1. Introduction
Bananas and plantains are important food crops and daily sources of carbohydrates for at least 100 million people on the African continent [
1,
2,
3]. The importance of bananas as a staple food is exemplified by Uganda, where per capita consumption reaches approximately 223 kg per person per year. Of the 20 million tons of bananas produced in East and Southern Africa, only 1% is exported, mainly by Zimbabwe, Uganda and South Africa. Approximately 4% of the 11 million tons produced in West and Central Africa are exported, mainly by Côte d’Ivoire and Cameroon [
4,
5].
In Africa, the production of bananas and plantains is often hampered by biological and environmental constraints. Many pests and diseases have been introduced to the continent through infected planting material. With the expansion of the banana industry in African countries, these diseases and pests spread to several new areas. In many areas, disease-causing pathogens occur in epidemic proportions. The enormous costs involved in controlling some of these diseases make it almost impossible to continue banana production in severely affected areas [
6].
In Mozambique, bananas (
Musa spp.) are produced mainly on small farms; however, some large farms are found around the provinces of Maputo Province, Gaza, Nampula and Manica. These farms mainly cultivate Cavendish varieties and supply large cities such as Maputo and Beira and the markets of South Africa, Botswana and the Kingdom of eSwatini. Currently, banana plantations (farms) occupy 62,000 ha in Mozambique, with an estimated production of 470,000 thousand tons per year [
7].
Production on small farms in southern Mozambique (Maputo Province and Gaza Province) averages 45 tons per ha for a production area of 3216 ha. In addition to the various pests (nematodes and weevils) and diseases (Sigatoka, Fusarium disease or Mal de Panamá - Foc TR4 and currently bunchy top da Bananeira) that can affect banana crops, the lack of production knowledge, low performance, low soil fertility and long drought periods (particularly in the south of the country) are factors that hinder crop management [
7]. Among the diseases cited, Fusariosis or Panama disease (FocTR4) and
Banana bunchy top disease (BBTD) have been the most important.
BBTD, whose etiological agent is the
Banana bunchy top virus (BBTV), was introduced and reported for the first time in Sub-Saharan Africa (SSA) in the 1960s in the Democratic Republic of Congo [
8]. Since then, the spread of BBTV has been confirmed in 16 countries in the Central, Southern and West African regions, and it was first recorded in South Africa in 2015 [
9] and in Mozambique in June 2016, in the Chókwè irrigated perimeter, more specifically in the Primeira Zona locality, in Gaza province [
7].
BBTV belongs to the domain
Monodnariria, kingdom
Shotokuvirae, phylum
Cresdnaviricota, class
Arfiviricetes, order
Mulpavirales, family
Nanoviridae and genus
Babuvirus [
10]. BBTV affects only members of the genus
Musa and is transmitted in a persistent circulatory manner by its only known vector,
Pentalonia nigronervosa Coq. [
11,
12]. The BBTV genome has six circular single-stranded DNA (ssDNA) components (R-DNA, U3-DNA, S-DNA, M-DNA, C-DNA and N-DNA), each approximately 1 kilobase (kb) in size [
13]. Each component potentially encodes a protein, except for R-DNA, which encodes two proteins linked to viral replication [
14,
15].
S-DNA encodes the capsid protein (CP), which forms isometric particles and aids in genome packaging [
16]. C-, N- and M-DNA encode cell cycle binding proteins (Clink), nuclear transport proteins (NSPs) and movement proteins (MPs), respectively (17-19). The function of U3-DNA remains unknown for BBTV. Rep and CPs are evolutionarily more conserved than the other proteins and therefore serve as useful markers for the identification and classification of ssDNA viruses [
20].
The R gene has been used to classify isolates from different countries located in distinct geographical regions [
21]. There are two suggested groups, the Pacific-Indian Oceans (PIO) and South-east Asian (SEA) groups, characterized by an approximately 10% nucleotide difference, while the intragroup difference ranges from 1.9 to 10% [
21].
In Mozambique, the genetic characteristics of BBTV isolates occurring in producing fields located in the south of the country have not yet been reported. These data could allow inferences about the possible geographical origin of the viral isolates present and support preventive measures related to the control of viral spread in Africa. In this study, the R-DNA and S-DNA of 40 virus isolates, representing the 175 isolates previously collected in 23 locations of 11 farms located in the district of Chókwè, were sequenced and analyzed. The results obtained are presented and discussed here.
2. Materials and Methods
2.1. BBTV Isolates Analyzed
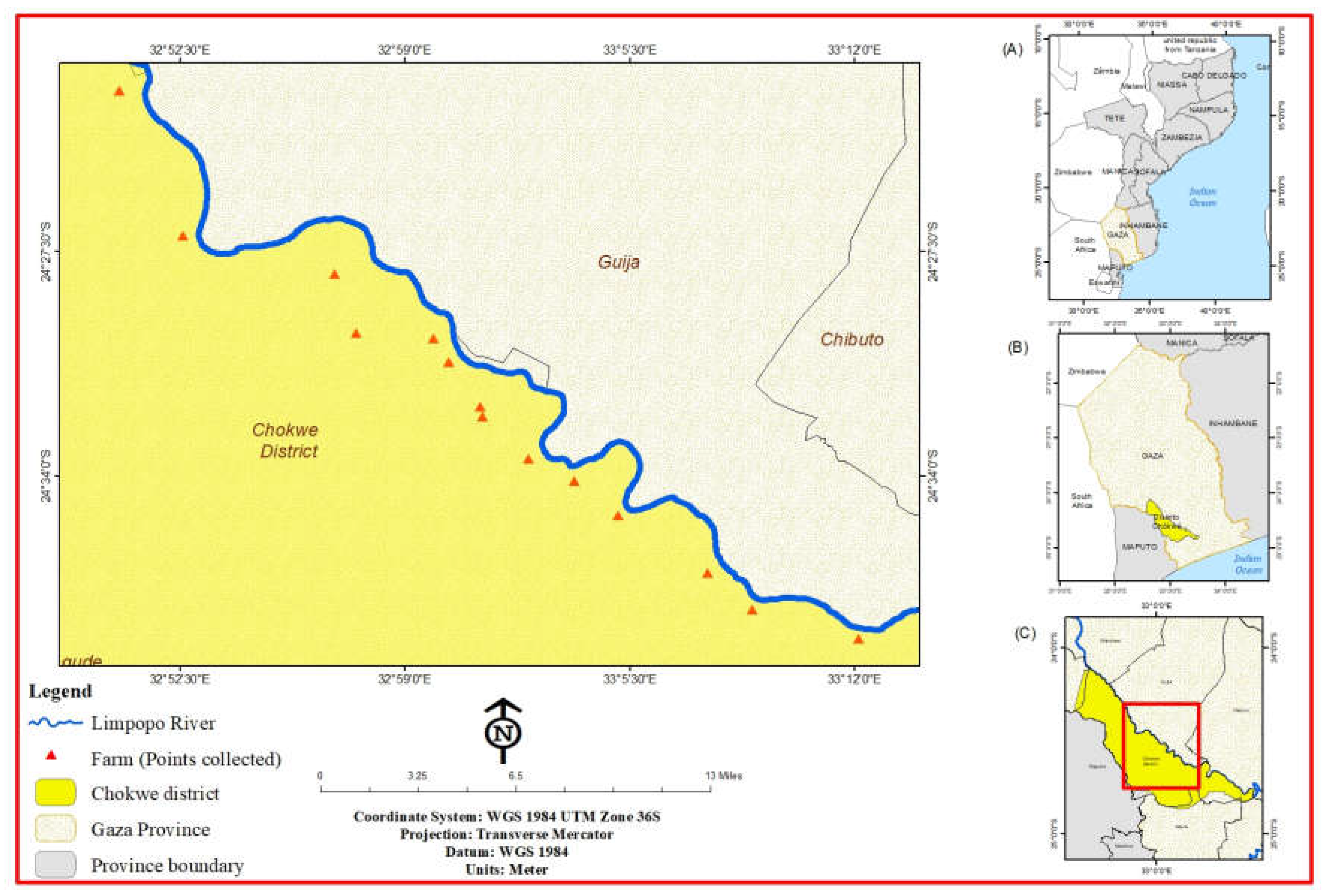
The 40 BBTV isolates used in this study were chosen proportionally to the number of isolates from 23 fields located on 11 farms in the 4 administrative posts of the Chókwè district (initial site of the outbreak), Gaza Province (
Figure 1b) in Mozambique (
Figure 1a), with the help of researchers from the Institute of Agricultural Research of Mozambique - IIAM and commercial and family farmers involved in banana production. Located in southern Gaza Province, the district of Chókwè (
Figure 1c) has the following geographical coordinates: 24°05′ and 24°48′ South latitude; 32°33′ and 33°35′ East Longitude [
22].
2.2. Amplification and Analysis of the S and R Genes
Total plant DNA was extracted from leaf tissue in the laboratory of the Biotechnology Center of the Eduardo Mondlane University of Mozambique (UEM), following the protocol of Lodhi et al. [
23]. For extraction, 0.35 g of fresh plant tissue was macerated in liquid nitrogen and then 1.5 mL of 2% CTAB buffer (100 mM Tris-HCl, pH 8.0; 20 mM EDTA; 1.4 M NaCl; 80 mM Na2SO3; 2% PVP-10 and 2% cetyl trimethyl ammonium bromide (CTAB) containing 0.2% β mercaptoethanol. After maceration, the tissue was transferred to a 1.5 mL sterile microcentrifuge tube. Then, 750 µl of extraction buffer with 2% PVP was added and incubated at 60°C for 30 minutes, and the tubes were mixed by inversion every ten minutes. A volume of 750 µl of chloroform:isoamyl alcohol solution (24:1) was added to the tubes and centrifuged at 12,000 rpm for 10 minutes at 23°C. The aqueous phase (supernatant) was transferred to new Eppendorf tubes, and the DNA was precipitated by adding 0.6 volumes of isopropanol and incubated at -20°C for one hour. After centrifugation at 12,000 rpm for 10 minutes, the supernatant was discarded, and the DNA was washed with 500 µl of 70% ethanol and resuspended in 100 µl of 1X TBE buffer. The tubes containing the extracted DNA were placed on dry ice and transported to Brazil to the Laboratory of Molecular Virology, Department of Plant Pathology, Federal University of Lavras (UFLA), for further studies.
In the DNA amplification reaction, primers specially designed to flank the S gene region and the R gene region were employed, as specified in
Table 1.
DNA amplification was performed in a 25 μl reaction consisting of 2.5 μl of 10X PCR buffer containing 15 mM MgCl2, 2.0 μl of 10 mM dNTPs, 1.0 μl of forward primer and 1.0 μl of reverse primer at a concentration of 10 μM, 0.25 μl of Taq DNA polymerase (Sigma, USA), 1.0 μl of DNA and enough water for a total reaction volume of 25 μl. Amplification was performed in a Master Cycler Thermocycler (Eppendorf, Germany) with the following program: 94°C for 2 minutes followed by 35 cycles of 94°C for 30 seconds, 49°C (DNA S) or 52°C (DNA R) for 20 seconds and 72°C for 60 seconds, with a final extension of 72°C for 5 minutes. The amplified products were analyzed by electrophoresis in a 1% agarose gel and counterstained with Gel Red (Biotium). The bands were visualized and documented using an Alpha Imager (Alpha Innotech Corp., USA).
2.3. Sequencing and Analysis of S and R DNA Sequences
The PCR products for each of the R-DNA and S-DNA (CP) fragments were purified and sent for sequencing at the Brazilian company FIOCRUZ. The sequence data were analyzed using the BioEdit software program (version 7.0.90) and NCBI (National Center for Biotechnology Information). The multiple alignments of the nucleotide and amino acid sequences of the studied isolates with other viral isolates available in GenBank (
Table 2), as well determination of the identity between them, were performed using the program CRUSTAL W2 (V.2.0). Isolates from the PIO group, including some isolates from sub-Saharan Africa, and from the SEA group were chosen for comparison. An isolate of pineapple bunchy top virus (ABTV). The phylogenetic trees were constructed by the neighbor-joining (NJ) method in the MEGA7.02 program [
24] using 2,000 bootstrap replicates.
3. Results
3.1. Amplification of the S and R Genes
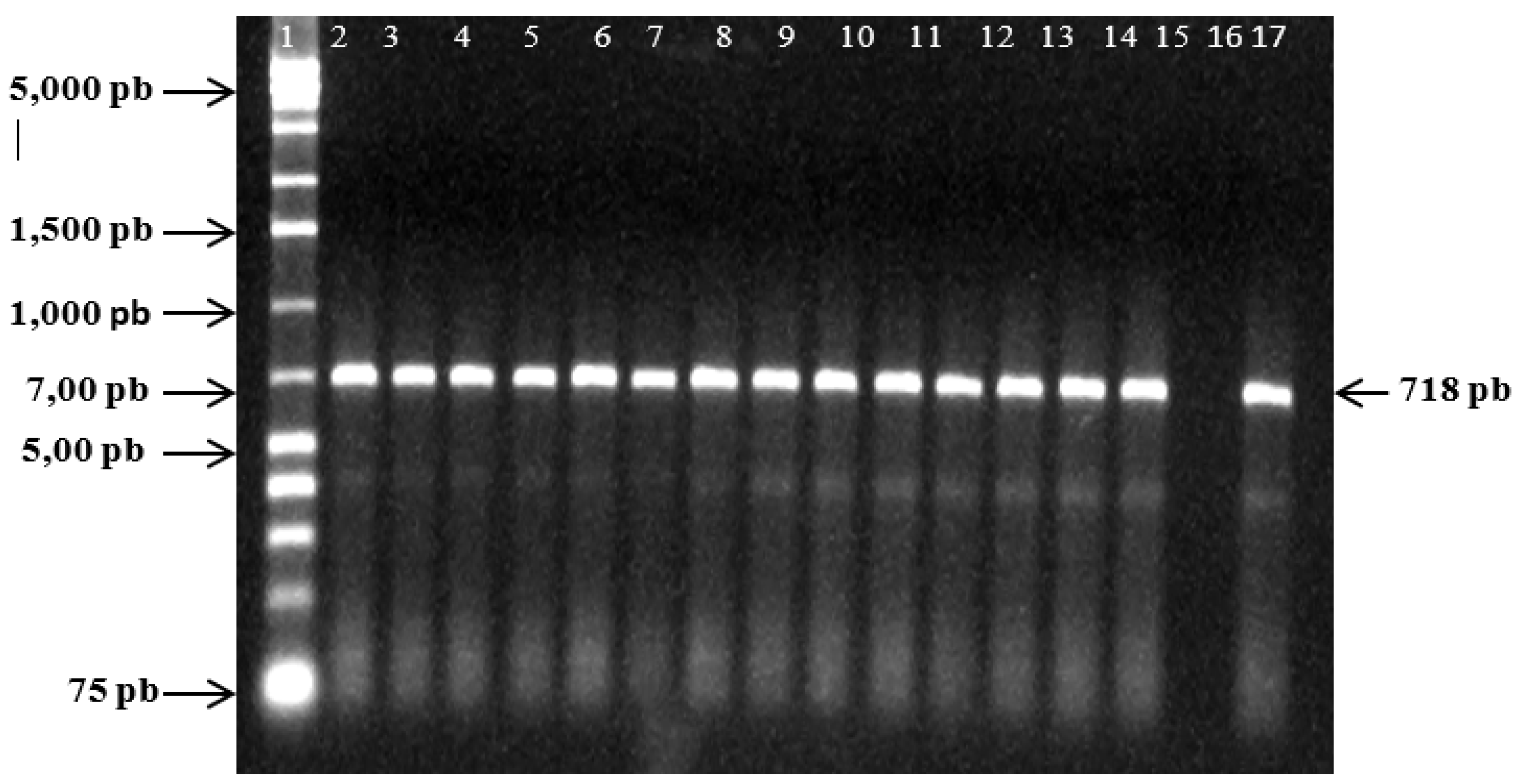
The 40 BBTV isolates from eleven farms of the four administrative posts of the district of Chóckwè, Gaza province, Mozambique, were sequenced and analyzed. The bands amplified from the S gene can be seen in
Figure 2 and those from the R gene in
Figure 3.
3.2. Analysis of the BBTV S-DNA Sequences
The nucleotide identities between the 40 isolates collected in Mozambique (
Table 3) and the isolates available in GenBank (
Table 2) showed that this gene is highly conserved, with more than 50% of the isolates having 100% identity, and the lowest identity was 98%. When compared to the GenBank isolates, the lowest identities observed (92-93%) occurred with isolates MT433376 and MT433375 from Indonesia, KM607468 from Taiwan, KM607469 from the Philippines, KM607536 and AF238876 from China and AB078023 from Japan. The highest identities occurred with isolates JQ820467 from Rwanda, KM607470 from Egypt, KM607505 from the Democratic Republic of Congo (DRC), and JF755980 and JQ820455 from Malawi, all from the African continent, although from different geographical regions. Only Malawi is found in the surrounding region, bordering western Mozambique. These isolates present in the African continent probably had a common geographical origin.
The amino acid identities observed among the Mozambican isolates were similar, with only seven isolates varying between 96 and 99% and the others showing 100% identity. When compared to isolates from GenBank, in addition to the higher identities already specified above, they also showed high identities with isolates from South and Southeast Asia and South Pacific: MK140621 from Pakistan, AB252642 from Miamar and KM607584 from Tonga. Notably, isolates BBTV_172_F11 from Mozambique and AB078023 from Japan showed 98% and 93% nucleotide identity, respectively, when compared to the other isolates. The amino acid identity of the Mozambican isolate was between 95 and 97%, indicating that the nucleotide substitutions were of the synonymous type, while the Japanese isolate presented amino acid identities between 95 and 98%, indicating that the substitutions were of the synonymous type.
Figure 4 and
Figure 5 illustrate the phylogenetic trees constructed based on the nucleotide and amino acid sequences, respectively, of the S-DNA. There was a clear subdivision of isolates into two clades, one with isolates from China, Japan, the Philippines, Taiwan and Indonesia and one with Mozambican isolates and the remaining isolates from
GenBank.
The Mozambican isolates were grouped into the same clade as the isolates from Myanmar, Malawi, Rwanda and DRC. Although isolates from Myanmar, Australia, Burundi, Egypt, Tonga, Pakistan and China were grouped in the same clade as the Mozambican isolates, they occupied different subclades.
In the tree constructed based on amino acid sequences, in addition to the isolates from Malawi and Rwanda, the isolates from Myanmar, Pakistan, Tonga and Egypt were also grouped with the Mozambican isolates. This must have occurred because the nucleotide substitutions are synonymous. In this tree, some isolates moved and occupied other subclades. Isolate BBTV_271_F1 occupied a subclade together with isolate AB252644 from Miami and isolate KM607505 from the DRC; isolate BBTV_292_F2 was placed in a subclade close to isolates from China, Japan, the Philippines, Indonesia and Taiwan, while isolates BBTV_286_F2 and BBTV_179_F11 were placed in different subclades and clades.
3.3. Analysis of the BBTV R DNA Sequences
Like the S-DNA, the R-DNA gene was also highly conserved. Except isolates BBTV_286_F2 and BBTV_273_F1, which presented identities between 93 and 96% when compared to each other, all the other isolates presented identities between 98% and 100%. When compared with the PIO isolates from GenBank, the identity of these two isolates ranged from 94% to 96%, while all other Mozambican isolates exhibited identities between 96% and 99%. On the other hand, when compared with the SEA isolates, the two isolates presented identities between 87 and 89%, while the others presented identities between 90 and 93%.
When comparing the identities between the PIO isolates from GenBank, it was observed that they have 97% to 99% identity among themselves and between 90 and 93% identities with the SEA isolates. These results allowed us to infer that the isolates that occur in Mozambique are of the PIO type, i.e., they belong to the Indian Pacific Ocean group.
The amino acid identities between the two isolates cited above (BBTV_273_F1 and BBTV_286_F2) and the other Mozambican isolates were lower than that of the nucleotides, between 91 and 93%, indicating that the nucleotide substitutions were nonsynonymous.
The same was observed with isolate 3, whose nucleotide identity ranged from 94% to 99% and amino acid identity ranged from 91% to 97%. Most isolates presented identities of 100% when compared to each other and from 97% to 100% when compared to the isolates from the PIO group and 91% to 95% when compared to the isolates from the SEA group.
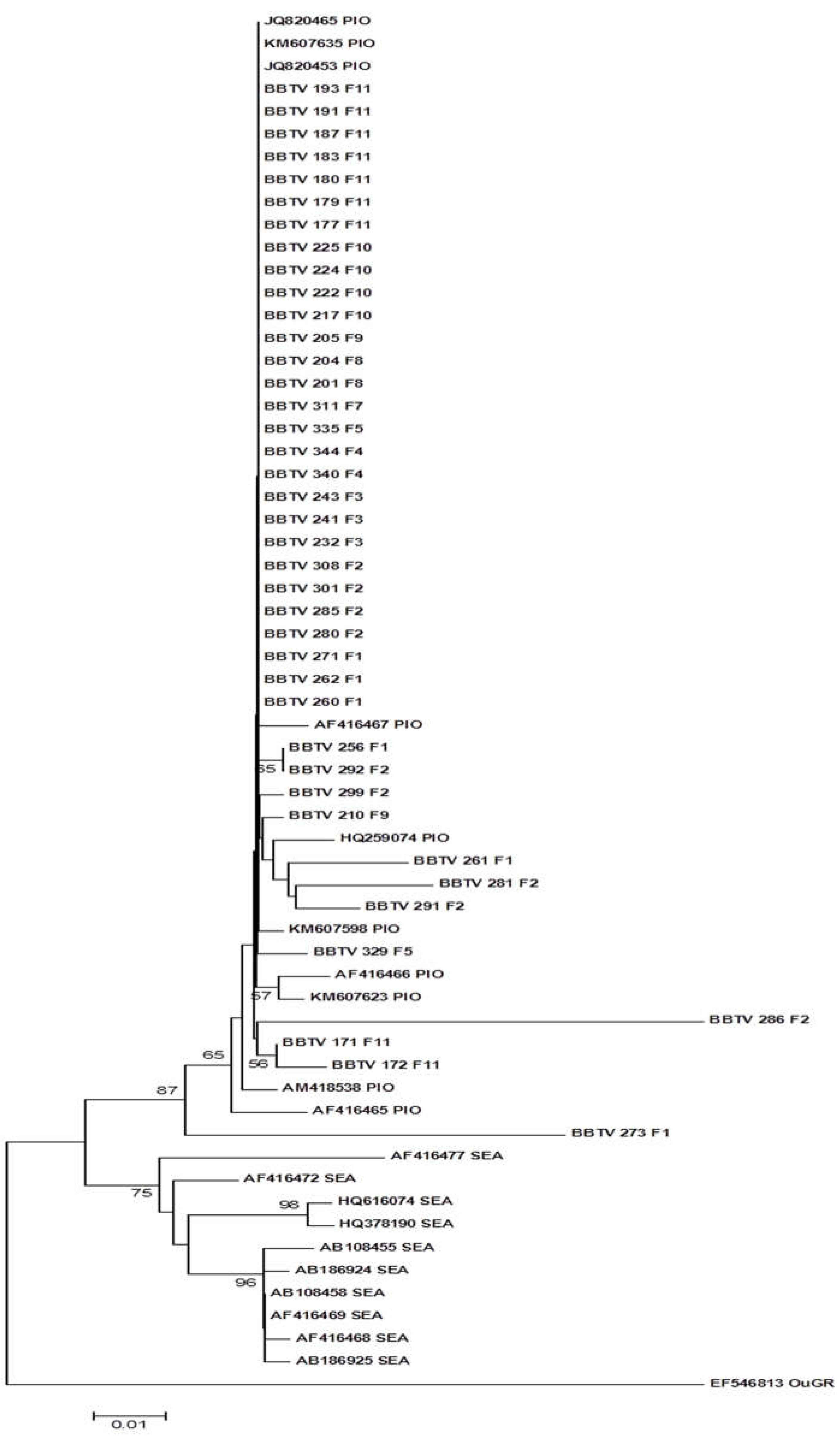
The phylogenetic tree based on the nucleotide sequences is illustrated in
Figure 6. The isolates were grouped into two distinct clades, one with the isolates of the PIO group and the other with the isolates of the SEA group. All Mozambican isolates were grouped into a subclade with the Malawi isolate JQ820453. The other isolates of the PIO group were grouped in the same clade but in a distinct subclade.
In the phylogenetic tree based on amino acid sequences (
Figure 7), the distribution was slightly different. There was also a separation of the isolates into two clades, one with the isolates of the PIO group and the other with the isolates of the SEA group. However, in addition to isolate JQ820453 from Malawi, isolates KM607635 from DRC, JQ820465 from Rwanda and, a little further away, AF416467 from Tonga also grouped together with most of the Mozambican isolates. Another eight isolates from Mozambique were mixed with isolates from different parts of the world in different combinations. Both the identity and phylogenetic results support the classification of the studied isolates within the PIO group.
4. Discussion
In this study, the R-DNA and S-DNA genes of Mozambican BBTV isolates from the Chókwè district of Gaza were sequenced and analyzed for the first time, revealing new information about their classification and possible geographical origin.
The nucleotide identities of the R-DNA among the 40 analyzed isolates revealed that 38 of them presented a difference of at most 2%, showing the high conservation of this gene, which has already been observed by authors from other regions of the world. Adegbola [
37] in Nigeria also found 100% identities among the local isolates. Another study conducted in Cameroon by Oben et al. [
38] showed a similar result, that is, 100% homology between nucleotide sequences of the analyzed isolates.
In phylogenetic studies performed by Karan et al. [
39] and reviewed by Yu et al. [
20], the classification of BBTV isolates, based on nucleotide sequences, into two groups, South Pacific group and the Asian group, was proposed. The Mozambican isolates showed higher identities with isolates from the PIO group, between 97% and 99%, and the lowest identities with the isolates from the SEA group, between 90% and 92%, indicating that the Mozambican isolates belong to the first group. These results were corroborated by the distribution of the isolates in the phylogenetic tree based on nucleotide sequences, which showed a clear subdivision of the isolates into two distinct clades, with all the Mozambican isolates grouped together with the isolates of the PIO group. Other authors studying isolates from different African countries also reported that the isolates belonged to the PIO group [
21,
37,
38,
40,
41,
42,
43].
Interestingly, the Mozambican isolates were very similar to isolate JQ820453 from Malawi, which borders Mozambique to the south and west. This demonstrates that there is a high probability that the Mozambican isolates came from this country. Kumar et al. [
8] had already considered Malawi as an area for the dissemination of BBTV through the exchange of planting material of preferred cultivars among farmers in bordering countries. However, it is necessary to consider that South Africa also has the presence of BBTV and shares a border with southern Mozambique. As there is no information on the complete gene sequences of the R-DNA in GenBank, it could not be included in this study. However, South Africa cannot be ruled out as the origin of the inoculants that reached Malawi and Mozambique, as it is geographically closer to southern Mozambique than Malawi.
Research on the introduction of BBTV in sub-Saharan Africa (SSA) indicated that the arrival of BBTV in Africa may have occurred in two ways: in the DRC in 1950, it was probably through infected propagules brought from southern Asia or the South Pacific [
44,
45,
46]; in Equatorial Guinea and Gabon, it may have been introduced by infected aphids (
Pentalonia nigronervosa) brought by migrant workers from the Philippines. Then, it is assumed that it has spread to other African countries (Rwanda, Burundi and Central African Republic) [
46,
47].
This virus is known to be widely prevalent in Central African countries and in Malawi, Southern Africa [
8]. To date, the occurrence of the disease has been reported in 16 countries of Central and Southern Africa (Angola, Benin, Egypt, Nigeria, Gabon, Burundi, Cameroon, Central African Republic, DRC, Congo, Equatorial Guinea, Malawi, Rwanda, Zambia, South Africa and Mozambique) [
48].
In the nucleotide sequence analysis of the S gene of the isolates from Mozambique, it was observed that it is also highly conserved, with more than 50% of the isolates being 100% identical, and the lowest identity was 98%. Additionally, in the case of the S gene, the greatest nucleotide identities were observed with the isolates of the PIO group, especially the African isolates JQ820467 from Rwanda, KM607470 from Egypt, KM607505 from the DRC, and JF755980 and JQ820455 from Malawi. The phylogenetic tree constructed based on nucleotide sequences, similar to that observed for the R gene, also indicated similarity to the isolates of the PIO group, especially isolates RJQ820467 from Rwanda and JQ820455 from Malawi.
Considering the danger that this disease poses to the banana industry and to food safety, urgent measures are needed to combat it, such as the installation of indexing laboratories and the availability of positive controls to increase confidence in the diagnosis based on PCR. It is also necessary to train the local team involved in teaching and extension activities and the farmers in the recognition and control of the virus. In this context, there is an urgent need to make farmers and government officials aware of the disease and the need to implement control measures, including (i) large-scale production and supply of virus free planting materials of the varieties preferred by farmers to rehabilitate banana production in the affected regions, preventing the use of infected planting material; (ii) raising awareness among farmers of the need to destroy infected material; (iii) greater phytosanitary surveillance and implementation of measures including restrictions on the movement of planting materials from regions affected by the disease, especially those bordering areas affected by BBTD; (iv) implementation of awareness programs among farmers, extensionists and regulatory agencies; (v) training to improve disease monitoring and diagnosis capacity; (vi) application of quarantine standards and integrated farming practices (ICP) aimed at reducing the spread of the disease and the aphid vector
Pentalonia niigronervosa; and (vii) promoting and developing robust research programs to identify resistant varieties, different strains of the virus and its origins as well as the genetic diversity existing in each banana production region in Mozambique. Such measures are also recommended by several researchers, such as James et al. [
31], Oben et al. [
38], Adegbola et al. [
37] and Ximba et al. [
49].
Considering that the presence of this virus in Mozambique and in neighboring countries constitutes a major threat to commercial crops and family banana producers, whose culture is considered subsistence for this segment of the population in Mozambique, these actions are essential and need to be implemented urgently and then supported by government agencies.
5. Conclusions
The results of this study support the classification of Mozambican isolates within the PIO group, as observed in Malawi. These and other results indicate that the introduction of BBTV in Mozambique from Malawi through the spread of infected material for planting is highly probable.
These results emphasize the need for intensive research to assess the extent of the geographical spread and severity of BBTV in Mozambique and for the implementation of quarantine and phytosanitary measures to prevent the internal and transboundary spread of this virus.
Author Contributions
All authors contributed to the study conception and design. The first draft of the manuscript was written by Sandra Carvalho I. Mussa Barros and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
The authors would like to thank the National Council for Scientific and Technological Development (CNPq), the Coordination for the Improvement of Higher Education Personnel (CAPES), Foundation for Research Support of the State of Minas Gerais (FAPEMIG) for their support in this project.
Data availability
Data are contained within the article.
Declaration
The authors declare that they do not have any potential technical or financial conflict of interest in relation to the entire content of the study presented.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dotto J, Matemu A, Ndakidemi P (2018) Potential of cooking bananas in addressing food security in East Africa. Int J Biosci 13(4):278-294. [CrossRef]
- Perrier X, De Langhe E, Donohue M, Lentfer C, Vrydaghs L, Bakry F, Carreel F, Hippolyte I, Horry J-P, Christophe J et al. (2011) Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc Natl Acad Sci 108(28):11311-11318.
- Sharrock S, Frison EA (1998) Musa production around the world-trends, varieties and regional importance. In: Network for the Improvement of Banana and Plantain. Montpellier: France.
- Karamura EB, Frison E, Karamura DA, Sharrock S (1999) Banana production Systems in Eastern and Southern Africa. In: Picq C, Fouré E, Frison EA (eds.). Bananas and Food security. International Network for the improvement of Banana an Plantain. France: Montpellier.
- Viljoen A, Kunert K, Kiggundu A, Escalant JV, Bornman CH (2004) Biotechnology for sustainable banana and plantain production in Africa: the South African contribution. S Afr J Bot 70(1):67–74. [CrossRef]
- Chabi MC, Dassou AG, Dossou-Aminon I, Ogouchoro D, Aman BO, Dansi A (2018) Banana and plantain production systems in Benin: ethnobotanical investigation, varietal diversity, pests, and implications for better production. J Ethnobiol Ethnomed 14(1):78. [CrossRef]
- MADER - Ministério de Agricultura e Desenvolvimento Rural (2018) Indústria da banana em Moçambique - situação de Foc TR4 e BBTV. Moçambique: MADER.
- Kumar PL, Hanna R, Alabi OJ, Soko MM, Oben TT, Vangu GHP et al. (2011) Banana bunchy top virus in sub-Saharan Africa: Investigations on virus distribution and diversity. Virus Res 159(2):171–182. [CrossRef]
- Jooste AEC, Wessels N, van der Merwe M (2016) First Report of Banana bunchy top virus in Banana (Musa spp.) from South Africa. Plant Dis 100(6):1251. [CrossRef]
- ICTV - International Committee on Taxonomy of Viruses (2021) International Committee on Taxonomy of Viruses 9th Report. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/ssdna-viruses 2011/w/ssdna_viruses/150/nanoviridae-figures Accessed 19 July 2022.
- Bell KE, Dale JL, Ha CV, Vu MT, Revill PA (2002) Characterisation of Rep-encoding components associated with banana bunchy top nanovirus in Vietnam. Arch Virol 147(4):695–707. [CrossRef]
- Hu JS, Wang M, Sether D, Xie W, Leonhardt KW (1996) Use of polymerase chain reaction (PCR) to study transmission of banana bunchy top virus by the banana aphid (Pentalonia nigronervosa). Ann Appl Biol 128(1):55–64.
- Burns TM, Harding RM, Dale JL (1995) The genome organization of banana bunchy top virus: analysis of six ssDNA components. J Gen Virol 76(6):1471–1482. [CrossRef]
- Herrera-Valencia VA, Dugdale B, Harding RM, Dale JL (2006) An iterated sequence in the genome of Banana bunchy top virus is essential for efficient replication. J Gen Virol 87(11):3409–3412. [CrossRef]
- Horser CL, Karan M, Harding RM, Dale JL (2001) Additional Rep-encoding DNAs associated with Banana bunchy top virus. Arch Virol 146(1):71–86. [CrossRef]
- Ji X, Yu N, Qu L, Li B, Liu Z (2019) Banana bunchy top virus (BBTV) nuclear shuttle protein interacts and re-distributes BBTV coat protein in Nicotiana benthamiana. 3 Biotech 9(4). [CrossRef]
- Amin I, Ilyas M, Qazi J, Bashir R, Yadav JS, Mansoor S et al. (2010) Identification of a major pathogenicity determinant and suppressors of RNA silencing encoded by a South Pacific isolate of Banana bunchy top virus originating from Pakistan. Virus Genes 42(2):272–281. [CrossRef]
- Yu N, Xie H, Yu Liang Zhang, Wang J, Zhou X, Zhi Xin Liu (2019) Independent modulation of individual genomic component transcription and a cis-acting element related to high transcriptional activity in a multipartite DNA virus. BMC Genomics 20(1). [CrossRef]
- Zhuang J, Lin W, Coates CJ, Shang P, Tàiyún Wèi, Wu Z et al. (2019) Cleavage of the Babuvirus Movement Protein B4 into Functional Peptides Capable of Host Factor Conjugation is Required for Virulence. Virol Sin 34(3):295–305. [CrossRef]
- Yu NT, Zhang YL, Feng TC, Wang JH, Kulye M, Yang WJ et al. (2012) Cloning and sequence analysis of two Banana bunchy top virus genomes in Hainan. Virus Genes 44(3):488–494. [CrossRef]
- Karan M, Harding RM, Dale JL (1994) Evidence for two groups of Banana bunchy top virus isolates. J Gen Virol 75(12):3541–3546. [CrossRef]
- UNDP - United Nations Development Programme (2007) MDG Achievement Fund – Environment and Climate Change Thematic Window. MDG-F Terms of Reference. Madrid: MDG-F https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.462.1402&rep=rep1&type=pdf Accessed 18 July 2021.
- Lodhi MA, Ye GN, Weeden NF, Reisch BI (1994) A simple and efficient method for DNA extraction from grapevine cultivars andVitis species. Plant Mol Biol Rep 12(1):6–13. [CrossRef]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33(7):1870–1874. [CrossRef]
- Islam, MN, Naqvi A, Jan A, Haq Q (2010) Genetic diversity and possible evidence of recombination among Banana Bunchy Top Virus (BBTV) isolates. Int Res J Microbiol 1(1):1-12.
- James AP (2011) Viruses of Banana in East Africa: Queensland University of Technology.
- Amin I, Qazi J, Mansoor S, Ilyas M, Briddon RW (2008) Molecular characterisation of Banana bunchy top virus (BBTV) from Pakistan. Virus Genes 36(1):191–198. [CrossRef]
- Stainton D, Martin DP, Muhire BM, Lolohea S, Halafihi M, Lepoint P et al. (2015) The global distribution of Banana bunchy top virus reveals little evidence for frequent recent, human-mediated long distance dispersal events. Virus Evol 1(1):1-15. [CrossRef]
- Furuya N, Kawano S, Natsuaki KT (2005) Characterization and genetic status of Banana bunchy top virus isolated from Okinawa, Japan. J Gen Plant Pathol 71(1):68–73. [CrossRef]
- Sharman M, Thomas JE, Skabo S, Holton TA (20078) Abacá bunchy top virus, a new member of the genus Babuvirus (family Nanoviridae). Arch Virol 153(1):135–147. [CrossRef]
- James AP, Soko MM, Mugini JA, Geijskes RJ, Dale JL, Harding RM (2012) Molecular characterisation of Banana bunchy top virus isolates from Malawi and Rwanda. Centre for Tropical Crops and Biocommodities. https://www.ncbi.nlm.nih.gov/nuccore/394326320 Accessed 15 July 2021.
- Bashir S, Naqvi SMS, Muhammad A, Hussain İ, Ali K, Khan MR et al. (2022) Banana bunchy top virus genetic diversity in Pakistan and association of diversity with recombination in its genomes. Plos One 17(3). [CrossRef]
- He Z et al. (2001) Cloning and sequencing of DNA component 1 of two BBTV strains. J Agric Biotechnol 9(2):145-148.
- Kawabe K, Onuki M (2002) Banana bunchy top virus coat protein (CP) gene. https://www.ncbi.nlm.nih.gov/nuccore/394326320 Accessed 10 Jan 2023.
- Rahayuniati RF, Subandiyah S, Hartono S et al. (2021) Recent distribution and diversity analysis on banana bunchy top virus of banana and alternative host in Indonesia. Trop Plant Pathol 46(5):506–517. [CrossRef]
- Furuya N, Natsuaki KT (2008) Identification of banana viruses in Myanmar. https://www.ncbi.nlm.nih.gov/nuccore/394326320. Accessed 21 Jan 2023.
- Adegbola RO, Ayodeji O, Awosusi OO, Atiri GI, Kumar PL (2013) First Report of Banana bunchy top virus in Banana and Plantain (Musa spp.) in Nigeria. Plant Dis 97(2):290–290. [CrossRef]
- Oben TT, Hanna R, Ngeve J, Alabi OJ, Naidu RA, Kumar PL (2009) Occurrence of Banana Bunchy Top Disease Caused by the Banana bunchy top virus on Banana and Plantain (Musa sp.) in Cameroon. Plant Dis 93(10):1076. [CrossRef]
- Karan M (1995) Sequence diversity of DNA fragments associated with Banana bunchy top virus. PhD thesis - Queensland University of Technology, Australia.
- Abdel-Salam SM, Dahot MU, Sadik AS (2012) Molecular comparative analysis of component 1 (DNA-R) of an Egyptian isolate of banana bunchy top nanovirus isolated from banana aphid (Pentalonia nigronervosa). J Genet Eng Biotechnol 10(1):55–65. [CrossRef]
- Karan M, Harding RM, Dale JL (1997) Association of Banana bunchy top virus DNA components 2 to 6 with bunchy top disease. Mol Plant Pathol 1-7.
- Niyongere C, Lepoint P, Losenge T, Blomme G (2015) Towards understanding the diversity of banana bunchy top virus in the Great Lakes region of Africa. Afr J Agric Res 10(7):702–709.
- Wanitchakorn R, Harding RM, Dale JL (2000) Sequence variability in the coat protein gene of two groups of banana bunchy top isolates. Arch Virol 145(3):593–602. [CrossRef]
- Dale JL (1987) Banana Bunchy Top: An Economically Important Tropical Plant Virus Disease. Adv Virus Res 33:301–325. [CrossRef]
- Fouré E, Manser PD (1982) Note sur l’apparition au Gabon d’une grave maladie virale des bananiers et plantains: le bunchy top. Fruits 37(6): 409-414.
- Wardlaw CW (1961) Banana diseases including plantain and abaca. London: Longmans, Green.
- Jeger MJ, Eden-Green SJ, Thresh JM, Johanson A, Waller JM, Brown AE (1995) Banana Diseases. In: Gowen S (ed.). Bananas and Plantains. London: Chapman and Hall.
- EPPO – European and Mediterranean Plant Protection Organization (2021) Global Database. Banana bunchy top vírus (BBTV00). https://gd.eppo.int/taxon/BBTV00/distribution Accessed 15 July 2021.
- Ximba SPF, Tshabalala J, Gubba A, Jooste AEC (2022) Monitoring the distribution of Banana bunchy top virus in South Africa: a country-wide survey. Arch Virol 167(6):1433–1441. [CrossRef]
Figure 1.
Geographic distribution of BBTV isolates collected in the Chókwè district, Gaza Province, Mozambique (indicated with red dots).
Figure 1.
Geographic distribution of BBTV isolates collected in the Chókwè district, Gaza Province, Mozambique (indicated with red dots).
Figure 2.
Electrophoretic analysis of PCR products generated by the DNA S gene of BBTV isolates in fourteen different samples: 1: 1Kb marker; line of 2 to 15 samples that tested positive among those analyzed; 16: negative control; 17: positive control.
Figure 2.
Electrophoretic analysis of PCR products generated by the DNA S gene of BBTV isolates in fourteen different samples: 1: 1Kb marker; line of 2 to 15 samples that tested positive among those analyzed; 16: negative control; 17: positive control.
Figure 3.
Electrophoretic analysis of PCR products amplified from the R DNA of BBTV isolates in fourteen different samples. 1: 1Kb marker; 2 to 15: band pattern of the amplified isolates; 16: negative control; 17: positive control.
Figure 3.
Electrophoretic analysis of PCR products amplified from the R DNA of BBTV isolates in fourteen different samples. 1: 1Kb marker; 2 to 15: band pattern of the amplified isolates; 16: negative control; 17: positive control.
Figure 4.
Phylogenetic tree constructed based on the nucleotide sequence of the S DNA of BBTV isolates collected in the district of Chókwè – Mozambique and isolates from GenBank. Bootstrap values were obtained using the MEGA 7.0 and Neighbor Joining program with 2,000 repetitions.
Figure 4.
Phylogenetic tree constructed based on the nucleotide sequence of the S DNA of BBTV isolates collected in the district of Chókwè – Mozambique and isolates from GenBank. Bootstrap values were obtained using the MEGA 7.0 and Neighbor Joining program with 2,000 repetitions.
Figure 5.
Phylogenetic tree constructed based on the amino acid sequence of the S DNA of BBTV isolates collected in the district of Chókwè – Mozambique and isolates from GenBank. Bootstrap values were obtained using the MEGA 7.0 and Neighbor Joining program with 2,000 repetitions.
Figure 5.
Phylogenetic tree constructed based on the amino acid sequence of the S DNA of BBTV isolates collected in the district of Chókwè – Mozambique and isolates from GenBank. Bootstrap values were obtained using the MEGA 7.0 and Neighbor Joining program with 2,000 repetitions.
Figure 6.
Phylogenetic tree constructed based on the nucleotide sequence of the R DNA of BBTV isolates collected in the district of Chókwè – Mozambique and isolates from GenBank. Bootstrap values were obtained using the MEGA 7.0 and Neighbor Joining program with 2,000 repetitions.
Figure 6.
Phylogenetic tree constructed based on the nucleotide sequence of the R DNA of BBTV isolates collected in the district of Chókwè – Mozambique and isolates from GenBank. Bootstrap values were obtained using the MEGA 7.0 and Neighbor Joining program with 2,000 repetitions.
Figure 7.
Phylogenetic tree constructed based on the amino acid sequence of the R DNA of BBTV isolates collected in the district of Chókwè – Mozambique and isolates from GenBank. Bootstrap values were obtained using the MEGA 7.0 and Neighbor Joining program with 2,000 repetitions.
Figure 7.
Phylogenetic tree constructed based on the amino acid sequence of the R DNA of BBTV isolates collected in the district of Chókwè – Mozambique and isolates from GenBank. Bootstrap values were obtained using the MEGA 7.0 and Neighbor Joining program with 2,000 repetitions.
Table 1.
Primers used for amplification of the R and S Genes with the respective annealing temperatures and amplified fragment.
Table 1.
Primers used for amplification of the R and S Genes with the respective annealing temperatures and amplified fragment.
| Primer |
Sequence
(5′-3′) |
Gene |
Temp
(°C) |
Size
(bp) |
| BBTV 67SF |
CGTTTAGATGGGTTTGGG |
DNA S |
49 |
718 |
| BBTV 784SR |
CGTTTAGATGGGTTTGGG |
| BBTV 32RF |
CGGGACGGGACATTTGC |
DNA R |
52 |
1007 |
| BBTV 1038SR |
CGTTAGGATAGCACAATC |
Table 2.
BBTV isolates of R DNA and S DNA and the number of accessions available in GenBank, used for comparison with the Mozambican isolates.
Table 2.
BBTV isolates of R DNA and S DNA and the number of accessions available in GenBank, used for comparison with the Mozambican isolates.
BBTV
Group * |
DNA R |
DNA S |
| Origin |
GenBank |
Reference |
Origin |
GenBank |
Reference |
| PIO |
Egypt |
HQ259074 |
lslam et al. [25] |
Egypt |
KM607471 |
Stainton et al. [28] |
| PIO |
Egypt |
AF416465 |
Horser et al. [15] |
Egypt |
KM607470 |
Stainton et al. [28] |
| PIO |
Fiji |
AF416466 |
Horser et al. [15] |
Tonga |
KM607584 |
Stainton et al. [28] |
| PIO |
Malawi |
JQ820453 |
James et al. [26] |
Malawi |
JF755980 |
Kumar et al. [8] |
| PIO |
Rwanda |
JQ820465 |
James et al. [26] |
Rwanda |
JQ820467 |
James et al. [31] |
| PIO |
Tonga |
AF416467 |
Horser et al. [15] |
Tonga |
KM607550 |
Stainton et al. [28] |
| PIO |
Pakistan |
AM418538 |
Amin et al. [27] |
Pakistan |
MK140621 |
Bashir et al. [32] |
| PIO |
DRC |
KM607635 |
Stainton et al. [28] |
DRC |
KM607505 |
Stainton et al. [28] |
| PIO |
Australia |
KM607623 |
Stainton et al. [28] |
Australia |
KM607489 |
Stainton et al. [28] |
| PIO |
Burundi |
KM607598 |
Stainton et al. [28] |
Burundi |
KM607456 |
Stainton et al. [28] |
| SEA |
China |
HQ616074 |
Yu et al. [20] |
China |
KM607536 |
Stainton et al. [28] |
| SEA |
China |
HQ378190 |
Yu et al. [20] |
China |
AF238876 |
He et al. [33] |
| SEA |
Japan |
AB108458 |
Furuya, Kawano and Natsuaki [29] |
Japan |
AB078023 |
Kawabe and Onuki [34] |
| SEA |
Indonesia |
AB186924 |
Furuya, Kawano and Natsuaki [29] |
Indonesia |
MT433375 |
Rahayuniati et al. [35] |
| SEA |
Vietnam |
AF416477 |
Bell et al. [11] |
Myanmar |
AB252644 |
Furuya and Natsuaki [36] |
| SEA |
Vietnam |
AF416472 |
Bell et al. [11] |
Myanmar |
AB252642 |
Furuya, Kawano and Natsuaki [29] |
| SEA |
Japan |
AB108455 |
Furuya, Kawano and Natsuaki [29] |
Malawi |
JQ820455 |
James et al. [31] |
| SEA |
Taiwan |
AF416468 |
Horser et al. [15] |
Taiwan |
KM607468 |
Stainton et al. [28] |
| SEA |
Filipinas |
AF416469 |
Horser et al. [15] |
Filipinas |
KM607469 |
Stainton et al. [28] |
| SEA |
Indonesia |
AB186925 |
Furuya, Kawano and Natsuaki [29] |
Indonesia |
MT433376 |
Rahayuniati et al. [35] |
| OuGR |
Malaysia |
EF546813 |
Sharman et al. [30] |
Malaysia |
NC_010316 |
Sharman et al. [30] |
Table 3.
Names of the BSV isolates collected Chokwe district, Gaza Province, Mozambique, with their respective accession number in GEnBAnk.
Table 3.
Names of the BSV isolates collected Chokwe district, Gaza Province, Mozambique, with their respective accession number in GEnBAnk.
| BSV Isolate Name |
GenBank Accession Number |
| DNA R |
DNA S |
| BBTV 252 F1 |
PP097007 |
PP096947 |
| BBTV 260 F1 |
PP097009 |
PP096949 |
| BBTV 261 F1 |
PP097010 |
PP096950 |
| BBTV 262 F1 |
PP097011 |
PP096951 |
| BBTV 271 F1 |
PP097012 |
PP096952 |
| BBTV 273 F1 |
PP097013 |
PP096953 |
| BBTV 280 F2 |
PP096991 |
PP096931 |
| BBTV 281 F2 |
PP096992 |
PP096932 |
| BBTV 285 F2 |
PP096995 |
PP096935 |
| BBTV 286 F2 |
PP096996 |
PP096936 |
| BBTV 291 F2 |
PP096999 |
PP096939 |
| BBTV 292 F2 |
PP097000 |
PP096940 |
| BBTV 299 F2 |
PP097001 |
PP096941 |
| BBTV 301 F2 |
PP097003 |
PP096943 |
| BBTV 308 F2 |
PP097005 |
PP096945 |
| BBTV 232 F3 |
PP096986 |
PP096926 |
| BBTV 241 F3 |
PP096988 |
PP096928 |
| BBTV 243 F3 |
PP096989 |
PP096929 |
| BBTV 340 F4 |
PP096983 |
PP096923 |
| BBTV 344 F4 |
PP096984 |
PP096924 |
| BBTV 329 F5 |
PP096980 |
PP096920 |
| BBTV 335 F5 |
PP096982 |
PP096922 |
| BBTV 311 F7 |
PP096979 |
PP096919 |
| BBTV 201 F8 |
PP096977 |
PP096917 |
| BBTV 204 F8 |
PP096978 |
PP096918 |
| BBTV 205 F9 |
PP096975 |
PP096915 |
| BBTV 210 F9 |
PP096976 |
PP096916 |
| BBTV 217 F10 |
PP096971 |
PP096911 |
| BBTV 222 F10 |
PP096972 |
PP096912 |
| BBTV 224 F10 |
PP096973 |
PP096913 |
| BBTV 225 F10 |
PP096974 |
PP096914 |
| BBTV 171 F11 |
PP096954 |
PP096894 |
| BBTV 172 F11 |
PP096955 |
PP096895 |
| BBTV 177 F11 |
PP096959 |
PP096899 |
| BBTV 179 F11 |
PP096961 |
PP096901 |
| BBTV 180 F11 |
PP096962 |
PP096902 |
| BBTV 183 F11 |
PP096963 |
PP096903 |
| BBTV 187 F11 |
PP096967 |
PP096907 |
| BBTV 191 F11 |
PP096968 |
PP096908 |
| BBTV 193 F11 |
PP096969 |
PP096909 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).