Submitted:
18 April 2024
Posted:
19 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
Gut Microbiome
Age
Genetics
Environment
Lifestyle
Microbiome Characterization Techniques
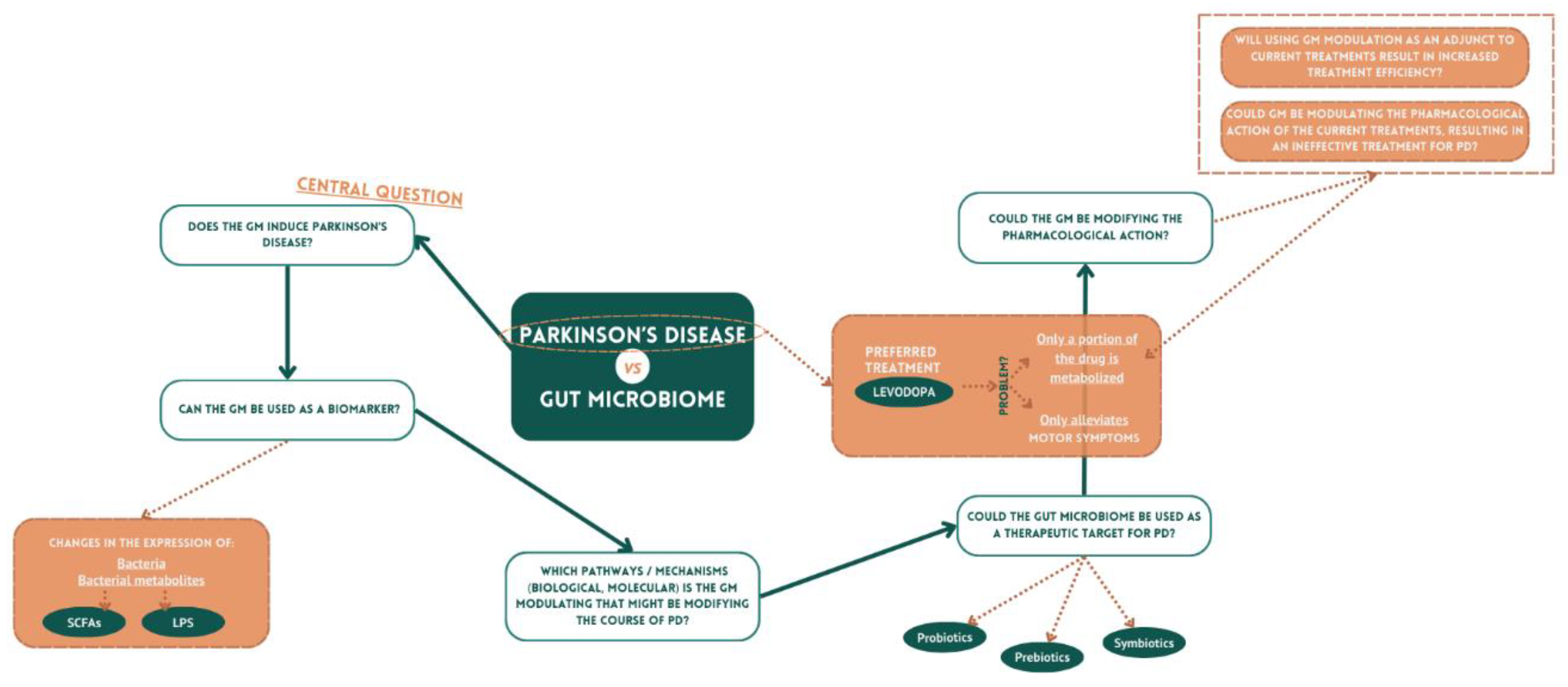
Microbiome and PD: From Diagnosis to Treatment
Microbiome vs αSyn – Potential Contributions of the Microbiome to PD: Braak’s Hypothesis
Gut Microbiome and Neuroinflammation
Molecular Mediators: Toll-Like Receptors (TLR)
Molecular Mediators: LPS and Lipopolysaccharide-Binding Protein (LBP)
Metabolic Mediators: Short-Chain Fatty Acids (SCFAs)
Microbiome: A New Source for PD Biomarkers?
Microbiome: Are We Looking for a New Therapeutic Target?
Gut Microbiome-Drug Interaction for the Treatment of PD
Future Perspectives
Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balestrino, R.; Schapira, A. H. V. Parkinson Disease. Eur. J. Neurol. 2020, 27, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Parnetti, L.; Gaetani, L.; Eusebi, P.; Paciotti, S.; Hansson, O.; El-Agnaf, O.; Mollenhauer, B.; Blennow, K.; Calabresi, P. CSF and Blood Biomarkers for Parkinson’s Disease. Lancet. Neurol. 2019, 18, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, B.; Lou, P.; Dai, T.; Chen, Y.; Zhuge, A.; Yuan, Y.; Li, L. The Relationship Between the Gut Microbiome and Neurodegenerative Diseases. Neurosci. Bull. 2021, 37, 1510–1522. [Google Scholar] [CrossRef] [PubMed]
- Lubomski, M.; Tan, A. H.; Lim, S.-Y.; Holmes, A. J.; Davis, R. L.; Sue, C. M. Parkinson’s Disease and the Gastrointestinal Microbiome. J. Neurol. 2020, 267, 2507–2523. [Google Scholar] [CrossRef] [PubMed]
- 5. World Health Organization. Parkinson disease, 20 April.
- Pavan, S.; Prabhu, A. N.; Prasad Gorthi, S.; Das, B.; Mutreja, A.; Shetty, V.; Ramamurthy, T.; Ballal, M. Exploring the Multifactorial Aspects of Gut Microbiome in Parkinson’s Disease. Folia Microbiol. (Praha). 0123. [Google Scholar] [CrossRef]
- Keshavarzian, A.; Engen, P.; Bonvegna, S.; Cilia, R. The Gut Microbiome in Parkinson’s Disease: A Culprit or a Bystander? Prog. Brain Res. 2020, 252, 357–450. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, S.; Chutna, O.; Bousset, L.; Aldrin-Kirk, P.; Li, W.; Björklund, T.; Wang, Z.-Y.; Roybon, L.; Melki, R.; Li, J.-Y. Direct Evidence of Parkinson Pathology Spread from the Gastrointestinal Tract to the Brain in Rats. Acta Neuropathol. 2014, 128, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh, K.; Rahimmi, A. Oxidative Stress and Neuroinflammation in the Story of Parkinson’s Disease: Could Targeting These Pathways Write a Good Ending? J. Cell. Physiol. 2018, 234, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Sun, M. F.; Shen, Y. Q. Dysbiosis of Gut Microbiota and Microbial Metabolites in Parkinson’s Disease. Ageing Res. Rev. 2018, 45, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Lim, K. L.; Tan, J. M. M. Role of the Ubiquitin Proteasome System in Parkinson’s Disease. BMC Biochem. 2007, 8 (SUPPL. 1), 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Benito, M.; Granado, N.; García-Sanz, P.; Michel, A.; Dumoulin, M.; Moratalla, R. Modeling Parkinson’s Disease With the Alpha-Synuclein Protein. Front. Pharmacol. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wüllner, U.; Borghammer, P.; Choe, C. un; Csoti, I.; Falkenburger, B.; Gasser, T.; Lingor, P.; Riederer, P. The Heterogeneity of Parkinson’s Disease. J. Neural Transm. 2023, 130, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Greenland, J. C.; Williams-Gray, C. H.; Barker, R. A. The Clinical Heterogeneity of Parkinson’s Disease and Its Therapeutic Implications. Eur. J. Neurosci. 2019, 49, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Mahlknecht, P.; Foltynie, T.; Limousin, P.; Poewe, W. How Does Deep Brain Stimulation Change the Course of Parkinson’s Disease? Mov. Disord. 2022, 37, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Hariz, M.; Blomstedt, P. Deep Brain Stimulation for Parkinson’s Disease. J. Intern. Med. 2022, 292, 764–778. [Google Scholar] [CrossRef] [PubMed]
- Virel, A.; Dudka, I.; Laterveer, R.; Af Bjerkén, S. (1)H NMR Profiling of the 6-OHDA Parkinsonian Rat Brain Reveals Metabolic Alterations and Signs of Recovery after N-Acetylcysteine Treatment. Mol. Cell. Neurosci. 2019, 98, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J. C.; Ursell, L. K.; Parfrey, L. W.; Knight, R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Diaz Heijtz, R.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M. L.; Forssberg, H.; Pettersson, S. Normal Gut Microbiota Modulates Brain Development and Behavior. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kasper, L. H. The Role of Microbiome in Central Nervous System Disorders. Brain. Behav. Immun. 2014, 38, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J. F.; O’Riordan, K. J.; Sandhu, K.; Peterson, V.; Dinan, T. G. The Gut Microbiome in Neurological Disorders. Lancet. Neurol. 2020, 19, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Foster, J. A.; McVey Neufeld, K.-A. Gut-Brain Axis: How the Microbiome Influences Anxiety and Depression. Trends Neurosci. 2013, 36, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Cox, M. J.; Cookson, W. O. C. M.; Moffatt, M. F. Sequencing the Human Microbiome in Health and Disease. Hum. Mol. Genet. 2013, 22, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tan, Y.; Cheng, H.; Zhang, D.; Feng, W.; Peng, C. Functions of Gut Microbiota Metabolites, Current Status and Future Perspectives. Aging Dis. 2022, 13, 1106–1126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Liu, X.; Ye, Y.; Yan, X.; Cheng, Y.; Zhao, L.; Chen, F.; Ling, Z. Gut Microbiota: A Novel Therapeutic Target for Parkinson’s Disease. Front. Immunol. 2022, 13, 937555. [Google Scholar] [CrossRef] [PubMed]
- Bull, M. J.; Plummer, N. T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. (Encinitas). 2014, 13, 17–22. [Google Scholar] [PubMed]
- Shannon, K. M. Gut-Derived Sterile Inflammation and Parkinson’s Disease. Front. Neurol. 2022, 13, 831090. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Hayes, R. B. Environmental Influences on the Human Microbiome and Implications for Noncommunicable Disease. Annu. Rev. Public Health 2021, 42, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; Lugli, G. A.; Rodriguez, J. M.; Bode, L.; de Vos, W.; Gueimonde, M.; Margolles, A.; van Sinderen, D.; Ventura, M. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- Reeve, A.; Simcox, E.; Turnbull, D. Ageing and Parkinson ’ s Disease : Why Is Advancing Age the Biggest Risk Factor ? Ageing Res. Rev. 2014, 14, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Wang, P.; Jankovic, J. The Genetics of Parkinson Disease. Ageing Res. Rev. 2018, 42, 72–85. [Google Scholar] [CrossRef]
- Biswas, S.; Bagchi, A. Study of the Effects of Nicotine and Caffeine for the Treatment of Parkinson’s Disease. Appl. Biochem. Biotechnol. 2022, 639–654. [Google Scholar] [CrossRef]
- Nuytemans, K.; Theuns, J.; Cruts, M.; Van Broeckhoven, C. Genetic Etiology of Parkinson Disease Associated with Mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 Genes: A Mutation Update. Hum. Mutat. 2010, 31, 763–780. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.-X.; Gulam, M. Y.; Chia, N. S. J.; Feng, L.; Rotzschke, O.; Tan, E.-K. Gut-Brain Axis: Potential Factors Involved in the Pathogenesis of Parkinson’s Disease. Front. Neurol. 2020, 11, 849. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lenardo, M. J. The Role of LRRK2 in Inflammatory Bowel Disease. Cell Res. 2012, 22, 1092–1094. [Google Scholar] [CrossRef] [PubMed]
- Vizziello, M.; Borellini, L.; Franco, G.; Ardolino, G. Disruption of Mitochondrial Homeostasis: The Role of Pink1 in Parkinson’s Disease. Cells 2021, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Hernán, M. A.; Takkouche, B.; Caamaño-Isorna, F.; Gestal-Otero, J. J. A Meta-Analysis of Coffee Drinking, Cigarette Smoking, and the Risk of Parkinson’s Disease. Ann. Neurol. 2002, 52, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Hill-Burns, E. M.; Debelius, J. W.; Morton, J. T.; Wissemann, W. T.; Lewis, M. R.; Wallen, Z. D.; Peddada, S. D.; Factor, S. A.; Molho, E.; Zabetian, C. P.; Knight, R.; Payami, H. Parkinson’s Disease and Parkinson’s Disease Medications Have Distinct Signatures of the Gut Microbiome. Mov. Disord. 2017, 32, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Prabhu, A. N.; Prasad Gorthi, S.; Das, B.; Mutreja, A.; Shetty, V.; Ramamurthy, T.; Ballal, M. Exploring the Multifactorial Aspects of Gut Microbiome in Parkinson’s Disease. Folia Microbiol. (Praha). 2022, 67, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Blesa, J.; Phani, S.; Jackson-Lewis, V.; Przedborski, S. Classic and New Animal Models of Parkinson’s Disease. J. Biomed. Biotechnol. 2012, 2012. [Google Scholar] [CrossRef]
- Reichmann, H.; Csoti, I.; Koschel, J.; Lorenzl, S.; Schrader, C.; Winkler, J.; Wüllner, U. Life Style and Parkinson’s Disease. J. Neural Transm. 2022, 129, 1235–1245. [Google Scholar] [CrossRef]
- Leevven-, A.; Obfervatioiis, M. An Ab/Lratt of A.
- Power, A.; Dobell, C. $ection of Tbe Lbtitor. of !Hebicine. The Discovery of the Intestinal Protozoa of Man.’. Proc R Soc Med.
- Ursell, L. K.; Metcalf, J. L.; Parfrey, L. W.; Knight, R. Defining the Human Microbiome. Nutr. Rev. 2012, 70 (SUPPL. 1). [Google Scholar] [CrossRef]
- Grice, E. A.; Segre, J. A. Published in Final Edited Form as: The Human Microbiome: Our Second Genome. Annu. Rev. Genomics Hum. Genet. 2012, 13, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Hollister, E. B.; Gao, C.; Versalovic, J. Compositional and Functional Features of the Gastrointestinal Microbiome and Their Effects on Human Health. Gastroenterology 2014, 146, 1449–1458. [Google Scholar] [CrossRef]
- Blum, H. E. The Microbiome: A Key Player in Human Health and Disease. J. Healthc. Commun. 2017, 02, 1–5. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z. X.; Chen, X. Y.; Wang, J. Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J. B.; Wei, L.; Li, J.; Chen, Z. S. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7. [Google Scholar] [CrossRef] [PubMed]
- Afzaal, M.; Saeed, F.; Shah, Y. A.; Hussain, M.; Rabail, R.; Socol, C. T.; Hassoun, A.; Pateiro, M.; Lorenzo, J. M.; Rusu, A. V.; Aadil, R. M. Human Gut Microbiota in Health and Disease: Unveiling the Relationship. Front. Microbiol. 2022, 13, 1–14. [Google Scholar] [CrossRef]
- Song, E. J.; Lee, E. S.; Nam, Y. Do. Progress of Analytical Tools and Techniques for Human Gut Microbiome Research. J. Microbiol. 2018, 56, 693–705. [Google Scholar] [CrossRef]
- Fraher, M. H.; O’Toole, P. W.; Quigley, E. M. M. Techniques Used to Characterize the Gut Microbiota: A Guide for the Clinician. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 312–322. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Ahmadi Bashirzadeh, D.; Nazemalhosseini Mojarad, E.; Asadzadeh Aghdaei, H.; Norouzinia, M.; Shahrokh, S. Signature of Gut Microbiome by Conventional and Advanced Analysis Techniques: Advantages and Disadvantages. Middle East J. Dig. Dis. 2020, 12, 5–11. [Google Scholar] [CrossRef]
- Cani, P. D. Gutmicrobiota and Obesity: Lessons from the Microbiome. Brief. Funct. Genomics 2013, 12, 381–387. [Google Scholar] [CrossRef]
- Scales, B. S.; Huffnagle, G. B. The Microbiome in Wound Repair and Tissue Fibrosis. J. Pathol. 2013, 229, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Suau, A.; Bonnet, R.; Sutren, M.; Godon, J. J.; Gibson, G. R.; Collins, M. D.; Doré, J. Direct Analysis of Genes Encoding 16S RRNA from Complex Communities Reveals Many Novel Molecular Species within the Human Gut. Appl. Environ. Microbiol. 1999, 65, 4799–4807. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Sakamoto, M.; Benno, Y. Phylogenetic Analysis of the Human Gut Microbiota Using 16S RDNA Clone Libraries and Strictly Anaerobic Culture-Based Methods. Microbiol. Immunol. 2002, 46, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; Okada, H.; Bono, H.; Nakagawa, S.; Hirota, K. Full-Length 16S RRNA Gene Amplicon Analysis of Human Gut Microbiota Using MinIONTM Nanopore Sequencing Confers Species-Level Resolution. BMC Microbiol. 2021, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Ghoshal, U.; Dhole, T. N.; Ghoshal, U. C. Fecal Microbiota in Patients with Irritable Bowel Syndrome Compared with Healthy Controls Using Real-Time Polymerase Chain Reaction: An Evidence of Dysbiosis. Dig. Dis. Sci. 2015, 60, 2953–2962. [Google Scholar] [CrossRef]
- Furet, J. P.; Firmesse, O.; Gourmelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Doré, J.; Corthier, G. Comparative Assessment of Human and Farm Animal Faecal Microbiota Using Real-Time Quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J. F.; Rôças, I. N.; Rosado, A. S. Application of Denaturing Gradient Gel Electrophoresis (DGGE) to the Analysis of Endodontic Infections. J. Endod. 2005, 31, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Imaeda, H.; Fujimoto, T.; Takahashi, K.; Kasumi, E.; Fujiyama, Y.; Andoh, A. Terminal-Restriction Fragment Length Polymorphism (T-RFLP) Analysis for Changes in the Gut Microbiota Profiles of Indomethacin-and Rebamipide-Treated Mice. Digestion 2012, 86, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Namsolleck, P.; Thiel, R.; Lawson, P.; Holmstrøm, K.; Rajilic, M.; Vaughan, E. E.; Rigottier-Gois, L.; Collins, M. D.; De Vos, W. M.; Blaut, M. Molecular Methods for the Analysis of Gut Microbiota. Microb. Ecol. Health Dis. 2004, 16, (2–3). [Google Scholar] [CrossRef]
- Wang, W. L.; Xu, S. Y.; Ren, Z. G.; Tao, L.; Jiang, J. W.; Zheng, S. Sen. Application of Metagenomics in the Human Gut Microbiome. World J. Gastroenterol. 2015, 21, 803–814. [Google Scholar] [CrossRef]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef]
- Ojala, T.; Kankuri, E.; Kankainen, M. Understanding Human Health through Metatranscriptomics. Trends Mol. Med. 2023, 29, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Issa Isaac, N.; Philippe, D.; Nicholas, A.; Raoult, D.; Eric, C. Metaproteomics of the Human Gut Microbiota: Challenges and Contributions to Other OMICS. Clin. Mass Spectrom. 2019, 14, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Peters, D. L.; Wang, W.; Zhang, X.; Ning, Z.; Mayne, J.; Figeys, D. Metaproteomic and Metabolomic Approaches for Characterizing the Gut Microbiome. Proteomics 2019, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, Y.; Xiao, X.; Zheng, J.; Zhou, H. Metaproteomics: A Strategy to Study the Taxonomy and Functionality of the Gut Microbiota. J. Proteomics 2020, 219. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, K. S.; Maier, T. V.; Walker, A.; Heinzmann, S. S.; Forcisi, S.; Martinez, I.; Walter, J.; Schmitt-Kopplin, P. Challenges of Metabolomics in Human Gut Microbiota Research. Int. J. Med. Microbiol. 2016, 306, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Vernocchi, P.; Del Chierico, F.; Putignani, L. Gut Microbiota Profiling: Metabolomics Based Approach to Unravel Compounds Affecting Human Health. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, R. F.; Beggs, M. L.; Erickson, B. D.; Cerniglia, C. E. DNA Microarray Analysis of Predominant Human Intestinal Bacteria in Fecal Samples. Mol. Cell. Probes 2004, 18, 223–234. [Google Scholar] [CrossRef]
- Bikel, S.; Valdez-Lara, A.; Cornejo-Granados, F.; Rico, K.; Canizales-Quinteros, S.; Soberón, X.; Del Pozo-Yauner, L.; Ochoa-Leyva, A. Combining Metagenomics, Metatranscriptomics and Viromics to Explore Novel Microbial Interactions: Towards a Systems-Level Understanding of Human Microbiome. Comput. Struct. Biotechnol. J. 2015, 13, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Nearing, J. T.; Comeau, A. M.; Langille, M. G. I. Identifying Biases and Their Potential Solutions in Human Microbiome Studies. Microbiome 2021, 9, 1–22. [Google Scholar] [CrossRef]
- Madhogaria, B.; Bhowmik, P.; Kundu, A. Correlation between Human Gut Microbiome and Diseases. Infect. Med. 2022, 1, 180–191. [Google Scholar] [CrossRef]
- Vijay, A.; Valdes, A. M. Role of the Gut Microbiome in Chronic Diseases: A Narrative Review. Eur. J. Clin. Nutr. 2022, 76, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J. G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F. D.; Costello, E. K.; Fierer, N.; Peña, A. G.; Goodrich, J. K.; Gordon, J. I.; Huttley, G. A.; Kelley, S. T.; Knights, D.; Koenig, J. E.; Ley, R. E.; Lozupone, C. A.; McDonald, D.; Muegge, B. D.; Pirrung, M.; Reeder, J.; Sevinsky, J. R.; Turnbaugh, P. J.; Walters, W. A.; Widmann, J.; Yatsunenko, T.; Zaneveld, J.; Knight, R. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nature methods, 20 May; 10. [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E. M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; Wilkening, J.; Edwards, R. A. The Metagenomics RAST Server - a Public Resource for the Automatic Phylogenetic and Functional Analysis of Metagenomes. BMC Bioinformatics 2008, 9, 386. [Google Scholar] [CrossRef]
- Morgan, X. C.; Huttenhower, C. Meta’omic Analytic Techniques for Studying the Intestinal Microbiome. Gastroenterology 2014, 146, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Lyu, R.; Qu, Y.; Divaris, K.; Wu, D. Methodological Considerations in Longitudinal Analyses of Microbiome Data: A Comprehensive Review. Genes (Basel). 2023, 15, 51. [Google Scholar] [CrossRef]
- Custer, G. F.; Gans, M.; van Diepen, L. T. A.; Dini-Andreote, F.; Buerkle, C. A. Comparative Analysis of Core Microbiome Assignments: Implications for Ecological Synthesis. mSystems 2023, 8. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Joardar, N.; Sengupta, S.; Sinha Babu, S. P. Gut Microbes as Future Therapeutics in Treating Inflammatory and Infectious Diseases: Lessons from Recent Findings. J. Nutr. Biochem. 2018, 61, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Behrouzi, A.; Nafari, A. H.; Siadat, S. D. The Significance of Microbiome in Personalized Medicine. Clin. Transl. Med. 2019, 8. [Google Scholar] [CrossRef]
- Kashyap, P. C.; Chia, N.; Nelson, H.; Segal, E.; Elinav, E. Microbiome at the Frontier of Personalized Medicine. Mayo Clin. Proc. 2017, 92, 1855–1864. [Google Scholar] [CrossRef]
- Ji, B.; Nielsen, J. From Next-Generation Sequencing to Systematic Modeling of the Gut Microbiome. Front. Genet. 2015, 6, 1–9. [Google Scholar] [CrossRef]
- Benameur, T.; Porro, C.; Twfieg, M. E.; Benameur, N.; Panaro, M. A.; Filannino, F. M.; Hasan, A. Emerging Paradigms in Inflammatory Disease Management: Exploring Bioactive Compounds and the Gut Microbiota. Brain Sci. 2023, 13. [Google Scholar] [CrossRef]
- Krishnan, S.; Alden, N.; Lee, K. Pathways and Functions of Gut Microbiota Metabolism Impacting Host Physiology. Curr. Opin. Biotechnol. 2015, 36, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Chen, Q.; Zhang, Y. Multidirectional Associations between the Gut Microbiota and Parkinson ’ s Disease, Updated Information from the Perspectives of Humoral Pathway, Cellular Immune Pathway and Neuronal Pathway. 2023, No. December, 1–20. [CrossRef]
- Aho, V. T. E.; Houser, M. C.; Pereira, P. A. B.; Chang, J.; Rudi, K.; Paulin, L.; Hertzberg, V.; Auvinen, P.; Tansey, M. G.; Scheperjans, F. Relationships of Gut Microbiota, Short-Chain Fatty Acids, Inflammation, and the Gut Barrier in Parkinson’s Disease. Mol. Neurodegener. 2021, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Martin, C. R.; Osadchiy, V.; Kalani, A.; Mayer, E. A. The Brain-Gut-Microbiome Axis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Kubista, M.; Andrade, J. M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; Ståhlberg, A.; Zoric, N. The Real-Time Polymerase Chain Reaction. Mol. Aspects Med. 2006, 27, (2–3). [Google Scholar] [CrossRef] [PubMed]
- Considine, E.; Yin, L.; Hartmann, M. A Review of the Primary Nutritional and Environmental Factors Associated with Parkinson’s Disease. J. Student Res. 2021, 10, 1–14. [Google Scholar] [CrossRef]
- Sampson, T. R.; Debelius, J. W.; Thron, T.; Janssen, S.; Shastri, G. G.; Ilhan, Z. E.; Challis, C.; Schretter, C. E.; Rocha, S.; Gradinaru, V.; Chesselet, M. F.; Keshavarzian, A.; Shannon, K. M.; Krajmalnik-Brown, R.; Wittung-Stafshede, P.; Knight, R.; Mazmanian, S. K. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Stolzenberg, E.; Berry, D.; Yang, D.; Lee, E. Y.; Kroemer, A.; Kaufman, S.; Wong, G. C. L.; Oppenheim, J. J.; Sen, S.; Fishbein, T.; Bax, A.; Harris, B.; Barbut, D.; Zasloff, M. A. A Role for Neuronal Alpha-Synuclein in Gastrointestinal Immunity. J. Innate Immun. 2017, 9, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q. Q.; Haikal, C.; Li, W.; Li, J. Y. Gut Inflammation in Association With Pathogenesis of Parkinson’s Disease. Front. Mol. Neurosci. 2019, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bjørklund, G.; Dadar, M.; Anderson, G.; Chirumbolo, S.; Maes, M. Preventive Treatments to Slow Substantia Nigra Damage and Parkinson’s Disease Progression: A Critical Perspective Review. Pharmacol. Res. 2020, 161, 105065. [Google Scholar] [CrossRef]
- Visanji, N. P.; Brooks, P. L.; Hazrati, L. N.; Lang, A. E. The Prion Hypothesis in Parkinson’s Disease: Braak to the Future. Acta Neuropathol. Commun. 2014, 2, 1–12. [Google Scholar] [CrossRef]
- Rietdijk, C. D.; Perez-Pardo, P.; Garssen, J.; van Wezel, R. J. A.; Kraneveld, A. D. Exploring Braak’s Hypothesis of Parkinson’s Disease. Front. Neurol. 2017, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, C. H.; Del Tredici, K.; Braak, H. Parkinson’s Disease: A Dual-Hit Hypothesis. Neuropathol. Appl. Neurobiol. 2007, 33, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kwon, S. H.; Kam, T. I.; Panicker, N.; Karuppagounder, S. S.; Lee, S.; Lee, J. H.; Kim, W. R.; Kook, M.; Foss, C. A.; Shen, C.; Lee, H.; Kulkarni, S.; Pasricha, P. J.; Lee, G.; Pomper, M. G.; Dawson, V. L.; Dawson, T. M.; Ko, H. S. Transneuronal Propagation of Pathologic α-Synuclein from the Gut to the Brain Models Parkinson’s Disease. Neuron 2019, 103, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Borghammer, P.; Van Den Berge, N. Brain-First versus Gut-First Parkinson’s Disease: A Hypothesis. J. Parkinsons. Dis. 2019, 9, S281–S295. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Brockmann, K. Blood and Cerebrospinal Fluid Biomarkers of Inflammation in Parkinson’s Disease. J. Parkinsons. Dis. 2022, 12, S183–S200. [Google Scholar] [CrossRef] [PubMed]
- Di Vincenzo, F.; Del Gaudio, A.; Petito, V.; Lopetuso, L. R.; Scaldaferri, F. Gut Microbiota, Intestinal Permeability, and Systemic Inflammation: A Narrative Review. Intern. Emerg. Med. 2023, 19, 275–293. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Wang, J.; Deng, Y.; Ye, Z.; Ge, Y. Inflammatory Microbes and Genes as Potential Biomarkers of Parkinson’s Disease. npj Biofilms Microbiomes 2022, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, S. Z.; Yu, Y. J.; Adeli, K. Role of Gut Microbiota in Neuroendocrine Regulation of Carbohydrate and Lipid Metabolism via the Microbiota-Gut-Brain-Liver Axis. Microorganisms 2020, 8, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L. M. T. Gut Bacteria and Neurotransmitters. Microorganisms 2022, 10, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Heidari, A.; Yazdanpanah, N.; Rezaei, N. The Role of Toll-like Receptors and Neuroinflammation in Parkinson’s Disease. J. Neuroinflammation 2022, 19, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Z.; Lv, A.; Fan, C. How Toll-like Receptors Influence Parkinson’s Disease in the Microbiome–Gut–Brain Axis. Front. Immunol. 2023, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Isik, S.; Yeman Kiyak, B.; Akbayir, R.; Seyhali, R.; Arpaci, T. Microglia Mediated Neuroinflammation in Parkinson’s Disease. Cells 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Tansey, M. G.; Wallings, R. L.; Houser, M. C.; Herrick, M. K.; Keating, C. E.; Joers, V. Inflammation and Immune Dysfunction in Parkinson Disease. Nat. Rev. Immunol. 2022, 22, 657–673. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-F.; Shen, Y.-Q. Dysbiosis of Gut Microbiota and Microbial Metabolites in Parkinson’s Disease. Ageing Res. Rev. 2018, 45, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.; Rietschel, E. T. Bacterial Lipopolysaccharides and Innate Immunity. J. Endotoxin Res. 2001, 7, 167–202. [Google Scholar] [CrossRef] [PubMed]
- Carvey, P. M.; Chang, Q.; Lipton, J. W.; Ling, Z. Prenatal Exposure to the Bacteriotoxin Lipopolysaccharide Leads to Long-Term Losses of Dopamine Neurons in Offspring: A Potential, New Model of Parkinson’s Disease. Front. Biosci. 2003, No. 10, 826–837. [Google Scholar] [CrossRef]
- Jangula, A.; Murphy, E. J. Lipopolysaccharide-Induced Blood Brain Barrier Permeability Is Enhanced by Alpha-Synuclein Expression. Neurosci. Lett. 2013, 551, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D. Q. H.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ji, X.; Song, Z.; Song, W.; Huang, Q.; Yu, T.; Shi, D.; Wang, F.; Xue, X.; Guo, J. Butyrate Inhibits the Mitochondrial Complex Ι to Mediate Mitochondria-Dependent Apoptosis of Cervical Cancer Cells. BMC Complement. Med. Ther. 2023, 23, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bienenstock, J.; Kunze, W.; Forsythe, P. Microbiota and the Gut-Brain Axis. Nutr. Rev. 2015, 73, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Gubert, C.; Kong, G.; Renoir, T.; Hannan, A. J. Exercise, Diet and Stress as Modulators of Gut Microbiota: Implications for Neurodegenerative Diseases. Neurobiol. Dis. 2020, 134. [Google Scholar] [CrossRef] [PubMed]
- Yemula, N.; Dietrich, C.; Dostal, V.; Hornberger, M. Parkinson’s Disease and the Gut: Symptoms, Nutrition, and Microbiota. J. Parkinsons. Dis. 2021, 11, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Gaßner, H.; Steib, S.; Klamroth, S.; Pasluosta, C. F.; Adler, W.; Eskofier, B. M.; Pfeifer, K.; Winkler, J.; Klucken, J. Perturbation Treadmill Training Improves Clinical Characteristics of Gait and Balance in Parkinson’s Disease. J. Parkinsons. Dis. 2019, 9, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Postuma, R. B.; Lang, A. E.; Munhoz, R. P.; Charland, K.; Pelletier, A.; Moscovich, M.; Filla, L.; Zanatta, D.; Romenets, S. R.; Altman, R.; Chuang, R.; Shah, B. Caffeine for Treatment of Parkinson Disease: A Randomized Controlled Trial. Neurology 2012, 79, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Gallo, V.; Vineis, P.; Cancellieri, M.; Chiodini, P.; Barker, R. A.; Brayne, C.; Pearce, N.; Vermeulen, R.; Panico, S.; Bueno-De-Mesquita, B.; Vanacore, N.; Forsgren, L.; Ramat, S.; Ardanaz, E.; Arriola, L.; Peterson, J.; Hansson, O.; Gavrila, D.; Sacerdote, C.; Sieri, S.; Kühn, T.; Katzke, V. A.; Van Der Schouw, Y. T.; Kyrozis, A.; Masala, G.; Mattiello, A.; Perneczky, R.; Middleton, L.; Saracci, R.; Riboli, E. Exploring Causality of the Association between Smoking and Parkinson’s Disease. Int. J. Epidemiol. 2019, 48, 912–925. [Google Scholar] [CrossRef] [PubMed]
- Lotankar, S.; Prabhavalkar, K. S.; Bhatt, L. K. Biomarkers for Parkinson’s Disease: Recent Advancement. Neurosci. Bull. 2017, 33, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Le, W.; Dong, J.; Li, S.; Korczyn, A. D. Can Biomarkers Help the Early Diagnosis of Parkinson’s Disease? Neurosci. Bull. 2017, 33, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, E.; Garrido, A.; Scholz, S. W.; Poewe, W. Challenges in the Diagnosis of Parkinson’s Disease. Lancet Neurol. 2021, 20, 385–397. [Google Scholar] [CrossRef]
- Delenclos, M.; Jones, D. R.; McLean, P. J.; Uitti, R. J. Biomarkers in Parkinson’s Disease: Advances and Strategies. Park. Relat. Disord. 2016, 22, S106–S110. [Google Scholar] [CrossRef]
- Dos Santos, M. C. T.; Scheller, D.; Schulte, C.; Mesa, I. R.; Colman, P.; Bujac, S. R.; Bell, R.; Berteau, C.; Perez, L. T.; Lachmann, I.; Berg, D.; Maetzler, W.; Da Costa, A. N. Evaluation of Cerebrospinal Fluid Proteins as Potential Biomarkers for Early Stage Parkinson’s Disease Diagnosis. PLoS One 2018, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, P.; Lashuel, H. A. Opportunities and Challenges of Alpha-Synuclein as a Potential Biomarker for Parkinson’s Disease and Other Synucleinopathies. npj Park. Dis. 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E. H.; Tennagels, S.; Gold, R.; Gerwert, K.; Beyer, L.; Tönges, L. Update on CSF Biomarkers in Parkinson’s Disease. Biomolecules 2022, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Siderowf, A.; Concha-Marambio, L.; Lafontant, D. E.; Farris, C. M.; Ma, Y.; Urenia, P. A.; Nguyen, H.; Alcalay, R. N.; Chahine, L. M.; Foroud, T.; Galasko, D.; Kieburtz, K.; Merchant, K.; Mollenhauer, B.; Poston, K. L.; Seibyl, J.; Simuni, T.; Tanner, C. M.; Weintraub, D.; Videnovic, A.; Choi, S. H.; Kurth, R.; Caspell-Garcia, C.; Coffey, C. S.; Frasier, M.; Oliveira, L. M. A.; Hutten, S. J.; Sherer, T.; Marek, K.; Soto, C. Assessment of Heterogeneity among Participants in the Parkinson’s Progression Markers Initiative Cohort Using α-Synuclein Seed Amplification: A Cross-Sectional Study. Lancet Neurol. 2023, 22, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Meng, L. B.; Chen, L. J.; Shi, X.; Tu, L.; Zhou, Q.; Yu, J. L.; Liao, X.; Zeng, Y.; Yuan, Q. Y. The Role of the Microbiota-Gut-Brain Axis and Intestinal Microbiome Dysregulation in Parkinson’s Disease. Front. Neurol. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Claudino Dos Santos, J. C.; Oliveira, L. F.; Noleto, F. M.; Gusmão, C. T. P.; Brito, G. A. de C.; Viana, G. S. de B. Gut-Microbiome-Brain Axis: The Crosstalk between the Vagus Nerve, Alpha-Synuclein and the Brain in Parkinson’s Disease. Neural Regen. Res. 2023, 18, 2611–2614. [Google Scholar] [CrossRef] [PubMed]
- Weis, S.; Schwiertz, A.; Unger, M. M.; Becker, A.; Faßbender, K.; Ratering, S.; Kohl, M.; Schnell, S.; Schäfer, K.-H.; Egert, M. Effect of Parkinson’s Disease and Related Medications on the Composition of the Fecal Bacterial Microbiota. NPJ Park. Dis. 2019, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yu, Y.-B. Intestinal Microbiota and Chronic Constipation. Springerplus 2016, 5, 1130. [Google Scholar] [CrossRef] [PubMed]
- Salim, S.; Ahmad, F.; Banu, A.; Mohammad, F. Gut Microbiome and Parkinson’s Disease: Perspective on Pathogenesis and Treatment. J. Adv. Res. 2023, 50, 83–105. [Google Scholar] [CrossRef] [PubMed]
- Gorecki, A. M.; Preskey, L.; Bakeberg, M. C.; Kenna, J. E.; Gildenhuys, C.; MacDougall, G.; Dunlop, S. A.; Mastaglia, F. L.; Anthony Akkari, P.; Koengten, F.; Anderton, R. S. Altered Gut Microbiome in Parkinson’s Disease and the Influence of Lipopolysaccharide in a Human α-Synuclein over-Expressing Mouse Model. Front. Neurosci. 2019, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, S.; Le, W. Intestinal Permeability, Dysbiosis, Inflammation and Enteric Glia Cells: The Intestinal Etiology of Parkinson’s Disease. Aging Dis. 2022, 13, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Haikal, C.; Chen, Q.-Q.; Li, J.-Y. Microbiome Changes: An Indicator of Parkinson’s Disease? Transl. Neurodegener. 2019, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Feng, X.; Zhou, X.; Zhao, M.; Xiao, H.; Li, R.; Shen, H. Identification of Gut Metabolites Associated with Parkinson’s Disease Using Bioinformatic Analyses. Front. Aging Neurosci. 2022, 14, 1–14. [Google Scholar] [CrossRef]
- Qian, Y.; Yang, X.; Xu, S.; Huang, P.; Li, B.; Du, J.; He, Y.; Su, B.; Xu, L. M.; Wang, L.; Huang, R.; Chen, S.; Xiao, Q. Gut Metagenomics-Derived Genes as Potential Biomarkers of Parkinson’s Disease. Brain 2020, 143, 2474–2489. [Google Scholar] [CrossRef] [PubMed]
- Hashish, S.; Salama, M. The Role of an Altered Gut Microbiome in Parkinson’s Disease: A Narrative Review. Appl. Microbiol. 2023, 3, 429–447. [Google Scholar] [CrossRef]
- Zacharias, H. U.; Kaleta, C.; Cossais, F.; Schaeffer, E.; Berndt, H.; Best, L.; Dost, T.; Glüsing, S.; Groussin, M.; Poyet, M.; Heinzel, S.; Bang, C.; Siebert, L.; Demetrowitsch, T.; Leypoldt, F.; Adelung, R.; Bartsch, T.; Bosy-Westphal, A.; Schwarz, K.; Berg, D. Microbiome and Metabolome Insights into the Role of the Gastrointestinal–Brain Axis in Parkinson’s and Alzheimer’s Disease: Unveiling Potential Therapeutic Targets. Metabolites 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K. Y.; Bhoopatiraju, S.; Silverglate, B. D.; Grossberg, G. T. Biomarkers in Parkinson’s Disease: A State of the Art Review. Biomarkers in Neuropsychiatry 2023, 9. [Google Scholar] [CrossRef]
- Jankovic, J.; Aguilar, L. G. Current Approaches to the Treatment of Parkinson’s Disease. Neuropsychiatr. Dis. Treat. 2008, 4, 743–757. [Google Scholar] [CrossRef] [PubMed]
- Poewe, W.; Antonini, A.; Zijlmans, J. C.; Burkhard, P. R.; Vingerhoets, F. Levodopa in the Treatment of Parkinson’s Disease: An Old Drug Still Going Strong. Clin. Interv. Aging 2010, 5, 229–238. [Google Scholar] [CrossRef]
- Mulak, A.; Bonaz, B. Brain-Gut-Microbiota Axis in Parkinson’s Disease. World J. Gastroenterol. 2015, 21, 10609–10620. [Google Scholar] [CrossRef]
- Toledo, A. R. L.; Monroy, G. R.; Salazar, F. E.; Lee, J. Y.; Jain, S.; Yadav, H.; Borlongan, C. V. Gut–Brain Axis as a Pathological and Therapeutic Target for Neurodegenerative Disorders. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Gulliver, E. L.; Young, R. B.; Chonwerawong, M.; D’Adamo, G. L.; Thomason, T.; Widdop, J. T.; Rutten, E. L.; Rossetto Marcelino, V.; Bryant, R. V.; Costello, S. P.; O’Brien, C. L.; Hold, G. L.; Giles, E. M.; Forster, S. C. Review Article: The Future of Microbiome-Based Therapeutics. Aliment. Pharmacol. Ther. 2022, 56, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, N.; Kitano, S.; Puah, G. R. Y.; Kittelmann, S.; Hwang, I. Y.; Chang, M. W. Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem. Rev. 2023, 123, 31–72. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J. S.; Ng, S. C.; Schnabl, B. Promises of Microbiome-Based Therapies. J. Hepatol. 2022, 76, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Wong, A. C. New Approaches to Microbiome-Based Therapies ABSTRACT.
- Jackson, A.; Forsyth, C. B.; Shaikh, M.; Voigt, R. M.; Engen, P. A.; Ramirez, V.; Keshavarzian, A. Diet in Parkinson’s Disease: Critical Role for the Microbiome. Front. Neurol. 2019, 10, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Hegelmaier, T.; Lebbing, M.; Duscha, A.; Tomaske, L.; Tönges, L.; Holm, J. B.; Bjørn Nielsen, H.; Gatermann, S. G.; Przuntek, H.; Haghikia, A. Interventional Influence of the Intestinal Microbiome Through Dietary Intervention and Bowel Cleansing Might Improve Motor Symptoms in Parkinson’s Disease. Cells 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Singh, R. K.; Chang, H. W.; Yan, D.; Lee, K. M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T. H.; Bhutani, T.; Liao, W. Influence of Diet on the Gut Microbiome and Implications for Human Health. J. Transl. Med. 2017, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Purdel, C.; Margină, D.; Adam-Dima, I.; Ungurianu, A. The Beneficial Effects of Dietary Interventions on Gut Microbiota—An Up-to-Date Critical Review and Future Perspectives. Nutrients 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Leeming, E. R.; Johnson, A. J.; Spector, T. D.; Roy, C. I. L. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 1–28. [Google Scholar] [CrossRef]
- Duan, W. X.; Wang, F.; Liu, J. Y.; Liu, C. F. Relationship Between Short-Chain Fatty Acids and Parkinson’s Disease: A Review from Pathology to Clinic. Neurosci. Bull. 0123. [Google Scholar] [CrossRef]
- Suganya, K.; Koo, B. S. Gut–Brain Axis: Role of Gut Microbiota on Neurological Disorders and How Probiotics/Prebiotics Beneficially Modulate Microbial and Immune Pathways to Improve Brain Functions. Int. J. Mol. Sci. 2020, 21, 1–29. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Makki, K.; Deehan, E. C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Liu, X.; Ye, Y.; Yan, X.; Cheng, Y.; Zhao, L.; Chen, F.; Ling, Z. Gut Microbiota: A Novel Therapeutic Target for Parkinson’s Disease. Front. Immunol. 2022, 13, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pardo, P.; de Jong, E. M.; Broersen, L. M.; Wijk, N. van; Attali, A.; Garssen, J.; Kraneveld, A. D. Promising Effects of Neurorestorative Diets on Motor, Cognitive, and Gastrointestinal Dysfunction after Symptom Development in a Mouse Model of Parkinson’s Disease. Front. Aging Neurosci. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Luo, Y.; Ray Chaudhuri, K.; Reynolds, R.; Tan, E. K.; Pettersson, S. The Role of Gut Dysbiosis in Parkinson’s Disease: Mechanistic Insights and Therapeutic Options. Brain 2021, 144, 2571–2593. [Google Scholar] [CrossRef] [PubMed]
- Socała, K.; Doboszewska, U.; Szopa, A.; Serefko, A.; Włodarczyk, M.; Zielińska, A.; Poleszak, E.; Fichna, J.; Wlaź, P. The Role of Microbiota-Gut-Brain Axis in Neuropsychiatric and Neurological Disorders. Pharmacol. Res. 2021, 172. [Google Scholar] [CrossRef] [PubMed]
- Soundharrajan, I.; Kuppusamy, P.; Srisesharam, S.; Lee, J. C.; Sivanesan, R.; Kim, D.; Choi, K. C. Positive Metabolic Effects of Selected Probiotic Bacteria on Diet-Induced Obesity in Mice Are Associated with Improvement of Dysbiotic Gut Microbiota. FASEB J. 2020, 34, 12289–12307. [Google Scholar] [CrossRef] [PubMed]
- Alard, J.; Lehrter, V.; Rhimi, M.; Mangin, I.; Peucelle, V.; Abraham, A. L.; Mariadassou, M.; Maguin, E.; Waligora-Dupriet, A. J.; Pot, B.; Wolowczuk, I.; Grangette, C. Beneficial Metabolic Effects of Selected Probiotics on Diet-Induced Obesity and Insulin Resistance in Mice Are Associated with Improvement of Dysbiotic Gut Microbiota. Environ. Microbiol. 2016, 18, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, F.; Fiasca, F.; Minelli, M.; Maio, D.; Mattei, A.; Vergallo, I.; Cifone, M. G.; Cinque, B.; Minelli, M. The Effects of Low-Nickel Diet Combined with Oral Administration of Selected Probiotics on Patients with Systemic Nickel Allergy Syndrome (SNAS) and Gut Dysbiosis. Nutrients 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Lubomski, M.; Davis, R. L.; Sue, C. M. The Gut Microbiota: A Novel Therapeutic Target in Parkinson’s Disease? Park. Relat. Disord. 2019, 66, 265–266. [Google Scholar] [CrossRef] [PubMed]
- Rekdal, V. M.; Bess, E. N.; Bisanz, J. E.; Turnbaugh, P. J.; Balskus, E. P. Discovery and Inhibition of an Interspecies Gut Bacterial Pathway for Levodopa Metabolism. Science (80-. ). 2019, 364. [Google Scholar] [CrossRef] [PubMed]
- van Kessel, S. P.; Frye, A. K.; El-Gendy, A. O.; Castejon, M.; Keshavarzian, A.; van Dijk, G.; El Aidy, S. Gut Bacterial Tyrosine Decarboxylases Restrict Levels of Levodopa in the Treatment of Parkinson’s Disease. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Su, W.; Li, S.; Du, W.; Ma, X.; Jin, Y.; Li, K.; Chen, H. Eradication of Helicobacter Pylori Infection Might Improve Clinical Status of Patients with Parkinson’s Disease, Especially on Bradykinesia. Clin. Neurol. Neurosurg. 2017, 160, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Lolekha, P.; Sriphanom, T.; Vilaichone, R. K. Helicobacter Pylori Eradication Improves Motor Fluctuations in Advanced Parkinson’s Disease Patients: A Prospective Cohort Study (HP-PD Trial). PLoS One 2021, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hashim, H.; Azmin, S.; Razlan, H.; Yahya, N. W.; Tan, H. J.; Manaf, M. R. A.; Ibrahim, N. M. Eradication of Helicobacter Pylori Infection Improves Levodopa Action, Clinical Symptoms and Quality of Life in Patients with Parkinson’s Disease. PLoS One 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A.; Visanji, N. P.; Liu, L. W. C.; Lang, A. E.; Pfeiffer, R. F. Gastrointestinal Dysfunction in Parkinson’s Disease. Lancet Neurol. 2015, 14, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Boelens Keun, J. T.; Arnoldussen, I. A.; Vriend, C.; Van De Rest, O. Dietary Approaches to Improve Efficacy and Control Side Effects of Levodopa Therapy in Parkinson’s Disease: A Systematic Review. Adv. Nutr. 2021, 12, 2265–2287. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, P. A.; Tropea, T. F.; Dahodwala, N. Overcoming Barriers to Parkinson Disease Trial Participation: Increasing Diversity and Novel Designs for Recruitment and Retention. Neurotherapeutics 2020, 17, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yue, L.; Fang, X.; Wang, G.; Li, C.; Sun, X.; Jia, X.; Yang, J.; Song, J.; Zhang, Y.; Guo, C.; Ma, G.; Sang, M.; Chen, F.; Wang, P. Altered Gut Microbiota in Parkinson’s Disease Patients/Healthy Spouses and Its Association with Clinical Features. Park. Relat. Disord. 2020, 81, 84–88. [Google Scholar] [CrossRef]
- Fonseca, D. C.; Marques Gomes da Rocha, I.; Depieri Balmant, B.; Callado, L.; Aguiar Prudêncio, A. P.; Tepedino Martins Alves, J.; Torrinhas, R. S.; da Rocha Fernandes, G.; Linetzky Waitzberg, D. Evaluation of Gut Microbiota Predictive Potential Associated with Phenotypic Characteristics to Identify Multifactorial Diseases. Gut Microbes 2024, 16, 2297815. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Sutton, J. Personalized Medicine Could Transform Healthcare (Review). Biomed. Reports 2017, 7, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Naylor, S.; Chen, J. Y. Unraveling Human Complexity and Disease with Systems Biology and Personalized Medicine. Per. Med. 2010, 7, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Goetz, L. H.; Schork, N. J. Personalized Medicine: Motivation, Challenges, and Progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Chen, Y.; Huang, W.; Zhou, H.; Zhang, W. Drug-Microbiota Interactions: An Emerging Priority for Precision Medicine. Signal Transduct. Target. Ther. 2023, 8. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I. D.; Nicholson, J. K. Gut Microbiome Interactions with Drug Metabolism, Efficacy and Toxicity Europe PMC Funders Author Manuscripts The Gut Microbiota Have the Capability of Preforming a Wide Range of Metabolic Reactions On. Transl. Res. 2017, 179, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.; Griffin, B. T.; Clarke, G.; Hyland, N. P. Drug–Gut Microbiota Interactions: Implications for Neuropharmacology. Br. J. Pharmacol. 2018, 175, 4415–4429. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Zuo, T. Interplays between Drugs and the Gut Microbiome. Gastroenterol. Rep. 2022, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Lim, S. H. Bidirectional Interaction between Intestinal Microbiome and Cancer: Opportunities for Therapeutic Interventions. Biomark. Res. 2020, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Weersma, R. K.; Zhernakova, A.; Fu, J. Interaction between Drugs and the Gut Microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Misera, A.; Łoniewski, I.; Palma, J.; Kulaszyńska, M.; Czarnecka, W.; Kaczmarczyk, M.; Liśkiewicz, P.; Samochowiec, J.; Skonieczna-Żydecka, K. Clinical Significance of Microbiota Changes under the Influence of Psychotropic Drugs. An Updated Narrative Review. Front. Microbiol. 2023, 14, 1–16. [Google Scholar] [CrossRef]
- Enright, E. F.; Gahan, C. G. M.; Joyce, S. A.; Griffin, B. T. The Impact of the Gut Microbiota on Drug Metabolism and Clinical Outcome. Yale J. Biol. Med. 2016, 89, 375–382. [Google Scholar] [PubMed]
- Fan, H. X.; Sheng, S.; Zhang, F. New Hope for Parkinson’s Disease Treatment: Targeting Gut Microbiota. CNS Neurosci. Ther. 2022, 28, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Yue, Y.; He, T.; Huang, C.; Qu, B.; Lv, W.; Lai, H. Y. The Association Between the Gut Microbiota and Parkinson’s Disease, a Meta-Analysis. Front. Aging Neurosci. 2021, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzian, A.; Green, S. J.; Engen, P. A.; Voigt, R. M.; Naqib, A.; Forsyth, C. B.; Mutlu, E.; Shannon, K. M. Colonic Bacterial Composition in Parkinson’s Disease. Mov. Disord. 2015, 30, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Jiang, J.; Wang, X.; Jiang, K.; Cao, H. Exposure to Prescribed Medication in Early Life and Impacts on Gut Microbiota and Disease Development. eClinicalMedicine 2024, 68, 102428. [Google Scholar] [CrossRef] [PubMed]
- WADE, D. N.; MEARRICK, P. T.; MORRIS, J. L. Active Transport of L-Dopa in the Intestine. Nature 1973, 242, 463–465. [Google Scholar] [CrossRef] [PubMed]
- Ngwuluka, N.; Pillay, V.; Du Toit, L. C.; Ndesendo, V.; Choonara, Y.; Modi, G.; Naidoo, D. Levodopa Delivery Systems: Advancements in Delivery of the Gold Standard. Expert Opin. Drug Deliv. 2010, 7, 203–224. [Google Scholar] [CrossRef] [PubMed]
- Smith-Hicks, C.; Raymond, G. V. CHAPTER 11 - Developmental Disorders; Irani, D. N. B. T.-C. F. in C. P., Ed.; W.B. Saunders: Philadelphia, 2009. [Google Scholar] [CrossRef]
- Gonçalves, S.; Nunes-Costa, D.; Cardoso, S. M.; Empadinhas, N.; Marugg, J. D. Enzyme Promiscuity in Serotonin Biosynthesis, From Bacteria to Plants and Humans. Front. Microbiol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Gershanik, O. S. Improving L-Dopa Therapy: The Development of Enzyme Inhibitors. Mov. Disord. 2015, 30, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Jenner, P. Stalevo(®) : A Pioneering Treatment for OFF Periods in Parkinsons Disease. Eur. J. Neurol. 2023, 30 Suppl 2, 3–8. [Google Scholar] [CrossRef]
- Tambasco, N.; Romoli, M.; Calabresi, P. Levodopa in Parkinson’s Disease: Current Status and Future Developments. Curr. Neuropharmacol. 2017, 16, 1239–1252. [Google Scholar] [CrossRef] [PubMed]
- Tsunoda, S. M.; Gonzales, C.; Jarmusch, A. K.; Momper, J. D.; Ma, J. D. Contribution of the Gut Microbiome to Drug Disposition, Pharmacokinetic and Pharmacodynamic Variability. Clin. Pharmacokinet. 2021, 60, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Sheng, S.; Zhang, F. Relationship Between Gut Bacteria and Levodopa Metabolism. Curr. Neuropharmacol. 2022, 21, 1536–1547. [Google Scholar] [CrossRef] [PubMed]
- Jameson, K. G.; Hsiao, E. Y. A Novel Pathway for Microbial Metabolism of Levodopa. Nat. Med. 2019, 25, 1195–1197. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A. L. Mapping Human Microbiome Drug Metabolism by Gut Bacteria and Their Genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef] [PubMed]

| Technique | Description | References |
|---|---|---|
| Culture-based | Isolation/ Growing of bacteria on selective media | [51,52,53,54] |
| 16S rRNA (16S ribossomal RNA) gene sequencing based on cloning |
The process involves cloning the entire 16S rRNA amplicon, followed by Sanger sequencing and capillary electrophoresis for analysis. | [51,52,55,56] |
| Direct sequencing of 16S rRNA amplicons |
High-throughput parallel sequencing of partial 16S rRNA amplicons. | [51,52,57] |
| Quantitative real-time polymerase chain reaction (qPCR) | 16 S rRNA amplification and quantification. A substance in the reaction mixture exhibits fluorescence when is attached to a double-stranded DNA. | [51,52,58,59] |
| Denaturing gradient gel electrophoresis (DGGE) / Temperature Gradient Gel Electrophoresis (TGGE) | 16S rRNA amplicons separated on a gel using a denaturant/ temperature gradient | [51,52,60] |
| Terminal restriction fragment length polymorphism (T-RFLP) |
After the quantification of fluorescently labeled primers, the 16S rRNA amplicon undergoes digestion utilizing restriction enzymes. Subsequently, the resulting digested fragments are separated through gel electrophoresis. | [51,52,61] |
| Fluorescence in situ hybridization (FISH) |
Oligonucleotide probes, labeled with fluorescent markers, bind to sequences that are complementary to the target 16S rRNA. The fluorescence generated during this hybridization process can be quantified using flow cytometry. | [51,52,62] |
| Microbiome Shotgun sequencing | Extensive parallel sequencing of the entire genome. | [51,52] |
| Metagenomics | Exploring high-resolution profiling of gut microbiota genomes and characterizing gene structures of uncultivated microbiota. | [51,63] |
| Metatranscriptomics | High-resolution gene expression profiling is achieved through sequencing messenger RNA (mRNA) or complementary DNA (cDNA). This approach is used to explore differential microbial gene expression under various physiological or environmental conditions, providing insights into microbial adaptation and responses. | [64,65] |
| Metaproteomics | High-resolution protein monitoring and profiling involve the identification of proteins and peptides, enabling the examination of differential microbial protein production in diverse physiological or environmental conditions. | [66,67,68] |
| Metabolomics | Metabolites undergo analysis to profile the metabolic activity of microbial hosts. | [67,69,70] |
| DNA microarrays | Oligonucleotide probes labeled with fluorescent markers undergo hybridization with complementary nucleotide sequences. The resulting fluorescence is detected using a fluorescence laser detector. | [51,71] |
| Clinical Trial Title | Study Type | Sample Size | Study Phase | ClinicalTrials.gov Identifier |
|---|---|---|---|---|
| Ketogenic Diet Interventions in Parkinson's Disease: Safeguarding the Gut Microbiome (KIM) | Interventional | 50 | Not yet recruiting | NCT05469997 |
| Characterization of Fecal Microbiome Changes After Administration of PRIM-DJ2727 in Parkinson's Disease Patients | Interventional | 0 | Withdrawn | NCT03026231 |
| The Microbiome in Parkinson's Disease | Observational | 210 | Unknown status | NCT03129451 |
| Microbiome and Diet in Parkinson's Disease - a Randomized, Controlled Phase 2 Trial (PD-Diet) | Interventional | 40 | Not yet recruiting | NCT06207136 |
| Levodopa Response and Gut Microbiome in Patients With Parkinson's Disease | Observational | 38 | Completed | NCT04956939 |
| Prebiotics in the Parkinson's Disease Microbiome | Interventional | 20 | Completed | NCT04512599 |
| Observational Small Intestine and Blood Fingerprint (SmIle) Study in Parkinson's Disease | Observational | 100 | Not yet recruiting | NCT06003608 |
| Deep Brain Stimulation Therapy and Intestinal Microbiota | Observational | 30 | Unknown status | NCT04855344 |
| Single-center Pathophysiological Study of the Role of Inflammation, Changes in the Intestinal Epithelial Barrier and the Intestinal Microbiota in Parkinson's Disease | Interventional | 77 | Terminated | NCT04652843 |
| A Pilot Study to Explore the Role of Gut Flora in Parkinson's Disease | Observational | 100 | Unknown status | NCT04148326 |
| Study of the Genome, Gut Metagenome and Diet of Patients With Incident Parkinson's Disease | Observational | 138 | Terminated | NCT04119596 |
| Resistant Maltodextrin for Gut Microbiome in Parkinson's Disease: Safety and Tolerability Study | Interventional | 30 | Active, not recruiting | NCT03667404 |
| Microbiota Modification for the Treatment of Motor Complication of Parkinson´s Disease | Interventional | 14 | Completed | NCT04730245 |
| Constipation and Changes in the Gut Flora in Parkinson's Disease | Observational | 80 | Recruiting | NCT05787756 |
| AADC/TDC in Advanced Parkinson's Disease | Observational | 50 | Recruiting | NCT05558787 |
| A Trial of Fecal Microbiome Transplantation in Parkinson's Disease Patients | Interventional | 51 | Completed | NCT04854291 |
| Gut Microbiota and Parkinson's Disease | Observational | 50 | Unknown status | NCT03710668 |
| Determining the Microbiota Composition of the Middle Meatus in Parkinson's | Observational | 48 | Completed | NCT03336697 |
| Study of the Fecal Microbiome in Patients With Parkinson's Disease | Interventional | 15 | Completed | NCT03671785 |
| Parkinson's Disease Biomarkers in Nerve Cells in the Gut | Observational | 60 | Recruiting | NCT05347407 |
| Efficacy and Safety of Fecal Microbiota Transplantation in the Treatment of Parkinson's Disease With Constipation | Interventional | 30 | Recruiting | NCT04837313 |
| Increased Gut Permeability to Lipopolysaccharides (LPS) in Parkinson's Disease | Observational | 43 | Completed | NCT01155492 |
| Fecal Microbiota Transplantation for Parkinson's Disease | Interventional | 49 | Completed | NCT03808389 |
| Microbiota Intervention to Change the Response of Parkinson's Disease | Interventional | 86 | Recruiting | NCT03575195 |
| Microbiome Composition and Function Contributes to Cognitive Impairment and Neuroinflammation in Parkinson's Disease | Observational | 100 | Recruiting | NCT05419453 |
| Gut Microbiota in the Progression of Alpha-synucleinopathies | Observational | 490 | Recruiting | NCT05353868 |
| Gut Health and Probiotics in Parkinson's (SymPD) | Interventional | 60 | Active, not recruiting | NCT05146921 |
| Effects of Probiotics on Peripheral Immunity in Parkinson's Disease | Interventional | 88 | Enrolling by invitation | NCT05173701 |
| Gut Microbiota Across Early Stages of Synucleinopathy: From High-risk Relatives, REM Sleep Behavior Disorder to Early Parkinson's Disease | Observational | 441 | Completed | NCT03645226 |
| Meridian Activation Remedy System for Parkinson's Disease (MARS-PD) | Interventional | 88 | Recruiting | NCT05621772 |
| Mediterranean Diet Intervention to Improve Gastrointestinal Function in Parkinson's Disease a Randomized, Controlled, Clinical Trial (MEDI-PD) | Interventional | 46 | Completed | NCT04683900 |
| MOVIN' CARE for PD (Risk Management) (jcpdmcP) | Interventional | 308 | Active, not recruiting | NCT06147284 |
| Fecal Microbiota Transplantation As a Potential Treatment for Parkinson's Disease | Interventional | 10 | Completed | NCT03876327 |
| Efficacy of Probiotics for Parkinson Disease (PD) | Interventional | 300 | Not yet recruiting | NCT06118294 |
| MOVIN' CARE for PD (Mind-body Interventions) (jcpdmcI) | Interventional | 154 | Active, not recruiting | NCT06078046 |
| Establishment of a Human Tissue Bank for Studying the Microbial Etiology of Neurodegenerative Diseases | Observational | 0 | Withdrawn | NCT01954875 |
| The Sunnybrook Dementia Study (SDS) | Observational | 1600 | Recruiting | NCT01800214 |
| Metabolic Cofactor Supplementation in Alzheimer's Disease (AD) and Parkinson's Disease (PD) Patients | Interventional | 120 | Completed | NCT04044131 |
| N-DOSE: A Dose Optimization Trial of Nicotinamide Riboside in Parkinson's Disease | Interventional | 80 | Recruiting | NCT05589766 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





