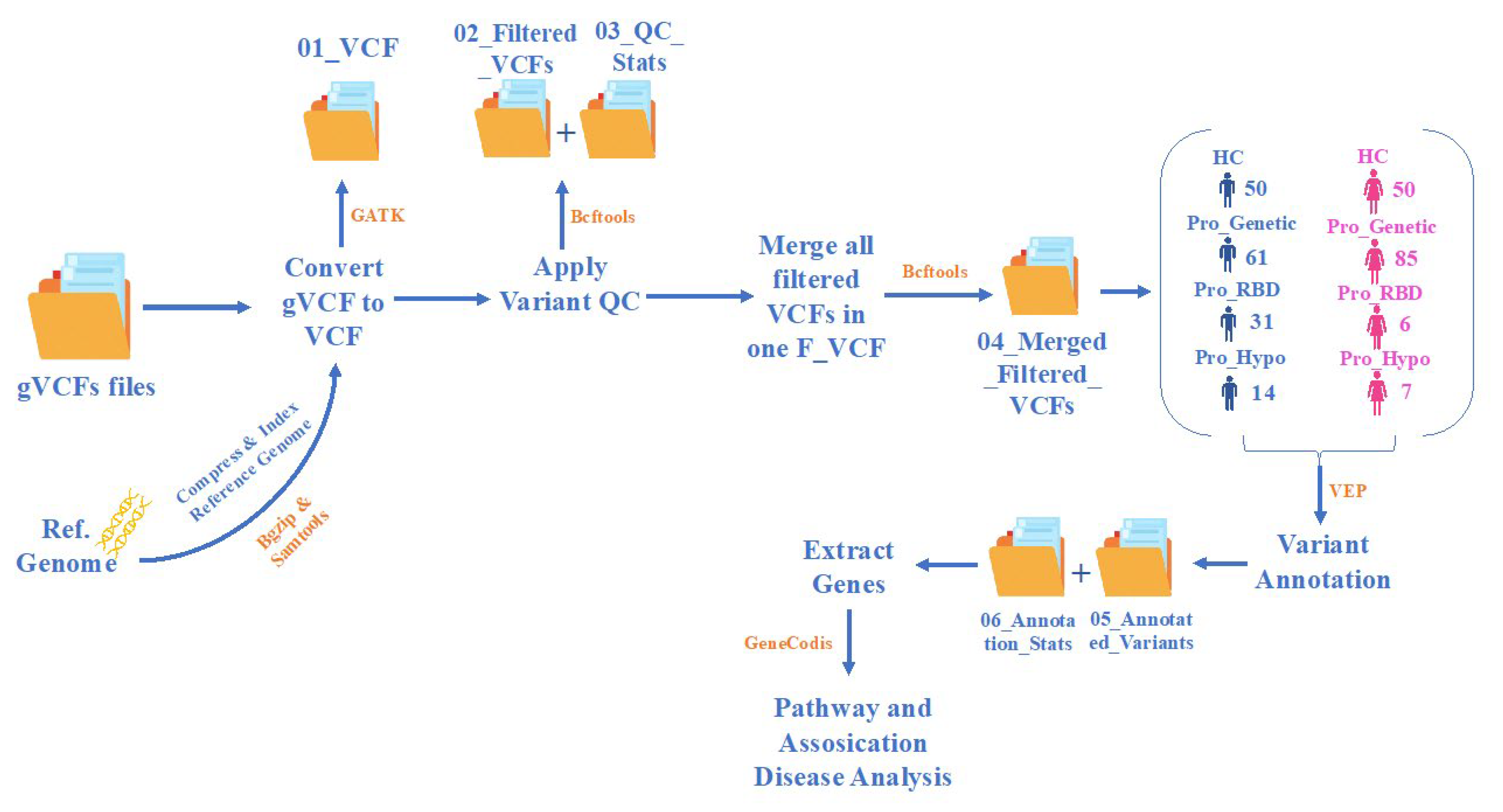
Introduction
Parkinson’s disease (PD) is a neurodegenerative disease that is pathologically defined as the death of dopaminergic neurons in the midbrain and the inclusion of Lewy bodies in the brain [
1]. It has become obvious that PD has a prodromal stage, which is the period before the beginning of neurodegeneration without detecting motor signs by classical diagnosis. The basis of the nonmotor prodromal stage is that the pathological process could not yet occur in the substantia nigra pars compacta (SNpc) [
2]. The classical diagnosis of PD relies on the loss of mesodiencephalic dopaminergic (mdDA) neurons in the substantia nigra pars compacta (SNpc) and the development of Lewy bodies in some surviving neurons [
3].
Early investigations focused on the role of genetic factors in Parkinson’s disease to identify rare mutations linked to familial disease [
4]. Moreover, the past decade has shown the great role of genetics in sporadic disease [
5]. The identification of novel variants and genes for the early diagnosis of the prodromal and/or PD stages is receiving increased amounts of attention [
6]. Variant Call Format (VCF) is becoming a community standard for reporting variations in genetic data acquired from medical genetic diagnostics and research [
7].
In this study, we analyzed gVCF data acquired from the Parkinson’s Progression Markers Initiative (PPMI) for healthy participants and prodromal PD patients. The prodromal PD subgroups involved in this study were genetic, RBD, and hyposmia. Sleep behavior disorder (RBD), rapid eye movement (REM), is a parasomnia condition characterized by complex abnormal motor movements in the REM state during sleep [
8]. RBD is mainly associated with abnormal movement behaviors, nightmares, and loss of normal skeletal atonia in the REM state [
9]. Moreover, hyposmia is an olfactory dysfunction that leads to a loss of smell ability and is the most common nonmotor symptom of PD [
10].
This study was based on annotation, biological pathway, and disease association analyses of prodromal patients with PD. We detected the percentages and types of variation in each population with their percentile on each chromosome. The gene lists were generated and are presented as gene-annotation network clusters and bar charts, which enabled us to detect novel genes with counts in prodromal stages identified recently in PD patients, as illustrated in
Table 2.
High Percentage of Intronic and Intergenic Variants
The Ensembl Variant Effect Predictor (VEP) is a powerful tool for analyzing genomic variation in coding and noncoding regions. It also provides access to a very extensive collection of genomic annotations [
15].
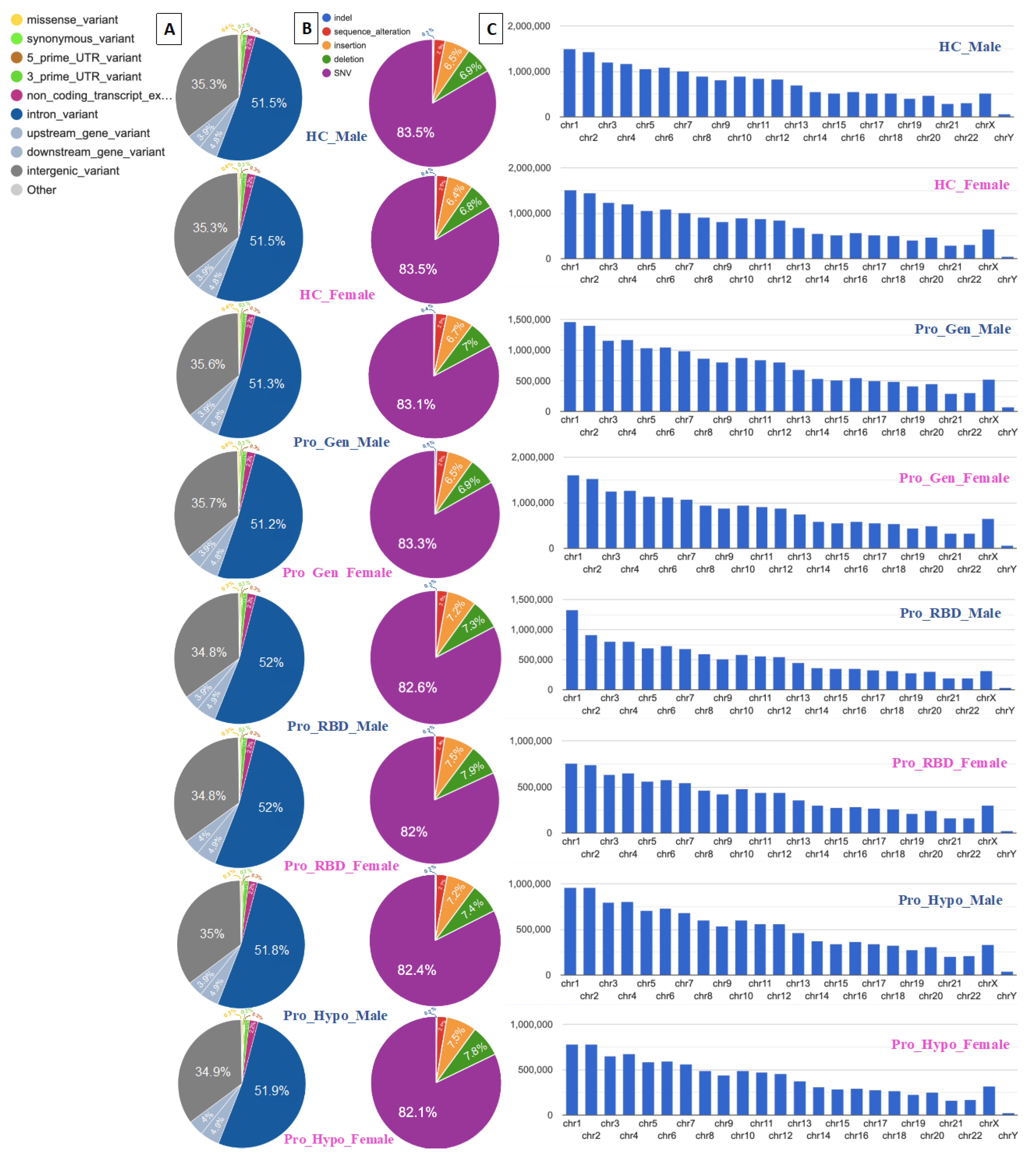
In this study, VEP was used for annotation analyses and detection of genomic variations and their associations. The highest two percentages of variation were detected for intronic and intergenic variants, as illustrated in
Figure 3A. Similarly, 51.5% and 35.3% of the healthy control male and female populations, respectively, exhibited the same percentages of intron variants and intergenic variants. Among those with a prodromal genetic male population, 51.3% had intron variants and 35.6% had intergenic variants, while among those with a prodromal genetic female population, 51.2% had intron variants and 35.7% had intergenic variants. Prodromal RBD males and females had the same percentages of intron variants and intergenic variants (52% and 34.8%, respectively). Among the males with prodromal hyposmia, 51.8% had intron variants and 35% had intergenic variants; moreover, among the females with prodromal hyposmia, 51.9% had intron variants and 34.9% had intergenic variants.
The detection of variants in intronic and intergenic regions is common across the entire human genome, as these noncoding regions make up half of the human noncoding genome and can play important regulatory roles [
16]. The presence of intronic and intergenic variants in the studied healthy population and prodromal populations suggests that these variants are not specifically associated with prodromal PD. However, these findings likely represent background genetic variation.
A single-nucleotide variant (SNV), also called a single-nucleotide polymorphism (SNP), is a variant of a specific single nucleotide and occurs at a specific position in the genome. Moreover, SNVs are the most common type of genetic variation [
17]. This clearly explains the high percentages of SNVs detected in all the healthy and prodromal populations presented in
Figure 3B. All the populations presented ≥ 82% of the SNVs.
Deletion mutations were detected as the second highest number of elements after SNVs. The two highest percentages of deletions were detected in the RBD females and hyposmia females (7.9% and 7.8%, respectively). These deletion mutations may be associated with genetic factors involved in the development of hyposmia and RBD in individuals, specifically females. Moreover, these deletions could contain genes or regulatory regions relevant to olfactory function and sleep regulation. A sex difference could be related to the association between high deletion percentages and prodromal symptoms in females. This could suggest potential sex-specific genetic risk factors for PD. Notably, all female participants in this study had a Y chromosome. It is suspected to be due to androgen insensitivity syndrome (AIS), which is characterized by evidence of feminization of the external genitalia at birth and abnromal sexual development with a 46,XY karyotype [
18]. Consequently, the two most common insertion mutations were also detected in 7.9% of the RBD females and 7.8% of the Hyposmia females, as shown in
Figure 2b. The detection of insertions in the RBD and Hyposmia populations suggested genetic variability within these groups. These high percentages of insertions could be associated with disease susceptibility or progression. Additionally, depending on their location within the genome, these insertions can have various functional consequences.
Chromosomes 1 and 2 are among the largest chromosomes in the human genome and contain a large number of genes and regulatory elements [
19]. Therefore, these genes may represent a greater number of variants simply because of their size and gene density. High percentages of variants were detected in all the analyzed samples, including those of the healthy controls. Healthy control males had variant counts of 1,500,086 on chr1 and 1,423,662 on chr2, while healthy control females had variant counts of 1,509,369 on chr1 and 1,441,362 on chr2 (
Figure 3C). Consequently, it could be normal to find these variants in prodromal populations. However, chromosomes 1 and 2 may influence biological processes relevant to PD, such as mitochondrial function, protein aggregation, the oxidative stress response, and neuroinflammation. Understanding how variants in these genomic regions affect molecular pathways associated with PD is crucial for providing insights into disease mechanisms. Gene-annotation network cluster and pathway analyses are shown in
Figure 4Figure 5 and
Figure 6.
Table 1.
General statistics of the variation analysis results for the healthy control (HC) (male and female), prodromal PD (genetic male and female), prodromal PD (RBD male and female), and prodromal PD (hyposmia male and female) populations.
Table 1.
General statistics of the variation analysis results for the healthy control (HC) (male and female), prodromal PD (genetic male and female), prodromal PD (RBD male and female), and prodromal PD (hyposmia male and female) populations.
| Population |
No. of samples in Population |
Lines of input read |
Processed Variants |
Novel/Existing Variants |
Overlapped Genes |
Overlapped Transcripts |
| HC_Male |
50 |
18191327 |
18191327 |
168623 (0.9%)/18022704 (99.1%) |
61987 |
250277 |
| HC_Female |
50 |
18387115 |
18387115 |
182458 (1.0%)/18204657 (99.0%) |
61663 |
249831 |
| Pro_Gen_Male |
61 |
17763968 |
17763968 |
179814 (1.0%)/17584154 (99.0%) |
62009 |
250316 |
| Pro_Gen_Female |
85 |
19371326 |
19371326 |
228369 (1.2%)/19142957 (98.8%) |
61655 |
249834 |
| Pro_RBD_Male |
31 |
12264624 |
12264624 |
63331 (0.5%)/12201293 (99.5%) |
61898 |
250173 |
| Pro_RBD_Female |
6 |
9568588 |
9568588 |
40093 (0.4%)/9528495 (99.6%) |
61542 |
249619 |
| Pro_Hypo_Male |
14 |
12081647 |
12081647 |
69229 (0.6%)/12012418 (99.4%) |
61924 |
250185 |
| Pro_Hypo_Female |
7 |
9942790 |
9942790 |
29510 (0.3%)/9913280 (99.7%) |
61565 |
249627 |
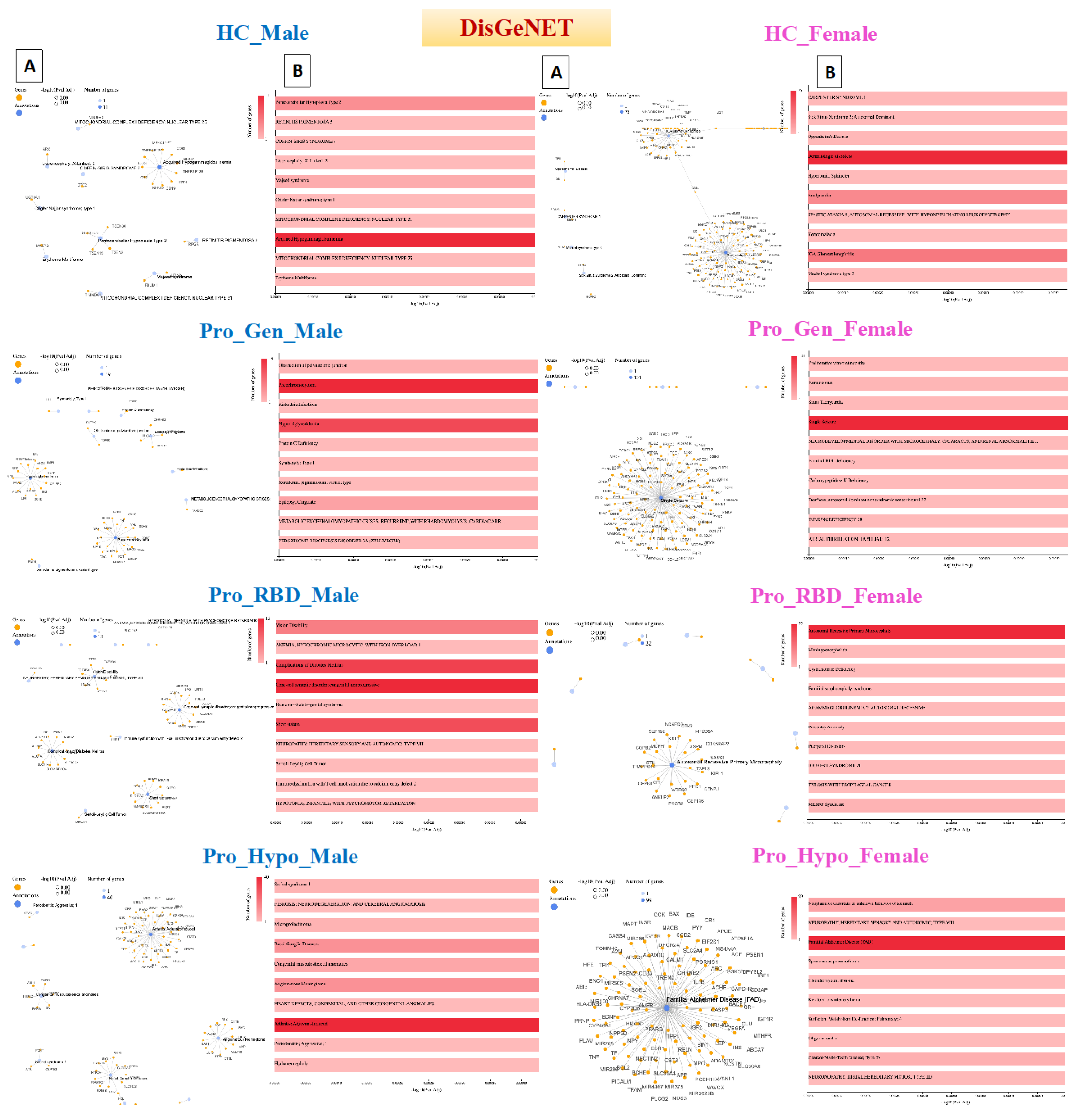
Disease Gene Network (DisGeNET) detection in prodromal PD populations
The disease gene network, known as DisGeNET, is a comprehensive knowledge base that integrates information on human disease-associated genes and variants from multiple sources [
20]. This database was accessed through the GeneCodis website, and annotation was carried out through this tool [
21].
Acquired hypogammaglobulinemia was detected in the heathy male population, in which 11 genes were associated with this disease. Acquired hypogammaglobulinemia is also known as secondary hypogammaglobulinemia and is a condition characterized by low levels of immunoglobulins (antibodies) in the blood. This condition can increase the risk of infections and can occur due to various factors, such as certain medications, underlying medical conditions, or environmental exposures [
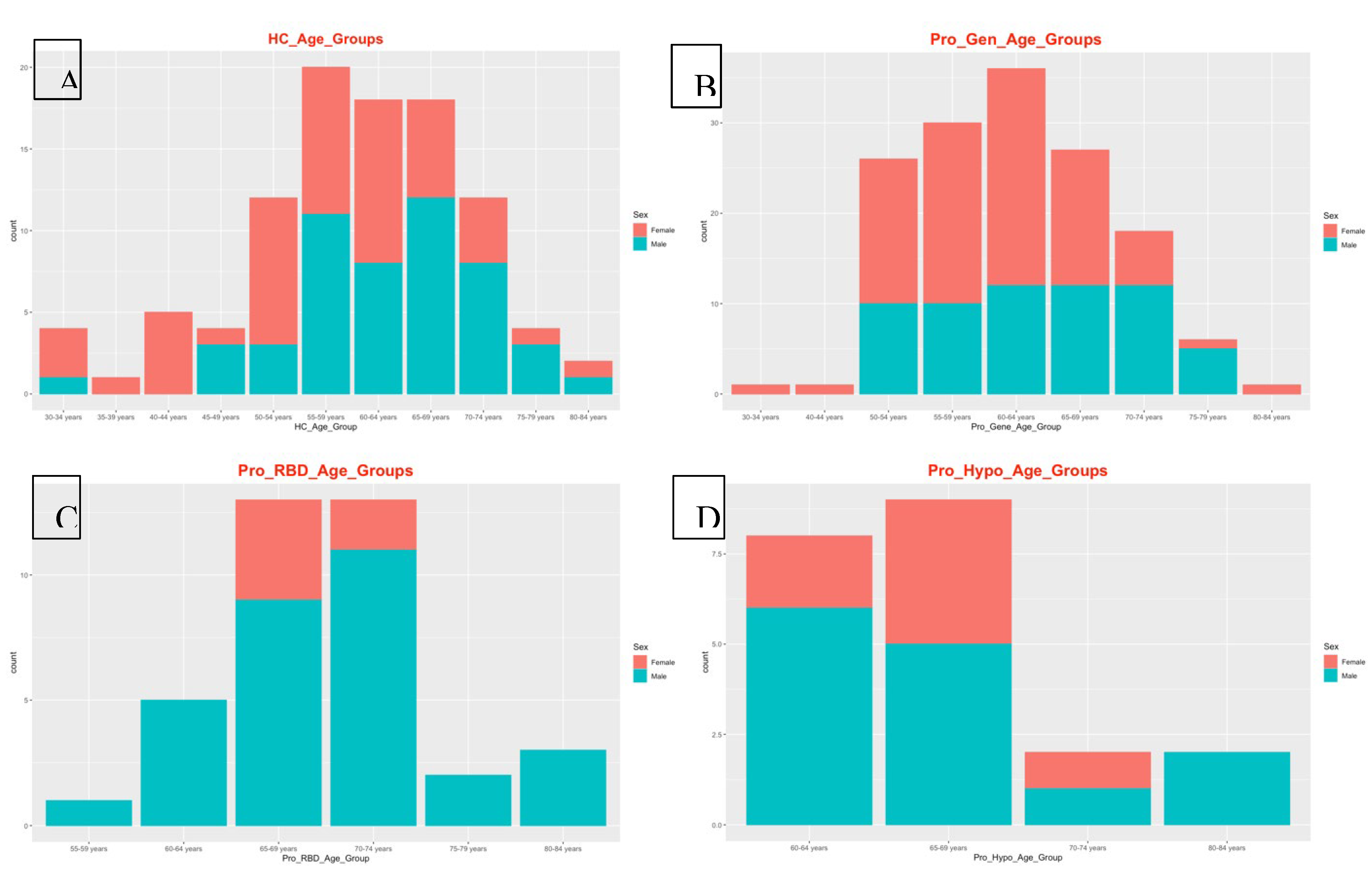
22]. Therefore, the detection of this disease in the healthy male population could be due to the age of the participants, as 49 participants were aged ≥ 45 years, as shown in
Figure 4. On the other hand, 75 genes were detected to be related to dermatological disorders in the healthy female population. Many dermatological disorders, including eczema, psoriasis, acne, and others, involve multiple genes and have complex genetic architectures. Variations in these genes can influence susceptibility to these conditions, and the involvement of 75 genes may indicate a polygenic inheritance pattern. Each gene may have a small effect on the overall risk of developing dermatological disorders. Environmental factors also contribute to disease susceptibility, and exposures to allergens, irritants, pollutants, UV radiation, and microbial agents can interact with genetic predispositions to trigger or exacerbate skin conditions.
Pheochromocytoma and hypertriglyceridemia were detected in the prodromal genetic male population with 19 and 15 genes, respectively. Pheochromocytoma (PCC) is a rare neuroendocrine tumor that arises from the adrenal glands and can also occur elsewhere in the sympathetic nervous system. It is characterized by the excessive production of catecholamines, such as noradrenaline and adrenaline, which can cause a variety of symptoms, including palpitations, hypertension, headache, anxiety, and sweating [
23]. The detection of pheochromocytoma in the prodromal genetic PD population is rare and unusual but possible. While there is no known direct genetic link between PCC and PD, it is important to note that both conditions can be influenced by genetic predispositions, environmental factors, and complex interactions between various biological pathways. Additionally, several genes associated with PD may have other roles in different cellular processes beyond the central nervous system.
Hypertriglyceridemia is an elevated level of triglycerides in the blood and is a lipid abnormality associated with an increased risk of cardiovascular disease [
24]. Hypertriglyceridemia is primarily influenced by lifestyle factors such as physical activity, diet, and obesity. Genetic factors can also play a role in lipid metabolism and contribute to elevated triglyceride levels. Investigating potential shared genetic predispositions between HTG and PD may provide insights into overlapping biological pathways or susceptibility genes. Furthermore, dysregulation of lipid metabolism and glucose homeostasis has been implicated in the pathogenesis of PD. Emerging evidence suggests potential links between metabolic dysfunction, insulin resistance, and neurodegeneration in PD patients. Detecting hypertriglyceridemia in individuals with prodromal genetic PD in the male population may raise questions about underlying metabolic disturbances and their implications for disease progression.
Among the prodromal genetic female population, 101 patients had a single seizure. These seizures can be triggered by fever (febrile seizures), head injury, metabolic disturbances, sleep deprivation, stress, alcohol, or drug withdrawal [
25]. While PD primarily affects dopaminergic neurons in the brain, there is evidence to suggest that individuals with PD, or in the prodromal stage, may have an increased susceptibility to seizures compared to the general population. Genetic factors, including mutations in genes associated with both PD and epilepsy, could contribute to this increased risk. The involvement of 101 genes may indicate a polygenic or multifactorial basis for the seizure phenotype, with variations in multiple genes contributing to the risk of seizures.
Cone-rod synaptic disorder (CRSD) was detected in 13 genes of the prodromal RBD male population. CRSD is a rare genetic disorder characterized by dysfunction of the synaptic connections between cone and rod photoreceptor cells in the retina. This leads to visual impairment, particularly affecting color vision, central vision, and visual acuity [
26]. Additionally, RBD is a rapid eye movement behavior disorder characterized by the loss of muscle atonia during REM sleep, leading to the enactment of dreams through vocalizations and movements. While CRSD primarily affects the retina, several genes associated with retinal function may have broader roles in neuronal health and function. Variants in these genes could contribute to neurodegenerative processes in conditions such as PD.
Autosomal recessive primary microcephaly (MCPH) was detected in the prodromal RBD female population with 22 genes. MCPH is a rare neurodevelopmental disorder characterized by significantly reduced head size (microcephaly) and intellectual disability. It is inherited in an autosomal recessive manner, meaning that both copies of the affected gene (one from each parent) must be mutated for the condition to manifest [
27]. While MCPH primarily affects brain development, several genes associated with neurodevelopmental disorders may have broader roles in neuronal health and function. Genetic variations in these genes may contribute to neurodegenerative processes in conditions such as prodromal RBD.
Adjuvant arthritis was detected in a prodromal hyposmia male population with 40 genes. In humans, a type of reactive arthritis occurs when the immune system reacts to a triggering event, such as an infection or exposure to certain substances. It typically presents with joint pain, swelling, and stiffness, similar to other forms of inflammatory arthritis. PD and autoimmune disorders such as rheumatoid arthritis (RA) have distinct etiologies, and there is a growing recognition of shared genetic susceptibility and environmental factors that may contribute to both conditions. However, when Arthritis was detected, adjuvant-induced Arthritis in a male population with prodromal hyposmia involving 40 genes suggested a complex interplay of genetic and environmental factors. More importantly, this arthritis occurs at older ages, and all the hyposmic male patients were aged ≥ 60 years. On the other hand, familial Alzheimer disease (FAD) was detected in 99 prodromal hyposmia females. FAD is associated with mutations in specific genes, including amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) [
28]. These mutations are typically inherited in an autosomal dominant manner, meaning that a single copy of the mutated gene is sufficient to cause the disease. While some genetic mutations may be associated with both Alzheimer's disease and Parkinson's disease, detecting FAD in a prodromal hyposmia female population involving 99 genes would need further in-depth investigations.
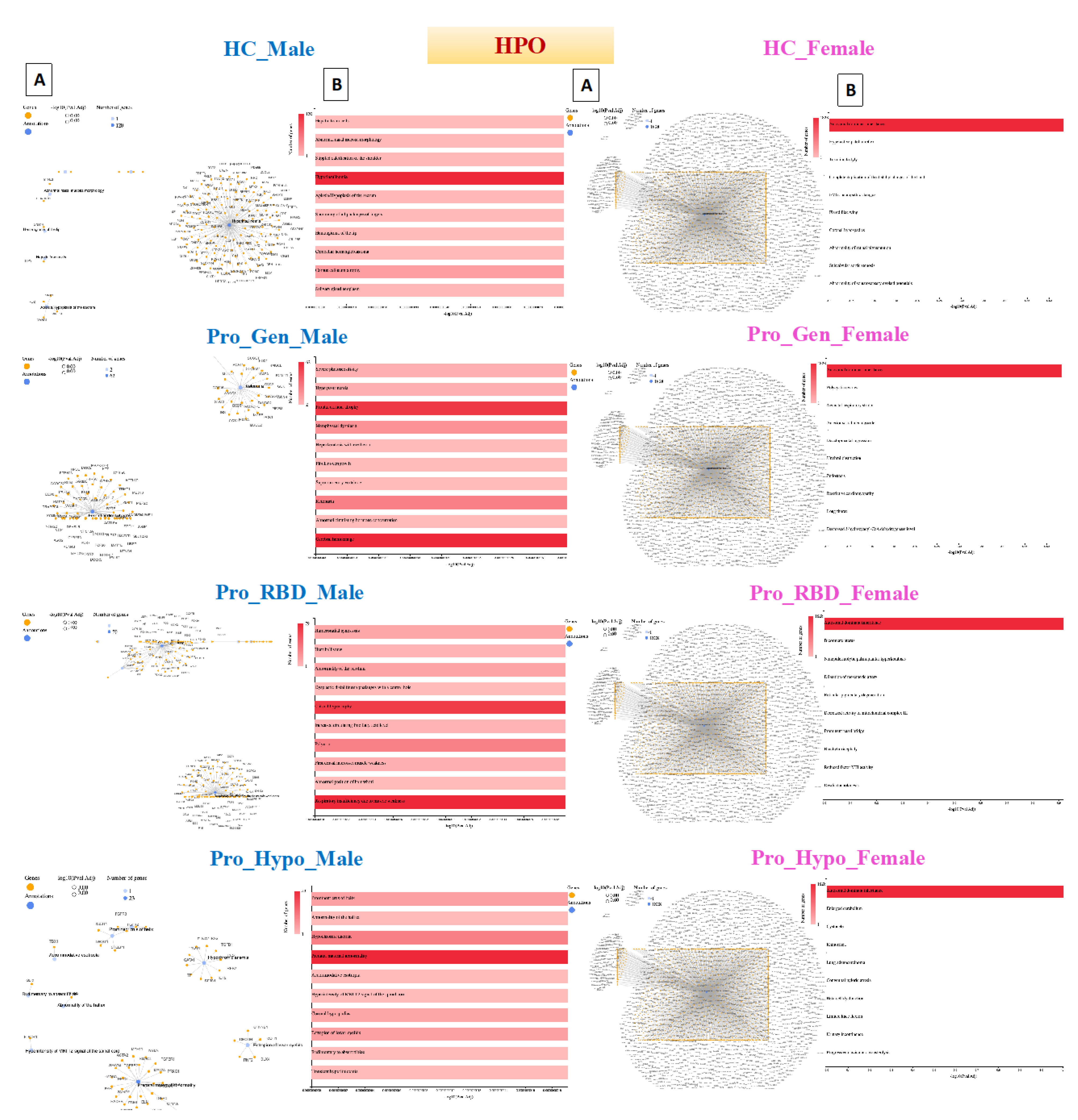
Detection of Human Phenotype Ontology (HPO) Data in Prodromal PD Populations
The HPO dataset is the Human Phenotype Ontology, which consists of phenotypic abnormalities encountered in human disease [
29].
Hyperinsulinemia was detected in a healthy male population with 120 genes. Hyperinsulinemia is a condition characterized by higher-than-normal levels of insulin in the blood. Insulin is a hormone produced by pancreatic bet cells that helps regulate glucose levels by facilitating the uptake of glucose into cells for energy or storage [
30]. The detection of hyperinsulinemia in the male population of HCs may suggest underlying metabolic abnormalities or insulin resistance. Moreover, hyperinsulinemia can occur as a compensatory response to insulin resistance and can be influenced by various factors, such as diet, physical activity, genetics, and medications. This could also be justified by the older ages of the healthy male population, as they are ≥ 45 years old
.
The HPO results revealed that all female populations, including the healthy population, had the same gene network of autosomal dominant inheritance, with 1828 genes being involved. Autosomal dominant inheritance is a pattern of inheritance for a trait or disorder determined by genes located on autosomal chromosomes (nonsex chromosomes). In other words, a single copy of the mutated gene, inherited from one parent, is sufficient to express the trait or disorder. This means that individuals who inherit the mutated gene from either parent will exhibit the trait or disorder. Examples of disorders with autosomal dominant inheritance include Parkinson's disease, Huntington's disease, familial hypercholesterolemia, Marfan syndrome, neurofibromatosis type 1, and some other forms of familial Alzheimer's disease [
31]. As mentioned in the chromosome analysis, all female samples had a Y chromosome (
Figure 3C).
Cerebral hemorrhage was detected in a prodromal genetic male population with 62 genes. Cerebral hemorrhage is a medical condition characterized by bleeding within brain tissues. It occurs when a blood vessel within the brain ruptures, leading to the leakage of blood into the surrounding brain tissue. This bleeding can cause damage to brain cells and disrupt normal brain function [
32]. In general, cerebral hemorrhage is not a common feature of prodromal PD. However, its detection in the prodromal genetic PD male population suggests a potential overlap or interaction between genetic factors predisposing patients to PD and cerebrovascular events.
Respiratory insufficiency due to muscle weakness was detected in 79 genes in the prodromal RBD male population. In this condition, the muscles involved in breathing are unable to adequately perform their function, leading to impaired respiratory function. This can occur because of various underlying causes, including neurological conditions, neuromuscular disorders, or muscular dystrophies [
33]. Detecting respiratory insufficiency due to muscle weakness in the prodromal RBD PD male population with 79 genes may suggest genetic predispositions or variants associated with neuromuscular or respiratory function. Moreover, respiratory insufficiency in PD patients is more commonly associated with factors such as upper airway obstruction, aspiration pneumonia, or respiratory muscle rigidity.
Prenatal maternal abnormalities were detected in the prodromal hyposmia male population with 23 genes. Prenatal maternal abnormalities are not linked to maternal health, but they may also occur because of genetic conditions or mutations carried by the father, which can be transmitted to the fetus and influence prenatal development and health outcomes. Epigenetic modifications, such as DNA methylation patterns or histone modifications, can reflect prenatal environmental exposures or maternal health conditions [
34]. Epigenetic changes could influence gene expression and neurodevelopmental processes relevant to PD risk.
Novel Gene Detection in Prodromal PD Populations
Interestingly, 12 potentially novel PD loci recently detected by Kim [
38] were found to be present in prodromal populations. The 12 potentially novel loci were MTF2, PIK3CA, ADD1, SYBU, IRS2, USP8, PIGL, FASN, MYLK2, USP25, EP300 and PPP6R2.
Table 2 shows the 12 novel genes and their counts in each prodromal population. This detection could lead to the use of this gene as a potential genetic biomarker for the early detection of prodromal PD patients.
Table 2.
Recently, 12 potentially novel PD genes were detected in the populations with prodromal PD (genetic male and female), prodromal PD (adolescent and female), and prodromal PD (hyposmia male and female).
Table 2.
Recently, 12 potentially novel PD genes were detected in the populations with prodromal PD (genetic male and female), prodromal PD (adolescent and female), and prodromal PD (hyposmia male and female).
| |
Population |
Pro_Gen_Male
Gene Count |
Pro_Gen_Female
Gene Count |
Pro_RBD_Male
Gene Count |
Pro_RBD_Female
Gene Count |
Pro_Hypo_Male
Gene Count |
Pro_Hypo_Female
Gene Count |
| Gene Name |
|
| MTF2 |
2985 |
3458 |
3065 |
1397 |
2035 |
1606 |
| ADD1 |
9095 |
10457 |
5535 |
3471 |
4404 |
3722 |
| PIK3CA |
2958 |
3072 |
1874 |
1390 |
1606 |
1830 |
| SYBU |
13897 |
15607 |
7991 |
5943 |
8012 |
6030 |
| IRS2 |
237 |
291 |
192 |
138 |
210 |
147 |
| USP8 |
5997 |
6524 |
4432 |
3358 |
4953 |
1953 |
| PIGL |
8277 |
9021 |
4350 |
2819 |
3902 |
2923 |
| FASN |
1184 |
1224 |
691 |
552 |
775 |
593 |
| MYLK2 |
265 |
296 |
183 |
132 |
111 |
107 |
| USP25 |
4081 |
4952 |
2672 |
753 |
2796 |
534 |
| EP300 |
3146 |
3510 |
1704 |
1256 |
1790 |
1398 |
| PPP6R2 |
5652 |
6393 |
3090 |
2693 |
3723 |
2561 |