Submitted:
11 April 2024
Posted:
16 April 2024
Read the latest preprint version here
Abstract
Keywords:
I. Introduction
A. Definition and Evolution of Computational Pharmaceutics
B. Significance of Integrating AI and Multi-Scale Modeling in Pharmaceutics
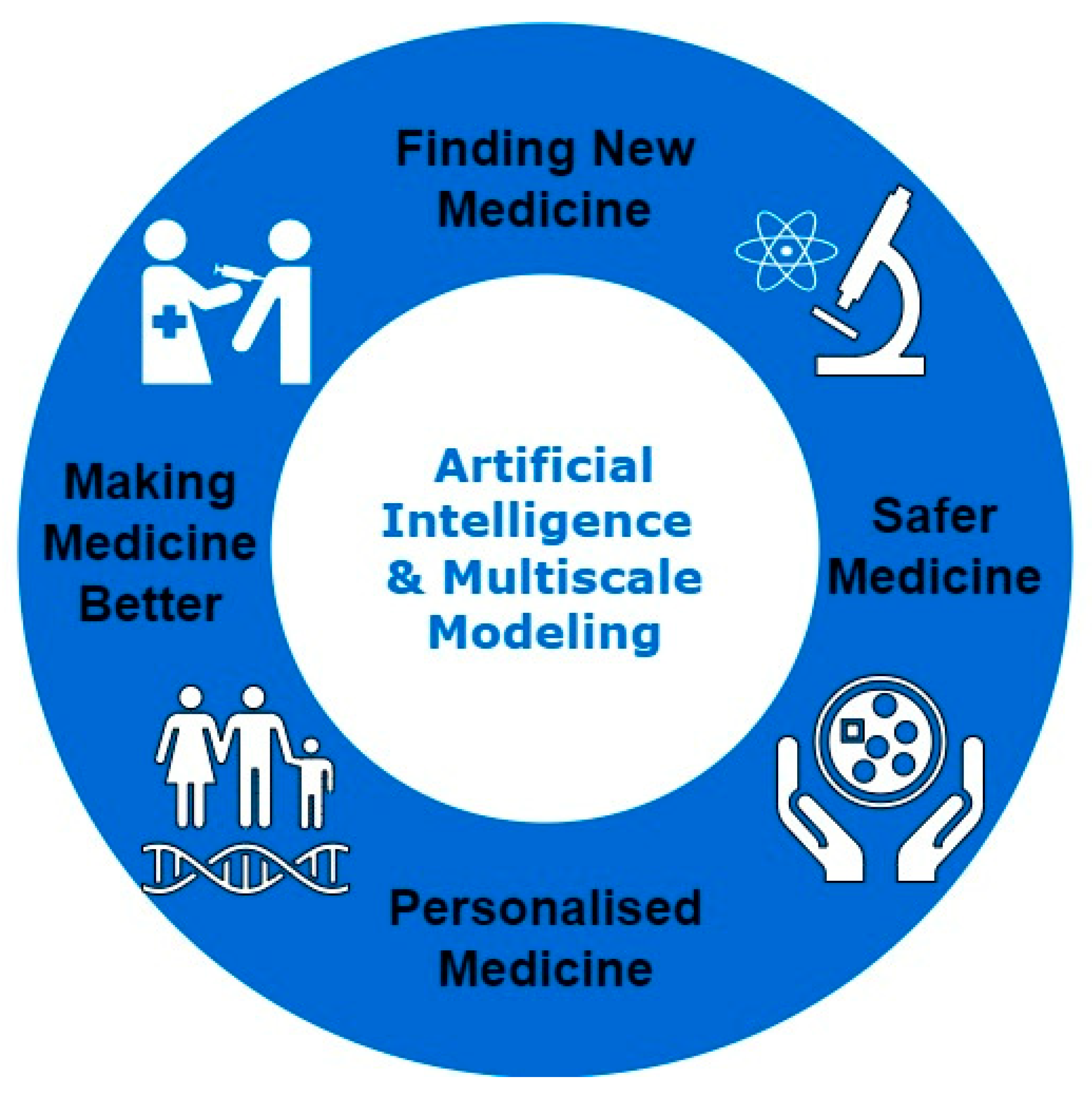
C. Overview of Computational Approaches in Pharmaceutics
| Computational Approach | Techniques/Tools | Impact on Pharmaceutical Science | Key References |
|---|---|---|---|
| Molecular Dynamic Simulations | Molecular dynamics software (e.g., GROMACS, AMBER, NAMD). | Facilitates the rational design of drug delivery systems, enhances understanding of drug behavior in complex environments. | Abd-Algaleel et al. (2021) |
| Docking Studies | Molecular docking software (e.g., Autodock, GOLD, Glide). | Expedites drug discovery processes, aids in hit identification and lead optimization. | Ashfaq et al. (2022); Chauhan et al. (2023); Sadybekov & Katritch (2023) |
| Quantum Phase Estimation Calculations | Quantum chemistry software (e.g., Gaussian, NWChem, Q-Chem). | Improves accuracy in quantum mechanical calculations, aids in understanding complex chemical systems. | Izsák et al. (2023) |
| Virtual Screening Methods | Virtual screening software (e.g., Schrödinger Suite, OpenEye, MOE). | Expedites lead discovery, minimizes costs, aids in drug repurposing efforts. | Abramov et al. (2022); Adhikary & Basak (2020); Sadybekov & Katritch (2023) |
| Machine Learning | Machine learning algorithms (e.g., neural networks, random forests, support vector machines). | Enhances predictive accuracy, facilitates data-driven drug discovery, aids in identifying novel chemical entities. | Sadybekov & Katritch (2023) |
| Bioinformatics Tools | Bioinformatics software (e.g., PyMOL, Chimera, BLAST). | Enables better understanding of biological systems, aids in drug-target interaction studies, facilitates personalized medicine. | Ashfaq et al. (2022); Chauhan et al. (2023) |
| Role in Drug Development | Computational chemistry techniques, in silico modeling, data mining approaches. | Accelerates drug development timelines, reduces failure rates, enables precision medicine approaches. | Adhikary & Basak (2020); Ashfaq et al. (2022); Chauhan et al. (2023); Sadybekov & Katritch (2023) |
| Impact on Pharmaceutical Science | Enhanced understanding of molecular interactions, expedited drug development timelines, improved drug safety profiles. | Facilitates the development of safer and more effective medications, enables precision medicine, reduces development costs. | Abd-Algaleel et al. (2021); Hussain et al. (2023); Sadiku et al. (2019); Ouyang & Smith (2015); Douroumis et al. (2015) |
| Applications | Computational modeling of physiological parameters, drug-drug interactions, toxicity prediction, dose-response modeling. | Enhances drug efficacy and safety profiles, aids in personalized medicine approaches, optimizes therapeutic regimens. | Abd-Algaleel et al. (2021); Hussain et al. (2023); Sadiku et al. (2019); Ouyang & Smith (2015); Douroumis et al. (2015) |
II. Past Achievements in Computational Pharmaceutics
A. Role of Quantum Mechanics (QM) in Predicting Molecular Properties
B. Molecular Dynamics Simulation for Understanding Physical Motion
C. Molecular Modeling in Investigating Structural and Energetic Aspects
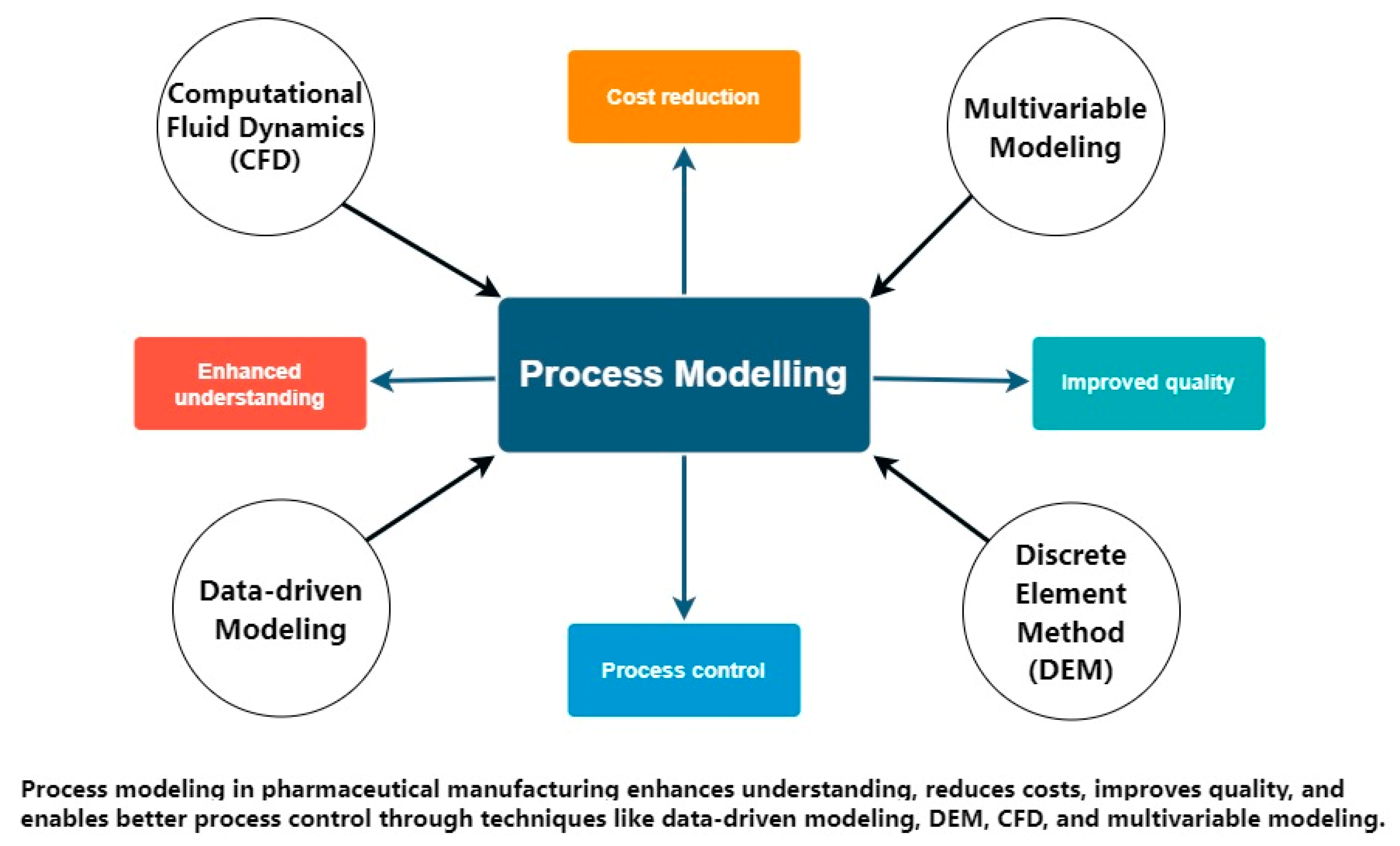
D. Process Modeling for Numerical Simulation in Manufacturing
E. Physiologically Based Pharmacokinetic (PBPK) Modeling for Predicting PK/PD
F. Contribution of Artificial Intelligence (AI) and Machine Learning Algorithms
III. Current Problems in AI Application in Pharmaceutics
A. Lack of Sufficient Data and Challenges in Data Sharing
B. Need for Interpretable Machine Learning Methods
C. Addressing the High Cost and Lengthy Research Time in Pharmaceutical Experiments
IV. Future Scenarios in Computational Pharmaceutics
A. Paradigm Shift in Drug Delivery Development with QbD Strategy
B. Acceleration of Drug Production through Continuous Manufacturing
C. Integration of Modeling Methods for Enhanced Process Understanding
D. Challenges and Strategies: Multi-Scale Modeling, Data Sharing, Experimental Methods, Talent Training, and Cultural Change
Conclusion
References
- Abd-Algaleel, S. A., Abdel-Bar, H. M., Metwally, A. A., & Hathout, R. M. (2021). Evolution of the Computational Pharmaceutics Approaches in the Modeling and Prediction of Drug Payload in Lipid and Polymeric Nanocarriers. Pharmaceuticals , 14(7). [CrossRef]
- Ab, R., & Ambos, M. (2000). New industrial training program for interns and postdoctoral fellows. Journal of Molecular Graphics & Modelling, 18(6), 615.
- Abramov, Y. A., Sun, G., & Zeng, Q. (2022). Emerging Landscape of Computational Modeling in Pharmaceutical Development. Journal of Chemical Information and Modeling, 62(5), 1160–1171. [CrossRef]
- Adhikary, T., & Basak, P. (2020). Interdisciplinary approaches incorporating computational intelligence in modern pharmacognosy to address biological problems. In Lecture Notes in Electrical Engineering (pp. 11–19). Springer Singapore.
- Albalwy, F., McDermott, J. H., Newman, W. G., Brass, A., & Davies, A. (2022). A blockchain-based framework to support pharmacogenetic data sharing. The Pharmacogenomics Journal, 22(5-6), 264–275. [CrossRef]
- Allen, C. L., VanGelder, K. F., & Maguire, C. K. (2022). Implementation of high throughput experimentation across medicinal chemistry, process chemistry and materials science. In ACS Symposium Series (pp. 23–33). American Chemical Society.
- Arastoopour, H., Gidaspow, D., & Lyczkowski, R. W. (2022). Application of multiphase flow simulation in pharmaceutical processes. In Mechanical Engineering Series (pp. 299–316). Springer International Publishing.
- Arya, H., & Bhatt, T. K. (2021). Molecular dynamics simulations. In The Design & Development of Novel Drugs and Vaccines (pp. 65–81). Elsevier.
- Ashfaq, U. A., Shahid, F., & Munir, S. (2022). Computational approaches for drug-metabolizing enzymes: Concepts and challenges. In Biochemistry of Drug Metabolizing Enzymes (pp. 29–72). Elsevier.
- Atz, K., Isert, C., Böcker, M. N. A., Jiménez-Luna, J., & Schneider, G. (2022). Δ-Quantum machine-learning for medicinal chemistry. Physical Chemistry Chemical Physics: PCCP, 24(18), 10775–10783. [CrossRef]
- Babar, A. V., Jain, G., Patel, J., Sharma, R., Khan, S., & Patel, R. (2022). Qbd approach in hplc method development and validation of tamoxifen. International Journal of Pharmaceutical Sciences and Medicine, 7(9), 90–105. [CrossRef]
- Balakrishnan, A., Kunhikatta, V., Nair, S., Khera, K., Viji, P. C., & Thunga, G. (2020). Training needs assessment for employability in pharmaceutical industries. Open Access Macedonian Journal of Medical Sciences, 8(A), 239–244. [CrossRef]
- Balakrishnan, A., Thunga, G., Nair, S., Kunhikatta, V., & Khera, K. (2018). Bridging the gap between industry and academia in pharmaceutical education. Indian Journal of Pharmaceutical Education, 52(4s), s7–s10. [CrossRef]
- Barrett, J. S. (2022). Data Sharing and Collaboration. In Fundamentals of Drug Development (pp. 431–443). Wiley. [CrossRef]
- Bisbal, P. (2019). Training computational scientists to build and package open-source software. The Journal of Computational Science Education, 10(1), 74–80. [CrossRef]
- Blunt, N. S., Camps, J., Crawford, O., Izsák, R., Leontica, S., Mirani, A., Moylett, A. E., Scivier, S. A., Sünderhauf, C., Schopf, P., Taylor, J. M., & Holzmann, N. (2022). A perspective on the current state-of-the-art of quantum computing for drug discovery applications. [CrossRef]
- Borkotoky, S., Joshi, A., Kaushik, V., & Nath Jha, A. (2022). Machine learning and artificial intelligence in therapeutics and drug development life cycle. In Drug Development Life Cycle. IntechOpen.
- Brigo, A., Naga, D., & Muster, W. (2022). Increasing the Value of Data Within a Large Pharmaceutical Company Through In Silico Models. Methods in Molecular Biology , 2425, 637–674.
- Bunker, A., & Róg, T. (2020). Mechanistic Understanding From Molecular Dynamics Simulation in Pharmaceutical Research 1: Drug Delivery. Frontiers in Molecular Biosciences, 7, 604770. [CrossRef]
- Butner, J. D., Dogra, P., Cristini, V., Deisboeck, T. S., & Wang, Z. (2023). Computational approaches for multiscale modeling. In Encyclopedia of Cell Biology (pp. 251–260). Elsevier.
- Chaudhari, Y., & Chaudhari, M. (2024). AI for Pharmacist. Pharmacy Infoline. [CrossRef]
- Chaudhari, Y., & Md, F. (2024). How Machine Learning Models Can Revolutionize Pharma and healthcare. Pharmacy Infoline. [CrossRef]
- Chauhan, S. S., Jamal, T., Singh, A., Sehrawat, A., & Parthasarathi, R. (2023). Structure-based virtual screening. In Cheminformatics, QSAR and Machine Learning Applications for Novel Drug Development (pp. 239–262). Elsevier.
- Chavda, V. P. (2023). Artificial intelligence and machine learning-based formulation and process development for drug products. In Bioinformatics Tools for Pharmaceutical Drug Product Development (pp. 183–195). Wiley. [CrossRef]
- Chen, Y., Li, J., Wang, F., Yue, K., Li, Y., Xing, B., Zhang, L., & Chen, L. (2023). DS2PM: A data-sharing privacy protection model based on blockchain and federated learning. IEEE Internet of Things Journal, 10(14), 12112–12125. [CrossRef]
- Chopard, B., Falcone, J.-L., Kunzli, P., Veen, L., & Hoekstra, A. (2018). Multiscale modeling: recent progress and open questions. Multiscale and Multidisciplinary Modeling Experiments and Design, 1(1), 57–68. [CrossRef]
- Conolly, R. P., Clewell, H. J., Moore, M. M., Campbell, J. L., Cheng, W., & Gentry, R. R. (2023). PBPK modeling to evaluate maximum tolerated doses: A case study with 3-chloroallyl alcohol. Frontiers in Pharmacology, 14, 1088011. [CrossRef]
- Conway, J. (1993). Cardiovascular research in the pharmaceutical industry: a personal appeal for rationalizing development. European Journal of Clinical Pharmacology, 45(5), 395–396. [CrossRef]
- Dankar, F. K. (2023). Practices and challenges in clinical data sharing. [CrossRef]
- Deepika, D., & Kumar, V. (2023). The Role of “Physiologically Based Pharmacokinetic Model (PBPK)” New Approach Methodology (NAM) in Pharmaceuticals and Environmental Chemical Risk Assessment. International Journal of Environmental Research and Public Health, 20(4). [CrossRef]
- Denninger, A., Becker, T., Westedt, U., & Wagner, K. G. (2023). Advanced In Vivo Prediction by Introducing Biphasic Dissolution Data into PBPK Models. Pharmaceutics, 15(7). [CrossRef]
- De Paola, G., & Danger, R. V. (2023, June 5). Efficient FDP optimization for AI enhanced decision making. Day 4 Thu, June 08, 2023. SPE EuropEC - Europe Energy Conference featured at the 84th EAGE Annual Conference & Exhibition, Vienna, Austria. [CrossRef]
- Ding, B., Zhou, J., Cheng, H., Zhicong, H., Xu, M., & Wang, T. (2020). Data sharing and data analytics implementing local differential privacy. https://typeset.io/papers/data-sharing-and-data-analytics-implementing-local-22zje4qt32.
- DiNuzzo, M. (2022). How artificial intelligence enables modeling and simulation of biological networks to accelerate drug discovery. Frontiers in Drug Discovery, 2. [CrossRef]
- Dong, Y., Yang, T., Xing, Y., Du, J., & Meng, Q. (2023). Data-driven modeling methods and techniques for pharmaceutical processes. Processes (Basel, Switzerland), 11(7), 2096. [CrossRef]
- Douroumis, D., Fahr, A., Siepmann, J., & Snowden, M. (2015). Computational pharmaceutics: application of molecular modeling in drug delivery. https://typeset.io/papers/computational-pharmaceutics-application-of-molecular-1nyslsuaae.
- D. Patil, H., B Patil, C., V. Patil, V., & S. Patil, P. (2023). An overview on quality by design. Asian Journal of Research in Pharmaceutical Science, 49–55.
- Đuriš, J., Kurćubić, I., & Ibrić, S. (2021). Review of machine learning algorithms’ application in pharmaceutical technology. Arhiv Za Farmaciju, 71(4), 302–317. [CrossRef]
- Edwards, T., Foloppe, N., Harris, S. A., & Wells, G. (2021). The future of biomolecular simulation in the pharmaceutical industry: what we can learn from aerodynamics modelling and weather prediction. Part 1. understanding the physical and computational complexity of in silico drug design. Acta Crystallographica. Section D, Structural Biology, 77(Pt 11), 1348–1356. [CrossRef]
- Elmokadem, A., Zhang, Y., Knab, T., Jordie, E., & Gillespie, W. R. (2023). Bayesian PBPK modeling using R/Stan/Torsten and Julia/SciML/Turing.Jl. CPT: Pharmacometrics & Systems Pharmacology, 12(3), 300–310. [CrossRef]
- Erhardt, E. M., Ursino, M., Biewenga, J., Jacobs, T., & Gasparini, M. (2019). Bayesian knowledge integration for an in vitro-in vivo correlation model. Biometrical Journal. Biometrische Zeitschrift, 61(5), 1104–1119. [CrossRef]
- Gao, L., & Guan, L. (2023). Interpretability of Machine Learning: Recent Advances and Future Prospects. http://arxiv.org/abs/2305.00537.
- Ghosh, S., Baranowski, E. S., Biehl, M., Arlt, W., Tino, P., & Bunte, K. (2022). Interpretable Models Capable of Handling Systematic Missingness in Imbalanced Classes and Heterogeneous Datasets. http://arxiv.org/abs/2206.02056.
- Gieschke, R., & Carr, R. (2022). Conceptual and organizational barriers to quantitative systems pharmacology modeling of pathophysiological systemic drug hypotheses. CPT: Pharmacometrics & Systems Pharmacology, 11(12), 1556–1559. [CrossRef]
- Goud, J. S., Elisetti, S. K., Kaur, N., Singh, R., Saini, K. S., & Arora, V. (2023). Artificial intelligence in formulation and product design of BCS class I and II drugs. In Micro-Electronics and Telecommunication Engineering (pp. 481–494). Springer Nature Singapore.
- Hariry, R. E., & Barenji, R. V. (2023). Embracing digital technologies in the pharmaceutical industry. In Control Engineering in Mechatronics (pp. 141–165). Springer Nature Singapore.
- Hathout, R. M., & Saharan, V. A. (2022). Computer-Aided Formulation Development. In Computer Aided Pharmaceutics and Drug Delivery (pp. 73–98). Springer Nature Singapore.
- Hie, B., Cho, H., & Berger, B. (2018). Realizing private and practical pharmacological collaboration. Science, 362(6412), 347–350. [CrossRef]
- Hocharoen, L., Noppiboon, S., & Kitsubun, P. (2021). Toward QbD Process Understanding on DNA Vaccine Purification Using Design of Experiment. Frontiers in Bioengineering and Biotechnology, 9, 657201. [CrossRef]
- Hussain, R., Haider, Z., Khalid, H., Fatmi, M. Q., Carradori, S., Cataldi, A., & Zara, S. (2023). Computational Medicinal Chemistry applications to cure Asian-prevalent strain of Hepatitis C Virus. In ChemRxiv. [CrossRef]
- Izsák, R., Riplinger, C., Blunt, N. S., de Souza, B., Holzmann, N., Crawford, O., Camps, J., Neese, F., & Schopf, P. (2023). Quantum computing in pharma: A multilayer embedding approach for near future applications. Journal of Computational Chemistry, 44(3), 406–421. [CrossRef]
- Jagtap, K., Chaudhari, B., & Redasani, V. (2022). Quality by Design (QbD) concept Review in Pharmaceuticals. Asian Journal of Research in Chemistry, 303–307. [CrossRef]
- Jiang, J., Ma, X., Ouyang, D., & Williams, R. O., 3rd. (2022). Emerging Artificial Intelligence (AI) Technologies Used in the Development of Solid Dosage Forms. Pharmaceutics, 14(11). [CrossRef]
- Jose, J., S, S., Mathew, B., & Parambi, D. G. T. (2022). Trial Approach for Biomedical Products: A Regulatory Perspective. Combinatorial Chemistry & High Throughput Screening, 25(12), 1991–2000. [CrossRef]
- Jupally, P., Damagundam, S., & Domaraju, P. (2023). Quality by design (QBD) tool for quality control in pharmaceutical industry. International Journal of Pharmaceutical Sciences and Nanotechnology(IJPSN), 16(2), 6480–6487. [CrossRef]
- Kargl, M., Regitnig, P., Müller, H., & Holzinger, A. (2020). Towards a better understanding of the workflows: Modeling pathology processes in view of future AI integration. In Artificial Intelligence and Machine Learning for Digital Pathology (pp. 102–117). Springer International Publishing.
- Katiyar, R. S., & Jha, P. K. (2018). Molecular simulations in drug delivery: Opportunities and challenges. Wiley Interdisciplinary Reviews. Computational Molecular Science, 8(4), e1358. [CrossRef]
- Kharb, M., & Rathore, K. S. (2022). QbD approach: A framework for integrating quality into pharmaceutical products. Journal of Pharmaceutical Research International, 1–11.
- Krstevska, A., Đuriš, J., Ibrić, S., & Cvijić, S. (2022). In-Depth Analysis of Physiologically Based Pharmacokinetic (PBPK) Modeling Utilization in Different Application Fields Using Text Mining Tools. Pharmaceutics, 15(1). [CrossRef]
- Kumari, R., MM College of Pharmacy, MM (DU), Mullana, Ambala, Haryana, Kumar, R., Bisht, A., Sharma, V., MM College of Pharmacy, MM (DU), Mullana, Ambala, Haryana, MM College of Pharmacy, MM (DU), Mullana, Ambala, Haryana, & MM College of Pharmacy, MM (DU), Mullana, Ambala, Haryana. (2023). A descriptive review on process analytical technology. Indian Journal of Health Care, Medical & Pharmacy Practice, 4(1), 1–12.
- Laky, D., Casas-Orozco, D., Rossi, F., Mackey, J. S., Reklaitis, G. V., & Nagy, Z. K. (2022). Determination of probabilistic design spaces in the hybrid manufacture of an active pharmaceutical ingredient using PharmaPy. In Computer Aided Chemical Engineering (pp. 2131–2136). Elsevier.
- Lancheros Porras, K. D., Alves, I. A., & Novoa, D. M. A. (2024). PBPK Modeling as an Alternative Method of Interspecies Extrapolation that Reduces the Use of Animals: A Systematic Review. Current Medicinal Chemistry, 31(1), 102–126. [CrossRef]
- Liang, L., Hu, J., Sun, G., Hong, N., Wu, G., He, Y., Li, Y., Hao, T., Liu, L., & Gong, M. (2022). Artificial Intelligence-Based Pharmacovigilance in the Setting of Limited Resources. Drug Safety: An International Journal of Medical Toxicology and Drug Experience, 45(5), 511–519. [CrossRef]
- Light, D. W., & Warburton, R. (2011). Demythologizing the high costs of pharmaceutical research. BioSocieties, 6(1), 34–50.
- Li, L., Zeng, L., Gao, Z., Yuan, S., Bian, Y., Wu, B., Zhang, H., Yu, Y., Lu, C., Zhou, Z., Xu, H., Li, J., Zhao, P., & Heng, P.-A. (2022). ImDrug: A benchmark for deep imbalanced learning in AI-aided drug discovery. [CrossRef]
- Lu, C., Liu, Q., Wang, C., Huang, Z., Lin, P., & He, L. (2019). Molecular Property Prediction: A Multilevel Quantum Interactions Modeling Perspective. arXiv: Computational Physics. https://typeset.io/papers/molecular-property-prediction-a-multilevel-quantum-3fm5z9ic6f.
- Mahesh, V., & Shijo, S. (2023). Accelerating drug discovery with quantum computing. In Evolution and Applications of Quantum Computing (pp. 175–181). Wiley. [CrossRef]
- Maithani, M., Chawla, V., & Chawla, P. A. (2022). Computers in Pharmaceutical Analysis. In Computer Aided Pharmaceutics and Drug Delivery (pp. 593–621). Springer Nature Singapore.
- McDermott, L. (2023). Process analytical technology (PAT) model lifecycle management. Spectroscopy , 9–13. [CrossRef]
- Mehrotra, S., Salwa, A., & Kumar, L. (2023). Implementation of Quality by Design in the Formulation and Development of Nanocarrier-Based Drug Delivery Systems. Critical Reviews in Therapeutic Drug Carrier Systems, 40(3), 1–46. [CrossRef]
- Michielan, L., & Moro, S. (2010). Pharmaceutical perspectives of nonlinear QSAR strategies. Journal of Chemical Information and Modeling, 50(6), 961–978. [CrossRef]
- Muqtadiroh, F. A., Purnomo, M. H., & Purnama, I. K. E. (2022). AI in the pharmaceutical industry promises cheaper, faster, better drugs (R. Ernunsari & S. Phillips (eds.)). Monash University. [CrossRef]
- Nainwal, N., Bahuguna, R., Banerjee, S., & Saharan, V. A. (2022). Historical developments on computer applications in pharmaceutics. In Computer Aided Pharmaceutics and Drug Delivery (pp. 39–72). Springer Nature Singapore.
- Nambiar, A. G., Singh, M., Mali, A. R., Serrano, D. R., Kumar, R., Healy, A. M., Agrawal, A. K., & Kumar, D. (2022). Continuous Manufacturing and Molecular Modeling of Pharmaceutical Amorphous Solid Dispersions. AAPS PharmSciTech, 23(7), 249. [CrossRef]
- Narayanan, H., Dingfelder, F., Condado Morales, I., Patel, B., Heding, K. E., Bjelke, J. R., Egebjerg, T., Butté, A., Sokolov, M., Lorenzen, N., & Arosio, P. (2021). Design of Biopharmaceutical Formulations Accelerated by Machine Learning. Molecular Pharmaceutics, 18(10), 3843–3853. [CrossRef]
- Niklas, J., Diaz Ochoa, J. G., Bucher, J., & Mauch, K. (2013). Quantitative Evaluation and Prediction of Drug Effects and Toxicological Risk Using Mechanistic Multiscale Models. Molecular Informatics, 32(1), 14–23. [CrossRef]
- Ouranidis, A., Davidopoulou, C., Tashi, R.-K., & Kachrimanis, K. (2021). Pharma 4.0 Continuous mRNA Drug Products Manufacturing. Pharmaceutics, 13(9). [CrossRef]
- Ouyang, D., & Smith, S. C. (2015). Introduction to computational pharmaceutics. In Computational Pharmaceutics (pp. 1–5). John Wiley & Sons, Ltd.
- Pandya, A., Howard, M. J., Zloh, M., & Dalby, P. A. (2018). An Evaluation of the Potential of NMR Spectroscopy and Computational Modelling Methods to Inform Biopharmaceutical Formulations. Pharmaceutics, 10(4). [CrossRef]
- Pareek, V., Sharma, L., Kumar, S., & Sharma, V. (2022). Need for artificial intelligence in pharmaceutical industry and its limitations. Journal of the Indian Academy of Geriatrics, 18(4), 217. [CrossRef]
- Patil, P., Kumar Nrip, N., Hajare, A., Hajare, D., K. Patil, M., Kanthe, R., & T. Gaikwad, A. (2023). Artificial intelligence and tools in pharmaceuticals: An overview. Journal of Advanced Pharmaceutical Technology & Research, 2075–2082.
- Pazhayattil, A. B., Sharma, S., Philip, J. P., Gischewski-Silva, M., & Ingram, M. (2023). Quality by design (QbD) process design. In AAPS Introductions in the Pharmaceutical Sciences (pp. 11–28). Springer International Publishing.
- Peletier, L. A., & Gabrielsson, J. (2015). Challenges in pharmacology modelling. Journal of Dynamics and Differential Equations, 27(3-4), 941–959. [CrossRef]
- Poduri, R. (2021). Pharmaceutical industry, academia, regulatory authorities and end user collaboration in successful drug discovery and development. In Drug Discovery and Development (pp. 465–471). Springer Singapore.
- Pokhriyal, P., Chavda, V. P., & Pathak, M. (2023). Future prospects and challenges in the implementation of AI and ML in pharma sector. In Bioinformatics Tools for Pharmaceutical Drug Product Development (pp. 401–416). Wiley. [CrossRef]
- Polasek, T. M., & Schuck, V. (2023). Improving the Efficiency of Clinical Pharmacology Studies. Clinical Pharmacology in Drug Development, 12(8), 771–774. [CrossRef]
- Pozzan, A. (2020). QM Calculations in ADMET Prediction. Methods in Molecular Biology , 2114, 285–305.
- Quazi, S., & Fatima, Z. (2023). Role of artificial intelligence and machine learning in drug discovery and drug repurposing. In Structural and Functional Aspects of Biocomputing Systems for Data Processing (pp. 182–197). IGI Global.
- Rischawy, F., Briskot, T., Schimek, A., Wang, G., Saleh, D., Kluters, S., Studts, J., & Hubbuch, J. (2022). Integrated process model for the prediction of biopharmaceutical manufacturing chromatography and adjustment steps. SSRN Electronic Journal. [CrossRef]
- Rizzuti, B. (2022). Molecular simulations of proteins: From simplified physical interactions to complex biological phenomena. Biochimica et Biophysica Acta: Proteins and Proteomics, 1870(3), 140757. [CrossRef]
- Russell, A., & Capece, M. (2022). Pharmaceutical Process Modeling. AAPS PharmSciTech, 23(4), 91. [CrossRef]
- Sadiku, M. N. O., Reeves, S. M., & Musa, S. M. (2019). The impact of computational pharmacology. European Scientific Journal, 15(9). [CrossRef]
- Sadybekov, A. V., & Katritch, V. (2023). Computational approaches streamlining drug discovery. Nature, 616(7958), 673–685. [CrossRef]
- Saharan, V. A., Banerjee, S., Penuli, S., & Dobhal, S. (2022). History and present scenario of computers in pharmaceutical research and development. In Computer Aided Pharmaceutics and Drug Delivery (pp. 1–38). Springer Nature Singapore.
- Saunders, R., Cardoso, S., Le Pouésard, M., Breslin, C., Myers, K., Swift, M., & Collins, T. (2020). Addressing essential skills gaps among participants in an OHS training program: a pilot study. Policy and Practice in Health and Safety, 18(2), 169–185. [CrossRef]
- Shanbhogue, M. H., Thirumaleshwar, S., Tegginamath, P. K., & Somareddy, H. K. (2021). Artificial Intelligence in Pharmaceutical Field - A Critical Review. Current Drug Delivery, 18(10), 1456–1466. [CrossRef]
- Sharma, A. K. (2022, November 9). A study on the applicability of AI in Pharmaceutical Industry. 2022 1st International Conference on Computational Science and Technology (ICCST). 2022 1st International Conference on Computational Science and Technology (ICCST), CHENNAI, India. [CrossRef]
- Shi, G., Lin, L., Liu, Y., Chen, G., Luo, Y., Wu, Y., & Li, H. (2021). Pharmaceutical application of multivariate modelling techniques: a review on the manufacturing of tablets. RSC Advances, 11(14), 8323–8345. [CrossRef]
- Song, W. (2023). The mechanism and design strategy of metal-organic frameworks in drug delivery. Applied and Computational Engineering, 7(1), 188–195. [CrossRef]
- Srivastava, R. (2022). Transformation of drug discovery towards artificial intelligence: An in silico approach. In Density Functional Theory - Recent Advances, New Perspectives and Applications. IntechOpen.
- Stevanovic, J. S. (2023). Process system integration. International Journal of Research and Innovation in Applied Science, VIII(IV), 78–82.
- Takemura, K., & Kitao, A. (2023). Molecular dynamics. In Plasma Membrane Shaping (pp. 431–443). Elsevier.
- Thakur, G., Masampally, V., Kulkarni, A., & Rathore, A. S. (2022). Process Analytical Technology (PAT) Implementation for Membrane Operations in Continuous Manufacturing of mAbs: Model-Based Control of Single-Pass Tangential Flow Ultrafiltration. The AAPS Journal, 24(4), 83. [CrossRef]
- Tielker, N., Eberlein, L., Hessler, G., Schmidt, K. F., Güssregen, S., & Kast, S. M. (2021). Quantum-mechanical property prediction of solvated drug molecules: what have we learned from a decade of SAMPL blind prediction challenges? Journal of Computer-Aided Molecular Design, 35(4), 453–472.
- Tugcu, G., Sipahi, H., Charehsaz, M., Aydın, A., & Saçan, M. T. (2023). Computational toxicology of pharmaceuticals. In Cheminformatics, QSAR and Machine Learning Applications for Novel Drug Development (pp. 519–537). Elsevier.
- Umashankar, V., & Gurunathan, S. (2011). In SilicoTools for Molecular Modeling. In General, Applied and Systems Toxicology. John Wiley & Sons, Ltd. [CrossRef]
- Vipanchi, V., Fatima, M., & Domaraju, P. (2023). Quality by Design approach for nanosystem based topical drug delivery. German Journal of Pharmaceuticals and Biomaterials, 2(1), 01–17. [CrossRef]
- Vora, L. K., Gholap, A. D., Jetha, K., Thakur, R. R. S., Solanki, H. K., & Chavda, V. P. (2023). Artificial Intelligence in Pharmaceutical Technology and Drug Delivery Design. Pharmaceutics, 15(7). [CrossRef]
- Wang, P.-H., Chen, J.-H., Yang, Y.-Y., Lee, C., & Tseng, Y. J. (2023). Recent advances in quantum computing for drug discovery and development. IEEE Nanotechnology Magazine, 1–5. [CrossRef]
- Wang, S., Di, J., Wang, D., Dai, X., Hua, Y., Gao, X., Zheng, A., & Gao, J. (2022). State-of-the-Art Review of Artificial Neural Networks to Predict, Characterize and Optimize Pharmaceutical Formulation. Pharmaceutics, 14(1). [CrossRef]
- Wang, Y., & Tuerhong, G. (n.d.). A Survey of Interpretable Machine Learning Methods. Retrieved April 11, 2024. [CrossRef]
- Wang, Z., & Ierapetritou, M. (2018). Global sensitivity, feasibility, and flexibility analysis of continuous pharmaceutical manufacturing processes. In Computer Aided Chemical Engineering (pp. 189–213). Elsevier.
- Wang, Z., Pan, Z., He, D., Shi, J., Sun, S., & Hou, Y. (2019). Simulation modeling of a pharmaceutical tablet manufacturing process via wet granulation. Complexity, 2019, 1–16. [CrossRef]
- Wanjul, P. B., Mourya, S. P., & Gangad, V. L. (2023). Future directions of AI in pharma : Innovation in pharmaceutical industry. International Journal For Multidisciplinary Research, 5(3). [CrossRef]
- Weinan, Lei, H., Xie, P., & Zhang, L. (2023). Machine learning-assisted multi-scale modeling. Journal of Mathematics and Physics, 64(7). [CrossRef]
- Williams, T., Kalinka, K., Sanches, R., Blanchard-Emmerson, G., Watts, S., Davies, L., Knevelman, C., McCloskey, L., Jones, P., Mitrophanous, K., Miskin, J., & Dikicioglu, D. (2023). Machine learning and metabolic modelling assisted implementation of a novel process analytical technology in cell and gene therapy manufacturing. Scientific Reports, 13(1), 834. [CrossRef]
- Xiong H.-S., Zhang Q., Zhang S.-N., Cai J.-Y., Su J., Zhu Y.-H., & Yan K.-J. (2023). [Methodology and application of process analytical technology (PAT) for traditional Chinese medicine manufacturing:a review]. Zhongguo Zhong yao za zhi = Zhongguo zhongyao zazhi = China journal of Chinese materia medica, 48(1), 22–29.
- Xu, B. (2023). The application of molecular dynamic simulations. Applied and Computational Engineering, 7(1), 147–151.
- Xu, J., Kalyani, D., Struble, T., Dreher, S., Krska, S., Buchwald, S. L., & Jensen, K. F. (2022). Roadmap to pharmaceutically relevant reactivity models leveraging High-Throughput Experimentation. In ChemRxiv. [CrossRef]
- Yang, H. (2022). Transforming pharma with data science, AI and machine learning. In Data Science, AI, and Machine Learning in Drug Development (pp. 1–17). Chapman and Hall/CRC.
- Yazdanpanah, N. (2022). Process development and integration by mathematical modeling and simulation tools. In Integration and Optimization of Unit Operations (pp. 273–292). Elsevier.
- Yeom, S. B., Ha, E.-S., Kim, M.-S., Jeong, S. H., Hwang, S.-J., & Choi, D. H. (2019). Application of the Discrete Element Method for Manufacturing Process Simulation in the Pharmaceutical Industry. Pharmaceutics, 11(8). [CrossRef]
- Zhang, H., Goedegebuure, S. P., Ding, L., Hawkins, W., DeNardo, D., Fields, R. C., Chen, Y., Payne, P., & Li, F. (2023). M3NetFlow: a novel multi-scale multi-hop multi-omics graph AI model for omics data integration and interpretation. In bioRxiv. [CrossRef]
- Zhou, H. (2023, January 6). High-efficiency drug design research based on virtual high-throughput screening. 2023 International Conference on Intelligent Supercomputing and BioPharma (ISBP). 2023 International Conference on Intelligent Supercomputing and BioPharma (ISBP), Zhuhai, China. [CrossRef]
- Zhou, M., Schultz, K., Wilson, J., Kruel, A., Thibert, S., Bracken, C., Orton, D., Gibbons, B., Chu, R., McNaughton, A., & Pasa-Tolic, L. (2022). High-throughput native mass spectrometry as experimental validation for in silico drug design. Office of Scientific and Technical Information (OSTI). [CrossRef]
- Zhou, W., Li, X., Han, L., & Fan, S. (2021). Application of network pharmacology based on artificial intelligence algorithms in drug development. In Network Pharmacology (pp. 35–73). Springer Singapore.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





