Submitted:
16 April 2024
Posted:
17 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
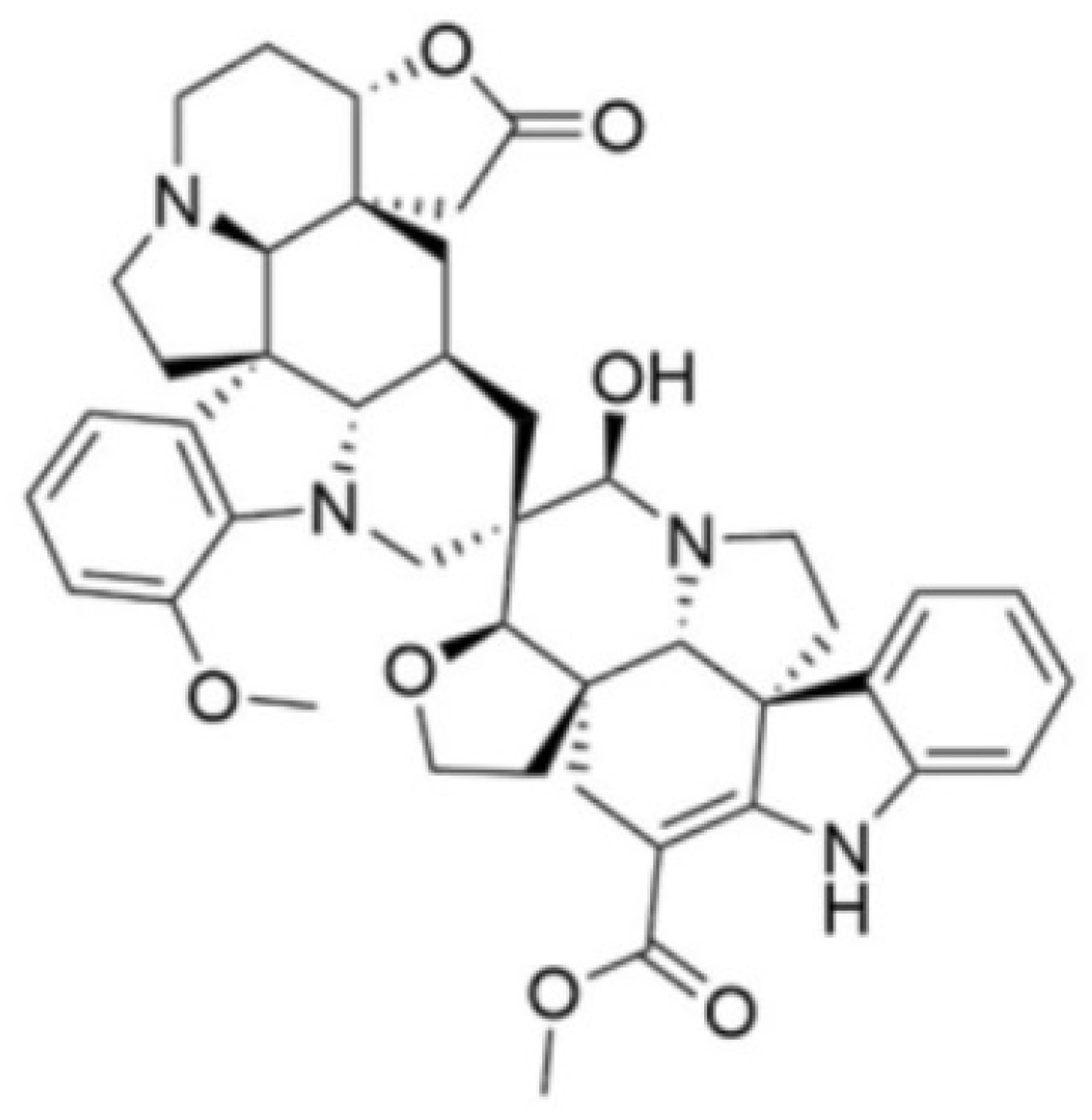
2.1. Test Compound
2.2. Colony-Forming Unit Viability Assay
2.3. MIC and MFC Determination
2.4. FAM-FLICA Poly-Caspase Assay
2.5. Molecular Docking against C. albicans Proteins
2.5.1. Ligand and Protein Preparation
2.5.2. Molecular Docking and Visualization of Interactions
2.6. Molecular Dynamics Simulations
3. Results
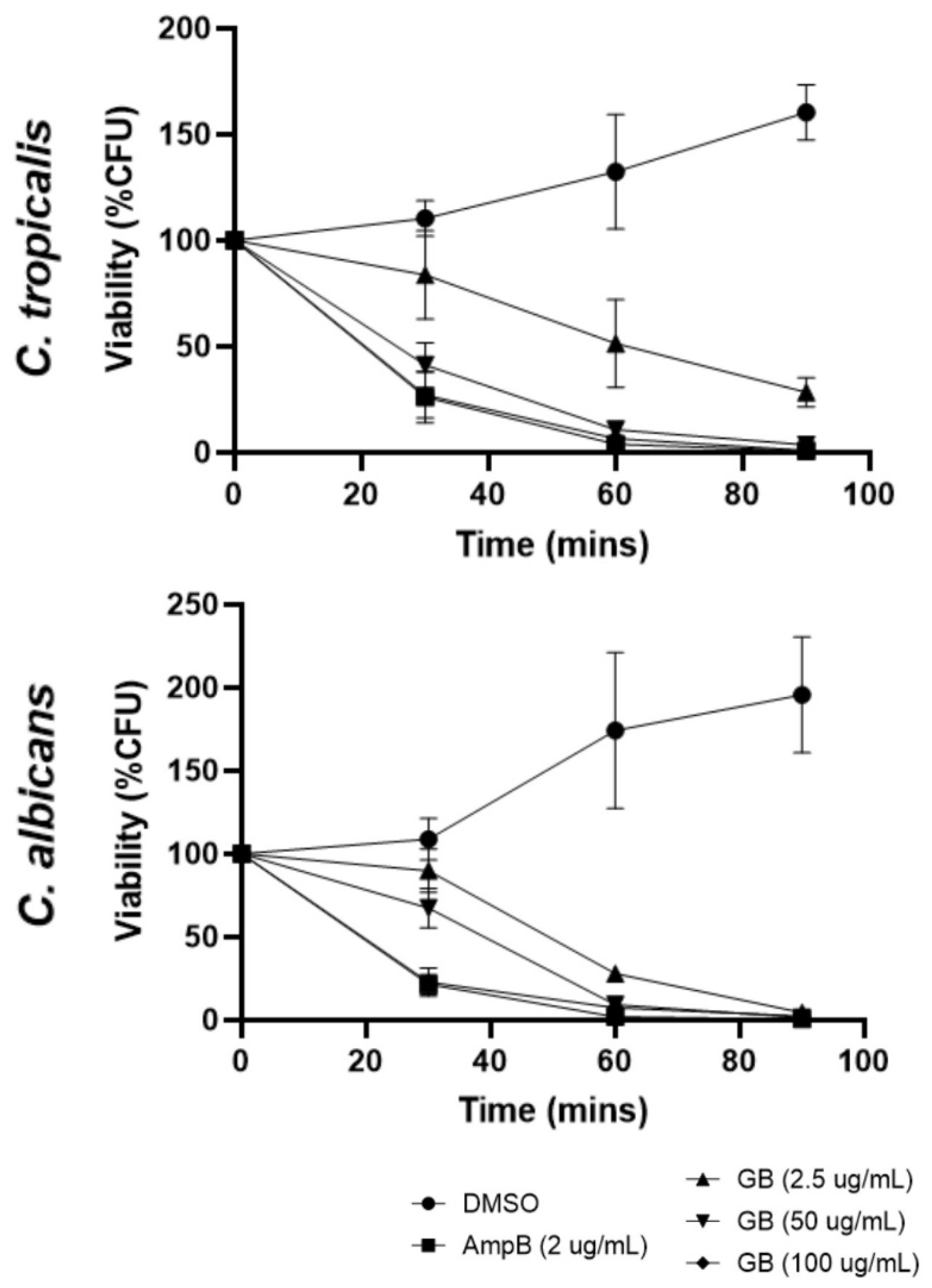
3.1. Effects of Globospiramine on C. albicans and C. tropicalis CFU Viability
3.2. MIC and MFC of Globospiramine versus C. albicans and C. tropicalis
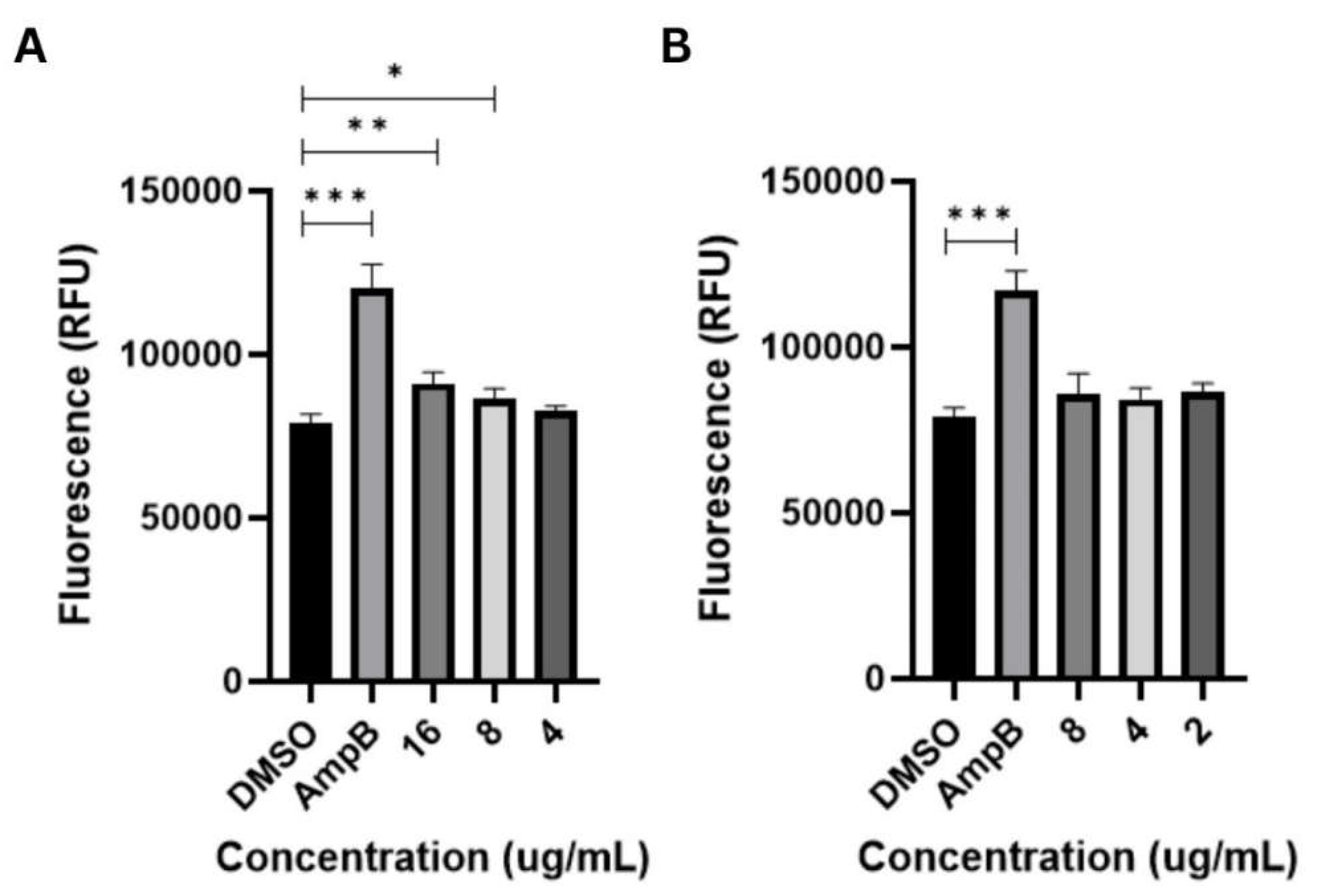
3.3. Apoptosis-Inducing Activities of Globospiramine vs C. albicans and C. tropicalis
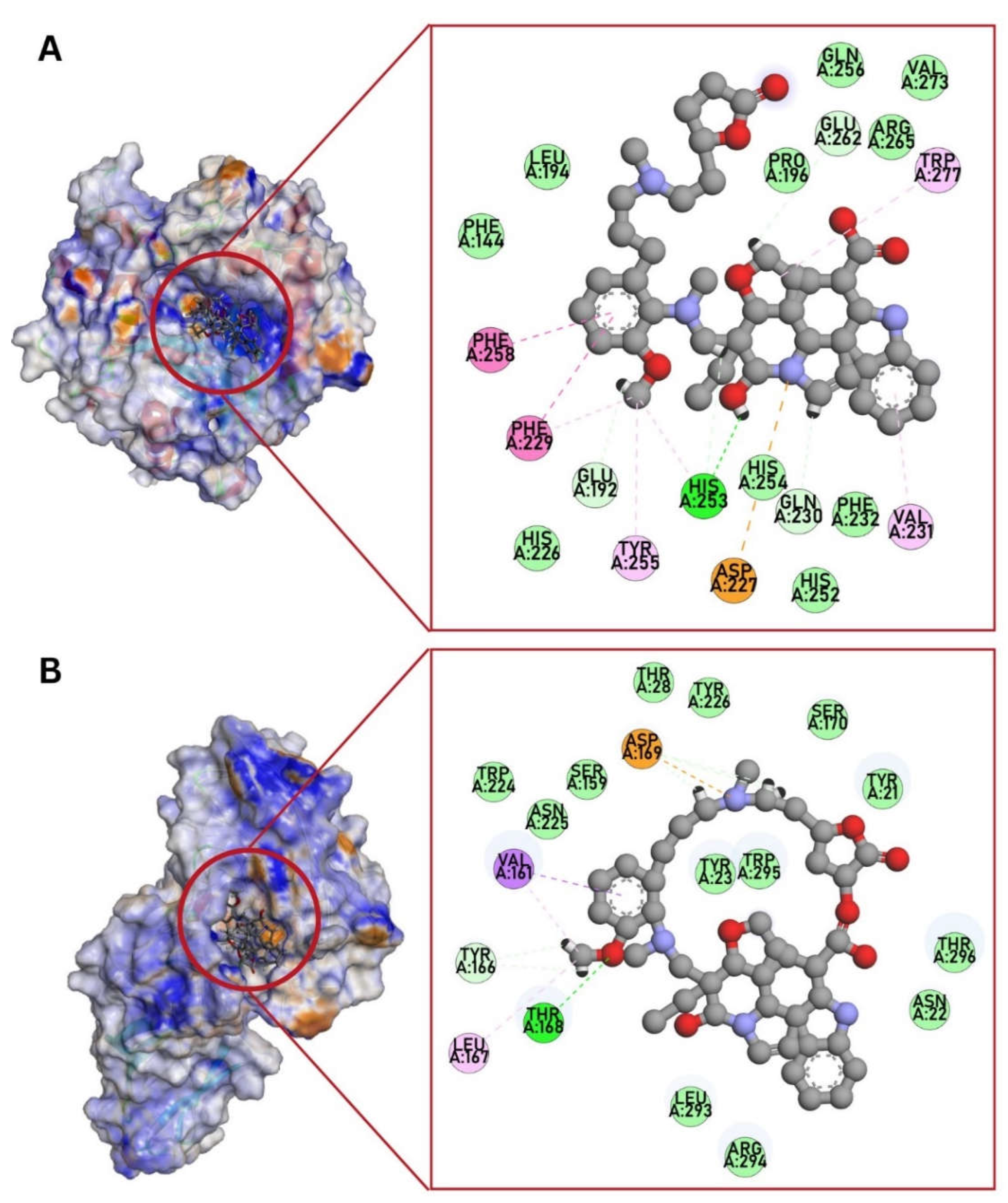
3.3. Molecular Docking against C. albicans Targets
3.4. Molecular Dynamics Simulations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vandeputte, P.; Ferrari, S.; Coste, A.T. Antifungal resistance and new strategies to control fungal infections. International Journal of Microbiology 2012, 1–26. [Google Scholar] [CrossRef]
- Chang, Y.-L.; Yu, S.-J.; Heitman, J.; Wellington, M.; Chen, Y.-L. New facets of antifungal therapy. Virulence 2016, 8, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Campoy, S.; Adrio, J.L. Antifungals. Biochemical Pharmacology 2017, 133, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A. Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. The American Journal of Medicine 2012, 125. [Google Scholar] [CrossRef] [PubMed]
- Tobudic, S.; Kratzer, C.; Presterl, E. Azole-resistant Candida spp. – emerging pathogens? Mycoses 2012, 55, 24–32. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Science Translational Medicine 2012, 4. [Google Scholar] [CrossRef]
- Miceli, M.H.; Díaz, J.A.; Lee, S.A. Emerging opportunistic yeast infections. The Lancet Infectious Diseases 2011, 11, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Eggimann, P.; Garbino, J.; Pittet, D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. The Lancet Infectious Diseases 2003, 3, 685–702. [Google Scholar] [CrossRef]
- Wächtler, B.; Citiulo, F.; Jablonowski, N.; Förster, S.; Dalle, F.; Schaller, M.; Wilson, D.; Hube, B. Candida albicans-epithelial interactions: Dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Martins, N.; Ferreira, I.C.; Barros, L.; Silva, S.; Henriques, M. Candidiasis: Predisposing factors, prevention, diagnosis and alternative treatment. Mycopathologia 2014, 177, 223–240. [Google Scholar] [CrossRef]
- Fuentefria, A.M.; Pippi, B.; Dalla Lana, D.F.; Donato, K.K.; de Andrade, S.F. Antifungals discovery: An insight into new strategies to combat antifungal resistance. Letters in Applied Microbiology 2018, 66, 2–13. [Google Scholar] [CrossRef] [PubMed]
- 8. Heard, S.C.; Wu, G.; Winter, J.M. Antifungal natural products. Current Opinion in Biotechnology 2021, 69, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Onishi, J.; Meinz, M.; Thompson, J.; Curotto, J.; Dreikorn, S.; Rosenbach, M.; Douglas, C.; Abruzzo, G.; Flattery, A.; Kong, L.; Cabello, A.; Vicente, F.; Pelaez, F.; Diez, M.T.; Martin, I.; Bills, G.; Giacobbe, R.; Dombrowski, A.; Schwartz, R.; Morris, S.; Harris, G.; Tsipouras, A.; Wilson, K.; Kurtz, M.B. Discovery of novel antifungal (1,3)-β-glucan synthase inhibitors. Antimicrobial Agents and Chemotherapy 2000, 44, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Long, S.-Y.; Li, C.-L.; Hu, J.; Zhao, Q.-J.; Chen, D. Indole alkaloids from the aerial parts of Kopsia fruticosa and their cytotoxic, antimicrobial and antifungal activities. Fitoterapia 2018, 129, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Yordanov, M.; Dimitrova, P.; Patkar, S.; Saso, L.; Ivanovska, N. Inhibition of Candida albicans extracellular enzyme activity by selected natural substances and their application in Candida infection. Canadian Journal of Microbiology 2008, 54, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.-B.; He, G.; Bai, H.-H.; Yang, T.; Zhang, G.-L.; Wu, L.-W.; Li, G.-Y. Indole alkaloids from Chaetomium globosum. Journal of Natural Products 2015, 78, 1479–1485. [Google Scholar] [CrossRef]
- Ahmed, A.; Li, W.; Chen, F.-F.; Zhang, J.-S.; Tang, Y.-Q.; Chen, L.; Tang, G.-H.; Yin, S. Monoterpene indole alkaloids from Rhazya stricta. Fitoterapia 2018, 128, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Anjum, K.; Kaleem, S.; Yi, W.; Zheng, G.; Lian, X.; Zhang, Z. Novel antimicrobial indolepyrazines A and B from the marine-associated Acinetobacter sp.. ZZ1275. Marine Drugs 2019, 17, 89. [Google Scholar] [CrossRef]
- Macabeo, A.P.; Vidar, W.S.; Chen, X.; Decker, M.; Heilmann, J.; Wan, B.; Franzblau, S.G.; Galvez, E.V.; Aguinaldo, Ma. A.; Cordell, G.A. Mycobacterium tuberculosis and cholinesterase inhibitors from Voacanga globosa. European Journal of Medicinal Chemistry 2011, 46, 3118–3123. [Google Scholar] [CrossRef]
- de Jesus, Ma.; Macabeo, A.; Ramos, J.; de Leon, V.; Asamitsu, K.; Okamoto, T. Voacanga globosa spirobisindole alkaloids exert antiviral activity in HIV latently infected cell lines by targeting the NF-ΚB CASCADE: In vitro and in silico investigations. Molecules 2022, 27, 1078. [Google Scholar] [CrossRef]
- Acebedo, A.R.; Amor, E.C.; Jacinto, S.D. Apoptosis-inducing activity of HPLC fraction from Voacanga globosa (Blanco) Merr. on the human colon carcinoma cell line, HCT116. Asian Pacific Journal of Cancer Prevention 2014, 15, 617–622. [Google Scholar] [CrossRef]
- Vital, P.G.; Rivera, W.L. Antimicrobial activity, cytotoxicity, and phytochemical screening of Voacanga globosa (Blanco) Merr. leaf extract (Apocynaceae). Asian Pacific Journal of Tropical Medicine 2011, 4, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Cascio, V.; Gittings, D.; Merloni, K.; Hurton, M.; Laprade, D.; Austriaco, N. S-adenosyl-L-methionine protects the probiotic yeast, Saccharomyces boulardii, from acid-induced cell death. BMC Microbiology 2013, 13. [Google Scholar] [CrossRef]
- Gardner, J. The Effect of Acute Heavy Metal (Cu And Cd) Toxicity on ROS Generation, Apoptosis, and Intracellular Glutathione Levels in Saccharomyces cerevisiae. Master’s Thesis, Georgia State University, Atlanta, Georgia, 2019. [Google Scholar]
- Poling, B.M. Differential Effects of Acute Cadmium, Copper, and Chromium Assault on Glutathione and Transcription Profiles in Saccharomyces cerevisiae. Master’s Thesis, Georgia State University, Atlanta, Georgia, 2021. [Google Scholar]
- da Nóbrega Alves, D.; Monteiro, A.F.; Andrade, P.N.; Lazarini, J.G.; Abílio, G.M.; Guerra, F.Q.; Scotti, M.T.; Scotti, L.; Rosalen, P.L.; Castro, R.D. Docking Prediction, antifungal activity, anti-biofilm effects on Candida spp., and toxicity against human cells of cinnamaldehyde. Molecules 2020, 25, 5969. [Google Scholar] [CrossRef] [PubMed]
- Gurgel do Amaral Valente Sá, L.; da Silva, C.R.; Neto, J.B.; do Nascimento, F.B.; Barroso, F.D.; da Silva, L.J.; Cabral, V.P.; Barbosa, A.D.; Silva, J.; Marinho, E.S.; de Moraes, M.O.; Rios, M.E.; Cavalcanti, B.C.; Lima, I.S.; Júnior, H.V. Antifungal activity of etomidate against growing biofilms of fluconazole-resistant Candida spp. strains, binding to mannoproteins and molecular docking with the ALS3 protein. Journal of Medical Microbiology 2020, 69, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Bouamrane, S.; Khaldan, A.; Hajji, H.; El-mernissi, R.; Alaqarbeh, M.; Alsakhen, N.; Maghat, H.; Ajana, M.A.; Sbai, A.; Bouachrine, M.; Lakhlifi, T. In silico identification of 1,2,4-triazoles as potential Candida albicans inhibitors using 3D-QSAR, molecular docking, molecular dynamics simulations, and ADMET profiling. Molecular Diversity 2022, 27, 2111–2132. [Google Scholar]
- Manzano, J.A.; Cruz, C.L.; Quimque, M.T.; Macabeo, A.P. In silico potentials of Alpinia galanga constituents against human placental aromatase vital in postmenopausal estrogen-dependent breast cancer pathogenesis. Philippine Journal of Science 2022, 151. [Google Scholar] [CrossRef]
- Manzano, J.A.; Llames, L.C.; Macabeo, A.P. Tetrahydrobisbenzylisoquinoline alkaloids from Phaeanthus ophthalmicus inhibit target enzymes associated with type 2 diabetes and obesity. Journal of Applied Pharmaceutical Science 2023. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Brogi, S.; Rossi, S.; Ibba, R.; Butini, S.; Calderone, V.; Campiani, G.; Gemma, S. In silico analysis of peptide-based derivatives containing bifunctional warheads engaging prime and non-prime subsites to covalent binding SARS-COV-2 main protease (mpro). Computation 2022, 10, 69. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Physical Review A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. The Journal of Chemical Physics 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Humphreys, D.D.; Friesner, R.A.; Berne, B.J. A multiple-time-step molecular dynamics algorithm for Macromolecules. The Journal of Physical Chemistry 1994, 98, 6885–6892. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. The Journal of Chemical Physics 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- da Silva, E.R.; Brogi, S.; Lucon-Júnior, J.F.; Campiani, G.; Gemma, S.; Maquiaveli, C. Dietary polyphenols rutin, taxifolin and quercetin related compounds target Leishmania amazonensis arginase. Food & Function 2019, 10, 3172–3180. [Google Scholar]
- Peng, F.; Hou, S.-Y.; Zhang, T.-Y.; Wu, Y.-Y.; Zhang, M.-Y.; Yan, X.-M.; Xia, M.-Y.; Zhang, Y.-X. Cytotoxic and antimicrobial indole alkaloids from an endophytic fungus Chaetomium sp. SYP-F7950 of Panax notoginseng. RSC Advances 2019, 9, 28754–28763. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, X.; Yuwen, H.-S.; Guo, L.-L.; Liu, J.-W.; Hao, X.-J. Alkaloids from Tabernaemontana divaricata combined with fluconazole to overcome fluconazole resistance in Candida albicans. Bioorganic Chemistry 2021, 107, 104515. [Google Scholar] [CrossRef]
- Lee, H.-S.; Yoon, K.-M.; Han, Y.-R.; Lee, K.J.; Chung, S.-C.; Kim, T.-I.; Lee, S.-H.; Shin, J.; Oh, K.-B. 5-hydroxyindole-type alkaloids, as Candida albicans isocitrate lyase inhibitors, from the tropical sponge Hyrtios sp. Bioorganic & Medicinal Chemistry Letters 2009, 19, 1051–1053. [Google Scholar]
- Leadsham, J.E.; Kotiadis, V.N.; Tarrant, D.J.; Gourlay, C.W. Apoptosis and the yeast actin cytoskeleton. Cell Death & Differentiation 2009, 17, 754–762. [Google Scholar]
- Al-Dhaheri, R.S.; Douglas, L.J. Apoptosis in Candida biofilms exposed to amphotericin B. Journal of Medical Microbiology 2010, 59, 149–157. [Google Scholar] [CrossRef]
- da Silva, C.R.; de Andrade Neto, J.B.; de Sousa Campos, R.; Figueiredo, N.S.; Sampaio, L.S.; Magalhães, H.I.; Cavalcanti, B.C.; Gaspar, D.M.; de Andrade, G.M.; Lima, I.S.; de Barros Viana, G.S.; de Moraes, M.O.; Lobo, M.D.; Grangeiro, T.B.; Nobre Júnior, H.V. Synergistic effect of the flavonoid catechin, quercetin, or epigallocatechin gallate with fluconazole induces apoptosis in Candida tropicalis resistant to fluconazole. Antimicrobial Agents and Chemotherapy 2014, 58, 1468–1478. [Google Scholar] [CrossRef]
- Soliman, S.; Alnajdy, D.; El-Keblawy, A.; Mosa, K.; Khoder, G.; Noreddin, A. Plants’ natural products as alternative promising anti-candida drugs. Pharmacognosy Reviews 2017, 11, 104. [Google Scholar] [CrossRef]
- Jia, C.; Zhang, J.; Yu, L.; Wang, C.; Yang, Y.; Rong, X.; Xu, K.; Chu, M. Antifungal activity of coumarin against Candida albicans is related to apoptosis. Frontiers in Cellular and Infection Microbiology 2019, 8. [Google Scholar] [CrossRef]
- Zhao, C.-R.; You, Z.-L.; Chen, D.-D.; Hang, J.; Wang, Z.-B.; Ji, M.; Wang, L.-X.; Zhao, P.; Qiao, J.; Yun, C.-H.; Bai, L. Structure of a fungal 1,3-β-glucan synthase. Science Advances 2023, 9. [Google Scholar] [CrossRef]
- Denning, D.W. Echinocandin antifungal drugs. The Lancet 2003, 362, 1142–1151. [Google Scholar]
- Odds, F.C.; Brown, A.J. P.; Gow, N.A. R. Antifungal agents: Mechanisms of action. Trends in Microbiology 2003, 11, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.; Latge, J.-P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiology Spectrum 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.L.; Colombo, A.L.; de Almeida Junior, J.N. Fungal cell wall: Emerging antifungals and drug resistance. Frontiers in Microbiology 2019, 10. [Google Scholar] [CrossRef]
- Lee, H.-S.; Kim, Y. Antifungal activity of Salvia miltiorrhiza against Candida albicans is associated with the alteration of membrane permeability and (1,3)-β-D-glucan synthase activity. Journal of Microbiology and Biotechnology 2016, 26, 610–617. [Google Scholar] [CrossRef]
- Perlin, D.S. Resistance to echinocandin-class antifungal drugs. Drug Resistance Updates 2007, 10, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.E.; Edlind, T.D. Topological and mutational analysis of Saccharomyces cerevisiae FKS1. Eukaryotic Cell 2012, 11, 952–960. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Filler, S.G. Candida albicans ALS3, a multifunctional adhesin and Invasin. Eukaryotic Cell 2011, 10, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Cota, E. Candida albicans agglutinin-like sequence (ALS) Family Vignettes: A review of ALS protein structure and function. Frontiers in Microbiology 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Kioshima, E.S.; Shinobu-Mesquita, C.S.; Abadio, A.K.; Felipe, M.S.; Svidzinski, T.I.; Maigret, B. Selection of potential anti-adhesion drugs by in silico approaches targeted to ALS3 from Candida albicans. Biotechnology Letters 2019, 41, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.R.; Sardi, J. de; Freires, I.A.; Silva, A.C.; Rosalen, P.L. In silico approaches for screening molecular targets in Candida albicans: A proteomic insight into drug discovery and development. European Journal of Pharmacology 2019, 842, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Cheng, S.; Clancy, C.J.; Nguyen, M.H. Caspofungin kills Candida albicans by causing both cellular apoptosis and necrosis. Antimicrobial Agents and Chemotherapy 2013, 57, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Zhang, B.; Li, J.; Zhang, B.; Wang, H.; Li, M. Endoplasmic reticulum-derived reactive oxygen species (ROS) is involved in toxicity of cell wall stress to Candida albicans. Free Radical Biology and Medicine 2016, 99, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Maurya, I.K.; Pathak, S.; Sharma, M.; Sanwal, H.; Chaudhary, P.; Tupe, S.; Deshpande, M.; Chauhan, V.S.; Prasad, R. Antifungal activity of novel synthetic peptides by accumulation of reactive oxygen species (ROS) and disruption of cell wall against Candida albicans. Peptides 2011, 32, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, J.; Sun, J.; Zhu, X.; Zhou, L.; Lu, Z.; Lu, Y. C16-Fengycin a affect the growth of Candida albicans by destroying its cell wall and accumulating reactive oxygen species. Applied Microbiology and Biotechnology 2019, 103, 8963–8975. [Google Scholar] [CrossRef]
- Phillips, A.J.; Sudbery, I.; Ramsdale, M. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proceedings of the National Academy of Sciences 2003, 100, 14327–14332. [Google Scholar] [CrossRef] [PubMed]
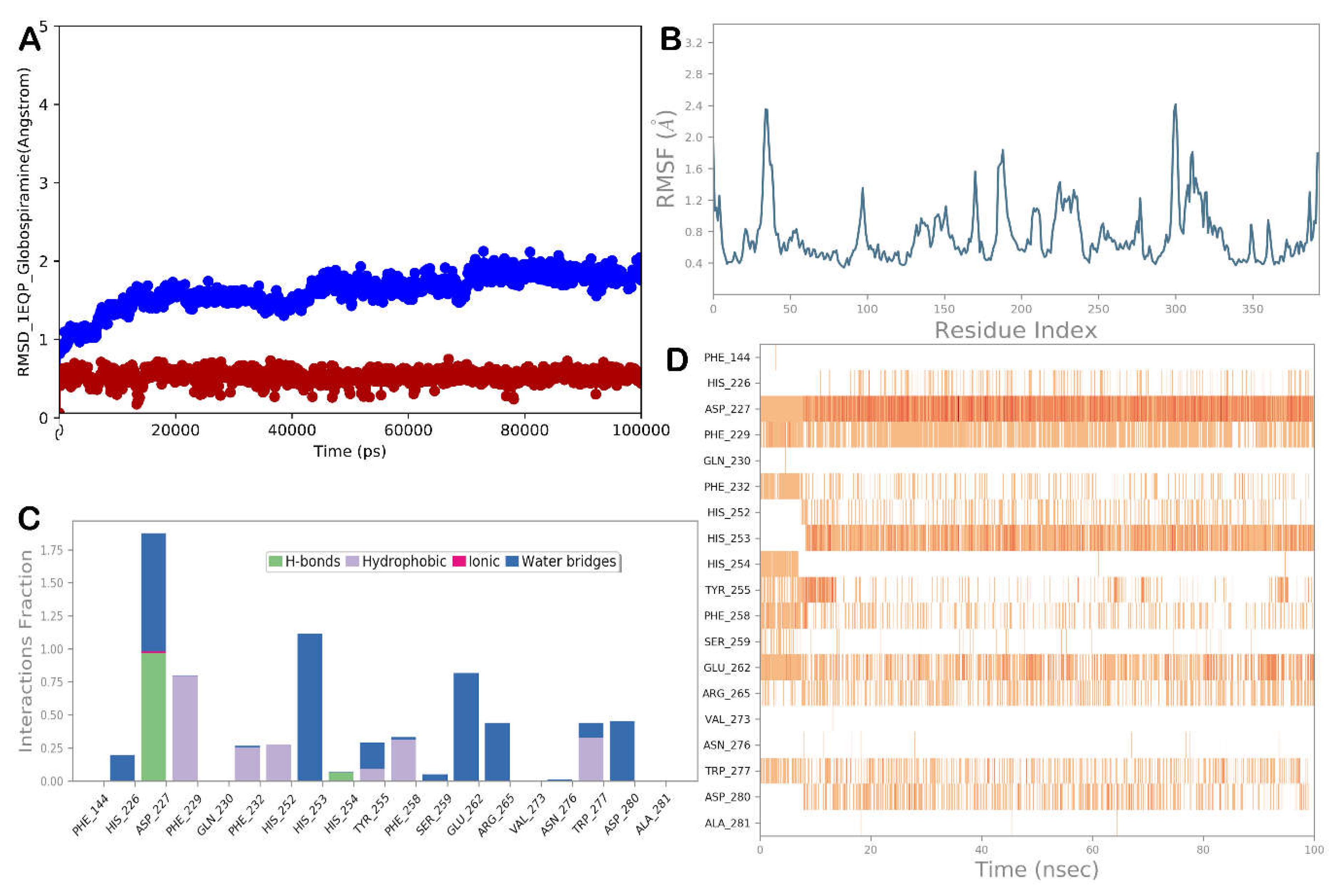
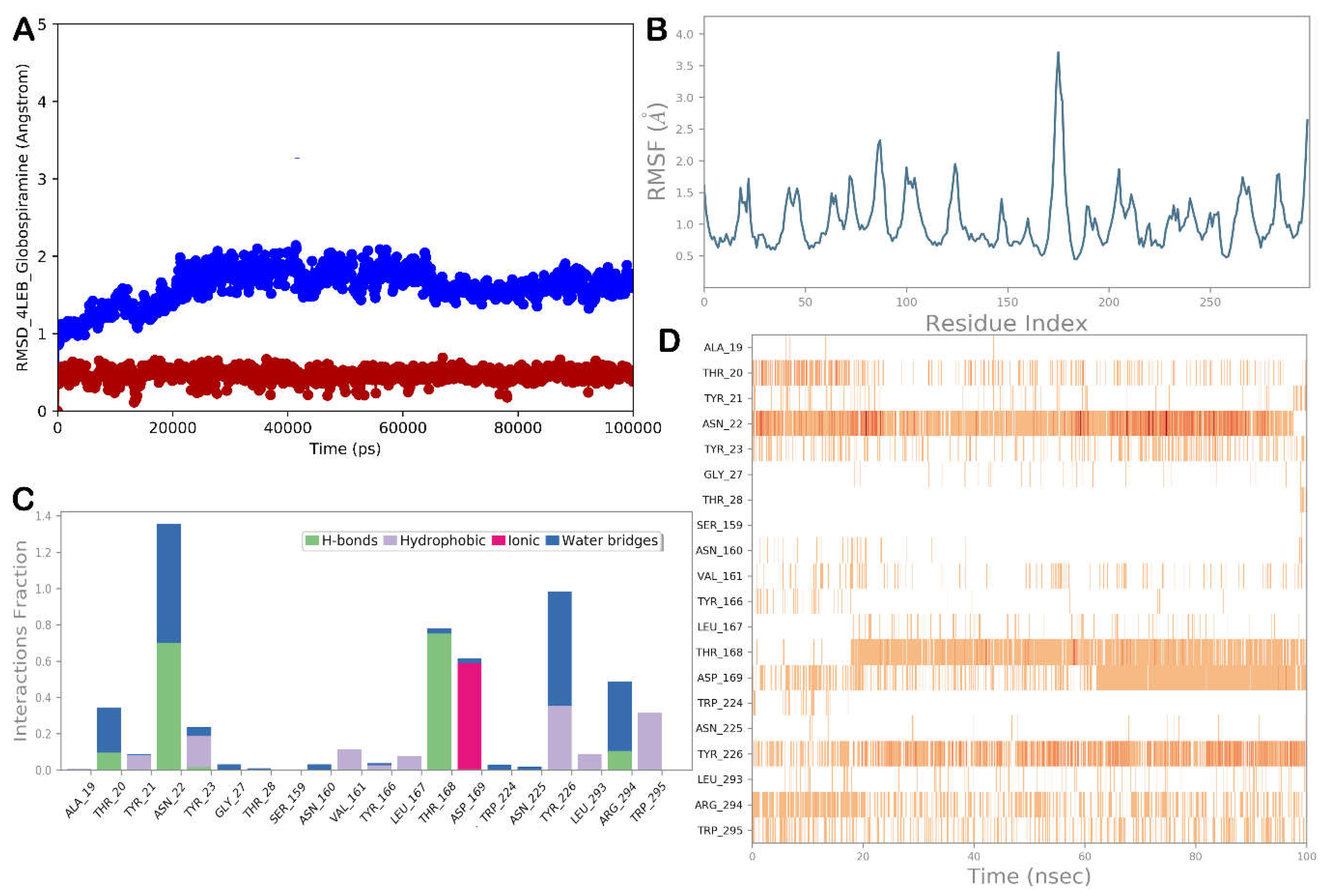
| Globospiramine | Amphotericin B | |
|---|---|---|
| MIC (µg/mL) | ||
| C. albicans | 8.0 | 0.50 |
| C. tropicalis | 4.0 | 0.50 |
| MFC (µg/mL) | ||
| C. albicans | 10.67 | 0.83 |
| C. tropicalis | >64.0 | 1.67 |
| PDB IDs | Globospiramine | Positive Controls | ||||||
|---|---|---|---|---|---|---|---|---|
| Caspofungin | Amphotericin B | Co-crystallized ligand / inhibitor | ||||||
| BE (kcal/ mol) | Interactions | BE (kcal/ mol) | Interactions | BE (kcal/ mol) | Interactions | BE (kcal/ mol) | Interactions | |
| 1EQP (1,3-β-glucan synthase) | -10.5 | His253 (H-bond), Phe258, Phe229 (pi-pi stacked), Trp277, Val231, Tyr255, Phe229, His253 (pi-alkyl), Glu192, His253, Gln230, Glu262 (C-H bond) | -8.0 | Trp277, Gln230 (H-bond), Asp227 (salt bridge), His254, Glu262 (C-H bond), Val273, Phe258, Phe144, Tyr255, Trp373 (alkyl, pi-alkyl), His253, Arg265 (unfavorable interaction) | -8.8 | Asn305, Asp151 (H-bond), Phe258, Phe144 (pi-alkyl), Tyr 153 (unfavorable donor-donor, C-H bond) | - | - |
| 4QUV (δ-14-sterol reductase) | -9.5 | Arg324, His320 (H-bond), Val96, His320 (pi-sigma), Leu253, Met99 (alkyl), Arg106, Arg323 (unfavorable positive-positive) | -7.2 | Arg106, Arg323, Arg324, Lys406 (H-bond), Tyr414, Trp352, Leu346, Cys403, Trp411, Lys319, Val96 (alkyl, pi-alkyl), His320 (pi-pi stacked), Gln97 (C-H bond), Arg324 (unfavorable positive-positive) | -7.5 | Gln97, Glu250, Arg323, Arg324, Gly343 (H-bond), Met99, Leu253 (alkyl), Arg106 (unfavorable positive-positive) | -9.5 | His248, Arg313, Thr254, Lys259, Lys319, Trp256, Arg395, Asn316, Thr255 (H-bond), Asp244, Asp399, Arg395 (attractive charge, pi-cation), Glu201 (C-H bond), Lys319 (unfavorable positive-positive), Tyr245 (pi-pi T-shaped), Arg398, Val252 (pi-alkyl, alkyl) |
| 5TZ1 (lanosterol 14-alpha demethylase or CYP51) | -7.4 | Arg469 (H-bond), Glu444 (attractive charge), Val452, Val454 (alkyl), Ser453, Lys451 (C-H bond) | -5.7 | Met508, Pro462, His468, Leu439, Leu471, Gly303 (H-bond), His468 (C-H bond), Ile304 (pi-sigma), Leu87, Phe233, Tyr64, Phe380, Phe228, Val509, Leu150, Ile304, Ile131, His377, Pro230, Leu88, Lys90 (pi-alkyl, alkyl), Arg381, Tyr132, Lys143 (unfavorable interactions) | -3.3 | Phe463 (H-bond), His468 (C-H bond), Tyr118 (pi-lone pair), Leu376, Ile379, Ala146, Ile304, Leu204, Phe475 (alkyl, pi-alkyl), Cys470, Ile379, Gly464, Arg381, Thr311, Phe475, Leu150, Ile471, Tyr132 (unfavorable bonds) | -10.6 | Gly303, Ile304 (C-H bond), Ser507, His377 (halogen), Tyr118, Tyr132 (pi-pi), Leu121, Phe233, Leu376, Pro230, Ile304, Ile131, Lys143 (alkyl, pi-alkyl) |
| 5UIV (thymidylate kinase) | -9.4 | Gly155, Asp91, Arg39 (H-bond), Asp13, Arg39, Glu159 (pi-cation / pi-anion / salt bridge), Glu159, Ser18 (C-H bond), Lys17 (pi-alkyl) | -8.2 | Ser18, Asp13, Asp91, Arg92, Lys17, Lys35, Arg39, Gly157 Gly155 (H-bond), Glu162, Glu159 (salt bridge, attractive charge), Asp13, Asp91, Lys35, Gly155 (C-H bond), Ile196, Arg153, Lys17, Arg39, Val199 (alkyl, pi-alkyl) | -7.7 | Arg92, Lys35, SerA (H-bond), Glu162, Gln159 (salt bridge, attractive charge), Pro37 (alkyl), Asp13 (C-H bond), Ser18 (unfavorable donor-donor) | -8.9 | Arg92, Lys17, Arg14, Ser18, Gly16 (H-bond), Glu159, Asp91, Asp13 (attractive charge, pi-anion), Lys35 (unfavorable donor-donor), Tyr100 (pi-pi), Leu51 (pi-alkyl) |
| 4LEB (Als3 adhesin) | -10.6 | Thr168 (H-bond), Asp169 (attractive charge), Asp169, Tyr166 (C-H bond), Val161 (pi-sigma), Val161, Leu167 (alkyl) | -6.5 | Thr168, Tyr226, Thr20, Asn22 (H-bond), Trp295 (pi-cation), Pro29, Arg171, Tyr21 (alkyl, pi-alkyl), Asn22 (pi-donor H-bond) | -7.7 | Asn22 (H-bond), Tyr226 (pi-alkyl), Arg294 (unfavorable positive-positive) | - | - |
| 2Y7L (Als9-2) | -8.1 | Thr293 (C-H bond), Trp294 (pi-cation), Tyr21, Pro160, Val161 (pi-alkyl) | -6.4 | Thr168 (H-bond), Arg171, Val22, Pro160, Val161, Ile167, Tyr23, Phe225, Pro29 (alkyl, pi-alkyl) | -7.1 | Glu86, Ser210, Asn213 (H-bond), Asn211 (C-H bond), Tyr261 (pi-alkyl) | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





