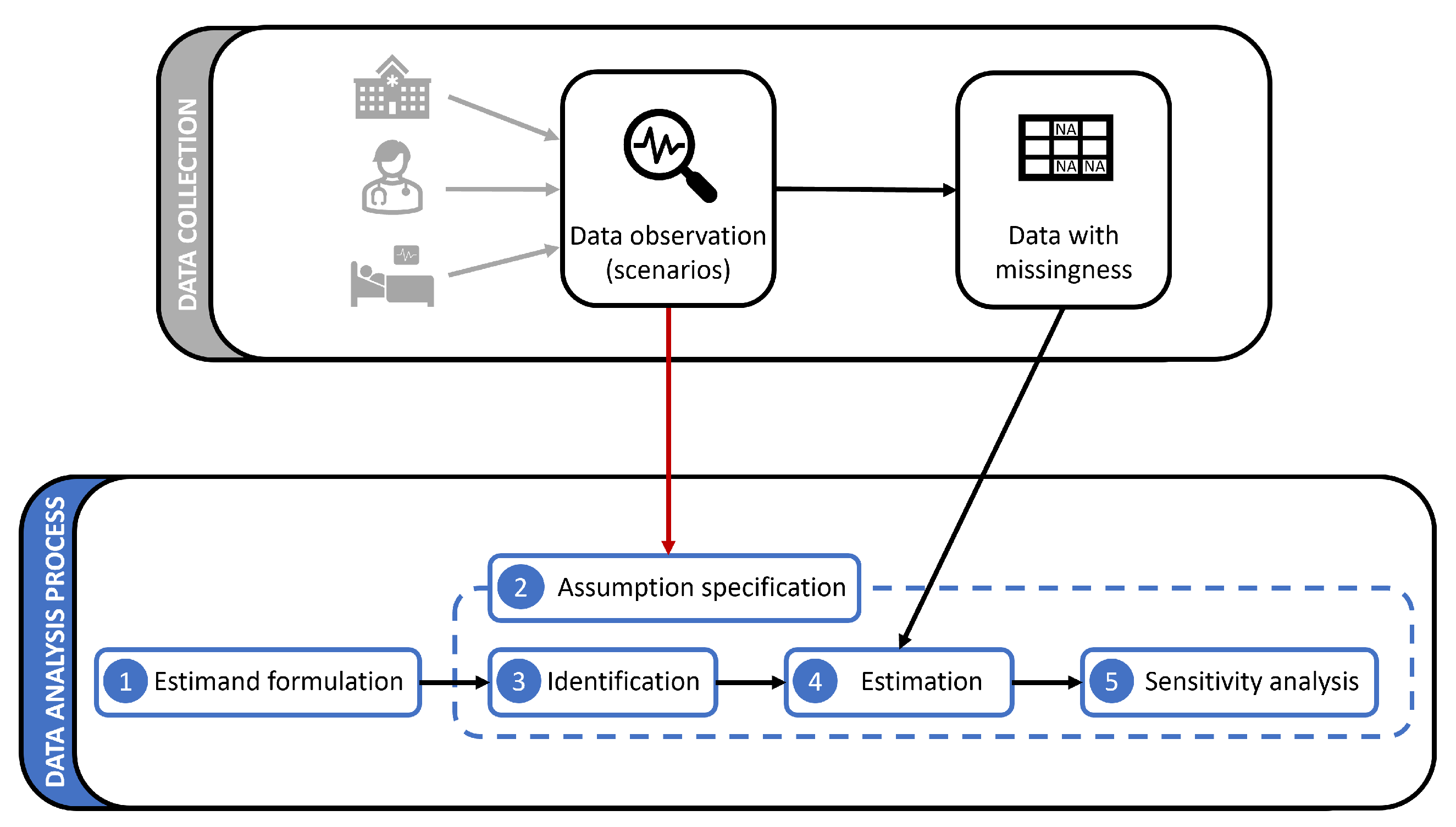
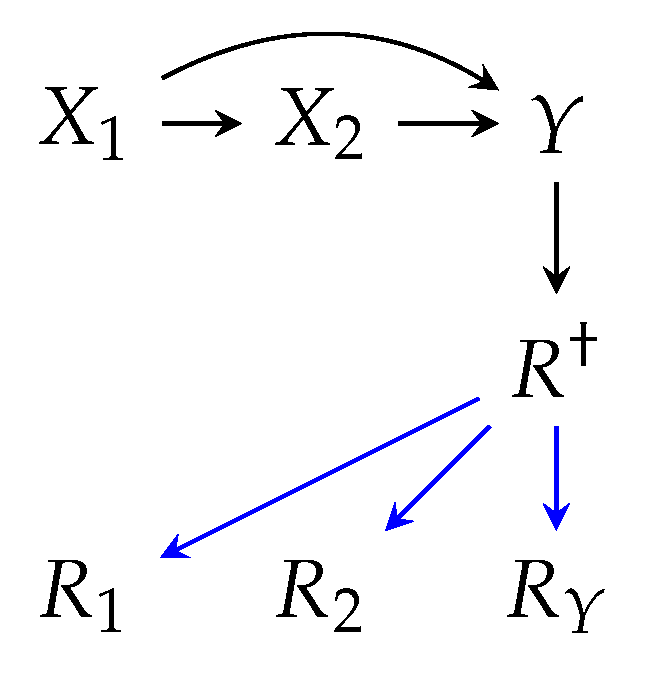3.1. Setting and Notation
Let the random vector comprise d study variables . For ease of reference, we denote a specific variable of interest beside X (e.g., the class labels in the ML classification problem) as Y. Furthermore, we denote independence between as . Independence by conditioning on a variable is denoted as .
In reality, the variable
is realized for all patients, though it may or may not always be available(i.e., observed, recorded, and present in the dataset). We therefore refer to
as a
counterfactual variable since this is what the data would have been if it had always been available, possibly contrary to reality. Corresponding to each
, we define a binary variable
, called the
missingness indicator, to express
’s availability: we set
when
is available, and
otherwise. The version of
which is masked by missingness is called
proxy variable, denoted as
where
represents the missing entries. By this definition, a proxy variable is modeled as
The distribution of
R is determined by the subset of scenarios from
Section 2, which describe the data observation and recording in a healthcare facility and dataset selection for analysis. A data availability policy
represents the union of scenarios, such that the missingness distribution follows the policy, i.e.,
. Subsequently, the resulting distribution given a policy
is denoted as
. We denote three special policies to reference in the paper: (1) the initial policy during data collection as
, (2) the full-availability policy as
, under which
X is always available, and (3) any other policy as
, which is neither
nor
. This notation yields
.
Example 1 (missingness under availability policies). Suppose a variable is realized for four patients, giving . If the fourth patient has missing values under a policy , we have . In this case, the mean estimations for X and are defined as and . Under a new policy where only the observations are available, we have
An availability policy for
is, in general, parameterized by all variables (including
itself) as well as other missingness indicators, i.e.,
. To encode these dependencies, we model the joint
distribution using m-graphs Mohan et al. [
2]. An m-graph under the availability policy
, denoted as
, is a causal directed acyclic graph (DAG) with the node set
. The edges in the structure
are deterministic, representing equation
1. While non-graphical approaches for missing data exist, we focus on m-graphs for their effectiveness and popularity in this paper.
Section 3.3 will provide more details about m-graphs.
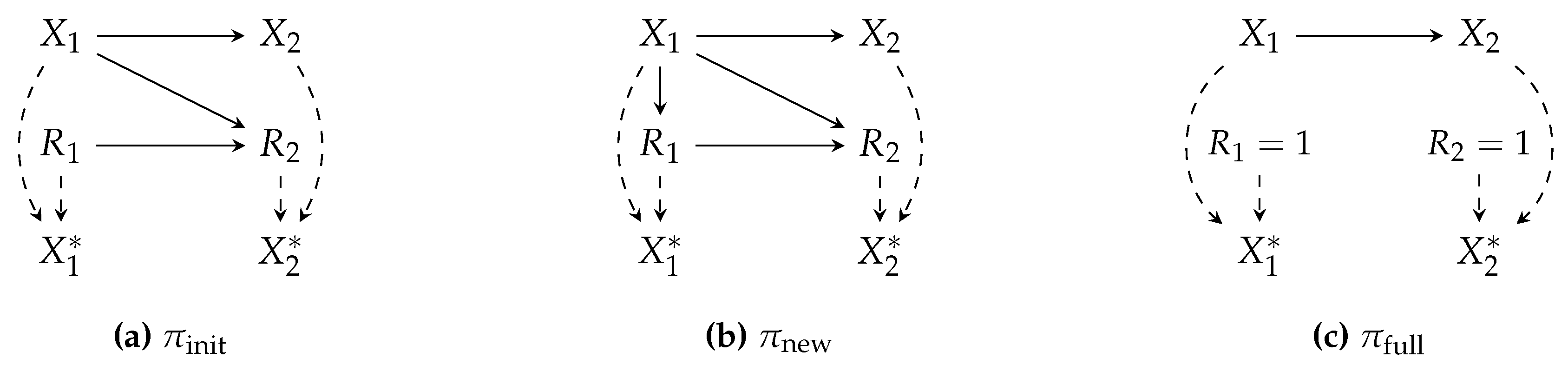
Example illustrations of m-graphs are depicted in
Figure 2, where three m-graphs model different policies for a similar
distribution.
3.2. Defining the Estimand
In the first step of data analysis, an objective must be set by the domain expert and the data scientist and translated into an estimand, which will be fitted to the data. Examples include finding the weights of a prediction model for patient morbidity or the mean value of a biomarker for a population. Based on the form of the estimand, and whether and how it depends on the unavailable data distribution under missingness, we may face diverse challenges.
A basic question of interest is the mean of an outcome variable
Y (e.g., the mean value of a test or the chance of recovery). If
Y is partially available under the policy
, one may formulate the question directly as
, which reads as "mean of
Y when it is available". However, we are often interested in estimating the entire population regardless of the missingness status "had
Y for all samples been available for analysis." This objective, referred to as the counterfactual mean estimation, is presented as
Example 2 (counterfactual mean LDL cholesterol level). As part of public health research, we aim to estimate the nationwide average LDL cholesterol level, denoted as Y. Available datasets are collected from a hospital where LDL levels are not available for all the patients.
gives the average observed value in the hospital.
As a possible new policy, gives the average value if the LDL level had been observed for all patients in the hospital.
gives the target estimand, i.e., the nationwide average LDL level.
As a more advanced objective, we may be interested in developing a prediction model for the outcome variable Y using the covariate vector X. i.e., , which reads as "conditional mean of Y given X". We often choose an ML model for estimation, such as a multi-layer perceptron neural network , parameterized by w. The weights of the network are learned by minimizing a loss function, e.g., the mean squared error (MSE): . Model performance at deployment can also be evaluated using the same formula.
Given a fully observed outcome and missing covariates, the estimand
formulates the MSE loss for the available
X. Estimand
3 suits the situation where the prediction model is to be deployed in an environment with the same observation policy, meaning that all missingness scenarios are the same during deployment as during the data collection stage. In estimand
3, we may use the information in
, e.g., we train (maximum)
separate sub-models
for each unique value (pattern) of
R [
20].
Example 3 (health status estimation at hospital discharge).
We aim to develop a prediction model for the 6-month outcome based on the observed variables during hospitalization, queried at discharge. The model deployment will not influence the physicians’ decisions. The fact that the hospitalization data is being analyzed retrospectively can justify the assumption that the observation and recording policy will not change at deployment. The MSE loss for this case is given by estimand 3.
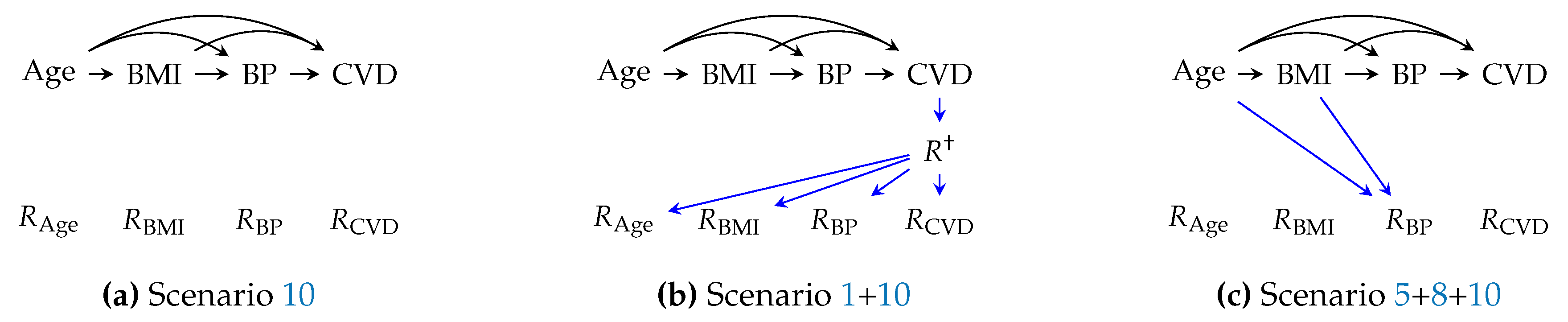
Alternatively, we may be interested in learning a prediction model that is deployed in healthcare facilities with different missingness scenarios, e.g., with varying guidelines of observation and protocols (scenario 5 and 6), for a different patient cohort (scenario 1 and 9), or in the same healthcare facility, but with a change of observation policy because the physicians would measure different variables to "feed" it to the prediction model. In particular, suppose a training dataset generated given the m-graph in
Figure 2 will be deployed in an environment modeled by the m-graph in
Figure 2. The estimand for such a case is
which reads as "MSE loss under new missingness scenarios at deployment," where
represents the new policy.
Example 4 (change of hospital discharge protocols).
Suppose the hospital in example 3 adopts a new discharge protocol mandating performing a medical test for all patients before discharge. The MSE loss under the newly adopted policy is given by estimand 4
A special case of estimand
4 is when the prediction model is expected to make predictions always using full covariates (
Figure 2c). The estimand for this case is
, with only one missingness pattern, the full-availability
. This objective is employed for most clinical prediction models (see Tsvetanova et al. [
8]). For more examples,
Appendix B presents the estimands for prediction using decision trees and feature importance.
Example 5 (clinical prediction model). Suppose a clinical prediction model is developed using an incomplete dataset. As a result of successful development, physicians will use the model while they actively collect all study variables every time to feed to the model. The MSE loss at deployment is given by .
3.3. Identification
As shown in the previous step, estimands may query different missingness distributions, while the only available distribution is given by the data collection policy
. If an estimand queries
, such as estimand
3, it can be computed directly using the training dataset
. On the other hand, estimands such as
2 and
4 query different distributions and hence are subjected to the distribution shift problem. In the identification step, we find a procedure that computes a consistent estimate of an estimand under a target distribution using the available
[
2].
To elaborate further, we consider an estimation approach under distribution shift, namely
importance sampling: for a functional
of the distribution at deployment
,
is estimated using the data collection distribution
as
where the fraction
is called the importance ratio. By equation
5, samples are drawn from
but re-weighted by their "importance" in reflecting
. Equation
5 states that a
estimation is possible given the
samples when
is known for all
over the support of
p.
The importance ratio can be re-written using the selection model factorization [
42], as
The conditional terms
and
in the fraction are the data collection and deployment availability policies, respectively. Assuming no additional counterfactual data distribution shift, i.e.,
, equation
6 is simplified as
, i.e., the ratio of missingness models at the data collection and deployment stages. When the availability policy does not change at deployment, the ratio is re-written as
and when a new policy is adopted at deployment, it is re-written as
While the following arguments are valid for equation
5 in general, we consider a special case where the full-availability policy
is running at deployment (e.g., estimand
2). In this case, we trivially have
for all incomplete data, since the numerator
is zero when
. This means that only the complete cases (R=1) are used for computation, for which
. The resulting estimator according to equation
5, is expressed (for estimation using
) as
for
N samples, where
selects only the complete cases. Equation
8 is known as the
inverse-probability weighting (IPW) estimator. The denominator in equation
8 is referred to as the
propensity score, often denoted as
.
The challenge of identification lies in the conditioning set of importance ratio terms, as they generally depend on the counterfactual distribution
, which is only partially available. As a solution, we assume an m-graph for the problem and seek independence properties among
variables that allow us to express the importance ratio in terms of factors that can be estimated using the available distribution
1 [
2,
43].
Example 6 (identification w.r.t. an m-graph).
Suppose a functional is to be estimated, given the data collection and deployment policies and , respectively. The propensity score for IPW estimator is which cannot be estimated using . Assuming the m-graph in Figure 2, we proceed as follows (we drop the distribution index for brevity):
factorize:
the assumed m-graph gives and . The propensity score is thus rewritten as
By the missingness definition in equation 1, we express the second term using the proxy variable and rewrite the propensity score as .
Both factors in the propensity score can be estimated using
In conclusion, identification in this manner requires an m-graph model, and within it, the causal relations of the missingness indicators are specifically important. It is therefore necessary to discover what kind of causal structures the missingness scenarios induce for
R. In particular, we specify the parents and ancestors (direct and indirect causes) for the
R nodes, as stated by inquiry 1. The causes of
R nodes are commonly referred to as the missingness mechanism.
Inquiry 1 (
missingness mechanism)
: Which causes for R nodes a scenario implies.
To facilitate identifying the causes, we categorize all potential causes to search for, in the following three categories:
(X and R components) First candidates for causes of
R are the study variables and their corresponding missingness indicators within the dataset. Examples can be found in
Figure 2a,b, where
X is a cause of indicator
R.
(latent/hidden confounders) Variables that have not been collected and available in the dataset may also causally influence R. More importantly, they may confound two or more study variables within the estimand, and may therefore hinder the identification process.
(exogenous causes) Other variables that may lie outside the dataset and do not confound the study variables of interest, are considered exogenous causes, having, in general, no identification implications.
Missingness in health-related variables such as lab test items are mainly caused by physicians under scenario 5 (missing due to diagnostic irrelevance), where they make measurement decisions based on the observed history. Therefore, in this case,
R indicators for health variables have incoming edges from the previous measurements (recorded or unrecorded). Other potential causes include the health status under scenarios 1 (patient complete non-visit) and 2 (missing follow-up visit due to health status). Examples of the latent/hidden confounders include socioeconomic variables as well as variables in secondary datasets with information about the non-visit population under scenario 1. As for the exogenous causes, many causes may be recognized, such as simply forgetting to enter the data for a patient, under scenario 8 (unrecorded observations). However, one should be cautious about treating all medically unrelated variables as exogenous causes, as they may still confound the study variables and missingness indicators. A detailed analysis of missingness scenarios with respect to inquiry 1 is presented in
Table A1.
Inquiry 1 explores the structural distribution shift caused by a change of m-graph between the data collection and deployment stages. Another possibility is that the m-graph stays invariant, but the causal relations are subjected to a parametric shift. For example, assume the m-graph in
Figure 2a holds for both data collection and deployment, but the missingness probability in
changes from
to
. As stated by inquiry 2, it is crucial to explore the potential parametric shift at deployment due to a change in observation and recording policies.
Inquiry 2 (
Missingness distribution shift)
: Whether a scenario is subjected to missingness parametric distribution shift at deployment.
Parametric shift may occur in scenario 5 (missing due to diagnostic irrelevance), if the definition of normal/abnormal ranges for a health marker changes. In this case, the results of primary tests still influence the performing decision of later tests, however, via different rules. As another example, a parametric shift may occur in scenario 7 (missing due to resource unavailability), if the monetary cost of a medical test decreases as a result of equipment upgrade or insurance plans, leading physicians to order the test more often. A detailed analysis of missingness scenarios with respect to inquiry 2 is presented in
Table A2.
Example 7 (parametric shift due to decreased test costs).
Consider a primary test and a secondary and more expensive test . Patients with abnormal primary test values () are more likely to give the test. After a cost reduction for the test, the overall frequency of the test () increases, such that now the relative number of tests for patients with abnormal values is ρ times larger than before; yet the association between and is retained. This statistics gives
which is the importance ratio for samples in equation 5. We leave it to the readers to calculate other importance ratios based on assumed statistics about this hypothetical problem.
So far, the described identification methodology has been based on the selection model factorization in equation
6 and the no-distribution-shift assumption for the counterfactual variables. However, there might exist missingness scenarios under which this assumption is violated. A case of violation is when the observation and measurement decisions directly affect the counterfactual variables. In terms of m-graphs, this translates to an
edge. The assumption that such a causal relation does not exist is referred to as
no-direct-effect (NDE) [
22], discussed in m-graph identifiability literature [
25]. Since violation of NDE influences the identification procedure, it is crucial to know whether the problem setting permits it, as stated by inquiry 3.
Inquiry 3 (
no-direct-effect assumption)
: Whether a scenario implies outgoing edges from missingness indicators to counterfactual variables.
A crucial case of NDE violation occurs when invasive tests such as biopsy affect the health status of patients. The effect of observation may be exerted on the corresponding counterfactual variable itself or other variables. This effect may also be exerted indirectly, e.g., through temporarily stopping a certain medication before a medical test. For example, due to the contraindication of radiology contrast agents and metformin, it is recommended for diabetic patients that the medication is stopped before performing angiography [
44]. Note that under violation of the NDE assumption, the problem definition stated in
Section 3.2 becomes ill-posed and requires further elaborations. An example of a problem definition under NDE violation is discussed in example 8. A detailed analysis of missingness scenarios with respect to inquiry 3 is presented in
Table A3.
Example 8 (problem definition under NDE violation). Assume the following m-graph , describing an dataset with fully observed Y, where measurement of X negatively influences Y. This problem cannot be analyzed similarly to example 1, as the counterfactual realizations cannot be described ignoring the missingness status. Assuming a hypothetical data generation mechanism, the relation follows in the absence of any measurement (). When X is measured (), Y distribution changes to . Therefore . Possible questions to pose with regard to a target quantity θ are:
if the observation policies remain unchanged,
if we begin to always observe X,
if we knew the value of X but without negative influences on Y, e.g., using a new testing technology.
Another common assumption for missing data problem is the
no-interference assumption, stating that the measurement decisions for one individual do not affect other individuals [
22]. This is similar to the i.i.d. assumption in general ML problems: having interfered measurements, the i.i.d. assumption cannot be made for the
R distribution. It is therefore important to check whether the no-interference assumption is permitted for observation scenarios, as stated by inquiry 4.
Inquiry 4 (
no-interference assumption)
: Whether a scenario causes interference among the availability status of data samples.
Similar to the NDE assumption, one may find realistic scenarios where the no-interference assumption is violated. In general, competing for limited resources under scenario 7 (unavailability or shortage of resources) or for available hospitalization services under scenario 1 and 2 (complete non-visit and missing follow-up) imply interference. A detailed analysis of missingness scenarios with respect to inquiry 4 is presented in
Table A4.
Finally, we discuss a unique case of missingness, where data samples are completely omitted from the dataset prior to any analysis. This case can be modeled in m-graphs via an
node that influences all
, such that if
, then
(
Figure 3). The risk in this situation lies in the fact that we cannot infer the occurrence of such omissions from a dataset without additional information, which may thus lead to the wrong conclusion that the dataset is complete and free of missingness. This case is commonly referred to as
selection bias in causal inference literature. Selection bias is argued in inquiry 5.
Inquiry 5 (
selection bias)
: Whether a scenario causes the omission of an entire data sample in the form of selection bias.
Clearly, sample omission can be a result of non-visit under scenario 1 and inclusion/exclusion criteria under scenario 9. Whether or not this should be conceived as a bias depends on whether the target parameter (e.g.
Y in Estimand
2) is believed to vary between the observed and the unobserved sub-populations. A detailed analysis of missingness scenarios with respect to inquiry 5 is presented in
Table A5.
3.4. Estimation
There are several methods for estimation with missing data, including likelihood-based methods such as Expectation-Maximization (EM) algorithm, multiple imputation (MI), IPW estimator, and outcome regression (OR) [
27,
42]. In the scope of this paper, we continue with the importance sampling approach in equation
5, in particular, the IPW estimator in equation
8 and the estimation of the propensity score.
Even though a successful identification step guarantees that the propensity score can be estimated using the available data, we still face some challenges, e.g., when the missingness pattern is
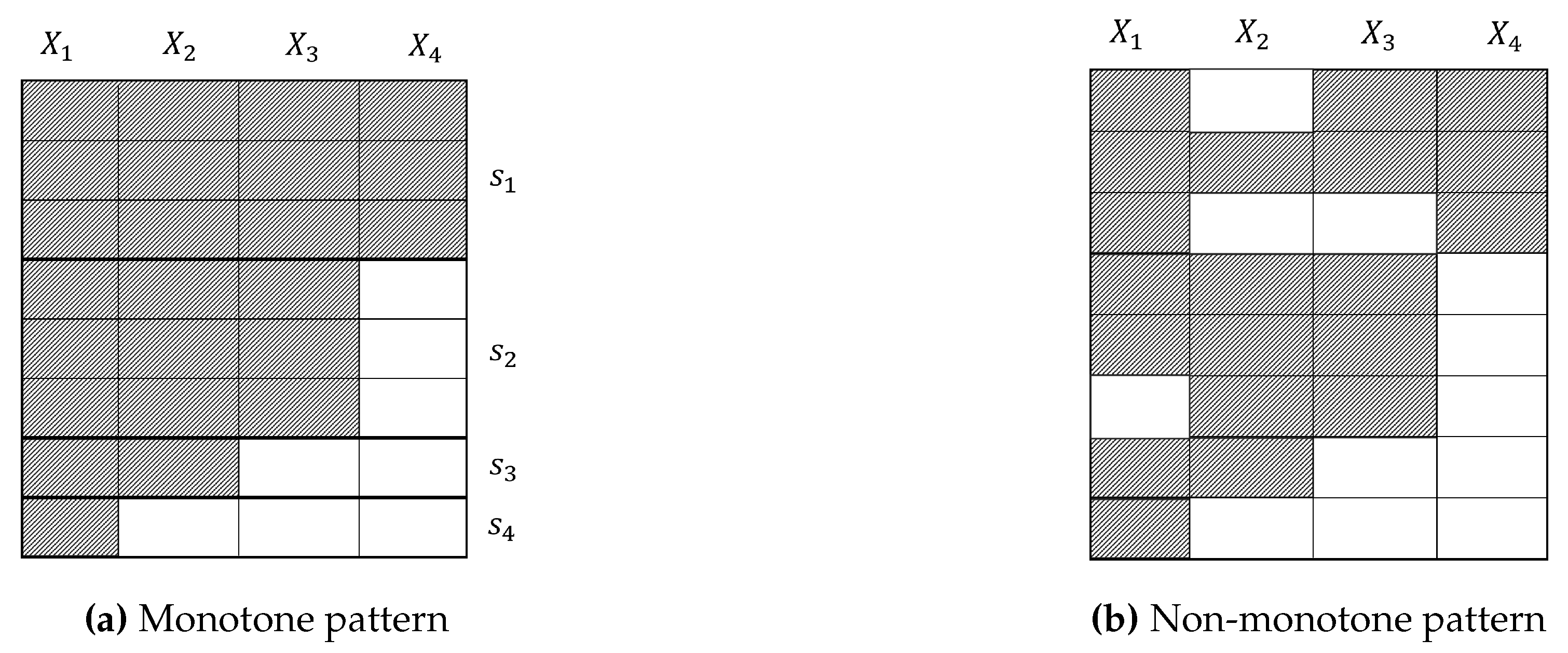
non-monotone. A missingness pattern is called monotone if there is at least one ordering of the variables, such that observing the
j-th variable ensures that all variables
in the ordering are all observed for all samples (
Figure 4a). Estimation of the propensity score has a straightforward solution for monotone patterns. Example 9 showcases propensity score estimation for identifiable monotone missingness.
Example 9 (Propensity score estimation for identifiable monotone missingness).
For the missingness in Figure 4a, we have , while . Assuming identifiability, each can be estimated using only the variables available in . As a result, the propensity score is estimated as
While methods have been developed for an effective estimation under non-monotone missingness [
27,
31], it is beneficial to adopt monotone solutions if applicable. In that regard, inquiry 6 argues whether a missingness scenario individually induces monotone missingness patterns.
Inquiry 6 (
monotonicity)
: Whether a scenario induces missingness with monotone patterns.
If an individual missingness scenario is active, monotonicity can be directly inferred from the emerged patterns, revealed by a simple sorting of the variables with respect to their missingness ratio (
Figure 4a). However, in practice, several scenarios influence a dataset. In such cases, the monotone pattern attributed to one scenario is broken by other scenarios. If we can attribute the emerged non-monotone pattern to a dominant monotone-inducing scenario along with less effective non-monotone scenarios (hypothetically in
Figure 4b), then methods exist based on resolving the missing entries up to recovery of the monotone pattern, e.g., via imputation, and proceeding with IPW estimation for monotone missingness [
45]. A noteworthy scenario likely inducing monotonicity is the sequential observations of physicians under scenario 5 (missing due to diagnostic irrelevance). Given a specific diagnostic flowchart, it is reasonable to assume that more specific secondary tests shall not be made unless primary tests are done. As said, many reasons may break this pattern, including more than one diagnostic flowchart being used, other scenarios such as 4 (patient’s refusal) or 7 (resource unavailability). A detailed analysis of missingness scenarios concerning inquiry 6 is presented in
Table A6.
3.5. Sensitivity Analysis
The assumptions made for handling missing data may not hold under all circumstances. They might be too strong for practical implementation, or we may expect the environment to undergo some perturbations that violate them. To ensure the robustness of the analysis, it is crucial to measure the sensitivity of results to departures from the assumptions and report the variation. Sensitivity analysis is usually done by perturbing the m-graph model.
In addition, it is possible that due to the nature of the problem, assumptions do not lead to a successful identification. In this case, we may impose stronger assumptions that lead to identifiability, model the departures from the actual assumptions, and finally measure the sensitivity to different degrees of magnitude of those departures.
Example 10 (sensitivity analysis for the unidentifiable self-masking missingness).
Consider an outcome variable Y that is subjected to missingness under the following mechanism . The estimand is unidentifiable under this mechanism, referred to as self-censoring [25] or self-masking [20].
We can assume the mean of the unobserved population is δ units away from the observed population, additively , or multiplicatively [6,33]. We then measure the variation of assuming a range of values for the sensitivity parameter δ.
For reliable meaningful sensitivity analysis results, it is crucial to interpret the sensitivity parameters based on meaningful real-world quantities. Inquiry 7 states that scenarios may carry valuable information for choosing meaningful parameters.
Inquiry 7 (
meaningful sensitivity parameters)
: Given a scenario, what are the meaningful units and ranges of parameters for sensitivity analysis.
Specific to the importance sampling approach and equation
8, the unidentifiable terms appear in the importance ratio. The importance ratio captures the difference in the levels of availability for different covariate strata. To make an informed guess about this quantity, we may refer to other research works or collaborations with health domain experts. For instance, Zamanian et al. [
36] suggest that the sensitivity parameters for physicians’ observations (scenario 5) is related to the odds of making an observation for relatively healthy or sick patients, which can be inferred based on the guidelines, protocols, and referring to the attending physicians. The sensitivity parameter for this case is formulated for the model in [
36], assuming a logistic model for missingness, by the following
Odds ratio term,
where
. Equation
10 follows the so-called exponential tilting model, where the multiplicative departure for
R is modeled as an exponential term [
34].
Likewise, the parameters for hospital visits (scenario 1) are related to the odds of visiting a healthcare facility for healthy and sick populations, formulated similarly as equation
10, for which some information can be extracted from epidemiologic studies and public health reports. Overall, the form of the sensitivity model depends on the estimand and the estimator. Yet, a similar ratio as in equation
10 often appears in the analysis, which must be specified. A detailed analysis of missingness scenarios concerning inquiry 7 is presented in
Table A7.