadi, J.; Rapp, D.; Dhawan, S.; Gupta, S.K.; Gupta, T.B.; Brightwell, G. Molecular detection and characterization of foodborne bacteria: Recent progresses and remaining challenges. Comprehensive Reviews in Food Science and Food Safety 2023, 22, 2433–2464. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ye, Q.; Jiang, A.; Zhang, J.; Shang, Y.; Li, F.; Zhou, B.; Xiang, X.; Gu, Q.; Pang, R.; et al. Pseudomonas aeruginosa Detection Using Conventional PCR and Quantitative Real-Time PCR Based on Species-Specific Novel Gene Targets Identified by Pangenome Analysis. Frontiers in Microbiology 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Murugan, N.; Malathi, J.; Therese, K.L.; Madhavan, H.N. Application of six multiplex PCR's among 200 clinical isolates of Pseudomonas aeruginosa for the detection of 20 drug resistance encoding genes. The Kaohsiung Journal of Medical Sciences 2017, 34, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; Casaburi, A.; Nasi, A.; Ferrocino, I.; Di Monaco, R.; Ferranti, P.; Mauriello, G.; Villani, F. Different molecular types of Pseudomonas fragi have the same overall behaviour as meat spoilers. Int J Food Microbiol 2010, 142, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Chen, S.; Zheng, Y.; Zheng, X.; Lin, J.-M. Droplet-based digital PCR (ddPCR) and its applications. TrAC Trends in Analytical Chemistry 2023, 158. [Google Scholar] [CrossRef]
- Zhang, L.; Parvin, R.; Fan, Q.; Ye, F. Emerging digital PCR technology in precision medicine. Biosensors and Bioelectronics 2022, 211. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Zou, Z.; Hu, Z.; Zhang, S.; Zhang, F.; Wang, B.; Lv, S.; Mu, Y. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab on a Chip 2020, 20, 979–986. [Google Scholar] [CrossRef]
- Zhao, G.; Shen, X.; Liu, Y.; Xie, P.; Yao, C.; Li, X.; Sun, Y.; Lei, Y.; Lei, H. Direct lysis-multiplex polymerase chain reaction assay for beef fraud substitution with chicken, pork and duck. Food Control 2021, 129. [Google Scholar] [CrossRef]
- Ruiz-Roldán, L.; Rojo-Bezares, B.; Lozano, C.; López, M.; Chichón, G.; Torres, C.; Sáenz, Y. Occurrence of Pseudomonas spp. in Raw Vegetables: Molecular and Phenotypical Analysis of Their Antimicrobial Resistance and Virulence-Related Traits. International Journal of Molecular Sciences 2021, 22. [Google Scholar] [CrossRef]
- Lee, C.S.; Wetzel, K.; Buckley, T.; Wozniak, D.; Lee, J. Rapid and sensitive detection of Pseudomonas aeruginosa in chlorinated water and aerosols targeting gyrB gene using real-time PCR. Journal of Applied Microbiology 2011, 111, 893–903. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Xue, P.; Zhan, Z.; Huang, Z.; Li, J.; Diao, B.; Kan, B. A duplex droplet digital PCR assay for Salmonella and Shigella and its application in diarrheal and non-diarrheal samples. International Journal of Infectious Diseases 2022, 120, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Li, J.; Yang, H.; Yu, J.; Wei, H.; Ledeboer, N.A. Accurate Detection of Methicillin-Resistant Staphylococcus aureus in Mixtures by Use of Single-Bacterium Duplex Droplet Digital PCR. Journal of Clinical Microbiology 2017, 55, 2946–2955. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Xia, L.; Zou, Z.; Zhuang, J.; Mu, Y. A direct and multiplex digital PCR chip for EGFR mutation. Talanta 2022, 250. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Luo, Y.; Wang, P.; Wu, L.; Cui, X.; Sun, B.; Li, G. Controlled Rehydration of Dried Reagents for Robust Multiplex Digital PCR. Analytical Chemistry 2022, 94, 13223–13232. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Kutsanedzie, F.Y.H.; Sun, H.; Wang, M.; Chen, Q.; Guo, Z.; Wu, J. Rapid Pseudomonas Species Identification from Chicken by Integrating Colorimetric Sensors with Near-Infrared Spectroscopy. Food Analytical Methods 2017, 11, 1199–1208. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Tian, D.; Phan, A.; Seididamyeh, M.; Alanazi, M.; Ping Xu, Z.; Sultanbawa, Y.; Zhang, R. Luminescent sensors for residual antibiotics detection in food: Recent advances and perspectives. Coordination Chemistry Reviews 2024, 498. [Google Scholar] [CrossRef]
- Liu, C.; Lu, C.; Tang, Z.; Chen, X.; Wang, G.; Sun, F. Aptamer-functionalized magnetic nanoparticles for simultaneous fluorometric determination of oxytetracycline and kanamycin. Microchimica Acta 2015, 182, 2567–2575. [Google Scholar] [CrossRef]
- Bolzon, V.; Bulfoni, M.; Pesando, M.; Nencioni, A.; Nencioni, E. Verification of a Rapid Analytical Method for the Qualitative Detection of Listeria spp. and Listeria monocytogenes by a Real-Time PCR Assay according to EN UNI ISO 16140-3:2021. Pathogens 2024, 13. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-Y.; Chen, Z.; Cao, X.; Ross, T.D.; Falbel, T.G.; Burton, B.M.; Venturelli, O.S. Programming bacteria for multiplexed DNA detection. Nature Communications 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Yan, L.; Zheng, X.-c.; Li, L.-z.; Liu, P.; Cao, W.-s. Rapid detection of Pseudomonas aeruginosa by cross priming amplification. Journal of Integrative Agriculture 2020, 19, 2523–2529. [Google Scholar] [CrossRef]
- Qin, M.; Ma, X.; Fan, S.; Wu, H.; Yan, W.; Tian, X.; Lu, J.; Lyu, M.; Wang, S. Rapid detection of Pseudomonas aeruginosa using a DNAzyme-based sensor. Food Science & Nutrition 2021, 9, 3873–3884. [Google Scholar]
- Zhao, T.; Zhang, J.; Han, X.; Yang, J.; Wang, X.; Vercruysse, M.; Hu, H.-Y.; Lei, X. A Pseudopaline Fluorescent Probe for the Selective Detection of Pseudomonas aeruginosa. CCS Chemistry 2021, 3, 2405–2417. [Google Scholar] [CrossRef]
- Wang, M.; Yang, J.; Gai, Z.; Huo, S.; Zhu, J.; Li, J.; Wang, R.; Xing, S.; Shi, G.; Shi, F.; et al. : Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk. International Journal of Food Microbiology 2018, 266, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Kim, J.-H.; Oh, S.-W. Combination of filtration and immunomagnetic separation based on real-time PCR to detect foodborne pathogens in fresh-cut apple. Journal of Microbiological Methods 2022, 201. [Google Scholar] [CrossRef]
- Maier, C.; Hofmann, K.; Huptas, C.; Scherer, S.; Wenning, M.; Lücking, G. Simultaneous quantification of the most common and proteolytic Pseudomonas species in raw milk by multiplex qPCR. Applied Microbiology and Biotechnology 2021, 105, 1693–1708. [Google Scholar] [CrossRef]
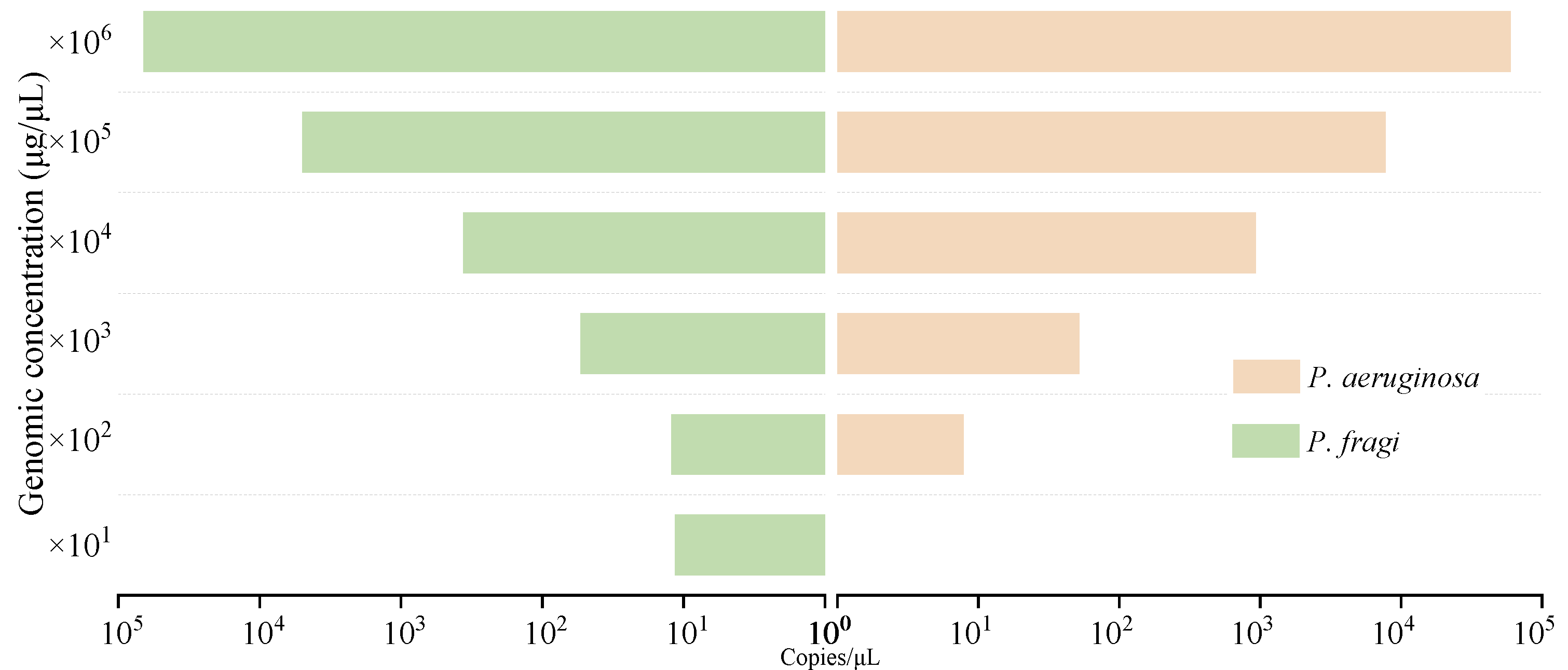
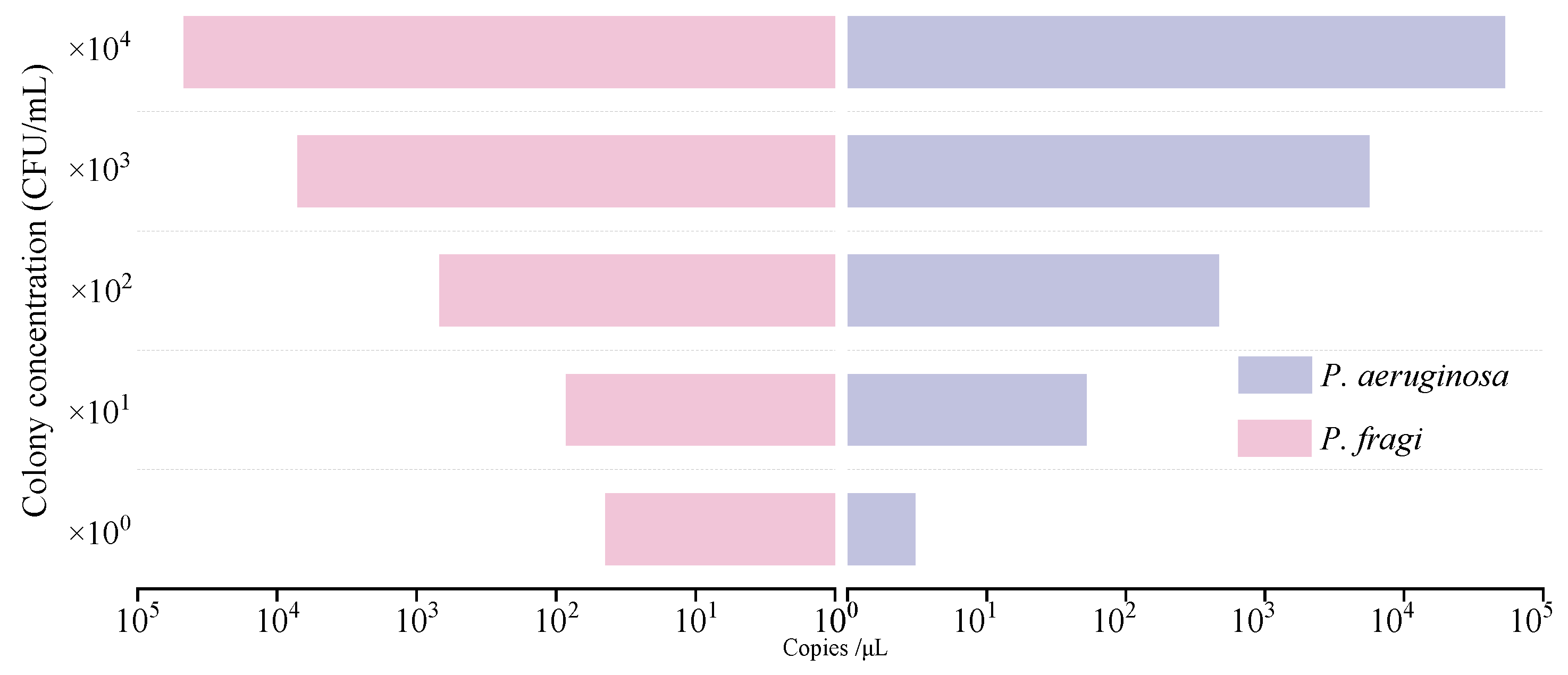
| Source | gene | Annotation | Primer | Sequence(5’-3’) | Source |
|---|---|---|---|---|---|
| P. fragi | RS22665 | Transcriptional regulatory protein | pf1-7 | ATAACGGCAAGAACACCA | In this study |
| CCAAACACGCCTCTGAAC | |||||
| RS22680 | NeuD/PglB/VioB family sugar acetyltransferase | pf1-18 | GGCACAAGTCAATGGTCG | ||
| CACAGTCAGGGCAAGGAT | |||||
| RS10890 | triacylglycerol lipase | Pf3-21 | CCTTGAATGCGCTTAACGCCCTGACCACC | ||
| CGTAGACCCGGTCCAGTAGGCGAGGCTGAT | |||||
| ribA | GTP cyclohydrolase II | Pf3-13 | CGATGTATTCGGGTCCAGACGCTGTGATT | ||
| ATAGTGGTAGTTGTCTTGGGACGGTAGGC | |||||
| P. aeruginosa | LasR | Transcriptional regulatory protein | lasR1 | CGAGAACGCCTTCATCGTCGGCAACTACC | In this study |
| GAAGAACTCGTGCTGCTTTCGCGTCTGGTA | |||||
| gyrB | DNA gyrase subunit B | gyrB2 | ATCCGCACCCTGCTGTTGACCTTCTTCTTCCG | ||
| TGATGTACTGCTCCTGCTTGCCACGCTTGACC | |||||
| rpoB | DNA-directed RNA polymerase beta chain | rpoB3 | TGCCCGATCGAAACCCCTGAAGGTCCGAA | ||
| ATCTCGTCGGTTACCAGGCTGTCCTTGACT |
| Bacterial strains | gene | primer | Sequences of primer(5’-3’) | PCR product | source |
|---|---|---|---|---|---|
| P. aeruginosa | LasR | Pa-2F | AGCCGGGAGAAGGAAGTGTT | 80 bp | In this study |
| Pa-2R | TCCGAGCAGTTGCAGATAACC | ||||
| Pa-2P | VIC-TGCGCCATCGGCAAGACCAGT-BHQ1 | ||||
| P. fragi | RS22680 | Pf-2F | GGCCGGCACGCAAGT | 59 bp | In this study |
| Pf-2R | CTTGGACAGTAGCGAAAAACGA | ||||
| Pf-2P | FAM-TGTCGAGAAGCCAGTCTCCGTGTTCC-BHQ1 |
| Component | Addition |
|---|---|
| 5× MIX | 4.5μL |
| Primer 1-F(10μM) | 1μM |
| Primer 1-R(10μM) | 1μM |
| Primer 2-F(10μM) | 1μM |
| Primer 2-R(10μM) | 1μM |
| Probe1(10μM) | 0.25μM |
| Probe2(10μM) | 0.25μM |
| ROX dye | 0.3μL |
| Enzyme | 0.2μL |
| Template 1 | 1μL |
| Template 2 | 1μL |
| Complemented by water to | 15μL |
| Bacterial strains | Source | Results | ||||||
|---|---|---|---|---|---|---|---|---|
| LasR | rpoB | gyrB | RS22665 | RS22680 | RS10890 | ribA | ||
| Pseudomonas fragi | SHBCC D24613 | - | - | - | + | + | + | + |
| Pseudomonas fragi | CGMCC1.3349 | - | - | - | + | + | + | + |
| Pseudomonas fragi | Laboratory isolates | - | - | - | + | + | + | - |
| Pseudomonas aeruginosa | ATCC 15442 | + | + | + | - | - | - | - |
| Pseudomonas aeruginosa | ATCC 27853 | + | + | + | - | - | - | - |
| Pseudomonas aeruginosa | DSM 939 | + | + | + | - | - | - | - |
| Pseudomonas aeruginosa | Laboratory isolates | + | + | + | - | - | - | - |
| Pseudomonas fluorescens | ATCC 13525 | - | - | - | + | - | + | - |
| Pseudomonas putida | ATCC 49128 | - | + | - | - | - | - | + |
| Pseudomonas pseudoalaligenes | CGMCC1.10611 | - | - | - | - | - | - | - |
| Pseudomonas mendocina | ATCC 25411 | - | + | - | - | - | - | + |
| Pseudomonas stutzeri | ATCC 17588 | - | - | - | - | - | - | - |
| Pseudomonas alcaligenes | ATCC 14909 | - | - | - | - | - | - | - |
| Pseudomonas cepacia | SHBCC D 14769 | - | - | - | - | - | - | - |
| Pseudomonas putida | ATCC 17485 | - | - | - | - | - | - | - |
| Pseudomonas fluorescens | GIM1.110 | - | - | - | - | - | + | - |
| Pseudomonas fluorescens | ATCC 17397 | - | - | + | - | - | - | - |
| Staphylococcus | CICC 10788 | - | - | - | - | - | - | - |
| Enterococcus avium | ATCC 14025 | - | - | - | - | - | - | - |
| Bacillus pumilus | CMCC 63202 | - | - | - | - | - | - | - |
| Listeria monocytogenes | CICC 21622 | - | - | - | - | - | - | - |
| salmonella enterica | CICC 21482 | - | - | - | - | - | - | - |
| Cronobacter sakazakii | CICC 21560 | - | - | - | - | - | - | - |
| Cronobacter universalis | NCTC 9529 | - | - | - | - | - | - | - |
| salmonella anatum | CICC 21498 | - | - | - | - | - | - | - |
| Escherichia coli | ATCC 25922 | - | - | - | - | - | - | + |
| Bacillus cereus | CICC 23384 | - | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





