Prioritized in the current study were all pertinent institutional, national, and/or international standards concerning the use and care of humans and animals. All study techniques involving humans and animals were approved by the Cairo University Ethics Committee for Human and Animal Handling (ECAHCU), located at the Faculty of Pharmacy, Cairo University, Egypt, in compliance with the Weatherall report's recommendations. This committee's approval number is P-9-12-2022. Every effort was made to minimize the number of participants and the suffering of the animals utilized in the research.
Screening experimental study.
This study was conducted from September 2022 to November 2023 at Cairo University's pharmacy faculty in Egypt.
The pharmacology and toxicology department of Cairo University's pharmacy faculty in Egypt procured and authorized animal models.
Fibrin blood clots are introduced into an adult male rabbit model that is overweight and weighs around 2 kg. The rabbits were given a week of adjustment before the testing. with a 12-hour light-dark cycle, 50% humidity, and a regulated temperature of 25°C. The rabbits had access to fresh grass.
Methods
Sample collection:
The samples were grassland soils, obtained at a depth of 11 to 20 cm, and randomly picked from different government sites. Samples were stored in sterile containers at 4℃ until processing.
Isolation of fibrinolytic producing bacterial isolates:
Each soil sample weighed one gram. Ninety nine milliliters of sterile distilled water were added to 250 millilitre Erlenmeyer flasks. The mixture was shaken for two minutes at 400 rpm using a gyrator shaker. In order to plate the dilutions from 10-1 to 10-6 on mineral Fibrinogen agar( MFA) medium, which contains 0.5 g KCl, 0.5 g Mgso4, 1 g KH2PO4, 0.1 g FeSO4, 0.1 g Znso4, 2 g Fibrinogen, and 2% Agar, the soil suspensions were serially diluted in sterile distilled water. At 37 ℃, the medium's pH was adjusted to 7.3. For 48 hours, the plates were incubated at 37 ℃. On the MFA medium, only microbes capable of using Fibrinogen as their only source of carbon and nitrogen could proliferate. Following two rounds of purification using the streak plate technique, colonies exhibiting growth were placed on nutrient agar slants and maintained at 4 ℃. It was possible to subculture the positive isolated colonies on sheep blood agar and determine whether or not beta-haemolysis was present. Every four weeks, the chosen isolate was routinely sub-cultured, and the slant culture was kept refrigerated.
Inoculum preparation:
The inoculum of the studied bacterial isolate was created in
250 ml Erlenmeyer flasks with
50 ml of
pH 7 nutrient broth liquid. The medium was autoclaved at
121℃ for
15 min and then infected with a loopful of culture from a nutritional agar slant that had been left overnight. The inoculum was the inoculated flasks, which were shaken for a whole day at
150 rpm. [
20]
Identification of fibrinolytic enzymes producing bacterial isolates was performed afterwards through Gram staining, biochemical reactions and 16S rRNA sequencing technique.
Gram stain:
It divided bacteria into two groups according to the composition of their cell walls. On a microscope slide, the bacteria were treated with a crystal violet solution and then iodine, causing the cells to become purple. Gram-positive bacteria retained the stain when coloured cells were treated with a solvent like acetone or alcohol, whereas gram-negative organisms lost the stain and became colourless. When the gram-negative, transparent bacteria were mixed with the counter-stain safranin, they became pink.
Spore shape:
The spore staining technique was used to find this. The slide was cleaned with a Kim-wipe and alcohol to remove any fingerprints. Two circles were drawn with a Sharpie on the bottom of the slide. Using an inoculation loop, two small droplets of water were added to each circle. Using an aseptic technique, a very little number of germs were removed from the culture tube. Microorganisms were present in the droplet of water on the slide. Air dried the slide fully. The slide was heat-fixed by passing it through the flame three or four times with the smear side up. The slide took a long time to cool entirely. The stains were covered up using a piece of paper towel that was positioned inside the slide's border. Above the slide was a beaker filled with heated water. The paper towel was drenched in a malachite green liquid and the slide was left to steam for three to five minutes. The stained paper towel was taken out and disposed of. Water was used to carefully wipe the slide in order to remove any loose paper towel particles.Safranin was used as the counter-stain for a minute. The slide's bottom was dried before it was placed on the microscope's stage and examined using the oil immersion lens.
Spore site:
During the Gram stain test, the spore location was discovered.
Cell shape:
During the Gram stain test, the cell shape was determined.
Blood haemolysis:
On blood agar media, the microorganism's capacity to haemolyze the blood was tried.
Motility test:
It was able to distinguish between germs that moved and those that did not.
The medium was punctured with a sterile needle up to 1 centimetre from the tube's bottom in order to identify a motile colony and assess its isolation. Without a doubt, the needle was held in place both throughout its insertion and withdrawal from the medium. It needed to be incubated for eighteen hours at 35°C, or until there was observable growth. In order to help identify the bacterial isolates that produced the fibrinolytic enzyme, biochemical experiments were carried out later.
Lecithinase test:
Several species were isolated and their differentiation was hypothesized based on lecithinase activity using the differentiated and enriched medium called egg yolk agar. One prevalent ingredient in egg yolks was lecithin. The enzyme lecithinase had the ability to break down lecithovitellin, a lipoprotein component of egg yolk agar, into phosphorylcholine and an insoluble diglyceride, which resulted in the formation of a precipitate in the medium. Lecithin was broken down by microorganisms that possessed the enzyme lecithinase into phosphorylcholine and insoluble diglyceride, which formed a white, opaque precipitation zone that extended outside the colony's boundaries. An opaque halo surrounded a colony that grew on egg yolk agar media, indicating a positive lecithinase activity test result. The test organism was streaked on the plate and then removed in a loop. The plate was checked for an opalescent halo surrounding the inoculates after 24 hours at 35–37°C.
Methyl red test:
Before adding the methyl red pH indicator to the MR broth contaminated tube, the Methyl Red test was conducted. When an organism employed the mixed acid fermentation pathway and generated stable acidic end products, the acids in the medium overcame the buffers, creating an acidic environment.
Catalase test:
To investigate if it might make catalase, a small inoculum of a particular bacterial strain was added to a 3% Hydrogen Peroxide solution. The quick development of oxygen bubbles was noticed.
Oxidase test:
A small piece of filter paper was treated with 1% Kovács oxidase reagent and let to air dry. Using a sterile loop, a well-isolated colony was removed from a newly cultured( 18–24 hours) bacterial plate and rubbed onto prepared filter paper. Changes in colour were observed.
Citrate utilization:
After autoclaving the Simmon Koser's citrate medium at 15 pounds for 15 minutes, five millilitres were extracted.The test tube holding the melted citrate medium was tilted to provide a distinct slant and butt. The designated microbe samples were injected on the inclined medium using labelled tubes and sterile wire. The tubes were incubated at 37°C for a whole day. The colour change of the medium was observed.
Starch hydrolysis:
The bacterial plates were injected for 48 hours at 37°C. The plates were sprayed with an iodine solution using a dropper for 30 seconds following incubation. After that, extra iodine was gushed out. The region around the bacterial growth line was examined.
Gelatin hydrolysis:
Agar medium was made using 1% gelatin. Using an inoculating loop, the provided microbe was applied to the gelatin agar plates, resulting in a single centre streak. The plates were incubated at 37 °C for a whole day. The plates were covered with a HgCl2 solution. The plates were inspected after a little period of time. Test result was positive; there was a noticeable halo around the injected area that indicated gelatin hydrolysis.
Growth at 45 0C:
On nutrient agar media, growth was observed to be possible at 45°C.
Indol test:
Five drops of the Kovács reagent were added straight to the test tube holding the inoculation microbe. An Indol test that was positive was indicated by the reagent layer turning pink to red( a cherry-red ring) a few seconds after the reagent was added to the medium.
Tolerance salinity:
Its capacity to develop on nutrient agar while being responsive to 5% and 7% NaCl was examined.
Voges-Proskauer(VP) test:
For the test, Voges-Proskauer broth, a glucose-phosphate broth loaded with microorganisms, was added to alpha-naphthol and Potassium hydroxide. A successful outcome was indicated by a cherry red tint, whereas an unfortunate outcome was indicated by a yellow-brown color.
Triple sugar Iorn test( TSI):
The triple-sugar iron agar( TSI agar) method was used to assess the fermentation of glucose, lactose, and sucrose, either with or without gas production. It also looked at whether hydrogen sulphide might be produced by amino acids. Phenol red served as the pH indicator in this test medium. The result was a TSIA slant. Using an inoculating needle, the indicated organism was injected into the butt of the TSIA slant. After the inoculating needle was taken out, TSIA zigzag-shaped lifted the surface of the slanted section of the tube. We let the slant incubate for a whole day. Following the incubation period, any alterations in the tube were recorded.
The lactose and/or sucrose fermentation was shown by the slant color. The butt color suggested that glucose was fermenting. Agar bubbles, a sign of gas production, were seen. The medium became dark, indicating the production of H2S.
Glucose fermentation test:
The fermentation reactions of glucose were investigated using glucose purple broth. Peptone and the PH indicator bromcresol purple made up the purple broth. A 1% concentration of glucose was added. Isolated colonies from a 24-hour pure culture of microorganisms were added to the glucose purple broth as an inoculant. Parallel to the inoculation of the glucose-based medium, a control tube of purple broth base was used. The inoculated medium was incubated aerobically for 3 days at a temperature of 35–37 ℃. The medium began to become yellow, which was a sign of a successful outcome. A poor carbohydrate fermentation response was indicated by the lack of yellow color development.
Fructose fermentation test:
The inoculum of a pure culture was aseptically added to a sterile tube with phenol red fructose broth. The infected tube was incubated at 35–37 ℃ for 18–24 hours. An acidic pH change, shown by a colour shift from red to yellow, was a positive reaction.
Maltose fermentation test:
Aseptic transfer of a pure culture inoculum was performed to a sterile tube filled with phenol red maltose broth. The infected tube was incubated at 35–37 ℃ for 18–24 hours. An acidic pH change, shown by a colour shift from red to yellow, was a positive reaction.
Sucrose fermentation test:
The inoculum of a pure culture was aseptically moved to a sterile tube filled with phenol red sucrose broth. The infected tube was incubated at 35–37 ℃ for 24 hours. An acidic pH change, shown by a hue shift from red to yellow, was a positive reaction.
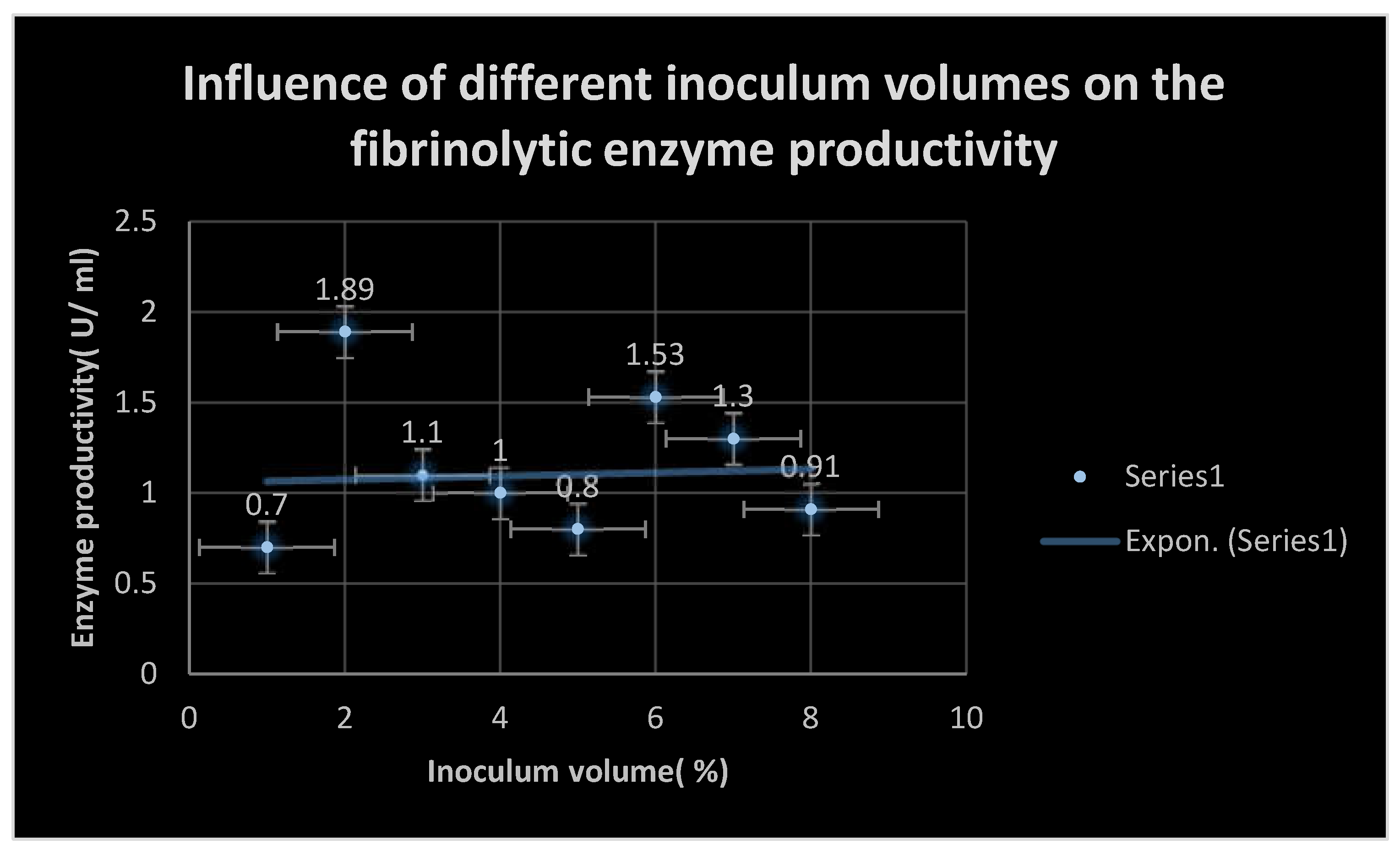
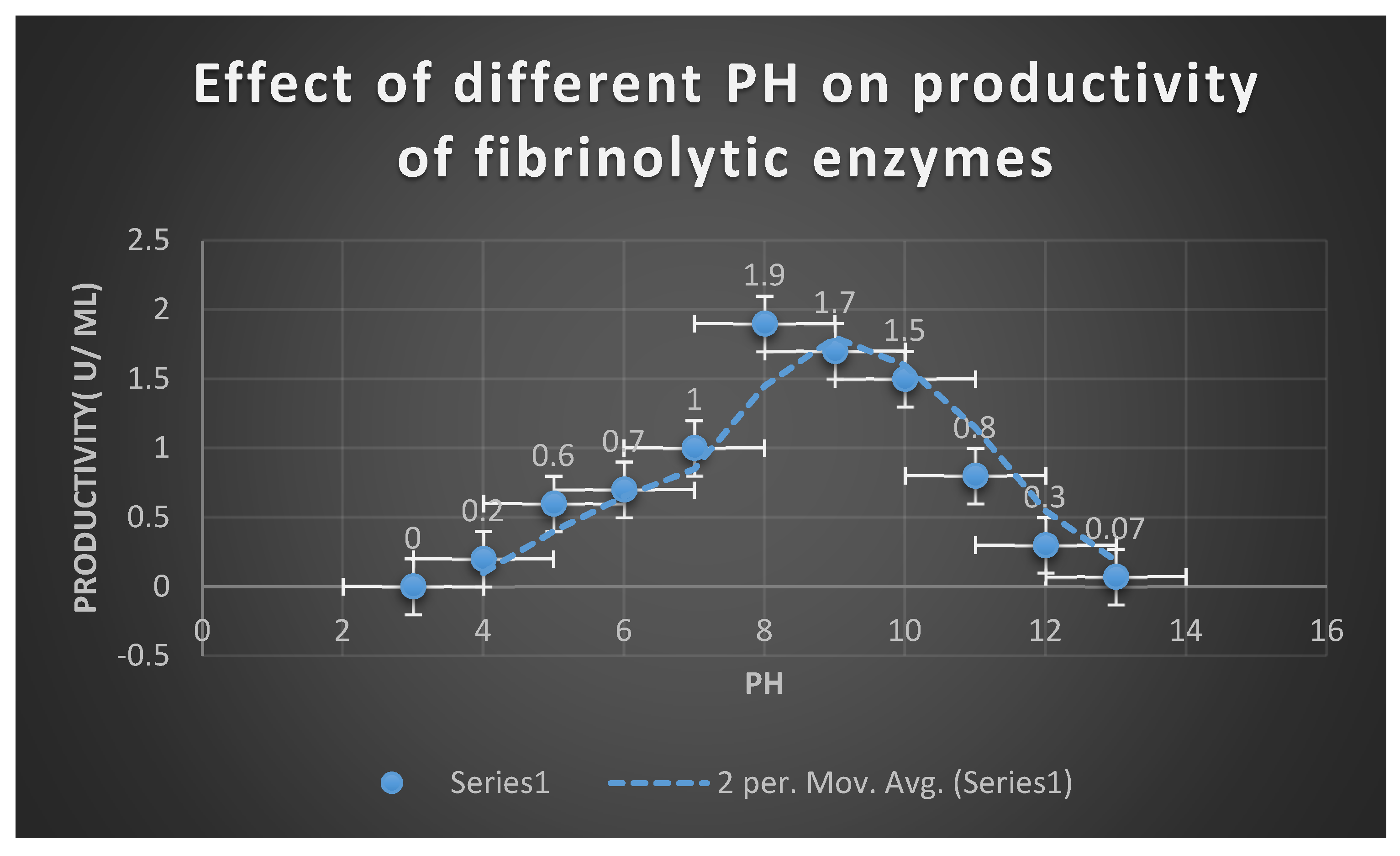
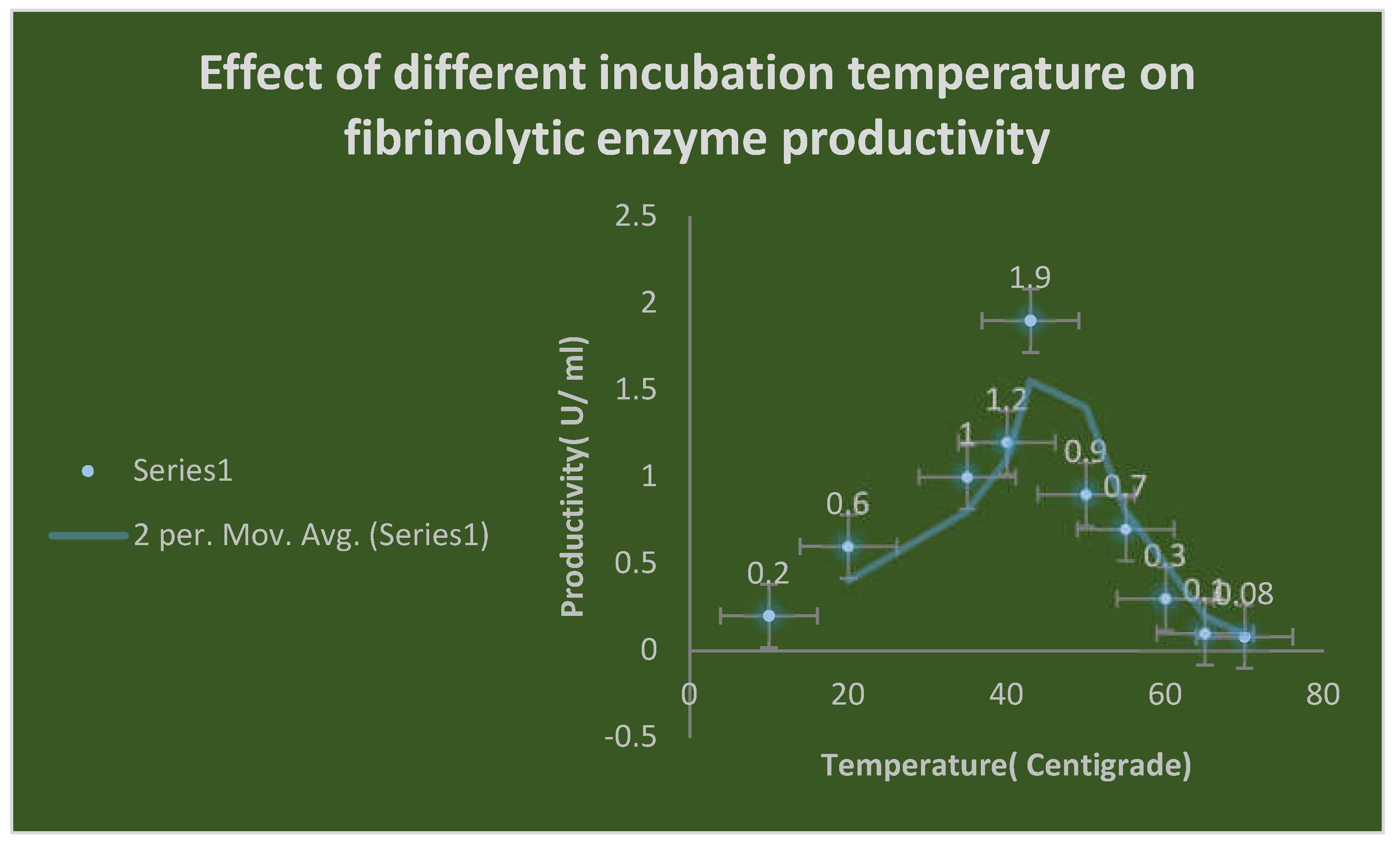
By cultivating multiple soil samples previously obtained on MFA, the ideal environmental and physiological factors affecting the growth of some selected bacterial isolates producing fibrinolytic enzymes, including metal ions such as Fe+2, Mg+2, Co+2, Mn+2, Ba+2, K+1, Na+1, and Ni+2 at a concentration of 10 mM, were determined. By measuring the fibrinolytic activity using the plasmin[ fibrin] plate test, the effects of different pH values[ 4–12], incubation temperatures[ 20–50 ℃], and inoculum quantities[ 1–10%] were also examined.
Optimization of soil bacterial fibrinolytic enzymes production:
The metal ions, nitrogen and carbon sources were obtained from Alnasr pharmaceutical chemical company, Abo-zabal, Alkhanka, Egypt.
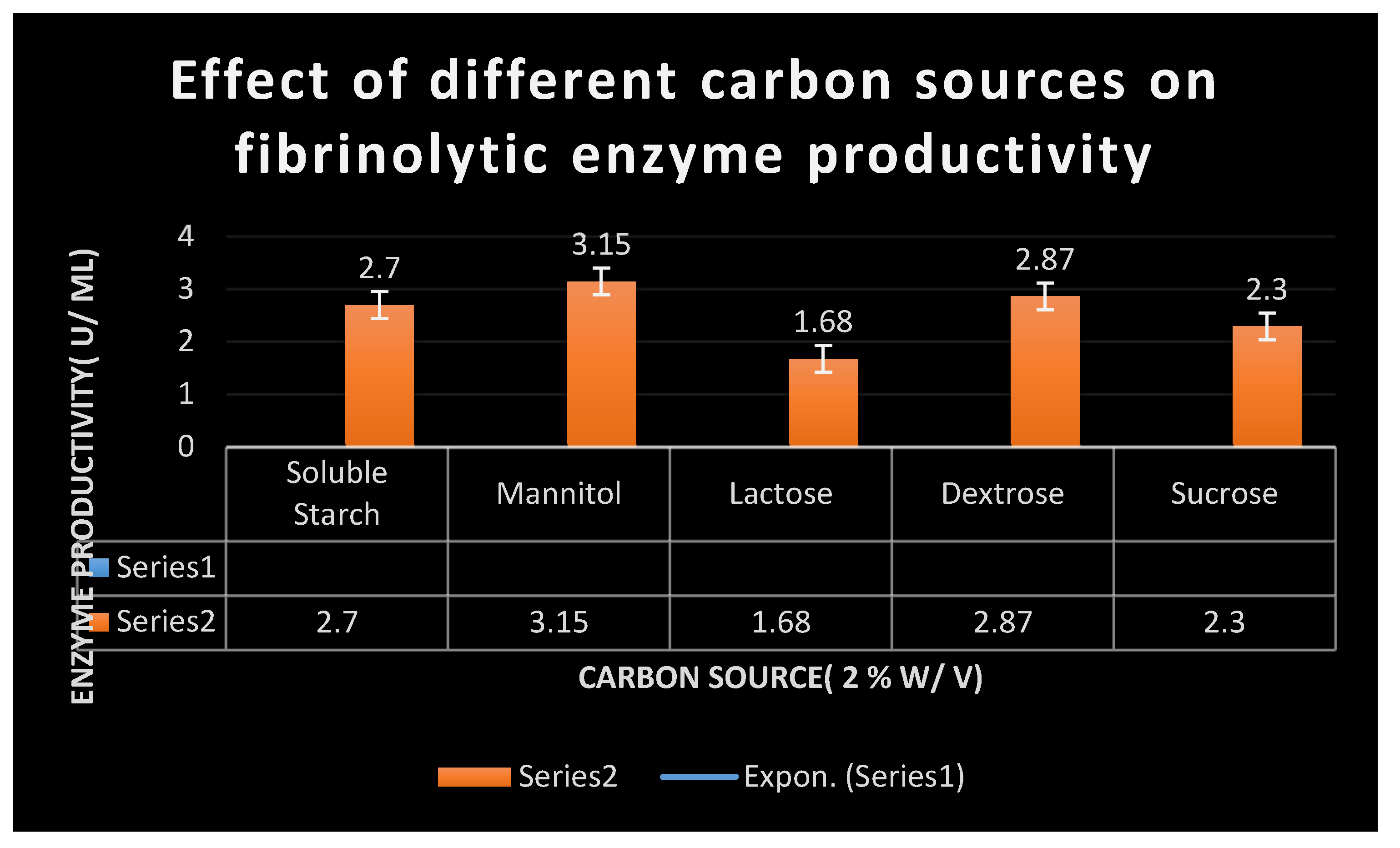
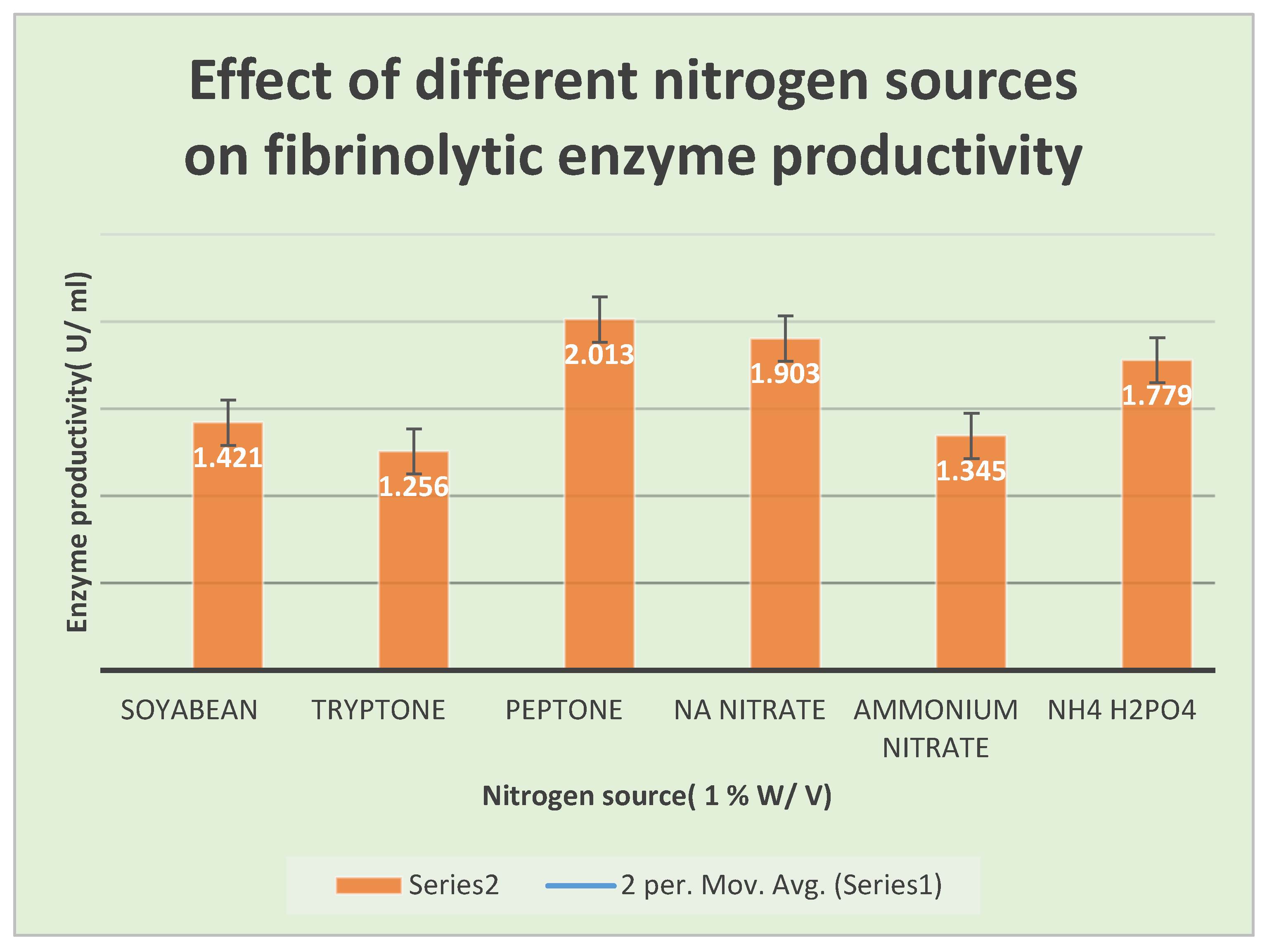
There have been investigations into several process variables that increase fibrinolytic enzymes yield. Peptone, Soyabean, and ammonium nitrate at a concentration of
1% W/V were used as nitrogen sources, and the effects of adding carbon sources such as sucrose, mannitol, dextrose and soluble starch at a concentration of
2% W/ V were also studied. The experiments were run in triplicate, and the mean results were presented. To determine the effective parameters and substrates, a series of fermentations were carried out in
500 ml Erlenmeyer flask containing
100 ml of
MFA medium with different carbon(
2% W/ V), nitrogen(
1% W/ V) and metal ion( 9 mM) sources as well as inoculation volumes(
1- 10%),
PH( 4-12) and temperatures(
20-500C). The carbon sources used were mannitol, dextrose, lactose and sucrose. The nitrogen sources were tryptone, peptone, soyabean, sodium nitrite, ammonium sulfate and ammonium nitrate. The salts of different metal ions including
CU +2, Ca +2, Mg +2, Fe +2, Mn +2, Zn +2, Co +2, Ni +2 and Ba +2 were utilized in the culture medium. [
21]
Molecular detection:
The predominant bacterial isolate with high fibrinolytic activity was identified using
16S rRNA sequencing and other biochemical tests. Nucleic acid was extracted from a swab by bead-beating in a buffered solution containing Phenol, Chloroform and Isoamyl alcohol. Variable region of
16S rRNA gene was then amplified from the resulting nucleic acid using
PCR. The genomic
DNA was extracted from 18 hours cultured cells using a
DNA purification kit[
PurreLinkTM Genomic DNA Mini Kit with Catalog number: K182002 was purchased from Invitrogen, USA] according to the protocol provided by the manufacturer of
DNA purification kit.
The 16S rRNA gene was amplified by
PCR[
PCR SuperMix kit was purchased from Invitrogen,
USA] using forward[
5-AGAGTTTGATCCTGGCTCAG-3-] and reverse[
5-GGTTACCTTGTTACGACTT-3-]
primers.
PCR amplicons from up to hundreds of samples were then combined and sequenced on a single run. The resulting sequences were matched to a reference database to determine relative bacterial abundances. Polymerase Chain Reaction(
PCR) is a powerful method for amplifying particular segments of
DNA. PCR uses the enzyme
PlatinumTM Taq DNA polymerase with catalog number
10966018[ purchased from Invitrogen,
USA] that directs the synthesis of
DNA from deoxynucleotide substrates on a single-stranded
DNA template.
DNA polymerase adds nucleotides to the 3` end of a custom-designed oligonucleotide when it is annealed to a longer template
DNA. Thus, if a synthetic oligonucleotide is annealed to a single-stranded template that contains a region complementary to the oligonucleotide,
DNA polymerase can use the oligonucleotide as a primer and elongate its 3` end to generate an extended region of double stranded
DNA. Denaturation is the initial
PCR cycle stage The
DNA template is heated to
94° C. This breaks the weak hydrogen bonds that hold
DNA strands together in a helix, allowing the strands to separate creating single stranded
DNA. Annealing is the second
PCR cycle. The mixture is cooled to anywhere from 50-70° C. This allows the primers to bind (anneal) to their complementary sequence in the template
DNA. Extension is the final step of PCR cycle. The reaction is then heated to 72° C, the optimal temperature for
DNA polymerase to act.
DNA polymerase extends the primers, adding nucleotides onto the primer in a sequential manner, using the target
DNA as a template. With one cycle, a single segment of double-stranded
DNA template is amplified into two separate pieces of double-stranded
DNA. These two pieces are then available for amplification in the next cycle. As the cycles are repeated, more and more copies are generated and the number of copies of the template is increased exponentially. The amplified
PCR product was sequenced using a genetic analyzer 3130XL[ purchased from Applied biosystems,
USA].
DNA sequence homology search analysis of the predominant bacterial isolate was achieved using Blast n algorithm at
NCBI website. [
22]
Upstream process:
Bacterial recombinant DNA production of fibrinolytic enzymes:
Primer for expression of FP,
Forward primer: TTCCAACAAAATGGACGTGA
Reverse primer: CAGGATAGCCGATTGTGCTT.
The expression vector was
pET-14b( purchased from Novagene in the United States). The expression vector was designed via GenSmart™ Design sortware. The promotor was
T7 Lac, and the tagged protein was
6x histidine linked to the C terminus of an molecule.
Escherichia coli BL21[ DE3] polys S served as the expression host. All of them were used in the manufacture of Fibrin-proteinase fibrinolytic enzyme using bacterial recombinant
DNA technology.
IPTG was the transcription process's inducer. The principal host for the plasmid synthesis and replication was
Escherichia coli DH5 [obtained from Stratagene corporation, USA]. Fibrin-proteinase genomic
DNA was isolated from
Bacillus cereus ATCC 14579 discovered in several Egypt soil conditions using restriction endonuclease type II enzymes[
DNA cutting enzymes]
Taq I and
Hae III. These cutting enzymes were bought from the German business Sigma-Aldrich. Furthermore, genomic
DNA was amplified using the polymerase chain reaction method before being sub-cloned to the prokaryotic expression vector
PET-14b using the same restriction endonuclease type II enzymes used to extract genomic
DNA from
Bacillus cereus ATCC 14579. This was followed by the transformation of pET-14b into the polys S expression host
Escherichia coli BL21[ DE3]. The addition of
IPTG stimulated transcription at the promotor site
T7 Lac, hence initiating gene expression. Luria agar and broth
[ LA, LB] were used for routine bacterial culture for 24 hours at 37
0C incubation temperature.
Ampicillin and/or
Kanamycin were added to the culture medium according to the manufacturer's guidelines. [
23]
Downstream process:
Purification was carried out using 500 ml of crude enzyme extract. For recombinant expression yield, a centrifuge tube was spun for 5 minutes at 4000 rpm. The expressed protein was extracted from the supernatant through salting out using 70% Ammonium sulfate.
Procedure of salting out using 70% Ammonium sulfate:
38 grams of ammonium sulfate were dissolved in
100 ml distilled water at
25 ℃ to prepare
70% saturated solution. The retentate was subjected to
70% W/ V Ammonium sulfate precipitation via stirring using a magnetic stirrer at
4 ℃ for over night; moreover the precipitate formed was centrifuged at
10000 rpm for
30 min in a centrifuge
4K15( purchased from Sigma-Aldrich, German) and the precipitate obtained was dissolved in a minimal quantity of
60 mM Tris-HCl buffer,
pH 7.2. Furthermore, the expressed protein was purified using
Ni-NTA Agarose Column affinity chromatography technique[
Ni-NTA Purification System with catalog number K95001 was purchased from Invitrogen, USA]. [
24]
Procedure of Ni-NTA Agarose Column affinity chromatography:
8 ml of cell lysate was added to a prepared purification column. To keep the resin suspended in the lysate solution, it was bound for
15–30 minutes at room temperature using gentle agitation on a rotating wheel. The resin was settled by low speed centrifugation at
500 rpm and carefully the supernatant was aspirated. The column was washed with
5 ml denaturing binding buffer by resuspending the resin and rocking for
3 minutes. The column was washed with
4 ml denaturing wash buffer(
pH 6.0) by resuspending the resin and rocking for two minutes; then the column was washed with
10 ml native wash buffer by resuspending the resin and rocking for
3 minutes. The column was clamped in a vertical position and the cap was snapped off on the lower end. The protein of interest was eluted with 9 ml native elution buffer.
An 1 ml of the supernatants after each wash were kept at
4º C for
SDS-PAGE analysis. [
25]
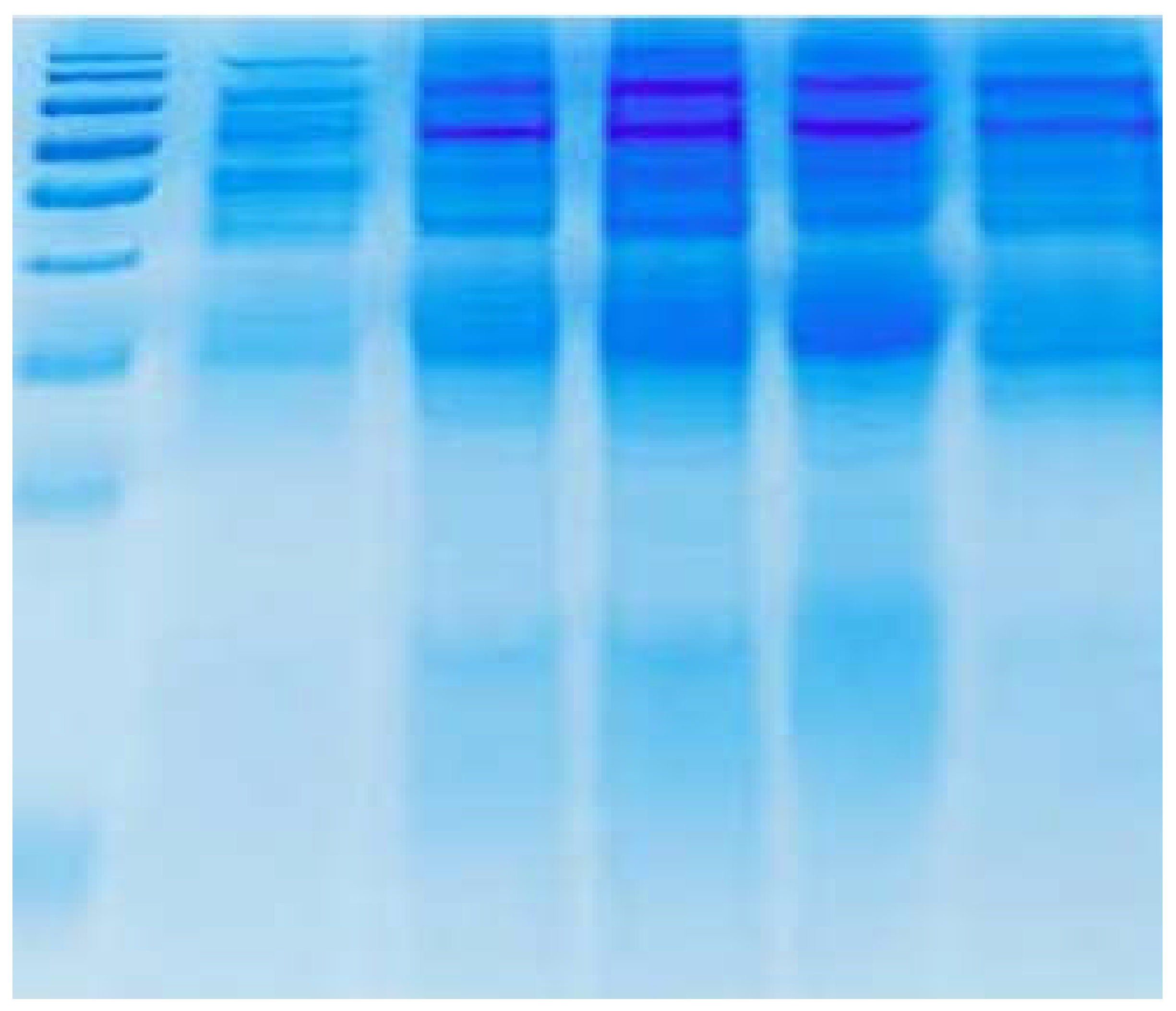
SDS-PAGE procedure:
SDS-PAGE Kit utilized during the present study was HiPer® SDS-PAGE Kit with Catalog number: HTP001 purchased from Hi-Media Laboratories, India. Procedure of SDS-PAGE was performed according to the manufacturer's instructions. The electrophoresis unit was assembled such that the glass plates were clamped to the unit along the spaces placed in-between them at two vertical edges. 1% Agarose( 0.5 g in 5 ml of distilled water) was prepared. The agarose was boiled to dissolve then a thin horizontal layer was poured the lower edge of the plates to seal the assembly. The agarose was allowed to solidify via allowing it to cool down for 5- 10 minutes. The preparation of 12% separating gel was carried out through the addition of the following:
The gel was poured in-between the plates and was allowed to solidify for an hour. Immediately after the gel was poured, the distilled water was added to level the gel. After one hour the water was poured off by inverting the casting assembly. 5% stacking gel was prepared through the addition of the following components:
After the addition of TEMED, the components were mixed gently by swirling the beaker. The stacking gel was poured was poured on the top of the separating gel and immediately the comb avoiding air bubbles was placed; as well as the gel was allowed to solidify for 30 minutes.
1X Tris-Glycine-SDS gel running buffer was poured in the unit such that the buffer connects the two electrodes, and hence completed the flow of current. The comb was removed from the stacking gel carefully. 5 µl of prestained protein ladder and 20 µl of samples immediately were loaded after the heat treatment in the wells created by the comb in the stacking gel. The power cord was connected to the electrophoertic power supply according to the conventions: red Anode and black Cathode. It was allowed to electrophorese at 100 volts and 10 mA until dye front reached 0.5 cm above the sealing gel. The gel was carefully removed from in-between the plates using spatula into the plastic tray containing distilled water. The gel was washed for 1 minute. The water was discarded and staining destaining procedure was proceeded. Staining and destaining of the gel included the addition of
50 ml of staining solution in the tray containing gel, till the bands were visible. The gel; afterwards was removed the staining solution. The staining solution could be reused
2-3 times; then the gel was washed by rinsing in distilled water till a considerable amount of stain was leached out from the gel. Changing the distilled water was kept for
3-4 times.
50 ml of destaining solution was added to the gel with constant moderate shaking. The destaining was continued till clear and distinct bands were observed. In the end of the process, the gel was removed from the destaining solution. After staining and destaining of the gel the molecular weights of samples were compared with that of the protein markers. Molecular weights were confirmed via a mass spectrometer. [
26]
Table 4a refers to the composition of
12 SDS-PAGE gel; while
Table 4b showed the components of
5% stacking gel.
Table 4a.
It demonstrates the components of 12% SDS-PAGE gel.
Table 4a.
It demonstrates the components of 12% SDS-PAGE gel.
| Component |
Volume |
| 30% Acrylamide-bis-acrylamide solution |
6 ml |
| Distilled water |
3 ml |
| 2.5 X Tris-SDS Buffer( PH 8.8) |
6ml |
| 10% APS solution |
125 µl |
| TEMED |
18 µl |
Table 4b.
It refers to the components of 5% stacking gel.
Table 4b.
It refers to the components of 5% stacking gel.
| Component |
Volume |
| 30% Acrylamide-bis-acrylamide solution |
1.3 ml |
| Distilled water |
5.1 ml |
| 5X Tris-SDS Buffer( PH 6.8) |
1.6 ml |
| 10% APS solution |
75 µl |
| TEMED |
10 µl |
Quantitative purified fibrinolytic protein estimation by lowry method:
The determination of of the unknown concentration of purified fibrinolytic enzyme in a sample was carried out using standard curve obtained through Lowry assay using bovine serum albumin as the standard, and the values were expressed as mg/ ml. [
27]
Determination of cellular position of fibrinolytic enzyme:
This was determined through spectrophotometric analysis of thrombolytic activity(
SATA) assay, standardized by tissue plasminogen activator(
tPA), which could quantitatively measure
in vitro thrombolytic activity. Blood clots were formed, uniformly, by mixing citrated whole blood with partial thromboplastin time(
PTT) reagent, together with calcium chloride. Then, designated concentrations of
tPA as a standard fibrinolytic drug, supernatants and cell lysates of different centrifuged soil samples containing test fibrinolytic enzymes at
500 rpm were added. The released red blood cells from each clot were quantified using spectrophotometry(
λmax=405nm) as an indicator of thrombolytic activity. [
28]
Assessment of secretion of antibodies to bacterial fibrinolytic enzyme:
The quantity of
IgG antibodies to the enzyme of interest in mouse serum was dictated via
ELISA.
100 µl of an emulsion containing
50 µg of
FP and an equal volume of complete Freund adjuvant[ purchased from Sigma Aldrich, USA) at 3 week intervals to 10 mice.Sera separated and used as primary antibodies. [
29]
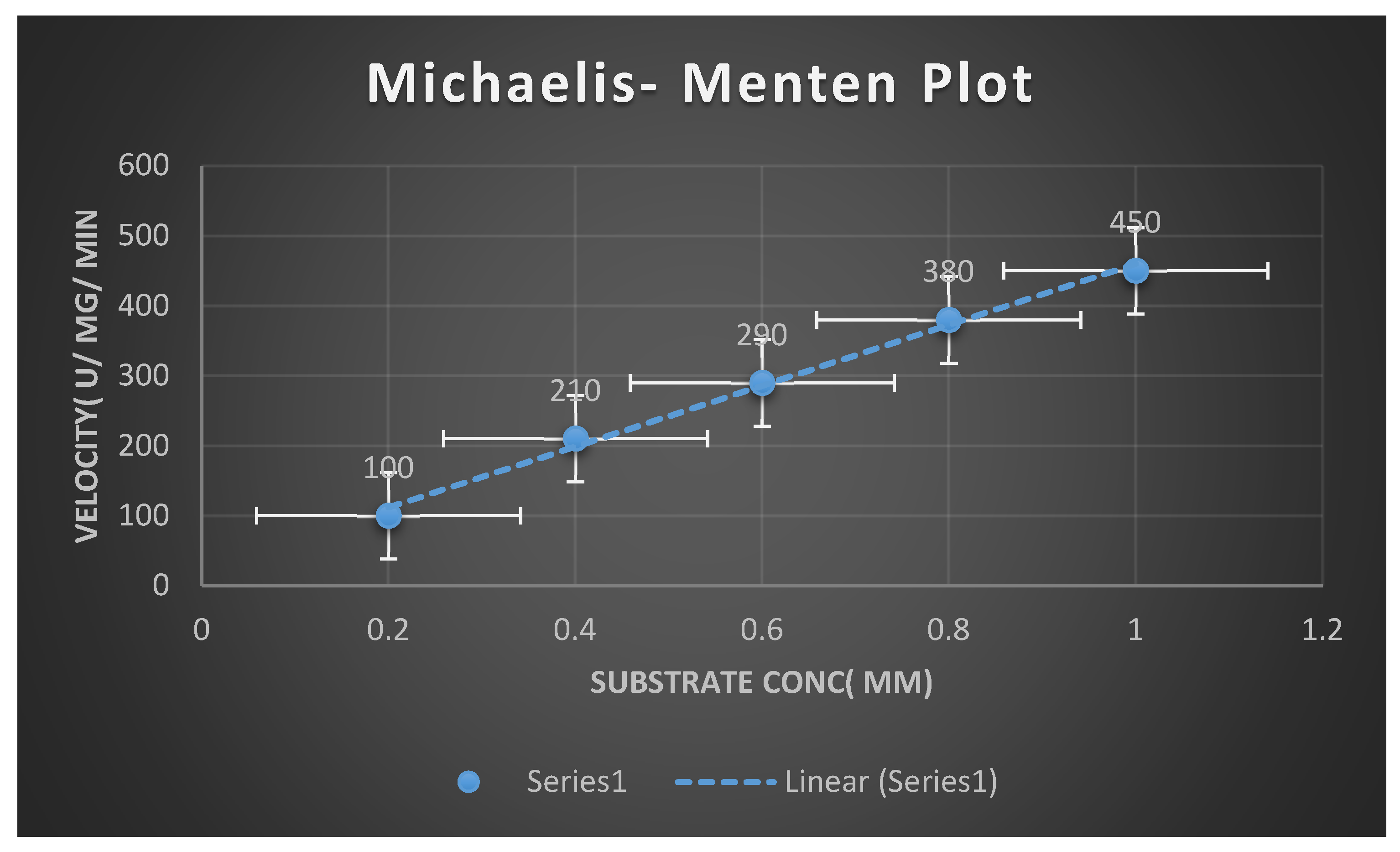
The kinetic parameters Km and Vmax determination:
The parameters of kinetic,
Michaelis–Menten constant(
Km) and maximum velocity (
Vmax) of purified fibrinolytic enzymes were ascertained with suited concentrations of fibrinogen(
2–11 mM) as the substrate. Data was connected to the nonlinear exponential stage union curve of regression. The outcome of fibrinolytic enzymes were computed aside measurement of the hydrolysis rate of fibrin clots below standard laboratory circumstances victimizing the equation of Michaelis-Menten. [
30]
Formulation of bacterial fibrinolytic enzymes:
The secreted fibrinolytic enzymes from the predominant bacterial isolate grown on SFA medium was formulated as a sterile solution intended for the utilization via parenteral routes of administration either
IV,
IM or
SC injections. Lyophphilized powder of recombinant
FP was obtained for preparation of a solution for injection of
50 units and
100 units in vials. The preparation was administered intravenously in the initial dose of
200 IU in
60 ml of isotonic solution of sodium chloride during
30 minutes(
25 drops per minute). Later, the administration of FP in the dose of
150 IU per hour was followed.
Table 5 demonstrates the composition of parenteral dosage form of
FP.
Compatibility studies of fibrinolytic enzyme and polymer:
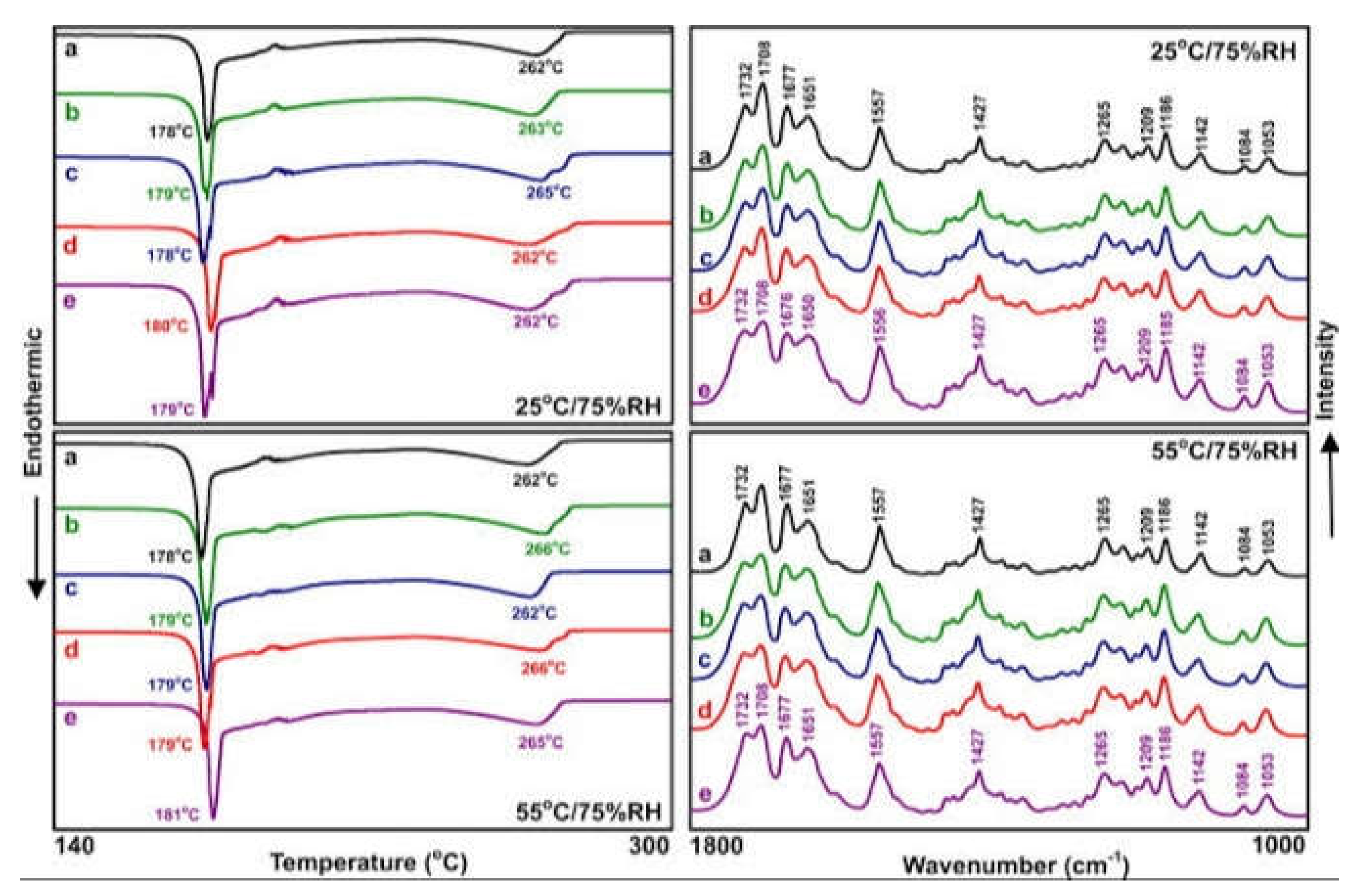
These were performed via FTIR spectroscopy and DSC. Recombinant FP and different excipients utilized in the preparation of intravenous and subcutaneous parenteral formulations were characterized by FT-IR( Perkins-Elmer 1600 FTIR spectrophotometer) spectroscopy and DSC( Shimadzu-DSC 50) to find the compatibility. The optimized formulations were blended with 100 mg KBr; then compressed into discs which were scanned at 5mm/ sec with a resolution of 1 cm-1 at a range of 4000-300 cm-1. Experiments of thermal analysis were carried out utilizing various scanning calorimeter( DSC). The samples of the optimized formulation were heated in hermetically sealed Aluminium pans at a temperature range of 0-4000 0C at a constant rate of 105 0C/minute under a purge of nitrogen( 40 ml/ min).
Testing the fibrinolytic activity of recombinant fibrinolytic enzymes:
This was achieved through the following calibration assays:
Fibrin plate method:
Sigma-Aldrich in the United States provided the human fibrinogen, plasmin, and thrombin. Using a fibrin plate test, fibrinolytic activity was ascertained. In a petri dish, agarose solution(
6 ml, 2% w/v, in 50 mM pH 7.4 sodium phosphate buffer), human fibrinogen (7 ml, 0.5% w/v, in 60 mM pH 7.4 sodium phosphate buffer), and
0.1 ml of
Thrombin(
5 NIH U/ml in 60 mM pH 7.4 sodium phosphate buffer) were combined. A fibrin clot layer formed after the solution was kept at room temperature for two hours. A
10 mm diameter well-punched fibrin plate was filled with
50 µl of the fibrinolytic enzyme-laden supernatant from various samples collected throughout the experiments. The plate was then incubated at
37°C for
24 hours. The lytic area was measured in order to quantify the enzyme activity. The quantity of enzyme in
30 µl of enzyme solution that created a clear zone measuring
1 mm2 at
pH 7.4 and
37°C for
18 hours is known as one unit of fibrinolytic enzyme activity. The
FEA of the samples was determined by creating a calibration plot with plasmin(
1000 U/ml) as the reference standard. [
31]
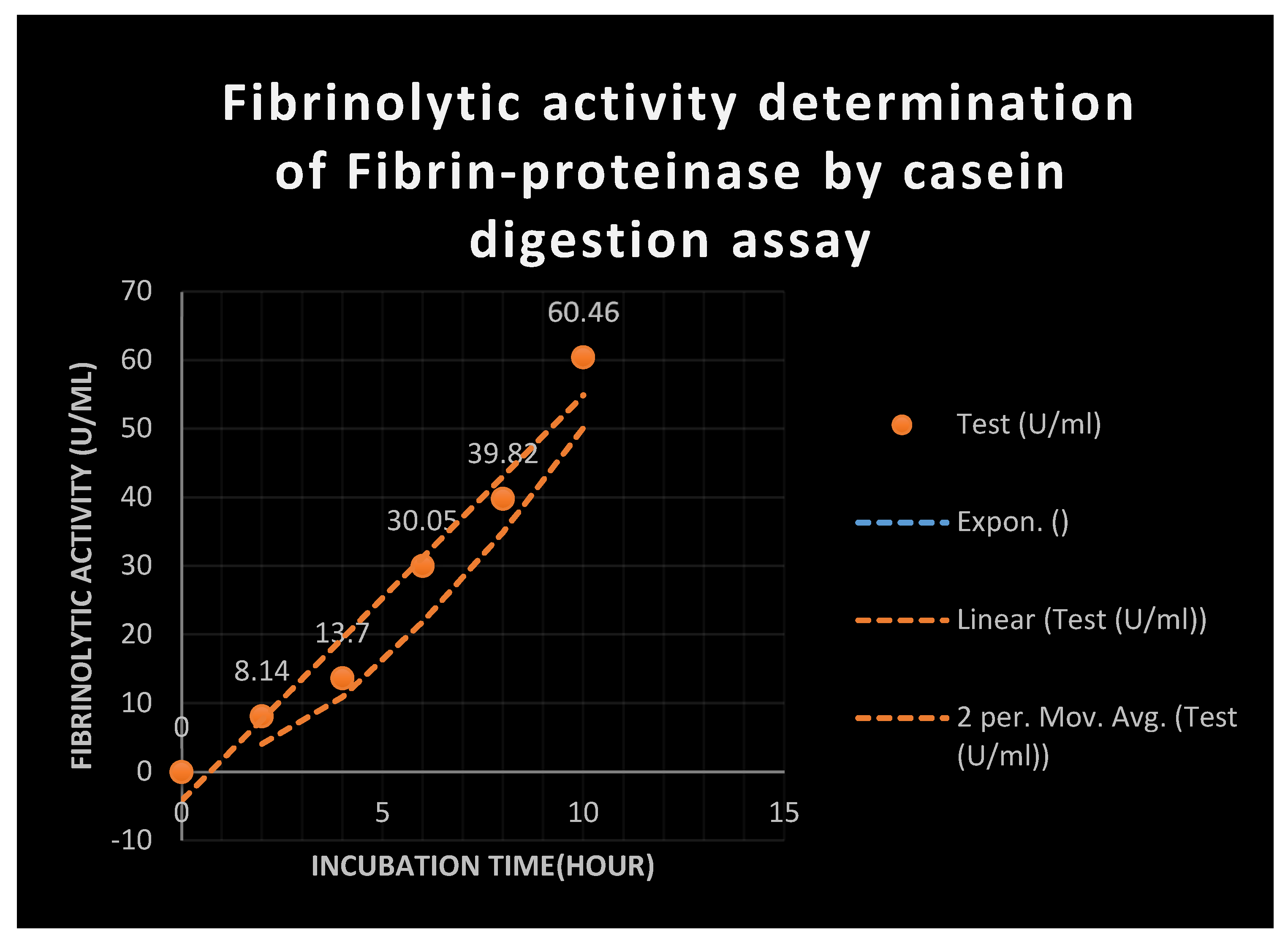
Casein digestion method:
Using the casein digestion method—which measures the amount of tyrosine produced from digested casein—fibrinolytic enzyme activity was ascertained. Utilizing the modified Sutar et al. technique, activity was confirmed to be 100% victimized (1986).
10 mg casein,
50 mM Tris-HCl,
pH 9.0, and 0.1 ml( or the suitable diluent) of supernatant were added to the reaction mixture(
3 ml). After
20 minutes at 37° C, the reactions were halted by adding
0.3 ml of
4.4 M HCl and
3.5 ml of 6% w/v trichloroacetic acid( TCA). After that, the reaction was placed on ice for thirty minutes, and then Whatman #1 filter paper was used for filtering. At
280 nm, the
TCA soluble fraction's absorbance was discussed. The standard plasmin curve was used to calculate the units of enzymatic activity. [
32]
Effect of plasminogen addition on fibrinolytic enzyme activity:
200 µl of 2% fibrin( pH 8
),
200 µl of 0.2 M Tris-HCl(
20 mM CaCl2, pH 7.8),
100 µl of the enzyme solution, and
100 µl of
plasminogen(
0.730 U) were all included in the incubation mixture. By employing the fibrin plate test, fibrinolytic activity was quantified and contrasted with the enzyme's activity in the absence of plasminogen. Five millilitres of 2.1% fibrin (pH 8) were added to four millilitres of
35 mM Tris-HCl( containing
0.80% agar and 0.20 M NaCl, pH 7.9) at
45°C to create
the fibrin plate. [
33]
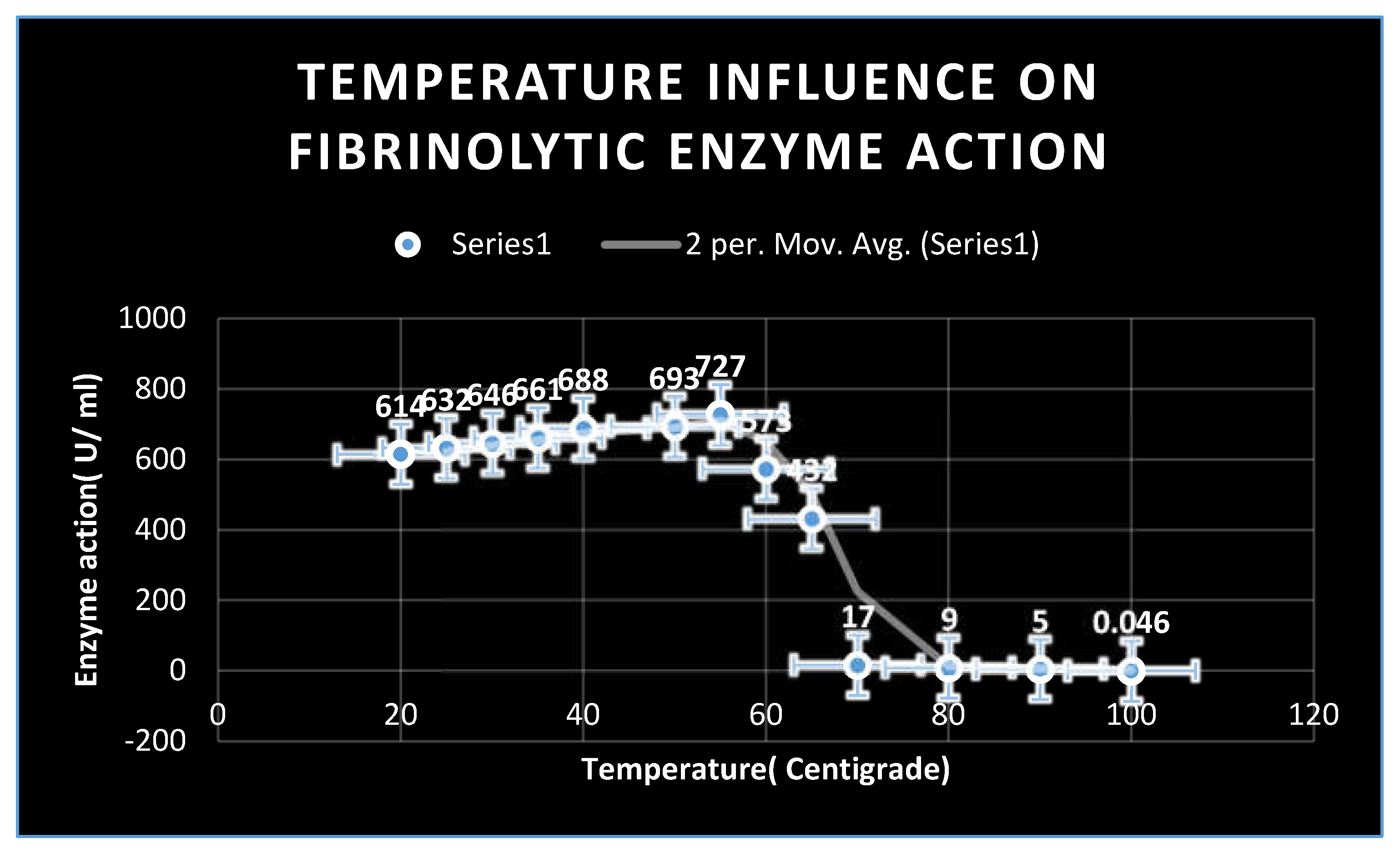
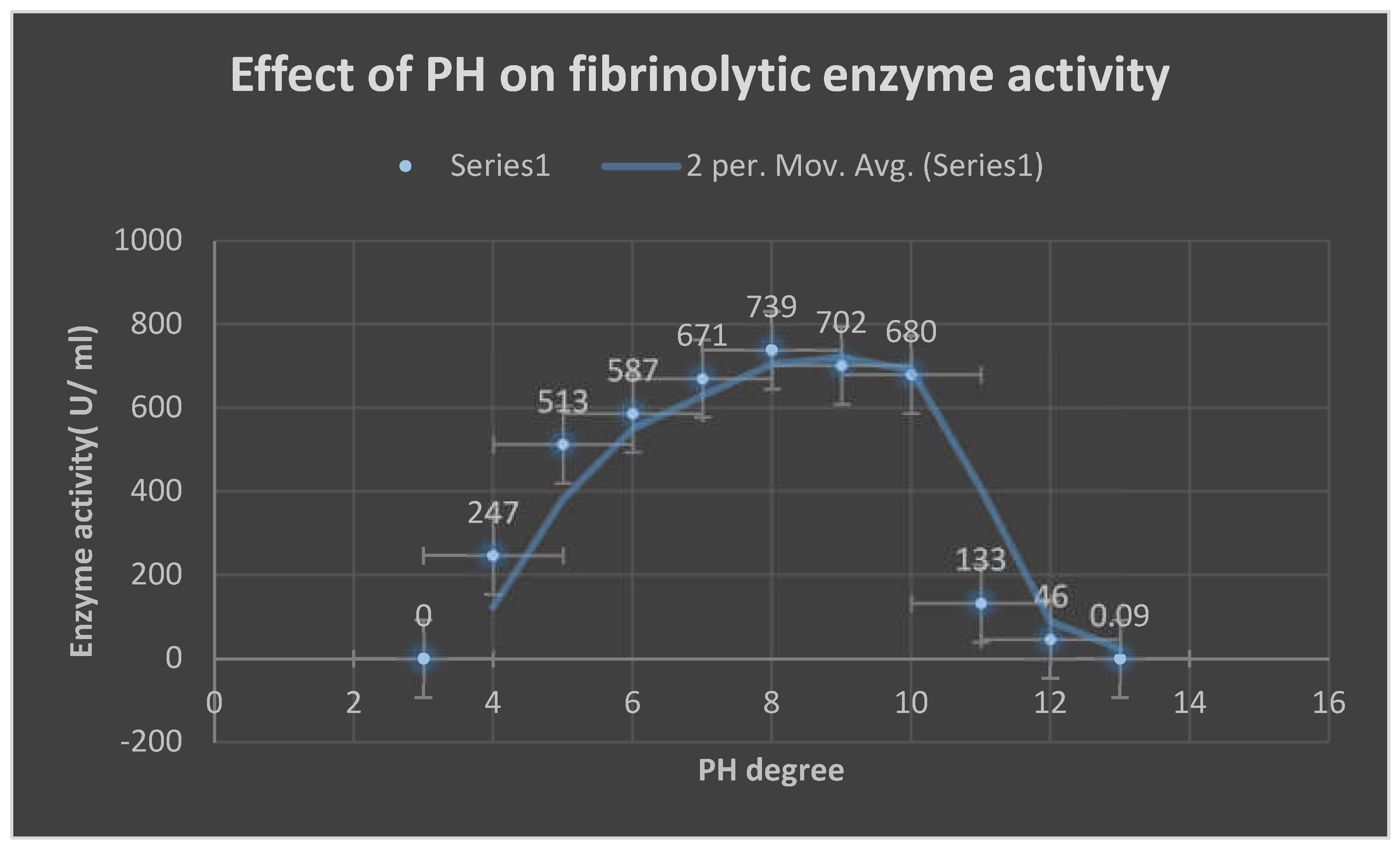
Effects of pH and temperature on enzyme activity:
The reaction mixture was incubated for two hours at temperatures ranging from 25 to 95 degrees Celsius in 605 millimolar phosphate buffer (pH 7.0) in order to establish the ideal temperature for the enzyme activity of extracellular fibrinolytic enzymes. The fibrin plate assay was used to measure enzyme activity. The pH range that was shown to be ideal for enzyme activity was pH 4.0 to pH 11.0. The enzyme was combined with several buffers in 65 mM sodium phosphate buffer (pH 7.0). The 0.2 M citrate-phosphate buffer(
pH 2.0-5.0), 65 mM sodium phosphate buffer (pH 6.0-7.0), Tris-Cl buffer(
pH 8.0-9.0), and
glycine-NaOH buffer(
pH 10.0-12.0) were the buffers that were employed. After the enzyme was incubated in
65 mM sodium phosphate buffer(
pH 6.5), the amount of activity that persisted was used to measure the thermostability of the enzyme activity. [
34]
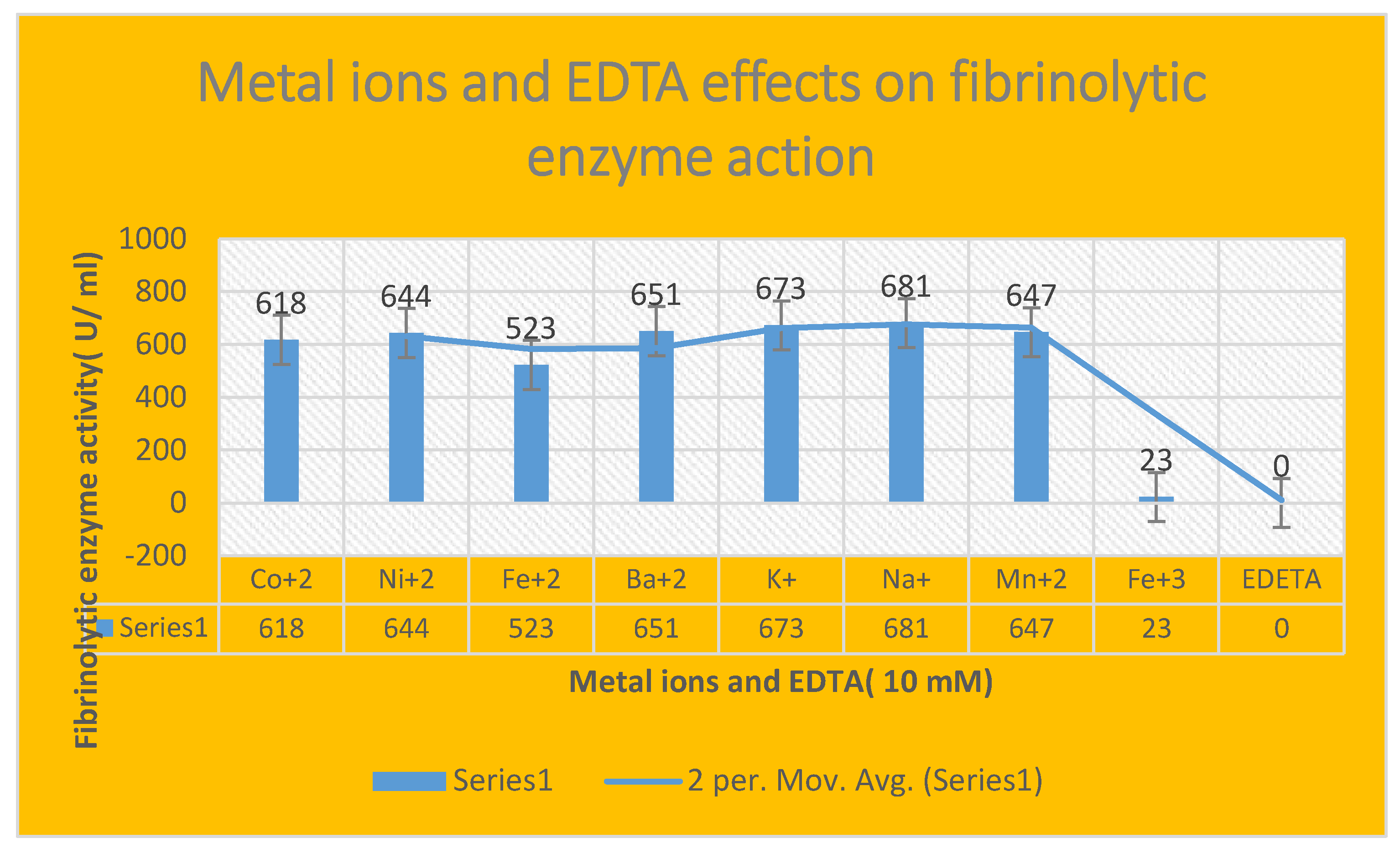
Effect of metallic ions on enzyme fibrinolytic activity:
The effect of the enzyme was investigated using
MgSO4, CoCl2, MnCl2, ZnCl2, FeSO4, CuSO4, and CaCl2. The partial purified protein was incubated in both the absence and the presence of metal ions including
Mg2+, Co2+, Mn2+, Zn2+, Fe2+, Cu2+ and Ca2+ with a final concentration of
10 mM in Tris buffer(
pH7.3) for
2 hrs at 37
o C and the fibrinolytic activity was assayed using fibrin plate assay. [
35]
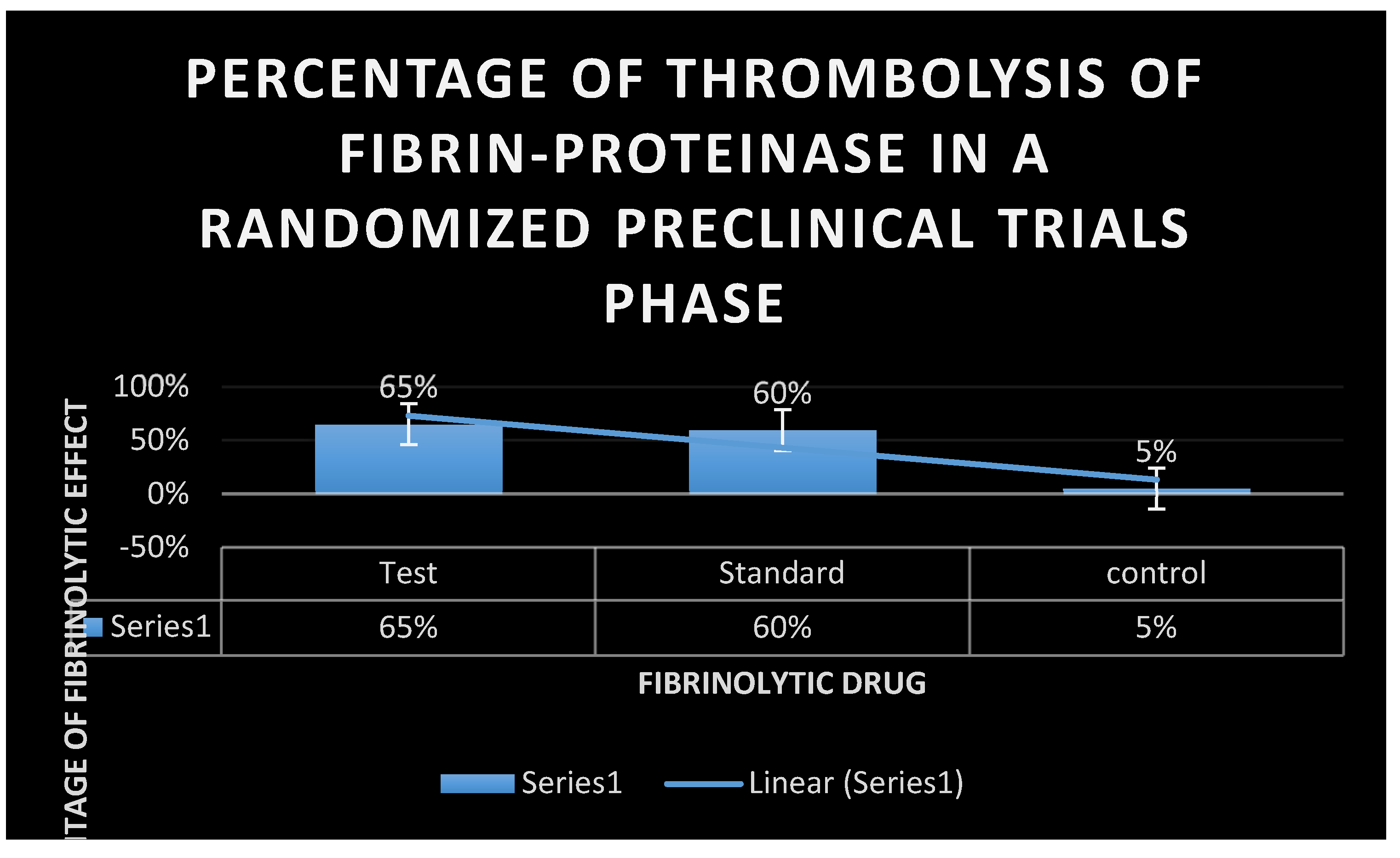
Experimental venous thrombosis Technique:
Biological activity of the test fibrinolytic enzyme was compared to standard activity in a rabbit model of experimental venous thrombosis.
300 male rabbit models weighing nearly
2 kg were collected and divided into three groups (test, standard, and control). Each group consisted of
100 of rabbit models. In test and control groups, thrombosis was induced by jugular vein stasis and injection of
10 mg/kg thromboplastin into the ear vein of each rabbit model. Animals were randomized to receive IV test fibrinolytic enzyme
0.2, 2.0, 4.0, or 9.0 mg/ kg or vehicle control and compared to standard intravenous plasmin and vehicle control. On the other hand, activated partial thromboplastin time(
APTT) test was performed to detect the anticoagulant effect of fibrinolytic enzymes; as well as the clotting time(
CT), thrombin time(
TT), Euglobulin lysis time(
ELT), fibrinogen(
FIG) and hemorheology conditions were determined. [
36]
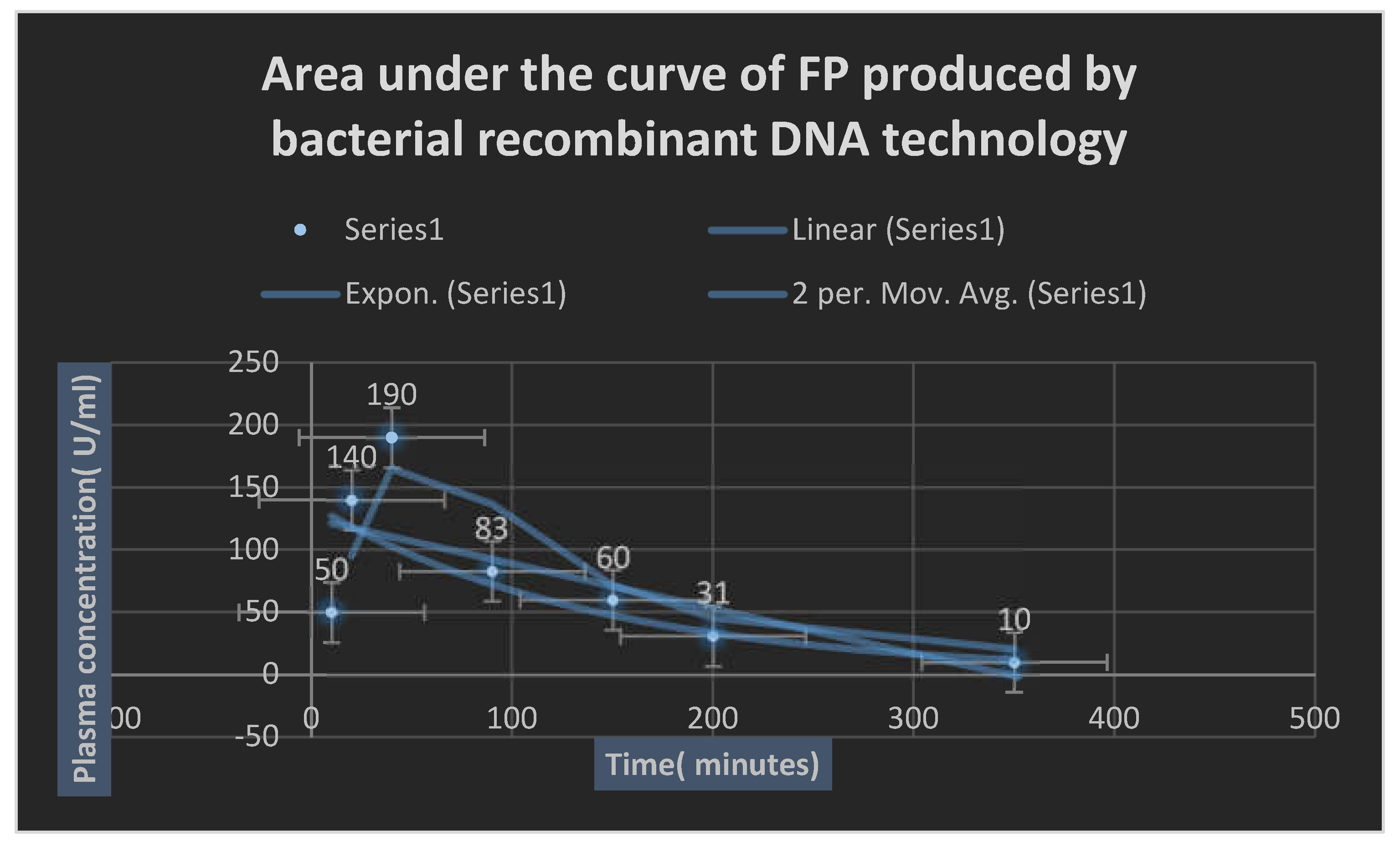
Determination of phamacokinetic profile:
In vitro fibrinolytic enzyme release and in vivo bioavailability study of optimized formulations were determined according to
British pharmacopoeia 2023. A
0.5- 1 ml rabbit animal model blood samples were withdrawn before dosing and immediately after dosing at
30, 60, 90, 120, 240 and 360 minutes; afterwards blood samples were refrigerated within 1 hour of sampling and centrifuged at
40C. Fibrin-proteinase concentrations were measured using HPLC.
HPLC analysis was performed on a reverse- phase column using phosphate buffer[
PH 4.3] and acteonitrile[
750: 250, V/V] as a mobile phase with a flow rate of
1 ml / min. The limit of the determination with
UV detection was at
225 nm. [
37]