Submitted:
15 April 2024
Posted:
17 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. DNA Extraction and Purification
2.3. DNA Quantification
2.4. Next-Generation Sequencing and Data Analysis
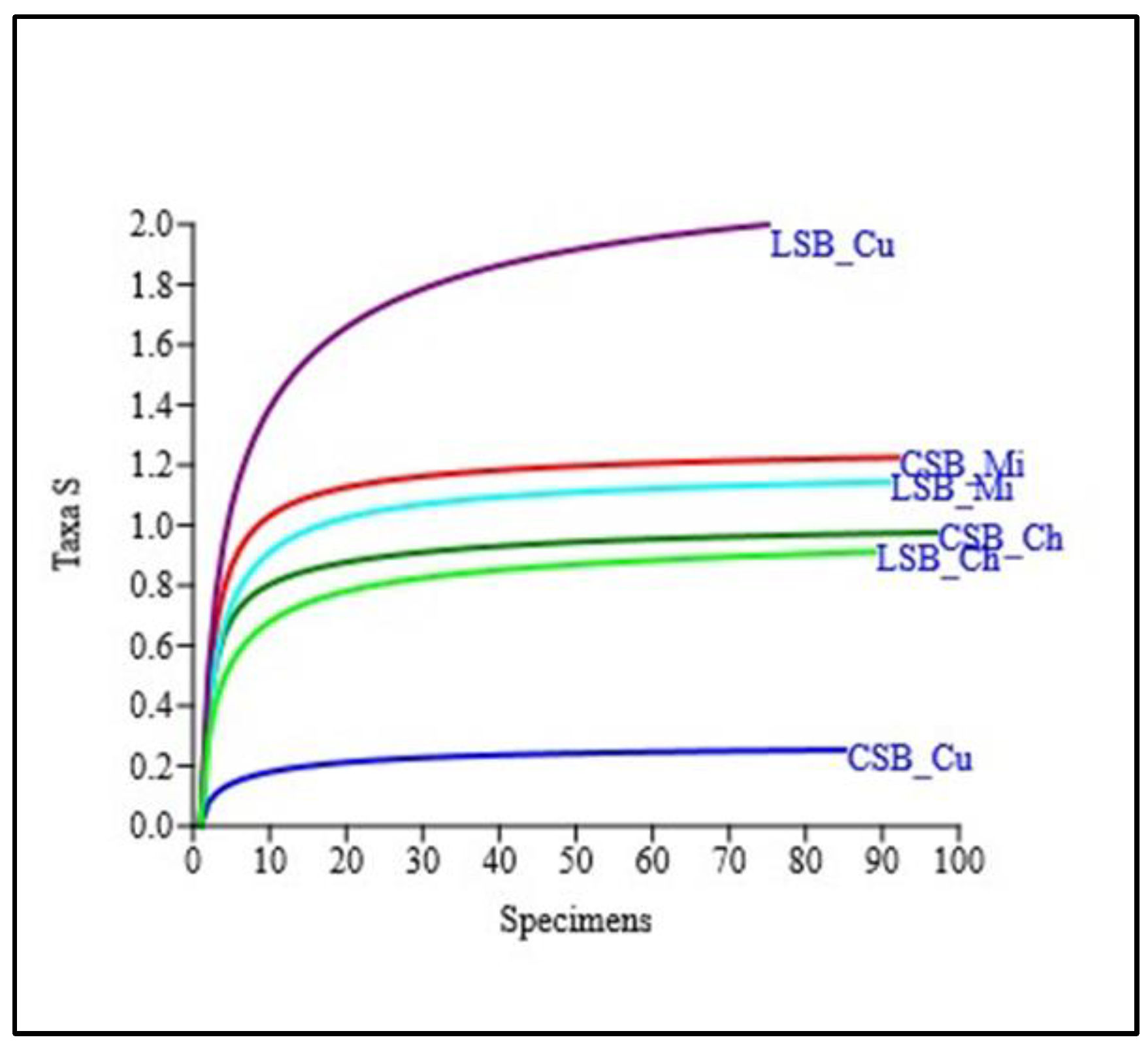
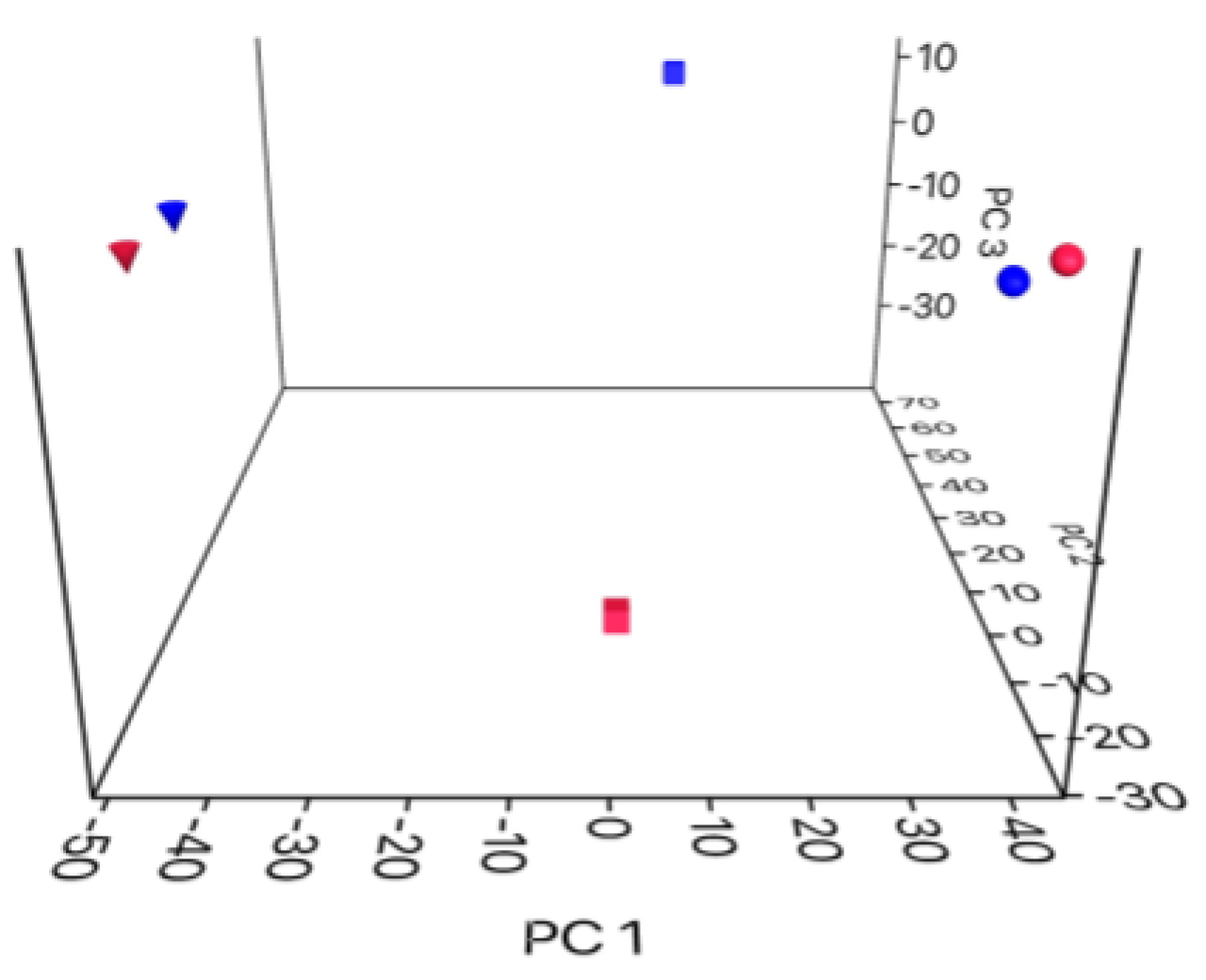
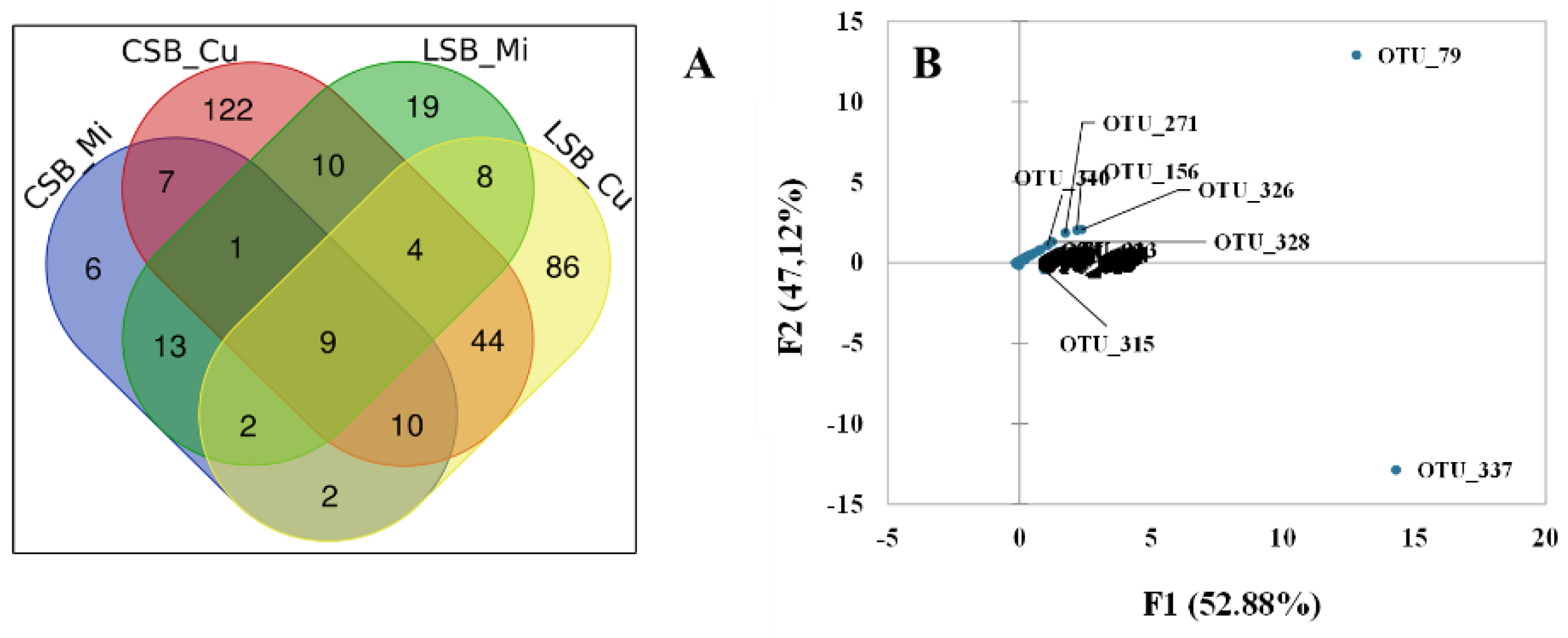
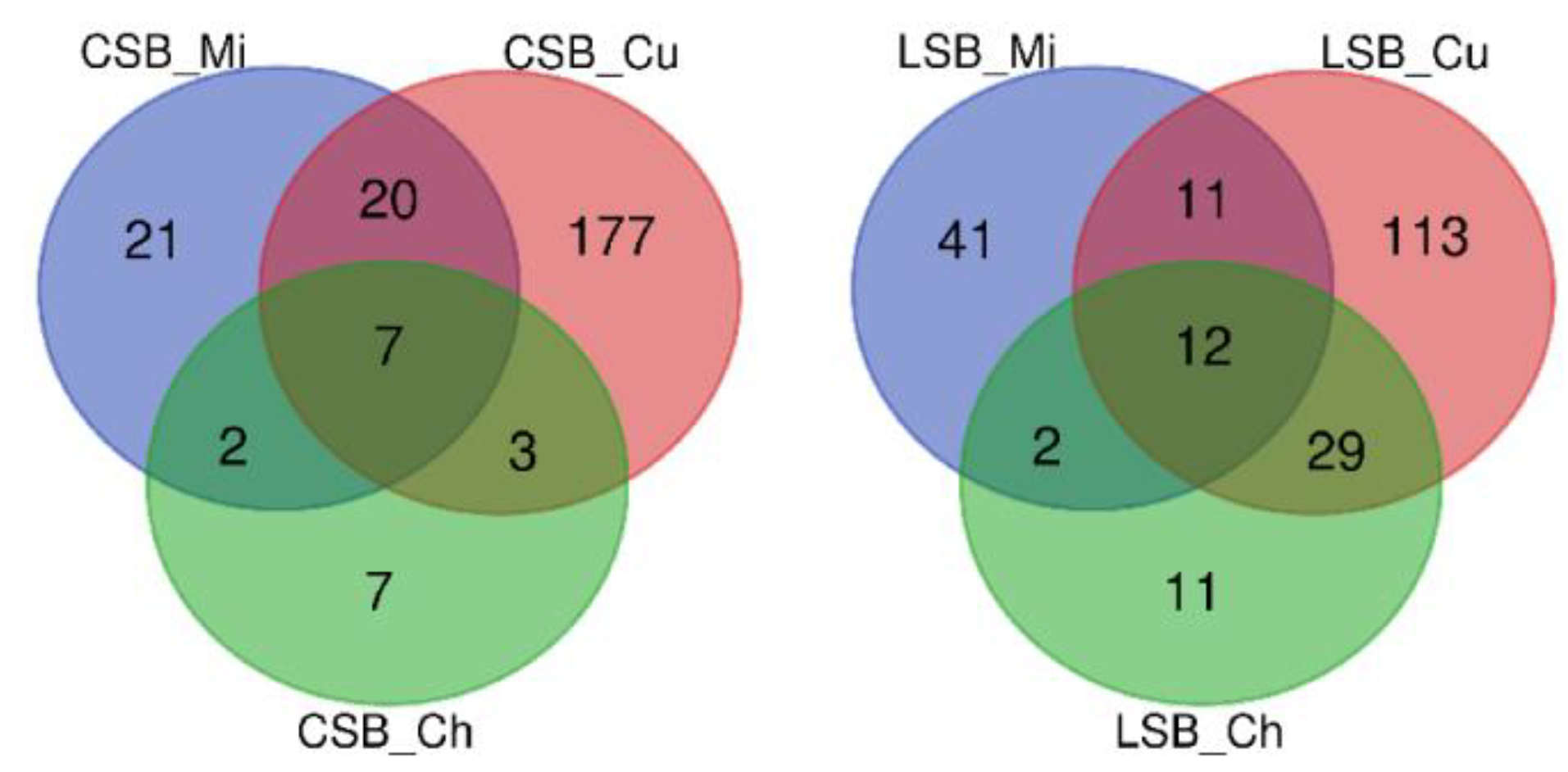
3. Results
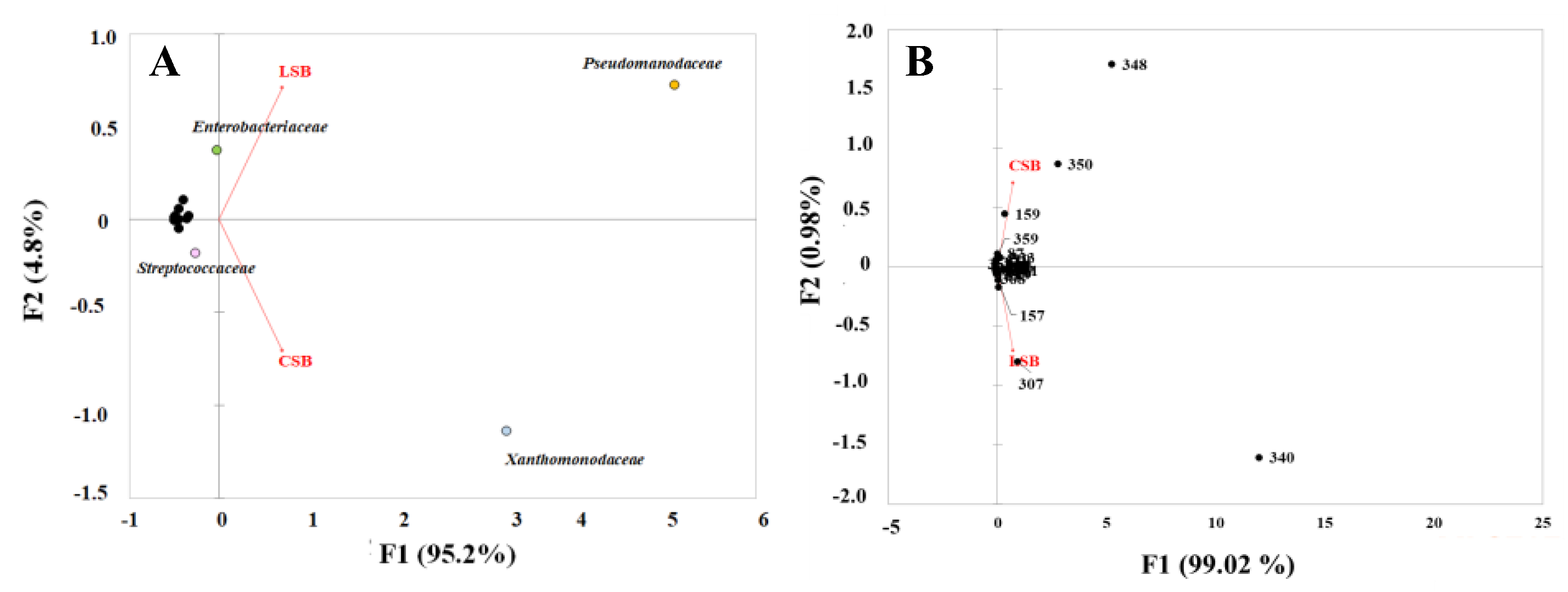
3.1. Sheep Milk Microbiota
3.2. Curd Microbiota
3.3. Cheese Microbiota
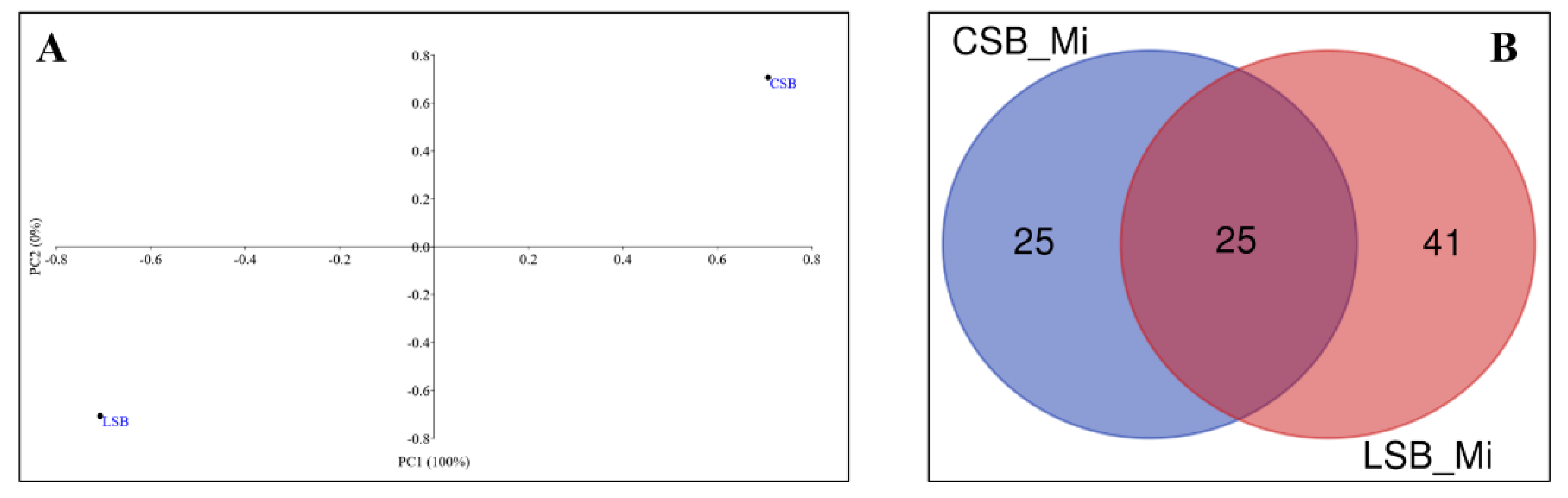
3.3.1. Influence of the Milk Microbiome
3.3.2. Influence of the Curd Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tamang, J.P.; Cotter, P.D.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; et al. Fermented foods in a global age: east meets West. Compr. Rev. Food Sci. Food Saf. 2020, 19, 184–217. [Google Scholar] [CrossRef] [PubMed]
- Chilton, S.N.; Burton, J.P.; Reid, G. Inclusion of fermented foods in food guides around the world. Nutrients 2015, 7, 390–404. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; et al. Health benefits of fermented foods: microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Kok, C.R.; Hutkins, R. Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr. Rev. 2018, 76, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Leeuwendaal, N.K.; Stanton, C.; O'Toole, P.W.; Beresford, T.P. Fermented foods, health and the gut microbiome. Nutrients 2022, 14, 1527. [Google Scholar] [CrossRef] [PubMed]
- Vinderola, G.; Cotter, P.D.; Freitas, M.; Gueimonde, M.; Holscher, H.D.; Ruas-Madiedo, P.; Salminen, S.; Swanson, K.S.; Sanders, M.E.; Cifelli, C.J. Fermented foods: a perspective on their role in delivering biotics. Front. Microbiol. 2023, 14, 1196239. [Google Scholar] [CrossRef]
- Shiby, V.K.; Mishra, H.N. Fermented milks and milk products as functional foods—a review. Crit. Rev. Food Sci. Nutr. 2013, 53, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Dadousis, C.; Pegolo, S.; Rosa, G.J.M.; Gianola, D.; Bittante, G.; Cecchinato, A. Pathway-based genome-wide association analysis of milk coagulation properties, curd firmness, cheese yield, and curd nutrient recovery in dairy cattle. J. Dairy Sci. 2017, 100, 1223–1231. [Google Scholar] [CrossRef]
- Dairy and Dairy Products (Chapter 7). in OECD/FAO (2021), OECD-FAO Agricultural Outlook 2021-2030, OECD Publishing, Paris. [CrossRef]
- Gobbetti, M.; Di Cagno, R. Chapter 32 - Extra-Hard Varieties. In Cheese, 4th ed.; McSweeney, P.L.H., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Academic Press, Cambridge, MA, USA, 2017, pp. 809–828, ISBN 9780124170124.
- Gobbetti, M.; Di Cagno, R. Extra-hard Varieties. In Encyclopedia of Dairy Sciences, 3rd ed.; McSweeney, P.L.H., McNamara, J.P., Eds; Academic Press, Cambridge, MA, USA, 2022; Volume 3, pp. 172–195, ISBN 9780128187678.
- Caridi, A.; Micari, P.; Caparra, P.; Curari, A.; Sarullo, V. Ripening and seasonal changes in microbial groups and in physico-chemical properties of the ewes’ cheese Pecorino del Poro. Int. Dairy J. 2003, 13, 191–200. [Google Scholar] [CrossRef]
- Di Cagno, R.; Banks, J.; Sheehan, L.; Fox, P.F.; Brechany, E.Y.; Corsetti, A.; Gobbetti, M. Comparison of the microbiological, compositional, biochemical, volatile profile and sensory characteristics of three Italian PDO ewes’ milk cheeses. Int. Dairy J. 2003, 13, 961–972. [Google Scholar] [CrossRef]
- Coda, R.; Brechany, E.; De Angelis, M.; De Candia, S.; Di Cagno, R.; Gobbetti, M. Comparison of the compositional, microbiological, biochemical, and volatile profile characteristics of nine Italian ewes' milk cheeses. J. Dairy Sci. 2006, 89, 4126–4143. [Google Scholar] [CrossRef]
- Ozbun, T. Production volume of Pecorino Romano PDO in Italy 2012-2022. (February 13, 2024) https://www.statista.com/statistics/551472/pecorino-romano-pdo-production-volume-in-italy/.
- Carloni, E.; Petruzzelli, A.; Amagliani, G.; Brandi, G.; Caverni, F.; Mangili, P.; Tonucci, F. Effect of farm characteristics and practices on hygienic quality of ovine raw milk used for artisan cheese production in central Italy. Anim. Sci J. 2016, 87, 591–599. [Google Scholar] [CrossRef]
- Fusté-Forné, F. Developing cheese tourism: a local-based perspective from Valle de Roncal (Navarra, Spain). J. Ethn. Foods. 2020, 7. [Google Scholar] [CrossRef]
- Montel, M.C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: rich and diverse microbiota with associated benefits. Int. J. Food Micro. 2014, 177, 136–154. [Google Scholar] [CrossRef] [PubMed]
- Fuka, M.M.; Wallisch, S.; Engel, M.; Welzl, G.; Havranek, J.; Schloter, M. Dynamics of bacterial communities during the ripening process of different croatian cheese types derived from raw ewe's milk cheeses. PLOS ONE. 2013, 8, e80734. [Google Scholar] [CrossRef] [PubMed]
- Hattem, H.E.; Taleb, A.T.; Manal, A.N.; Hanaa, S.S. Effect of pasteurization and season on milk composition and ripening of Ras cheese. J. Brew. Distilling. 2012, 3, 15–22. [Google Scholar] [CrossRef]
- Pasquali, F.; Valero, A.; Possas, A.; Lucchi, A.; Crippa, C.; Gambi, L.; Manfreda, G.; De Cesare, A. Occurrence of foodborne pathogens in Italian soft artisanal cheeses displaying different intra- and inter-batch variability of physicochemical and microbiological parameters. Front. Microbiol. 2022, 959648. [Google Scholar] [CrossRef] [PubMed]
- Condoleo, R.; Palumbo, R.; Mezher, Z.; Bucchini, L.; Taylor, R.A. Microbial risk assessment of Escherichia coli shiga-toxin producers (STEC) in raw sheep’s milk cheeses in Italy. Food Control 2022, 137, 108951. [Google Scholar] [CrossRef]
- Pinto, U.M.; De Dea, L.J. Editorial: Community series in microbiological safety and quality aspects of fermented dairy products, volume II. Front. Microbiol. 2023, 14, 1182373. [Google Scholar] [CrossRef]
- Yuan, H.; Han, S.; Zhang, S.; Xue, Y.; Zhang, Y.; Lu, H.; Wang, S. Microbial properties of raw milk throughout the year and their relationships to quality parameters. Foods 2022, 1, 3077. [Google Scholar] [CrossRef]
- Bettera, L.; Dreier, M.; Schmidt, R.S.; Gatti, M.; Berthoud, H.; Bachmann, H.P. Selective enrichment of the raw milk microbiota in cheese production: Concept of a natural adjunct milk culture. Front. Microbiol. 2023, 14, 1154508. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; La Storia, A.; Villani, F.; Ercolini, D. Exploring the sources of bacterial spoilers in beefsteaks by culture-independent high-throughput sequencing. PLoS One 2013, 8, e70222. [Google Scholar] [CrossRef] [PubMed]
- Lusk, T.S.; Ottesen, A.R.; White, J.R.; Allard, M.W.; Brown, E.W.; Kase, J.A. Characterization of microflora in Latin-style cheeses by next-generation sequencing technology. BMC microbiol. 2012, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 2013, 79, 3148–3155. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, I.; Di Cagno, R.; Buchin, S.; De Angelis, M.; Gobbetti, M. Microbial ecology dynamics reveal a succession in the core microbiota involved in the ripening of pasta filata Caciocavallo Pugliese cheese. Appl. Environ. Microbiol. 2014, 80, 6243–6255. [Google Scholar] [CrossRef] [PubMed]
- Kergourlay, G.; Taminiau, B.; Daube, G.; Vergès, M.C.C. Metagenomic insights into the dynamics of microbial communities in food. Int. J. Food Microbiol. 2015, 213, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Dugat-Bony, E.; Garnier, L.; Denonfoux, J.; Ferreira, S.; Sarthou, A.S.; Bonnarme, P.; Irlinger, F. Highlighting the microbial diversity of 12 French cheese varieties. Int. J. Food Microbiol. 2016, 238, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Ceccarani, C.; Curone, G.; Severgnini, M.; Pollera, C.; Bronzo, V.; Riva, F.; Addis, M.F.; Filipe, J.; Amadori, M.; et al. Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows. PLoS ONE 2018, 13, e0205054. [Google Scholar] [CrossRef]
- Esteban-Blanco, C.; Gutiérrez-Gil, B.; Puente-Sánchez, F.; Marina, H.; Tamames, J.; Acedo, A.; Arranz, J.J. Microbiota characterization of sheep milk and its association with somatic cell count using 16s rRNA gene sequencing. J. Anim. Breed Genet. 2020, 137, 73–83. [Google Scholar] [CrossRef]
- Luziatelli, F.; Melini, F.; Ficca, A.G.; Melini, V.; Nardilli, F.; Ruzzi, M. Core microbiome and bacterial diversity of the Italian Mediterranean river buffalo milk. Appl. Microbiol. Biotechnol. 2023, 107, 1875–1886. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinform. 2015, 31.21, 3476–3482. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Opens external link in new window. Nucl. Acids Res. 2014, 42, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. Paleontological Statistics (PAST) 2.06. University of Oslo, Oslo, Norway. 2015, Retrieved, 24.
- Secchi, G.; Amalfitano, N.; Carafa, I.; Franciosi, E.; Gallo, L.; Schiavon, S.; Sturaro, E.; Tagliapietra, F.; Bittante, G. Milk metagenomics and cheese-making properties as affected by indoor farming and summer highland grazing. J. Dairy Sci. 2023, 106, 96–116. [Google Scholar] [CrossRef]
- Yuan, L.; Sadiq, F.A.; Burmølle, M.; Wang, N.I.; He, G. Insights into psychrotrophic bacteria in raw milk: a review. J. Food Prot., 2019, 82, 1148–1159. [Google Scholar] [CrossRef]
- Esteban-Blanco, C.; Gutiérrez-Gil, B.; Marina, H.; Pelayo, R.; Suárez-Vega, A.; Acedo, A.; Arranz, J.-J. The Milk microbiota of the Spanish Churra sheep breed: new insights into the complexity of the milk microbiome of dairy species. Animals 2020, 10, 1463. [Google Scholar] [CrossRef]
- Elomari, M.; Coroler, L.; Hoste, B.; Gillis, M.; Izard, D.; Leclerc, H. DNA relatedness among Pseudomonas strains isolated from natural mineral waters and proposal of Pseudomonas veronii sp. nov. Int. J. Syst. Bacteriol. 1996, 4, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Nam, I.H.; Chang, Y.S.; Hong, H.B.; et al. A novel catabolic activity of Pseudomonas veronii in biotransformation of pentachlorophenol. Appl. Microbiol. Biotechnol. 2003, 62, 284–290. [Google Scholar] [CrossRef]
- Onaca, C.; Kieninger, M.; Engesser, K.H.; Altenbuchner, J. Degradation of alkyl methyl ketones by Pseudomonas veronii MEK700. J. Bacteriol. 2007, 10, 3759–3767. [Google Scholar] [CrossRef]
- Meng, L.; Liu, H.; Dong, L.; Zheng, N.; Xing, M.; Zhang, Y.; Zhao, S.; Wang, J. Identification and proteolytic activity quantification of Pseudomonas spp. isolated from different raw milks at storage temperatures. J. Dairy Sci., 2018, 101, 2897–2905. [Google Scholar]
- Kamal-Eldin, A.; Ayyash, M.; Sobti, B.; Nagy, P. Camel Milk. In Encyclopedia of Dairy Sciences, 3rd ed.; McSweeney, P.L.H., John, P. McNamara, J.P., Eds.; Academic Press, Cambridge, MA, USA, 2022; Volume 5, pp. 504–513, ISBN 9780128187678.
- Oikonomou, G.; Addis, M.F.; Chassard, C.; Nader-Macias, M.E.F.; Grant, I.; Delbès, C.; Bogni, C.I.; Le Loir, Y.; Even, S. Milk microbiota: what are we exactly talking about? Front. Microbiol. 2020, 11, 60. [Google Scholar] [CrossRef]
- McInnis, E.A.; Kalanetra, K.M.; Mills, D.A.; Maga, E.A. Analysis of raw goat milk microbiota: Impact of stage of lactation and lysozyme on microbial diversity. Food Microbiol. 2015, 46, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, H.; Fehr, K.B.; Sepehri, S.; Francoz, D.; De Buck, J.; Barkema, H.W.; Plaizier, J.C.; Khafipour, E. Invited review: Microbiota of the bovine udder: contributing factors and potential implications for udder health and mastitis susceptibility. J. Dairy Sci. 2018, 101, 10605–10625. [Google Scholar] [CrossRef]
- Kuehn, J.S.; Gorden, P.J.; Munro, D.; Rong, R.; Dong, Q.; et al. Bacterial community profiling of milk samples as a means to understand culture-negative bovine clinical mastitis. PLoS ONE 2013, 8, e61959. [Google Scholar] [CrossRef]
- Zeinhom, M.M.A.; Hassan, G.M.; Salem, H.A.M.; Corke, H. Prevalence and survival of Stenotrophomonas species in milk and dairy products in Egypt. Foodborne Pathog. Dis., 2021, 18, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Barchitta, M.; Cipresso, R.; Giaquinta, L.; Romeo, M.A.; Denaro, C.; Pennisi, C.; Agodi, A. Acquisition and spread of Acinetobacter baumannii and S. maltophilia in intensive care patients. Int. J. Hyg. Environ. Health. 2009, 212, 330–337. [Google Scholar] [CrossRef] [PubMed]
- De Vrankrijker, A.M.; Wolfs, T.F.; Van der Ent, C.K. Challenging and emerging pathogens in cystic fibrosis. Paediatr. Respir. Rev. 2010, 11, 246–254. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xu, W.L.; Li, C.D.; Wang, F.C.; Guo, Y.S.; Ya, M. Determination of the microbial community of traditional Mongolian cheese by using culture-dependent and independent methods. Food Sci. Nutr. 2022, 2, 828–837. [Google Scholar] [CrossRef]
- Lyerly, D.M.; Kreger, A.S. Importance of Serratia protease in the pathogenesis of experimental Serratia marcescens pneumonia. Infect. Immun. 1983, 40, 113–119. [Google Scholar] [CrossRef]
- Abreo, E.; Altier, N. Pangenome of Serratia marcescens strains from nosocomial and environmental origins reveals different populations and the links between them. Sci. Rep. 2019, 9, 46. [Google Scholar] [CrossRef]
- Crone, S.; Vives-Flórez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martínez-García, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; Whiteley, M.; Kolter, R.; Bjarnsholt, T. The environmental occurrence of Pseudomonas aeruginosa. APMIS 2020, 128, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Endres, C.M.; Castro, I.M.S.; Trevisol, L.D.; Severo, J.M.; Mann, M.B.; Varela, A.P.M.; Frazzon, A.P.G.; Mayer, F.Q.; Frazzon, J. Molecular characterization of the bacterial communities present in sheep's milk and cheese produced in South Brazilian Region via 16S rRNA gene metabarcoding sequencing. LWT 2021, 147, 111579. [Google Scholar] [CrossRef]
- Stellato, G.; De Filippis, F.; La Storia, A.; Ercolini, D. Coexistence of lactic acid bacteria and potential ppoilage microbiota in a dairy processing environment. Appl. Environ. Microbiol. 2015, 22, 7893–7904. [Google Scholar] [CrossRef] [PubMed]
- Stellato, G.; La Storia, A.; Cirillo, T.; Ercolini, D. Bacterial biogeographical patterns in a cooking centre for hospital foodservice. Int. J. Food Microbiol. 2015, 193, 99–108. [Google Scholar] [CrossRef]
- Ruta, S.; Murray, M.; Kampff, Z.; McDonnell, B.; Lugli, G.A.; Ventura, M.; Todaro, M.; Settanni, L.; van Sinderen, D.; Mahony, J. Microbial Ecology of Pecorino Siciliano PDO Cheese Production Systems. Fermentation 2023, 9, 620. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Button, J.E.; Santarelli, M.; Dutton, R.J. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 2014, 158, 422–433. [Google Scholar] [CrossRef]
- Tirloni, E.; Bernardi, C.; Stella, S. Pseudomonas spp.: are food grade organic acids efficient against these spoilage microorganisms in fresh cheeses? Foods 2021, 4, 891. [Google Scholar] [CrossRef] [PubMed]
- Todaro, M.; Francesca, N.; Reale, S.; Moschetti, G.; Vitale, F.; Settanni, L. Effect of different salting technologies on the chemical and microbiological characteristics of PDO Pecorino Siciliano cheese. Eur. Food Res. Technol., 2011, 233, 931–940. [Google Scholar] [CrossRef]
- Foulds, J.D.; Shemin, D. Properties and characteristics of a bacteriocin from Serratia marcescens. J. Bacteriol. 1969, 99, 655–660. [Google Scholar] [CrossRef]
- Enfedaque, J.; Ferrer, S.; Guasch, J.F.; Tomás, J.; Regué, M. Bacteriocin 28b from Serratia marcescens N28b: identification of Escherichia coli surface components involved in bacteriocin binding and translocation. Can. J. Microbiol. 1996, 42, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.A.; Kuo, C.H.; Lai, Y.K.; Graumann, P.L.; Tu, J. Phosphate limitation induces the intergeneric inhibition of Pseudomonas aeruginosa by Serratia marcescens isolated from paper machines. FEMS Microbiol. Ecol. 2013, 84, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sheoran, P.; Gupta, A.; Yadav, J.; Tiwari, S.K. Antibacterial property of bacteriocin produced by Lactobacillus plantarum LD4 isolated from a fermented food. Ann. Microbiol. 2016, 66, 1431–1440. [Google Scholar] [CrossRef]
- Younas, S.; Mazhar, B.; Liaqat, I.; Ali, S.; Tahir, H.M.; Ali, N.M. Bacteriocin production by Lactobacilli and their role as antibacterial tool against common pathogens. J. Oleo Sci. 2022, 4, 541–550. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




