Submitted:
09 April 2024
Posted:
10 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Molecular Analysis
2.2. Analysis of the Data Using Mathematical Formulas
2.3. The Support Vector Machine (SVM) Algorithm
2.4. Analysis of the Red Blood Count Indices and HPLC Results in the Two Most Common α Gene Defects
2.5. Data Analysis
2.6. Ethics
3. Results
3.1. Comparative Results of the Formulas
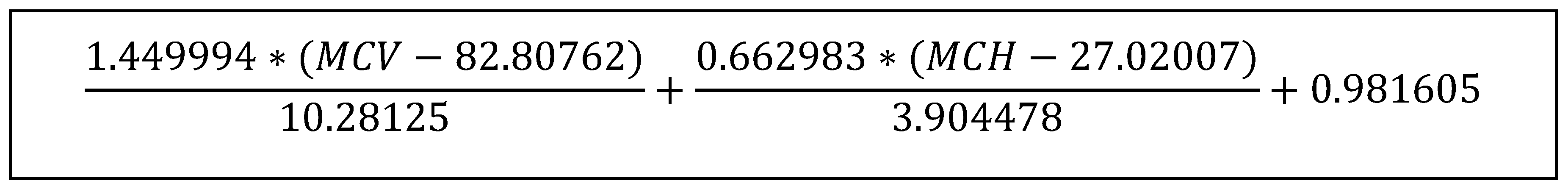
3.2. Analysis of Samples Suspected to Have a Diagnosis of Iron Deficiency Anemia
3.3. Comparison of the Results from the Two Common α Globin Mutations Found
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Higgs, D.R. The Molecular Basis of -Thalassemia. Cold Spring Harb Perspect Med. 2013, 3, a011718–a011718. [Google Scholar] [CrossRef] [PubMed]
- Koren, A. The continuing global challenges of treating patients with beta-thalassemia. Br J Haematol. 2023, 201, 183–4. [Google Scholar] [CrossRef] [PubMed]
- Koren, A.; Profeta, L.; Zalman, L.; Palmor, H.; Levin, C.; Zamir, R.B.; et al. Prevention of β Thalassemia in Northern Israel - A cost-benefit analysis. Mediterr J Hematol Infect Dis. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Musallam, K.M.; Lombard, L.; Kistler, K.D.; Arregui, M.; Gilroy, K.S.; Chamberlain, C.; et al. Epidemiology of clinically significant forms of alpha- and <scp>beta-thalassemia</scp> : A global map of evidence and gaps. Am J Hematol. 2023, 98, 1436–51. [Google Scholar] [PubMed]
- Koren, A.; Zalman, L.; Palmor, H.; Zarnir, R.B.; Levin, C.; Openheim, A.; et al. Sickle cell anemia in Northern Israel: Screening and prevention. Israel Medical Association Journal. 2009, 11. [Google Scholar]
- Koren, A.; Zalman, L.; Palmor, H.; Ekstein, E.; Schneour, Y.; Schneour, A.; et al. The prevention programs for beta thalassemia in the Jezreel and Eiron valleys: results of fifteen years experience. Harefuah. 2002, 141. [Google Scholar]
- Piel, F.B.; Weatherall, D.J. The α-Thalassemias. New England Journal of Medicine. 2014, 371, 1908–16. [Google Scholar] [CrossRef] [PubMed]
- Goh, L.P.W.; Chong, E.T.J.; Lee, P.C. Prevalence of Alpha(α)-Thalassemia in Southeast Asia (2010–2020): A Meta-Analysis Involving 83,674 Subjects. Int J Environ Res Public Health. 2020, 17, 7354. [Google Scholar] [CrossRef]
- Galanello, R.; Cao, A. Alpha-thalassemia. Genetics in Medicine. 2011, 13, 83–8. [Google Scholar] [CrossRef] [PubMed]
- Songdej, D.; Fucharoen, S. Alpha-Thalassemia: Diversity of Clinical Phenotypes and Update on the Treatment. Thalassemia Reports. 2022, 12, 157–72. [Google Scholar] [CrossRef]
- Dehbozorgian, J.; Moghadam, M.; Daryanoush, S.; Haghpanah, S.; Imani fard, J.; Aramesh, A.; et al. Distribution of alpha-thalassemia mutations in Iranian population. Hematology. 2015, 20, 359–62. [Google Scholar] [CrossRef] [PubMed]
- Keikhaei, B.; Slehi-fard, P.; Shariati, G.; Khosravi, A. Genetics of Iranian Alpha-Thalassemia Patients: A Comprehensive Original Study. Biochem Genet. 2018, 56, 506–21. [Google Scholar] [CrossRef] [PubMed]
- STEPHEN A. LIEBHABER, YUET WAI KAN. Differentiation of the mRNA Transcripts Originating from the al- and a2-Globin Loci in Normals and a-Thalassemics. J Clin Inwvest. 1981, 68, 439–46. [Google Scholar] [CrossRef] [PubMed]
- Oron, V.; Filon, D.; Oppenheim, A.; Rund, D. Severe thalassaemia intermedia caused by interaction of homozygosity for α-globin gene triplication with heterozygosity for β thalassaemia. Br J Haematol. 1994, 86, 377–9. [Google Scholar] [CrossRef] [PubMed]
- Amendolia, S.R.; Cossu, G.; Ganadu, M.L.; Golosio, B.; Masala, G.L.; Mura, G.M. A comparative study of K-Nearest Neighbour, Support Vector Machine and Multi-Layer Perceptron for Thalassemia screening. Chemometrics and Intelligent Laboratory Systems. 2003, 69, 13–20. [Google Scholar] [CrossRef]
- Getta, H.A.; Yasseen, H.A.; Said, H.M. Hi & Ha, are new indices in differentiation between Iron deficiency anemia and beta-Thalassaemia trait /A Study in Sulaimani City-Kurdistan/Iraq. Journal of Dental and Medical Sciences. 2015, 14, 67–72. [Google Scholar]
- Rahim, F.; Keikhaei, B. Better differential diagnosis of iron deficiency anemia from beta-thalassemia trait. Turk J Hematol. 2009, 26, 138–45. [Google Scholar]
- Bordbar, E.; Taghipour, M.; Zucconi, B.E. RELIABILITY OF DIFFERENT RBC INDICES AND FORMULAS IN DISCRIMINATING BETWEEN Β-THALASSEMIA MINOR AND OTHER CAUSES OF MICROCYTIC HYPOCHROMIC ANEMIA. Mediterr J Hematol Infect Dis. 2015, 7, e2015022. [Google Scholar] [CrossRef] [PubMed]
- Sirachainan, N.; Iamsirirak, P.; Charoenkwan, P.; Kadegasem, P.; Wongwerawattanakoon, P.; Sasanakul, W.; et al. New mathematical formula for differentiating thalassemia trait and iron deficiency anemia in thalassemia prevalent area: a study in healthy school-age children. Southeast Asian J Trop Med Public Health. 2014, 45, 174–82. [Google Scholar] [PubMed]
- Ehsani, M.A.; Shahgholi, E.; Rahimineja, M.S.; Seighali, F.; Rashidi, A. A New Index for Discrimination Between Iron Deficiency Anemia and Beta-Thalassemia Minor: Results in 284 Patients. Pakistan Journal of Biological Sciences. 2009, 12, 473–5. [Google Scholar]
- Sirdah, M.; Tarazi, I.; Al Najjar, E.; Al Haddad, R. Evaluation of the diagnostic reliability of different RBC indices and formulas in the differentiation of the beta-thalassaemia minor from iron deficiency in Palestinian population. Int J Lab Hematol. 2008, 30, 324–30. [Google Scholar] [CrossRef] [PubMed]
- Romero Artaza, J.; Carbia, C.D.; Ceballo, M.F.; Díaz, N.B. Red cell distribution width (RDW): its use in the characterization of microcytic and hypochromic anemias. Medicina (B Aires). 1999, 59, 17–22. [Google Scholar] [PubMed]
- d’Onofrio, G.; Zini, G.; Ricerca, B.M.; Mancini, S.; Mango, G. Automated measurement of red blood cell microcytosis and hypochromia in iron deficiency and beta-thalassemia trait. Arch Pathol Lab Med. 1992, 116, 84–9. [Google Scholar]
- Green, R.; King, R. A new red cell discriminant incorporating volume dispersion for differentiating iron deficiency anemia from thalassemia minor. Blood Cells. discussion 492-5.. 1989, 15, 481–91. [Google Scholar] [PubMed]
- Ricerca, B.M.; Storti, S.; d’Onofrio, G.; Mancini, S.; Vittori, M.; Campisi, S.; et al. Differentiation of iron deficiency from thalassaemia trait: a new approach. Haematologica. 1987, 72, 409–13. [Google Scholar] [PubMed]
- Shine, I.; Lal, S. A STRATEGY TO DETECT β-THALASSÆMIA MINOR. The Lancet. 1977, 309, 692–4. [Google Scholar] [CrossRef] [PubMed]
- Mentzer William, C. DIFFERENTIATION OF IRON DEFICIENCY FROM THALASSÆMIA TRAIT. The Lancet. 1973, 301, 882. [Google Scholar] [CrossRef] [PubMed]
- ENGLAND, J. DISCRIMINATION BETWEEN IRON-DEFICIENCY AND HETEROZYGOUS-THALASSqMIA SYNDROMES IN DIFFERENTIAL DIAGNOSIS OF MICROCYTOSIS. The Lancet. 1979, 313, 145–8. [Google Scholar] [CrossRef] [PubMed]
- England, J.M.; Fraser Patricia, M. DIFFERENTIATION OF IRON DEFICIENCY FROM THALASSÆMIA TRAIT BY ROUTINE BLOOD-COUNT. The Lancet. 1973, 301, 449–52. [Google Scholar] [CrossRef] [PubMed]
- Schriever Henry, G.; Srivastava, P.C. DIFFERENTIATION OF THALASSÆMIA MINOR FROM IRON DEFICIENCY. The Lancet. 1973, 302, 154–5. [Google Scholar] [CrossRef]
- Roth, I.L.; Lachover, B.; Koren, G.; Levin, C.; Zalman, L.; Koren, A. Detection of β-thalassemia carriers by red cell parameters obtained from automatic counters using mathematical formulas. Mediterr J Hematol Infect Dis. 2018, 10. [Google Scholar]
- Burges, C.J. A tutorial on support vector machines for pattern recognition. Data mining and knowledge discovery. 1998, 2, 121–67. [Google Scholar] [CrossRef]
- Koren, A.; Levin, C.; Zalman, L.; Palmor, H.; Filon, D.; Chubar, E.; et al. Hb TAYBE: clinical and morphological findings IN 43 patients. Eur J Haematol. 2016, 97. [Google Scholar] [CrossRef] [PubMed]
- Koren, A. The continuing global challenges of treating patients with beta-thalassemia. Br J Haematol. 2023. [CrossRef] [PubMed]
- Phirom, K.; Charoenkwan, P.; Shoombuatong, W.; Charoenkwan, P.; Sirichotiyakul, S.; Tongsong, T. DeepThal: A Deep Learning-Based Framework for the Large-Scale Prediction of the α+-Thalassemia Trait Using Red Blood Cell Parameters. J Clin Med. 2022, 11, 6305. [Google Scholar] [CrossRef] [PubMed]
- Hatton, C.; Wilkie, A.; Drysdale, H.; Wood, W.; Vickers, M.; Sharpe, J.; et al. Alpha-thalassemia caused by a large (62 kb) deletion upstream of the human alpha globin gene cluster. Blood. 1990, 76, 221–7. [Google Scholar] [CrossRef] [PubMed]
- Liebhaber, S.A.; Griese, E.U.; Weiss, I.; Cash, F.E.; Ayyub, H.; Higgs, D.R.; et al. Inactivation of human alpha-globin gene expression by a de novo deletion located upstream of the alpha-globin gene cluster. Proceedings of the National Academy of Sciences. 1990, 87, 9431–5. [Google Scholar] [CrossRef] [PubMed]
- Wilkie, A.O.M.; Lamb, J.; Harris, P.C.; Finney, R.D.; Higgs, D.R. A truncated human chromosome 16 associated with α thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature. 1990, 346, 868–71. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, D.M.; Sonati, M.F. . Regulation of human alpha-globin gene expression and alpha-thalassemia. Genet Mol Res. 2008, 7, 1045–53. [Google Scholar] [CrossRef] [PubMed]
- De Gobbi, M.; Viprakasit, V.; Hughes, J.R.; Fisher, C.; Buckle, V.J.; Ayyub, H.; et al. A Regulatory SNP Causes a Human Genetic Disease by Creating a New Transcriptional Promoter. Science (1979). 2006, 312, 1215–7. [Google Scholar] [CrossRef] [PubMed]
- Akhavan-Niaki, H.; Youssefi Kamangari, R.; Banihashemi, A.; Kholghi Oskooei, V.; Azizi, M.; Tamaddoni, A.; et al. Hematologic features of alpha thalassemia carriers. Int J Mol Cell Med. 2012, 1, 162–7. [Google Scholar] [PubMed]
- Oron-Karni, V.; Filon, D.; Shifrin, Y.; Fried, E.; Pogrebijsky, G.; Oppenheim, A.; et al. Diversity of ?-globin mutations and clinical presentation of ?-thalassemia in Israel. Am J Hematol. 2000, 65, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Shaulov, A.; Filon, D.; Rund, D. Haplotype analysis of α-thalassemia chromosomes reveals heterogeneity and multiple founders in Ashkenazi Jews. Eur J Med Genet. 2016, 59, 555–8. [Google Scholar] [CrossRef] [PubMed]
- Kattamis, A.C.; Camaschella, C.; Sivera, P.; Surrey, S.; Fortina, P. Human α-Thalassemia syndromes: Detection of molecular defects. Am J Hematol. 1996, 53, 81–91. [Google Scholar] [CrossRef]
- Embury, S.H.; Miller, J.A.; Dozy, A.M.; Kan, Y.W.; Chan, V.; Todd, D. Two different molecular organizations account for the single alpha-globin gene of the alpha-thalassemia-2 genotype. Journal of Clinical Investigation. 1980, 66, 1319–25. [Google Scholar] [CrossRef] [PubMed]
- Gilad, O.; Steinberg-Shemer, O.; Dgany, O.; Krasnov, T.; Noy-Lotan, S.; Tamary, H.; et al. Alpha-Thalassemia Carrier due to –α<sup>3.7</sup> Deletion: Not So Silent. Acta Haematol. 2020, 143, 432–7. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.T. Variable Clinical Phenotypes of α-Thalassemia Syndromes. The Scientific World JOURNAL. 2009, 9, 615–25. [Google Scholar] [CrossRef] [PubMed]
- Arnon, S.; Tamary, H.; Dgany, O.; Litmanovitz, I.; Regev, R.; Bauer, S.; et al. Hydrops fetalis associated with homozygosity for hemoglobin Taybe (α 38/39 THR deletion) in newborn triplets. Am J Hematol. 2004, 76, 263–6. [Google Scholar] [CrossRef] [PubMed]
- England, J.M.; Fraser Patricia, M. DIFFERENTIATION OF IRON DEFICIENCY FROM THALASSÆMIA TRAIT BY ROUTINE BLOOD-COUNT. The Lancet. 1973, 301, 449–52. [Google Scholar] [CrossRef]
| Diagnosis | Non β thalassemia trait Mean ± SD (range) |
β thalassemia trait Mean ± SD (range) |
α thalassemia trait and/or suspected α trait Mean ± SD (range) |
p value between α and β trait |
|---|---|---|---|---|
| Number of individuals (%) | 18572 (81.3) | 2936 (12.8) | 1334 (5.9) | |
| RBC (x 109/dl) | 4.2 ± 0.43 (2.17 – 7.67) | 5.42 ± 0.55 (3.21 – 7.81) |
4.87 ± 0.44 (3.48 – 6.34) |
<0.001 |
| Hgb (g/dl) | 11.75 ± 1.06 (9.00 – 19.2) |
10.65 ± 0.95 (9.00 – 15.4) |
11.51 ± 1.07 (9.0 – 15.6) |
<0.001 |
| MCV (fl) | 85.9 ± 6.84 (34 – 125.9) |
63.14 ± 5.76 (48 – 91.5) |
73.56 ± 4.52 (53.3 – 91.4) |
<0.001 |
| MCH (pg) | 28.16 ± 2.7 (16.2 – 40.7) |
19.71 ± 1.95 (14 – 31.3) |
23.71 ± 1.92 (16.5 – 29.9) |
<0.001 |
| MCHC (g/dl) | 32.75 ± 1.76 (12.6 – 45.7) |
31.23 ± 1.73 (17.5 – 65) |
32.17 ± 1.68 (19.2 – 36.2) |
<0.001 |
| RDW (%) | 14.96 ± 2.02 (10.1 – 36.4) |
16.39 ± 2.71 (12 – 22.8) |
15.16 ± 1.94 (12.1 – 28) |
<0.001 |
| Hgb F (%) | 0.34 ± 0.61 (0. – 14) |
2.12 ± 2.67 (0 – 38) |
0.4 ± 0.29 (0.1 – 1.9) |
<0.001 |
| Hgb A2 (%) | 1.12 ± 1.38 (0 – 3.4) |
5.6 ± 0.8 (3.5 – 8.8) |
2.6 ± 0.26 (2 – 3.3) |
<0.001 |
| Diagnosis | α thalassemia trait + mutation Mean ± SD (range) |
α thalassemia trait suspected Mean ± SD (range) |
α thalassemia trait normal sequence Mean ± SD (range) |
p value |
|---|---|---|---|---|
| Number of individuals (%) | 291 (21.8 %) | 962 (72.12%) | 81 (6.07 %) | |
| RBC (x 109/dl) | 4.93 ± 0.46 (3.79 – 6.34) |
4.87 ± 0.43 (3.48 – 6.27) |
4.67 ± 0.36 (3.7 – 5.69) |
<0.001 |
| Hgb (g/dl) | 11.58 ± 0.97 (9.0 – 14.8) |
11.51 ± 1.1 (9.0 – 15.6) |
11.39 ± 0.93 (9.1 – 13.5) |
NS |
| MCV (fl) | 72.95 ± 5.17 (53.3 – 83.9) |
73.55 ± 4.25 (57.8 – 91.4) |
75.1 ± 4.07 (62.2 – 85) |
NS |
| MCH (pg) | 23.61 ± 2.1 (16.5 – 27.1) |
23.66 ± 1.84 (16.5 – 29.9) |
24.43 ± 1.81 (18.3 – 28.6) |
NS |
| MCHC (g/dl) | 32.26 ± 1.72 (19.2 – 36.10) |
32.15 ± 1.33 (25.9 – 36.2) |
32.51 ± 1.38 (29.4 – 35.5) |
NS |
| RDW (%) | 14.91 ± 1.8 (12.3 – 23.4) |
15.19 ± 1.87 (12.1 – 28.0) |
15.89 ± 2.35 (12.9 – 26.5) |
0.004 |
| Hgb F (%) | 0.5 ± 0.52 (0.1 – 7.3) |
0.4 ± 0.29 (0.1 – 1.9) |
0.4 ± 0.34 (0 – 5.1) |
NS |
| Hgb A2 (%) | 2.6 ± 0.29 (1.3 – 3.6) |
2.6 ± 0.26 (2 – 3.3) |
2.6 ± 0.27 (0.8 – 3.3) |
NS |
| No. | Study (Reference) |
Formula | βThal cut-off | α thal PPV | α thal NPV | α thal specificity | α thal sensitivity | Percentile 75% | Percentile 95% (lower limit) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Srivastava [30] | MCH/RBC | <3.8 | 39.39 | 93.59 | 99.35 | 5.82 | 5.38 | 6.04 (5.97) |
| 2 | 1) England & Fraser – 1973 [49] 2)England & Fraser - 1979 [28] |
MCV-RBC-(5-Hb)-K* | <0 | 0.87 | 93.10 | 97.53 | 0.3 | 70.29 | 73.37 (72.96) |
| 3 | Mentzer [27] | MCV/RBC | <13 | 45.72 | 93.94 | 99.01 | 11.57 | 16.45 | 18.24 (18.09) |
| 4 | Shine & Lal [26] | MCV2 x MCH/100 | <1530 | 34.42 | 98.83 | 88.22 | 85.54 | 1466.58 | 1626.52 (1607.81) |
| 5 | Ricerca et al. [25] | RDW/RBC | <3.3 | 13.66 | 96.83 | 68.81 | 68.63 | 3.38 | 4.01 (3.95) |
| 6 | Green & King [24] | MCV2 x RDW/(Hb x 100) | <65 | 58.45 | 95.15 | 98.44 | 30.47 | 78.26 | 92.63 (90.12) |
| 7 | D’Onofrio et al. [23] | MCV / MCH | >0.9 | 6.74 | 0 | 0 | 100 | 3.18 | 3.34 (3.33) |
| 8 | Romero Artaza et al. [22] | RDW x MCV / RBC | <220 | 53.65 | 95.86 | 97.4 | 41.64 | 251.18 | 290.99 (285.58) |
| 9 | Sirdah et al. [21] | MCV-RBC-(3XHb) | <27 | 56.63 | 93.69 | 99.55 | 7.15 | 37.19 | 41.73 (41.11) |
| 10 | Ehsani et al. [20] | MCV-(10 x RBC) | <15 | 45.45 | 93.86 | 99.1 | 10.42 | 29.8 | 35.4 (34.8) |
| 11 | Sirachainan et al. [19] | 1.5 x Hb–0.05 x MCV | <14 | 6.08 | 91.95 | 32.69 | 60.39 | 14.58 | 16.01 (15.85) |
| 12 | Bordbar et al. [18] | [80-MCV]*[27-MCH] | >44.76 | 9.17 | 93.72 | 84.63 | 21.46 | 37.8 | 101.32 (93.44) |
| 13 | Rahim & Keikhaei [17] | HbxRDWx100 / RBC2 x MCHC | <21 | 63.44 | 95.2 | 98.72 | 30.89 | 25.14 | 29.3 (28.66) |
| 14 | Hisham index [16] | MCH x RDW / RBC | <67 | 55.85 | 94.99 | 98.4 | 28.13 | 80.87 | 94.52 (92.59) |
| 15 | Hameed index [16] | MCH x Hct x RDW / (RBC x Hb)2 | <220 | 6.73 | 0 | 0 | 100 | 4.99 | 6.52 (6.35) |
| 16 | Amendolia et al. – SVM [15] | SVM - (RBC, Hgb, Hct, MCV) | 98.75 | 13.59 | 98.99 | 57.99 | 1.65 | 1.81 (1.79) | |
| 17 | SVM [31] | SVM (MCV and MCH) (Fig 1) | <0 | 21.52 | 99.93 | 73.67 | 99.33 | -0.23 | 0.29 (0.22) |
| α globin genetics | αα/-α3.7kb | -α3.7kb/-α3.7kb | P* | αIVS I-1 /αα | αIVS I-1 / αIVS I-1 | P** | αIVS I-1 / -α3.7kb |
|---|---|---|---|---|---|---|---|
| Number of individuals (%) | 134 | 24 | 97 | 8 | 7 | ||
| RBC (x 109/dl) | 4.8 ± 0.4 (6.16 – 3.79) |
5.3 ± 0.49 (4.37 – 6.21) |
<0.001 | 4.9 ± 0.36 (4.1 – 6.1) |
5.4 ± 0.57 (4.52 – 5.96) |
0.002 | 5.5 ± 0.56 (4.72 – 6.14) |
| Hgb (g/dl) | 11.6 ± 0.96 (9.1 – 14.2) |
11.2 ± 1.01 (9.0– 13.3) |
NS | 11.8 ± 0.88 (9.4 – 14.8) |
10.5 ± 1.19 (9.0 – 11.7) |
<0.001 | 11.1 ± 0.66 (10.3 – 11.8) |
| MCV (fl) | 74.4 ± 4.35 (59 – 83.9) |
68.7 ±5.6 (58.7–77.1) |
<0.001 | 73.7 ± 4.36 (53.3– 82.2) |
64.9 ± 4.66 (59.1 – 73.6) |
<0.001 | 63.3 ± 1.41 (61.8 – 65.2) |
| MCH (pg) | 24.3 ± 1.75 (18 – 27.1) |
21.8 ± 2.11 (17.1 – 24.6) |
<0.001 | 23.9 ± 1.63 (16.5– 26.5) |
19.5 ± 2.15 (17.9 – 24.7) |
<0.001 | 20.2 ± 0.96 (19.2 – 21.9) |
| MCHC (g/dl) | 32.6 ± 1.18 (29.7 – 36.1) |
31.7 ± 1.52 (29.2 – 34.4) |
<0.001 | 32.4 ± 1.2 (28.9– 35.7) |
30.0 ± 2.2 (25.5 – 33.5) |
<0.001 | 28.7 ± 6.66 (19.2 – 35.4) |
| RDW (%) | 14.8 ± 1.65 (12.5 – 21.7) |
15.5 ± 2.56 (13.3 – 23.4) |
NS | 14.4 ± 1.29 (12.3– 20.1) |
16.3 ± 2.35 (12.6 – 20.5) |
0.001 | 18.8 ± 2.64 (16.5 – 22.6) |
| Hgb F (%) | 0.5 ± 0.67 (0.1 – 7.3) |
0.4 ± 0.38 (0.2 – 1.6) |
NS | 0.5 ± 0.35 (0.1 – 1.8) |
0.6 ± 0.26 (0.3 – 1.1) |
NS | 0.5 ± 0.46 (0.2 – 1.1) |
| Hgb A2 (%) | 2.7 ± 0.26 (1.7 – 3.6) |
2.6 ± 0.19 (2.2 – 2.9) |
0.09 | 2.7 ± 0.3 (1.6 – 3.1) |
2.3 ± 0.3 (1.7 – 2.7) |
0.002 | 2.8 ± 0.15 (2.5 – 2.9) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




