1. Introduction
Ageing is the process of becoming older and is correlated entirely to time, it is considered a natural part of the life cycle for all living organisms. Ageing is associated with physiological processes declining leading to an increased susceptibility to diseases, a decrease in physical and cognitive capabilities, and ultimately mortality. Whilst ageing is a reference of time passed, the rate at which physiological processes decline can vary due to genetic robustness and environmental factors [
1].
One hallmark of ageing has come from epigenetic change, epigenetic ageing analyses the changes in gene expression patterns that occur without altering the underlying DNA sequence, these changes are associated with the biological age of an organism [
2]. These changes are mediated by epigenetic mechanisms, including DNA methylation, histone modification, and the expression of non-coding RNAs, which regulate how genes are turned on and off in response to environmental and lifestyle factors [
2,
3]. One of the key aspects of epigenetic ageing is the development of epigenetic clocks. These predictive tools aim to estimate an individual’s biological age based on the pattern of DNA methylation at specific sites in the genome. The most widely recognized of these is the Horvath clock [
2], however other clocks have also been created including CheekAge [
4], GrimAge2 [
5] and DunedinPACE [
6]. These clocks have aimed to understand how epigenetic change occurs over time and how this influences the ageing process, with the aim of developing interventions that could delay ageing or prevent age-related diseases by modifying epigenetic marks.
Epigenetic clocks aim to recreate a linear regression in line with chronological age with little variability, the MuhdoAge clock has been created to have the capability of variability for environmental/pathologic impact but have a strong correlation to those who are disease free.
2. Materials and Methods
Muhdo Health is a commercial DTB/DTC genetic analysis and epigenetic analysis testing company that has been carrying out genetic and epigenetic investigation over the past 7 years across multiple Illumina (Illumina, Inc) arrays, some of which are combined IP. The company utilises a mobile application in both iOS and Android to deliver questionnaires (Supplemental information 1) and allows for tracking data (steps, HR etc.). Of the 23,589 individuals tested, 2109 had at least 2 MethylationEPIC array tests completed within <6months of each other (with the most recent being chosen for the study), agreed to their anonymised data being used (via. informed consent) for study and had filled out the in-app questionnaires fully. Saliva data collection was via. a 2ml passive drool saliva sample in a uniquely coded plastic tube (GeneFiX™ Saliva DNA/RNA Collection), prefilled with preservative liquid mix (non-toxic stabilization buffer) with analysis carried out at Eurofins Global Laboratory (Certification: ISO 17025:2005, ISO 17025:2017). Individuals with two tests were chosen for quality assurance purposes with comparative analysis done across sites to analyse parity.
Demographics
Of the 2109 participants, 1107 are stated as Male at birth, 1002 are stated as Female at birth. At the point of testing 991 are from Western Europe, 380 are from Eastern Europe, 486 are from Asia and 252 are from America (USA). The chronological age range is 14 – 97 years at point of testing.
Construction and methodology of the MuhdoAge Clock
2109 participants had completed a MethylationEPIC array consisting of an analysis of 850,000~ CpG sites, utilising the Tag.Bio (
https://www.tag.bio/) data analytics platform two separate cohorts were created from this data set.
Cohort 1 (Young) (14 – 20 years n=75)
Cohort 2 (Older) (75+ Years n=165)
All sites were analysed in an unbiased manner across the two cohorts to discover sites that had a significant mean change value of p=0.05 or below. From this analysis 662 CpG sites showed a significance level of p=0.05 or below (Supplemental information 2).
Of the 662 sites, 4 sites showed high failure rates (<90% call)
Table 1.
Removed sites due to higher failure rate.
Table 1.
Removed sites due to higher failure rate.
Mean methylation across the 658 sites was analysed across a linear regression on the 2109 participants to discover the coefficient of determination (R²), the data showed an R² of 0.238 indicating that 23.8% of the variance in methylation was determined by age.
To tighten the R² the significant mean was reduced to p=0.01 and below for the CpG sites analysed in the young cohort against the older cohort. This resulted in 237 CpG sites (supplemental information 3) being chosen for the MuhdoAge clock.
3. Results
Mean methylation was taken on 237 CpG which showed a significance of p = 0.01 or lower across the 2109 participants (supplemental information 4).
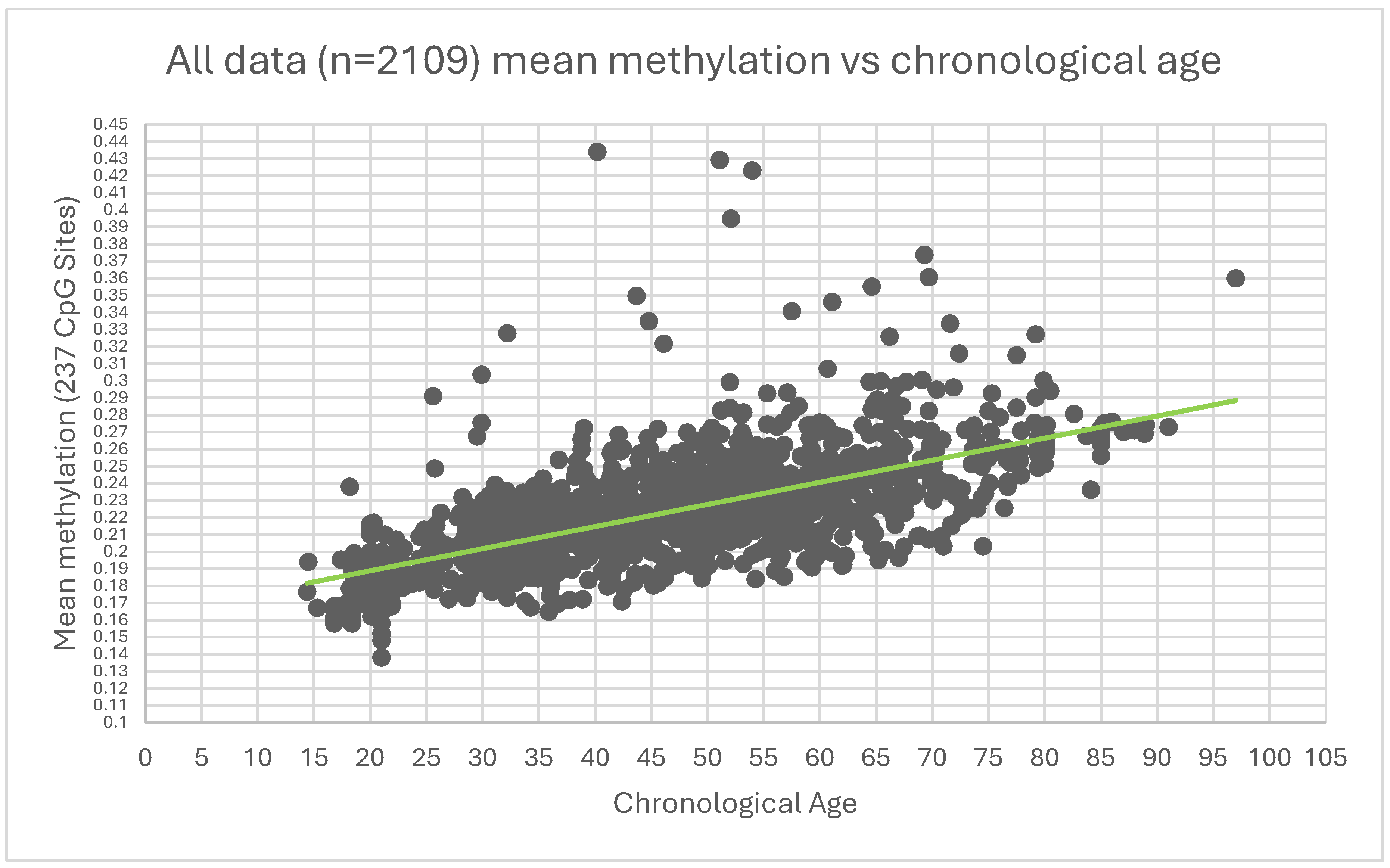
Figure 1.
Mean methylation of 237 CpG against age of the 2109 cohort.
Figure 1.
Mean methylation of 237 CpG against age of the 2109 cohort.
The data analysis showed an R² of 0.544 indicating an improvement over the 0.238 of the 658 sites that had a significance of 0.05 and lower between the young and older cohort.
Removal of outliers
The removal of outliers was considered to be important at this stage as these may skew the results of the analysis and lead to inaccurate clock. The method used to identify and remove outliers from the dataset is based on the Interquartile Range (IQR) considering that mean values may be affected by extreme outliers.
After analysis 18 participants were removed from the dataset meaning that the original dataset with 2109 participants was reduced to 2091.
Mean methylation was taken on 237 CpG which showed a significance of p = 0.01 or lower across the cleaned data of 2091 participants (supplemental information 5).
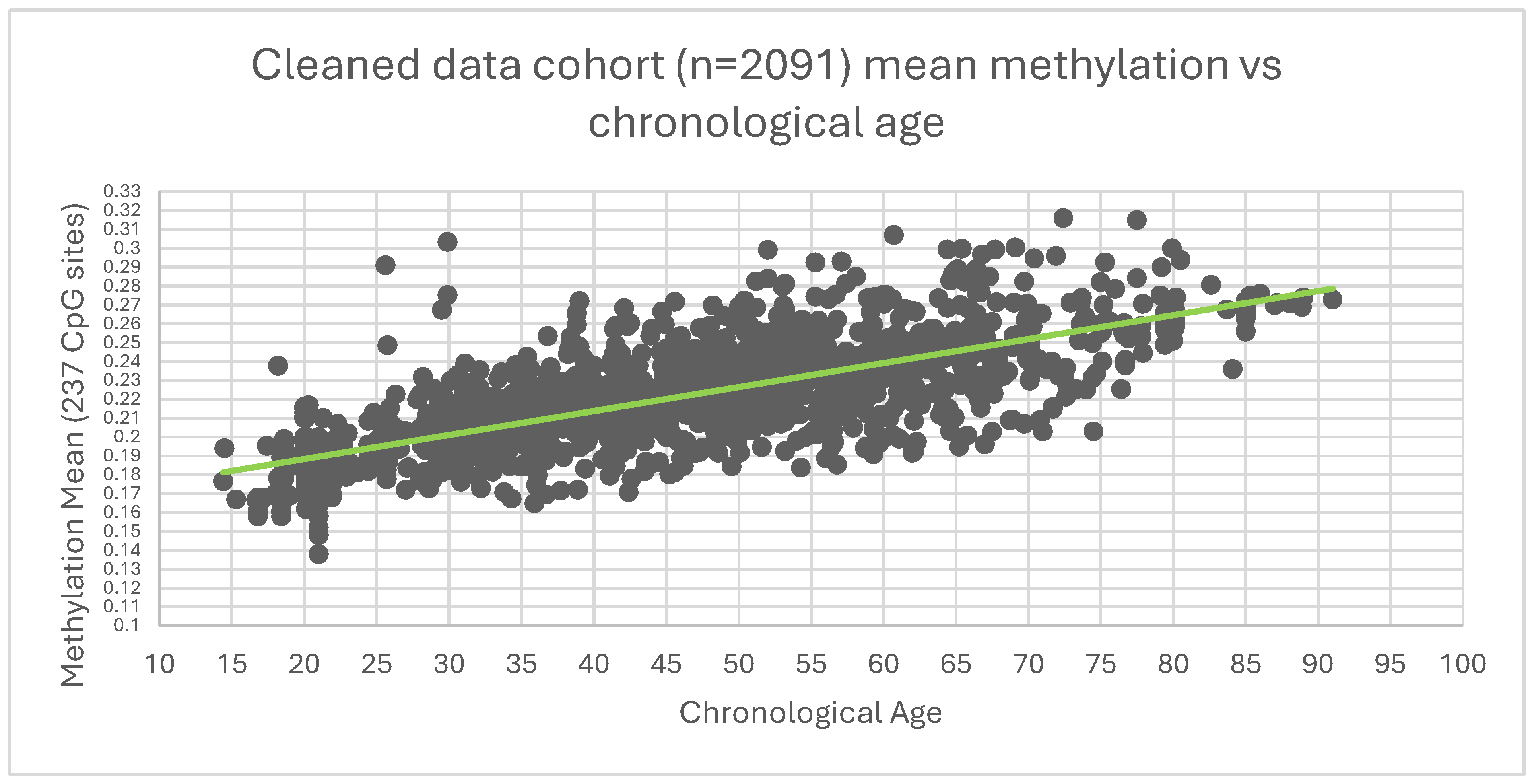
Figure 2.
Mean methylation of 237 CpG against age of the 2091 cohort.
Figure 2.
Mean methylation of 237 CpG against age of the 2091 cohort.
The data analysis showed an R² of 0.652 indicating an improvement over the non-cleaned data set.
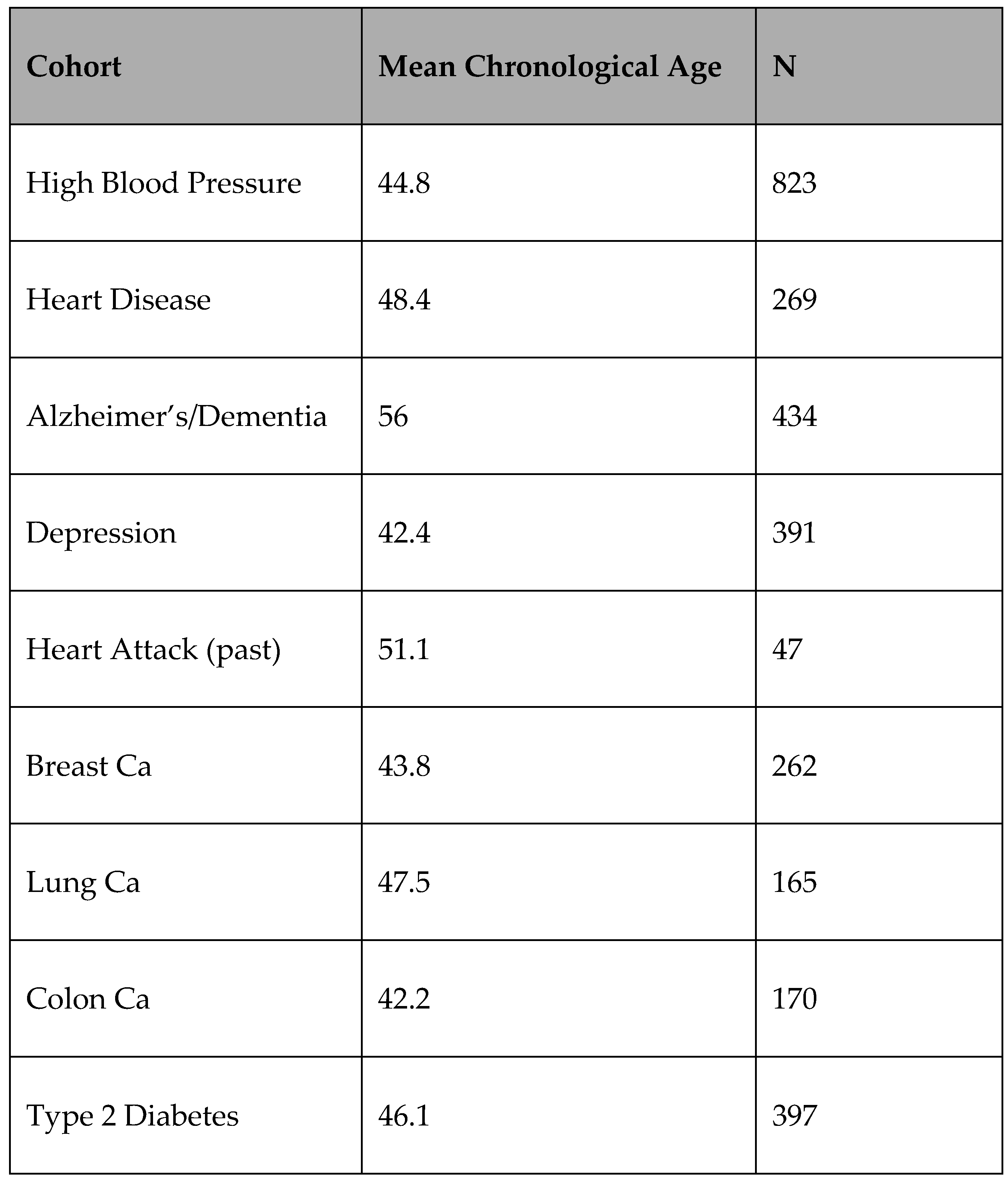
A training model was created initially utilising linear regression before moving towards random forest regression through analysing each CpG site individually in multiple of the Muhdo databases via. individuals that accepted anonymised data (included those with a single EPIC 850k array test to increase numbers) to be used for research purposes. This model was designed to analyse if any sites correlate with age without affect from variability from other sources vs sites that appear to be affected by environmental/other factors, i.e., sites that have no significant change besides via. time against sites that have significant change (p = <0.05) in any of the pathology/subjective/lifestyle or supplement cohorts with knowledge that there is cross-over across some cohorts.
Table 2.
Pathological Cohorts.
Table 2.
Pathological Cohorts.
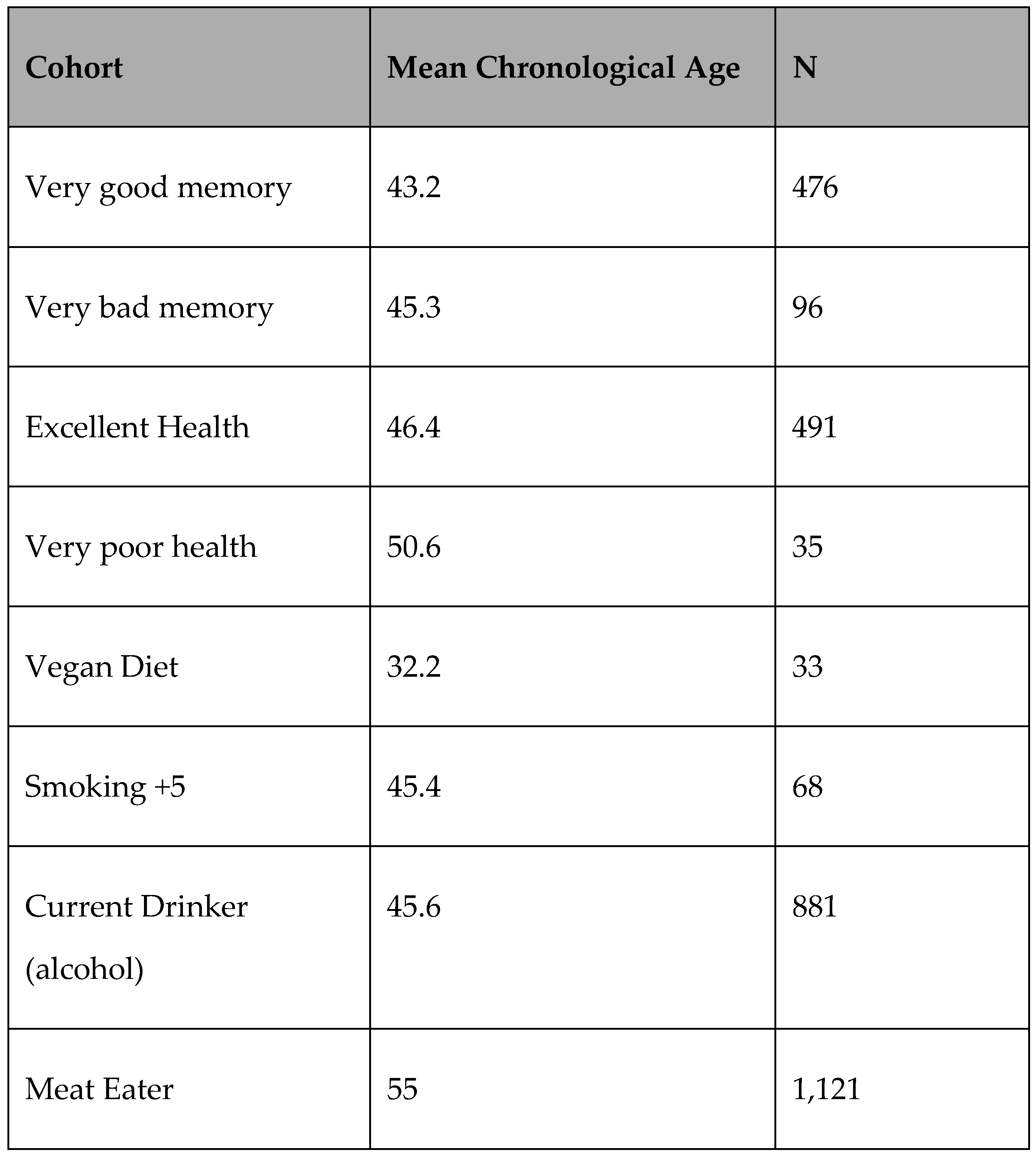
Table 3.
Subjective/lifestyle cohorts.
Table 3.
Subjective/lifestyle cohorts.
Table 4.
Supplemental Cohorts.
Table 4.
Supplemental Cohorts.
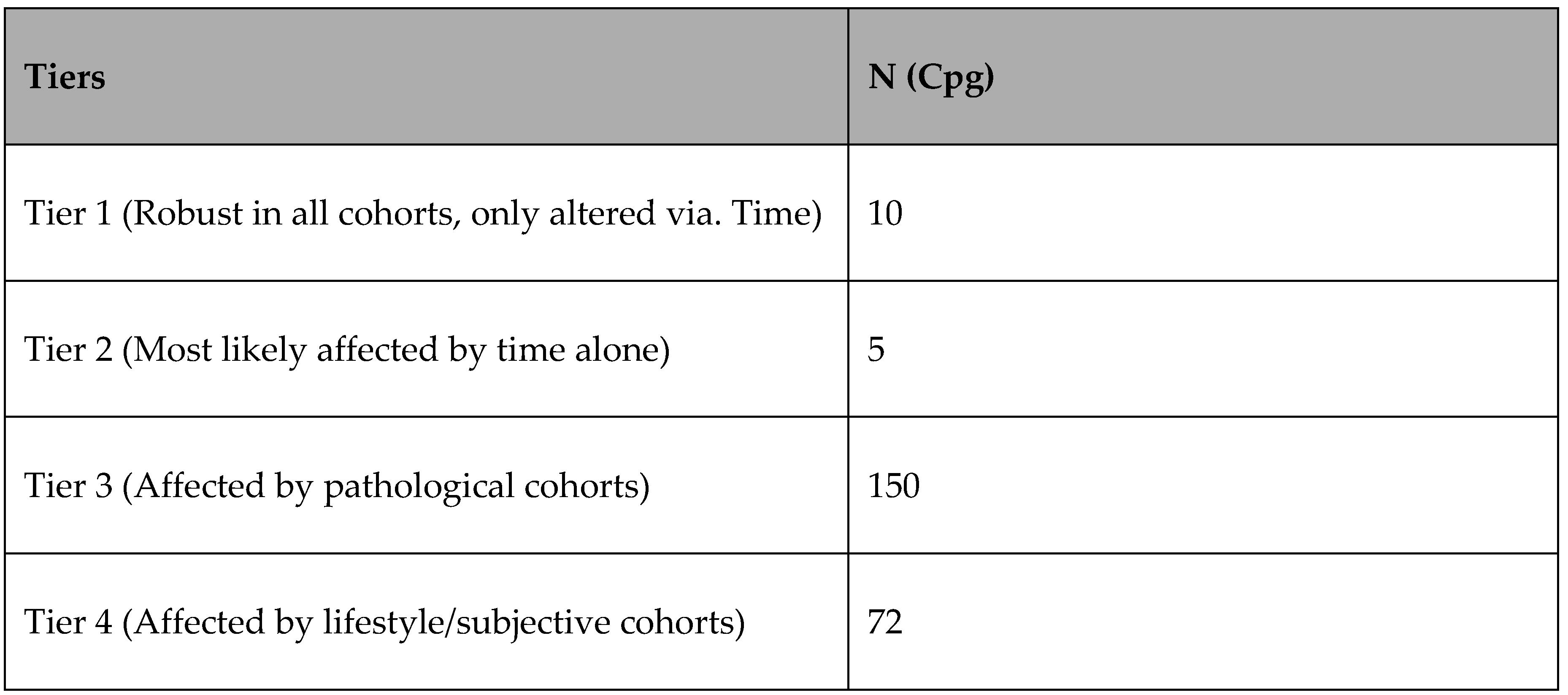
The analysis highlighted CpG sites that are robust to lifestyle, pathology and subjectivity, these sites hyper/hypo methylated with time regardless. The analysis then tiered sites in order of robustness (supplemental information 6) to the pathology/lifestyle/subjective cohorts:
Table 5.
Tiered CpG sites.
Table 5.
Tiered CpG sites.
With the prime directive of predicting age, each tier was weighted differently based on the training set of the original 2109 (including outliers) creating a simple weighting system:
M1+0.07M2+0.02M3+0.005M4=PA
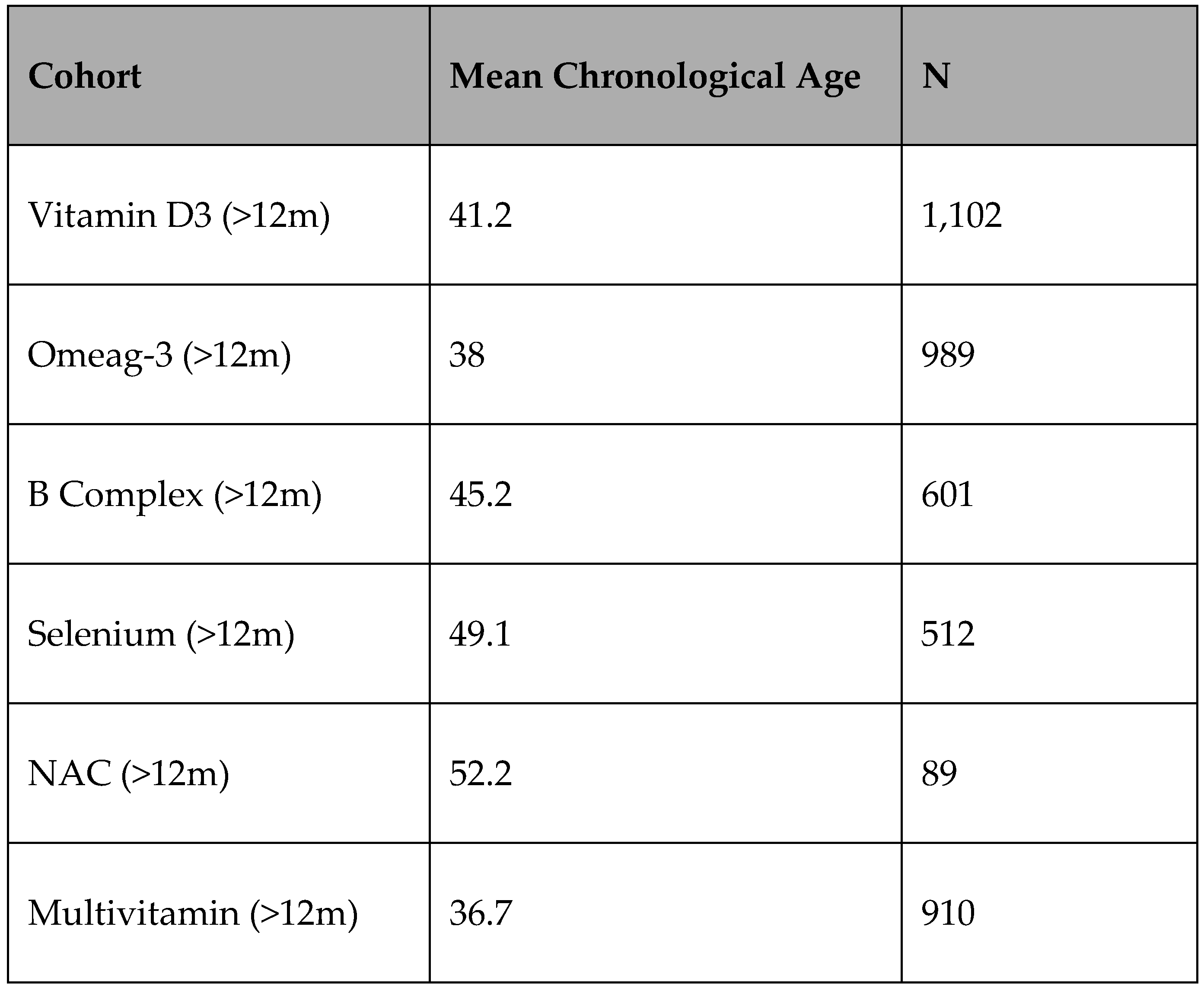
Muhdo Health went back to their client base to explore the potential of opening a larger cohort to use the new algorithm on, of the 4000~ asked (with two or more EPIC 850k array tests within <6 months), 2682 agreed (via. informed consent for their data to be anonymised and used) with some cross-over (41.3%/1108) of the original 2109. It was considered that there was limited value in using the algorithm on the exact same dataset as the training model, hence the new cohort was created.
Demographics of the new 2682 cohort
Of the 2682 participants, 1620 are stated as Male at birth, 1062 are stated as Female at birth. At the point of testing 1080 are from Western Europe, 400 are from Eastern Europe, 500 are from Asia and 702 are from America (USA). The chronological age range is 18 – 88 years at point of testing.
The algorithm as ran across the new dataset of 2682 (supplemental information 7).
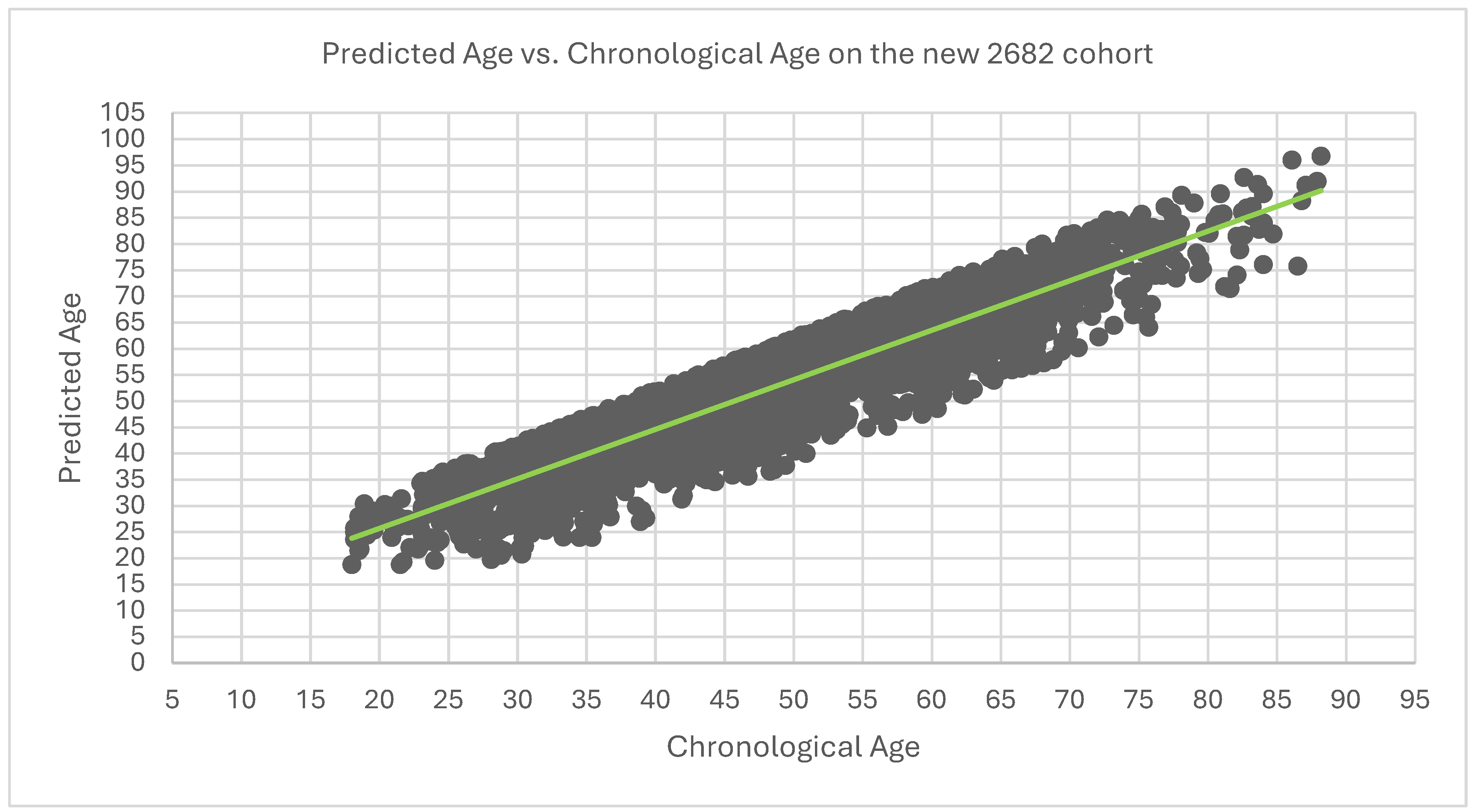
Figure 3.
Predicted Age vs. Chronological Age on the 2682 cohort.
Figure 3.
Predicted Age vs. Chronological Age on the 2682 cohort.
The R² for the dataset is 0.726
Mean Absolute Error (MAE) for the dataset is 5.97. Therefore, on average there is a sway of 5.97years of predicted age away from chronological age.
Mean Squared Error (MSE) for the dataset is 47.46.
Root Mean Squared Error (RMSE) for the dataset is 6.89.
Upon linear regression analysis there is an intercept of 6.79 and a slope of approximately 0.95. Indicating that for every year increase in chronological age there is a predicted increase in predicted age by 0.95 starting from the intercept of 6.79.
The new algorithm appears to have increased robustness (0.652 vs 0.726) towards pathology, subjective wellness/health, lifestyle and/or supplements.
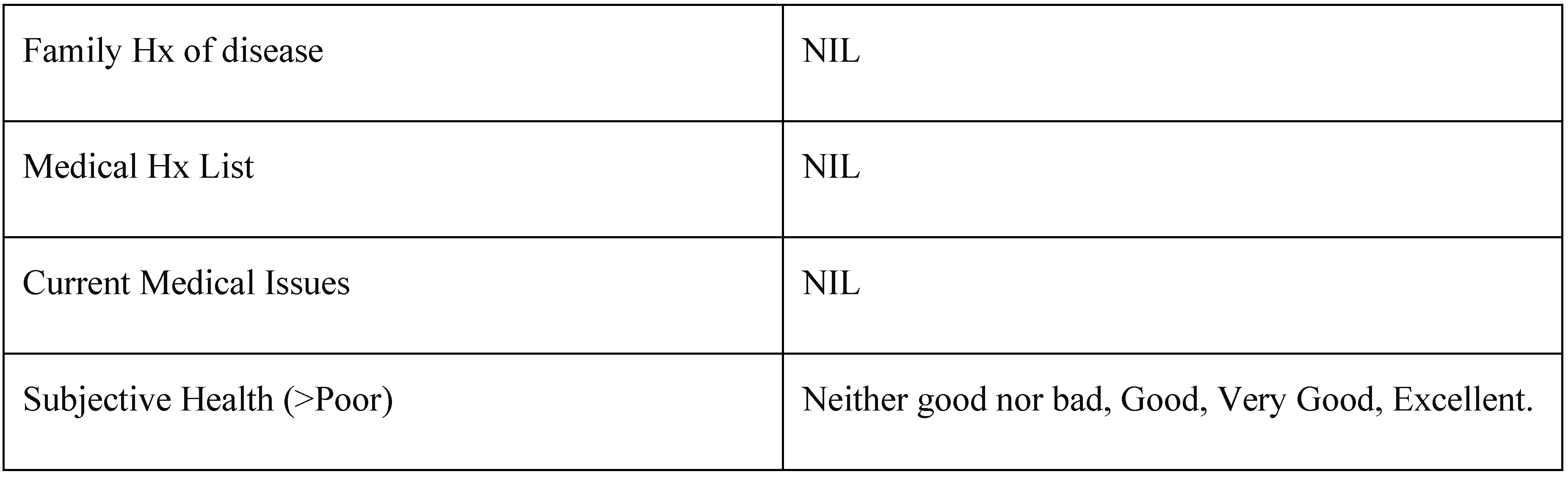
To analyse the theory that variability found in the MuhdoAge clock could/is caused by pathology a new cohort was created called cohort “Health” (Supplemental information 8):
Table 6.
Cohort Health Parameters.
Table 6.
Cohort Health Parameters.
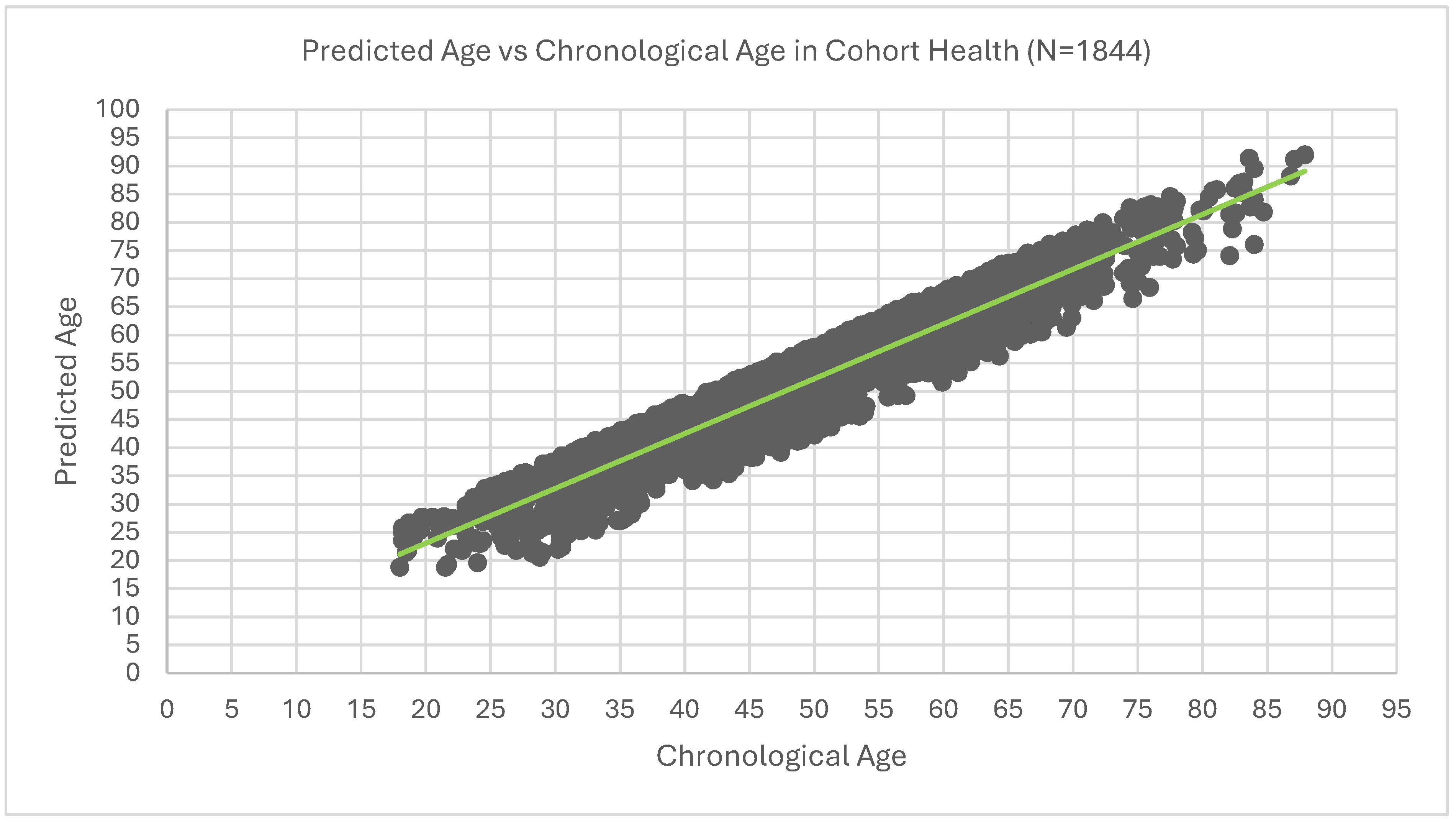
Figure 4.
Predicted Age vs Chronological Age in Cohort Heath.
Figure 4.
Predicted Age vs Chronological Age in Cohort Heath.
The R² for the dataset is 0.878
Mean Absolute Error (MAE) for the dataset is 3.49. Therefore, on average there is a sway of 3.49years of predicted age away from chronological age.
Mean Squared Error (MSE) for the dataset is 17.16.
Root Mean Squared Error (RMSE) for the dataset is 4.14.
Upon linear regression analysis there is an intercept of 3.61 and a slope of approximately 0.97.
When analysing the 2682 cohort against cohort Health we found:
Cohort Health:
Average Chronological Age: 47.96 years
Average Predicted Age: 50.25 years
Standard Deviation for Chronological Age: 13.34 years
Standard Deviation for Predicted Age: 13.62 years
Average Age Difference (Predicted - Chronological): +2.29 years
2682 Cohort:
Average Chronological Age: 47.65 years
Average Predicted Age: 51.83 years
Standard Deviation for Chronological Age: 13.16 years
Standard Deviation for Predicted Age: 13.57 years
Average Age Difference (Predicted - Chronological): +4.19 years
Both cohorts showed a similar average chronological age, with the original 2682 cohort being slightly younger on average (47.96 vs. 47.65). The original 2682 cohort had a higher average predicted age even though the chronological age was slightly lower compared to the Cohort Health (50.25 vs 51.83). The average difference between predicted and chronological ages is significantly higher in the original 2682 cohort (+4.19 years) than in the Cohort Health (+2.29 years), indicating that the Comparative Cohort’s predicted age is more elevated compared to its chronological age. The standard deviations for both chronological and predicted ages are similar across both cohorts, indicating a similar age distribution within each cohort. These statistics suggest that the 2682 cohort has a higher average predicted age compared to its chronological age than the Cohort Health which may back the usage of MuhdoAge showing disparity (accelerated age) in pathologic groups.
4. Discussion
This study introduced the MuhdoAge clock, a novel epigenetic clock based on saliva samples, demonstrating a significant correlation with biological age in a healthy, disease-free cohort (
R² 0.878). Our findings show that the MuhdoAge clock can accurately predict age compared with chronological age but shows variability to health, offering a robust tool for aging research and potential clinical applications. These results are particularly relevant considering the increasing interest in understanding the biological rather than chronological determinants of aging [
2,
3,
4]. MuhdoAge could be used in public health strategies to monitor and manage the ageing population more effectively. By identifying populations that are ageing faster due to environmental or lifestyle factors, targeted interventions can be developed to improve the overall health and longevity of these groups.
Comparison with Existing Epigenetic Clocks
The MuhdoAge clock shows a strong predictive capability in a non-adapted cohort (R² 0.726) and a very strong predictive capability in an adapted healthy cohort (R²0.878) this aligns closely with established epigenetic clocks like the Horvath clock (R² 0.96 across all tissues), GrimAge (R² 0.75 predicting point of death), Hannuman (R² 0.81), Cheek Age (R² 0.93) [
2,
4,
5,
7]. Unlike many alternative clocks, MuhdoAge leverages saliva over other tissues and blood which makes a viable test for mass testing and home based direct to consumer testing, highlighting its non-invasive advantage. This form of testing is particularly significant for longitudinal studies and the monitoring of ageing interventions where frequent sampling is required [
8].The use of saliva offers a non-invasive, easily accessible, and cost-effective means of collecting DNA for epigenetic analysis. Saliva has been increasingly recognised for its rich biomarker potential, reflecting both local and systemic health states [
9]. This study further underscores the utility of saliva in epigenetic aging research, providing a more accessible avenue for widespread application in both research and clinical settings.
The MuhdoAge clock embraced the rich data from health databases to account for alterations in predicted age offering new insights into the epigenetic mechanisms of ageing. This aligns with growing evidence that lifestyle interventions can modulate epigenetic aging rates [
10,
11,
12]. This study backs up the concept that health-based intervention may reduce biological ageing, also highlighting the predictive ability of DNA methylation for pathological change.
Limitations
The MuhdoAge study marks a step forward in the usage of saliva as a method of predicting age and that ageing may be increased in pathologic groups, however limitations exist. The cohort, although large and diverse, is limited geographically and ethnically, the database comes primarily from a direct to consumer/business working model which may pose a risk of mistakes being made by individuals filling out questionnaires. A particular type of person may have also been interested in purchasing a health-based DNA/epigenetic test and this demographic disposition was not studied during the study, however the clients would have required to have a level of disposable income to purchase a test.
Future Study
Future study will focus on longitudinal studies to validate the predictive accuracy of MuhdoAge over time and explore the causal relationships between lifestyle factors and epigenetic age acceleration. Moreover, integrating multi-omics data, such as transcriptomic or proteomic profiles, could enhance the understanding of the biological underpinnings of the epigenetic changes observed.
Muhdo Health aims to leverage health databases to create pathology specific prediction tests based on methylation that appears independent of ageing. During the processes of this study particular CpG sites were identified that appeared to methylate differently based on pathology alone and were independent of time, further analysis into these sites may allow for earlier prediction tools for these conditions.
5. Conclusions
The MuhdoAge clock represents an advancement in the field of epigenetic research, offering a practical, non-invasive tool for assessing biological age. This study not only contributes to the understanding of the epigenetic landscape of ageing but also opens new avenues for preventive health strategies aimed at extending health span.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, supplemental information 1-8,
Figure 1,
Figure 2,
Figure 3 and
Figure 4.
Funding
This research received no external funding
Institutional Review Board Statement
Ethical review and approval were waived for this study due to the study being conducted via a GDPR compliant company on individuals wishing for their data to be utilized for research purposes via. Informed and signed consent.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Data in anonymized.
Acknowledgments
Consultation was sought from Dr. Richard Siow from King’s College London
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. Hallmarks of aging: an expanding universe. Cell. 2023;186:243–78.
- Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14(10), R115.
- Jung, M., & Pfeifer, G. P. (2015). Aging and DNA methylation. BMC Biology, 13, 7.
- Shokhirev, M.N., Torosin, N.S., Kramer, D.J. et al. CheekAge: a next-generation buccal epigenetic aging clock associated with lifestyle and health. GeroScience (2024).
- Lu AT, Binder AM, Zhang J, Yan Q, Reiner AP, Cox SR, Corley J, Harris SE, Kuo PL, Moore AZ, et al. DNA methylation GrimAge version 2. Aging (Albany NY). 2022;14:9484–549.
- Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, Baccarelli A, Chamarti K, Gao X, Hannon E, et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife. 2022;11.
- Hannum, G., Guinney, J., Zhao, L., Zhang, L., Hughes, G., Sadda, S. V. R., Klotzle, B., Bibikova, M., Fan, J., Gao, Y., Deconde, R., Chen, M., Rajapakse, I., Friend, S. H., Ideker, T., & Zhang, K. (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular Cell, 49(2), 359–367.
- Leigh, S. J., & Morris, M. J. (2020). The role of epigenetic change in aging and age-related disease. Ageing Research Reviews, 64, 101174.
- Zhang CZ, Cheng XQ, Li JY, Zhang P, Yi P, Xu X, Zhou XD. Saliva in the diagnosis of diseases. Int J Oral Sci. 2016 Sep 29;8(3):133-7. [CrossRef] [PubMed] [PubMed Central]
- Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, Baccarelli A, Chamarti K, Gao X, Hannon E, Harrington HL, Houts R, Kothari M, Kwon D, Mill J, Schwartz J, Vokonas P, Wang C, Williams BS, Moffitt TE. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife. 2022 Jan 14;11:e73420. [CrossRef] [PubMed] [PubMed Central]
- Waki, T., Tamura, G., Sato, M., and Motoyama, T. (2003). Age-related methylation of tumor suppressor and tumor-related genes: An analysis of autopsy samples. Oncogene 22 (26), 4128–4133. [CrossRef]
- Marioni, R. E., Shah, S., McRae, A. F., Ritchie, S. J., Muniz-Terrera, G., Harris, S. E., et al. (2015). The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int. J. Epidemiol. 44 (4), 1388–1396. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).