Submitted:
19 April 2024
Posted:
22 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction:
2. Materials and Methods
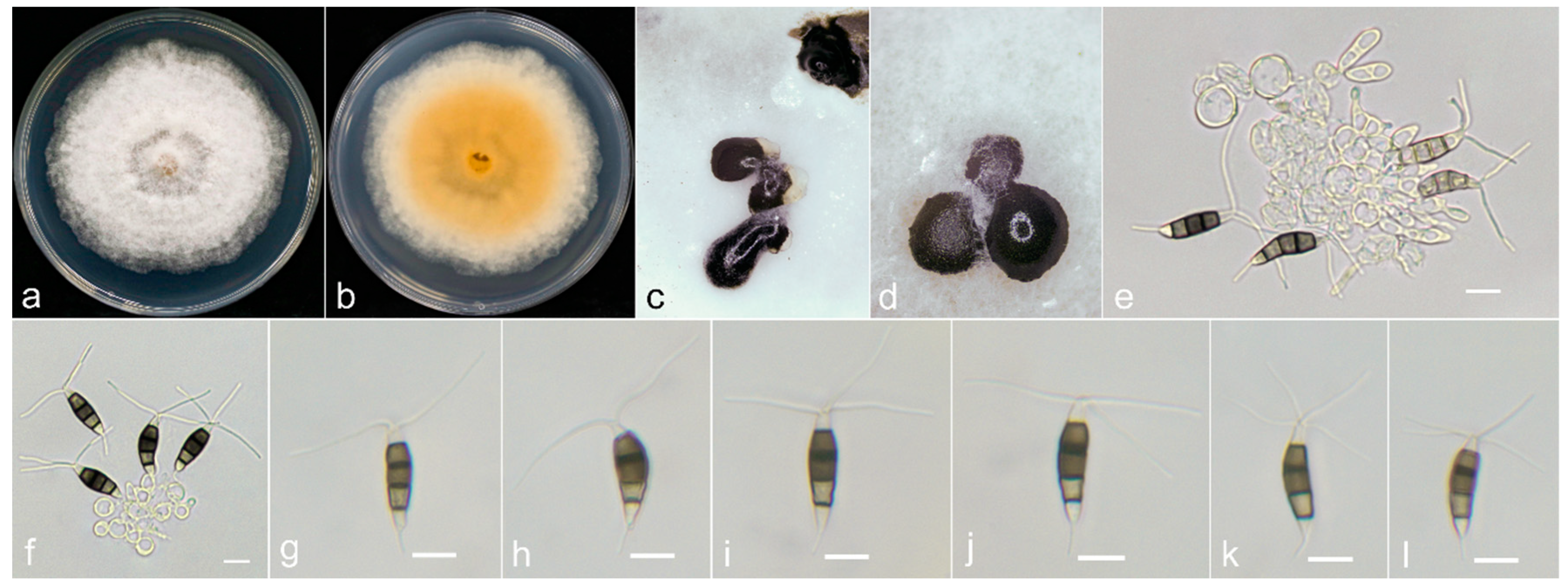
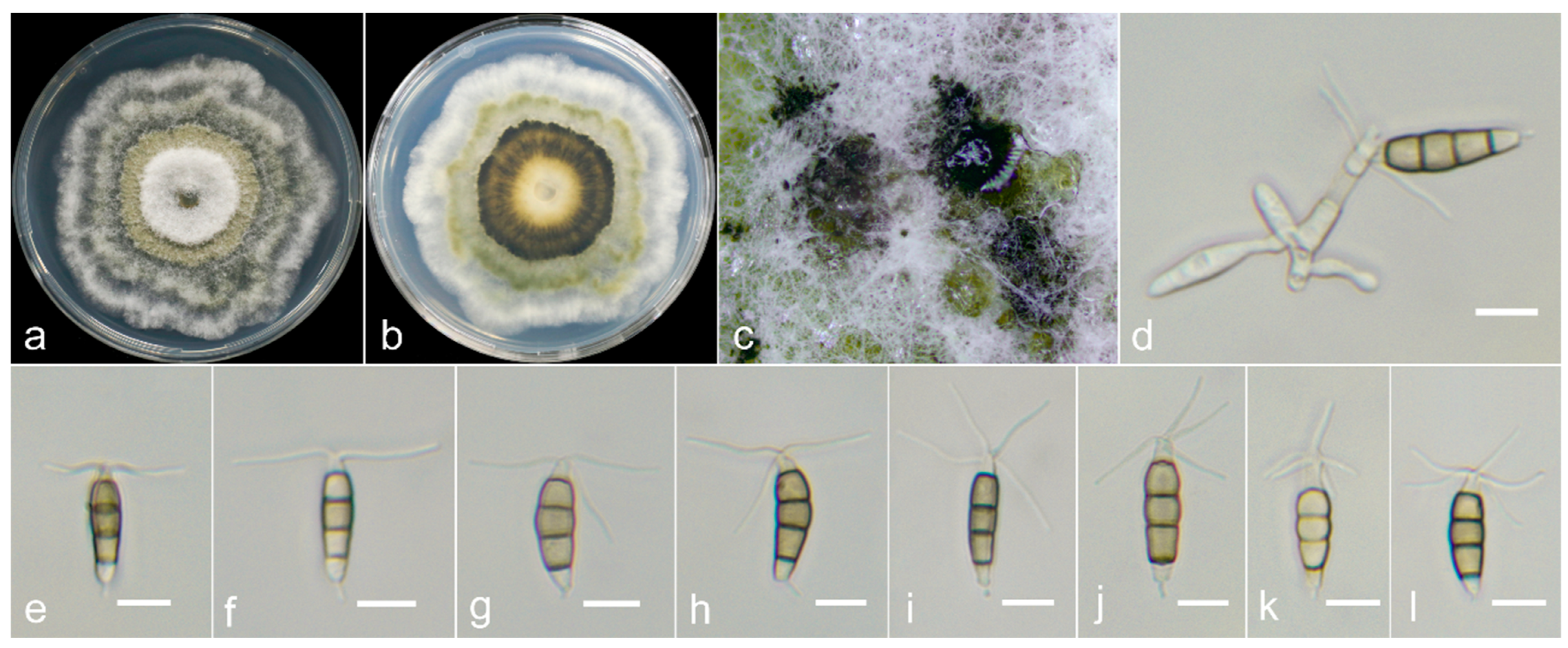
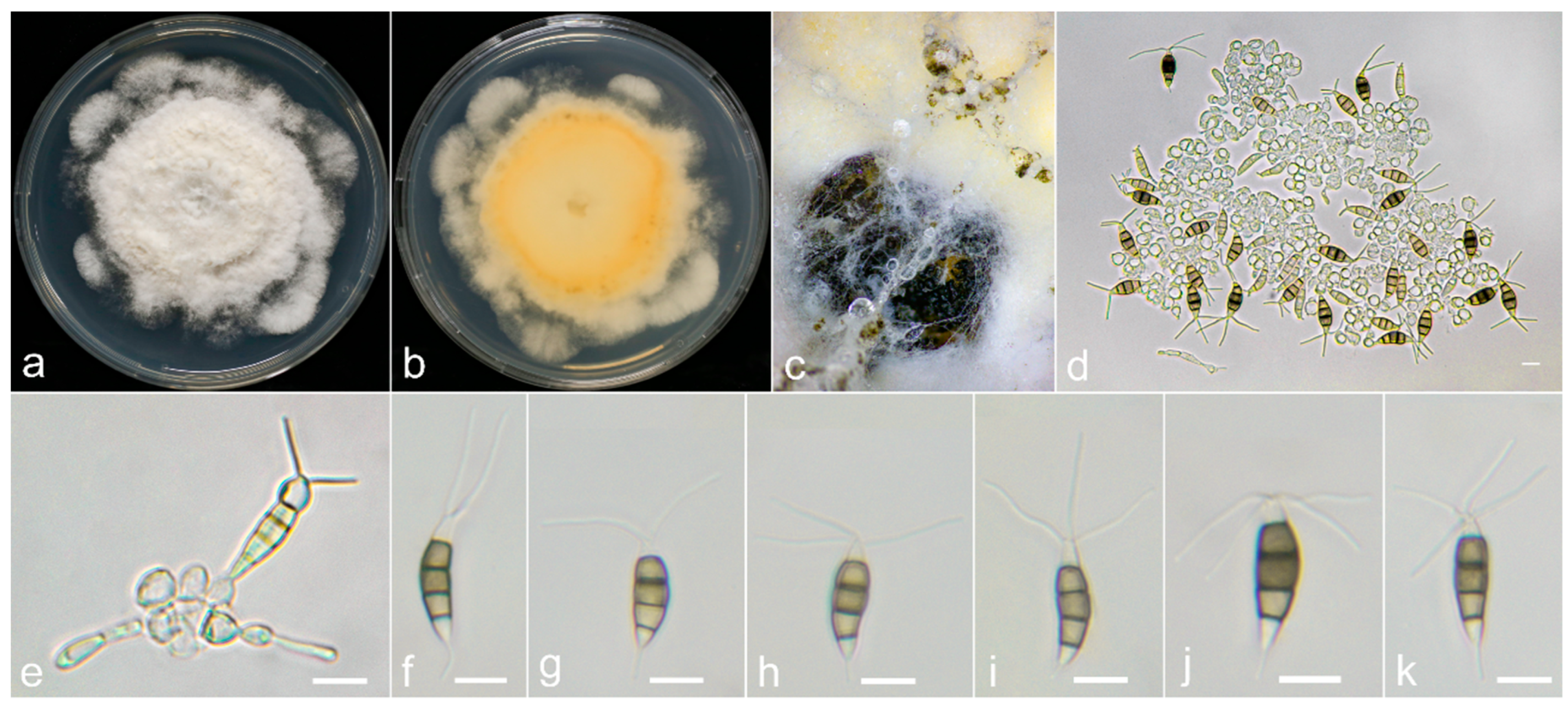
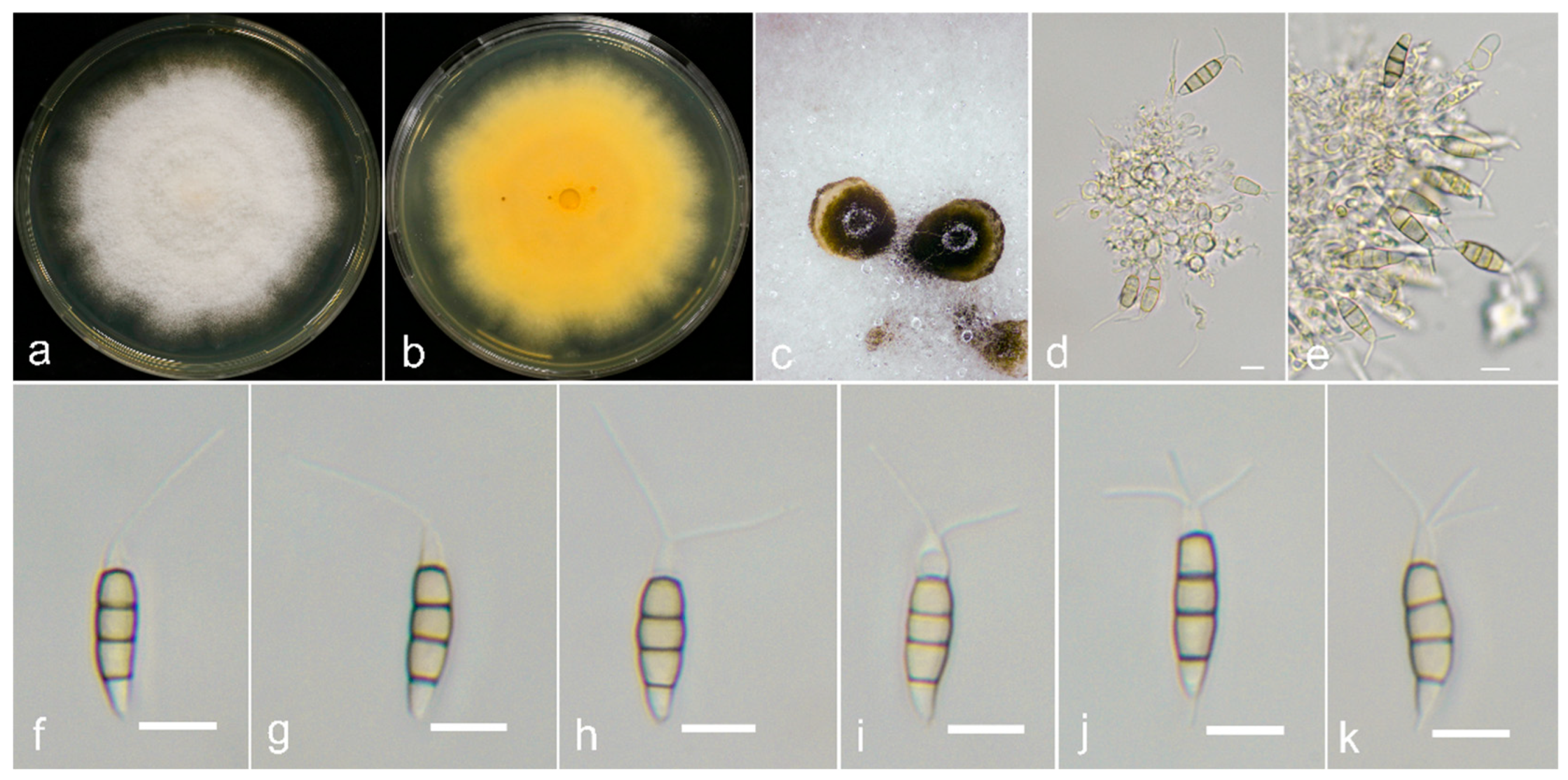
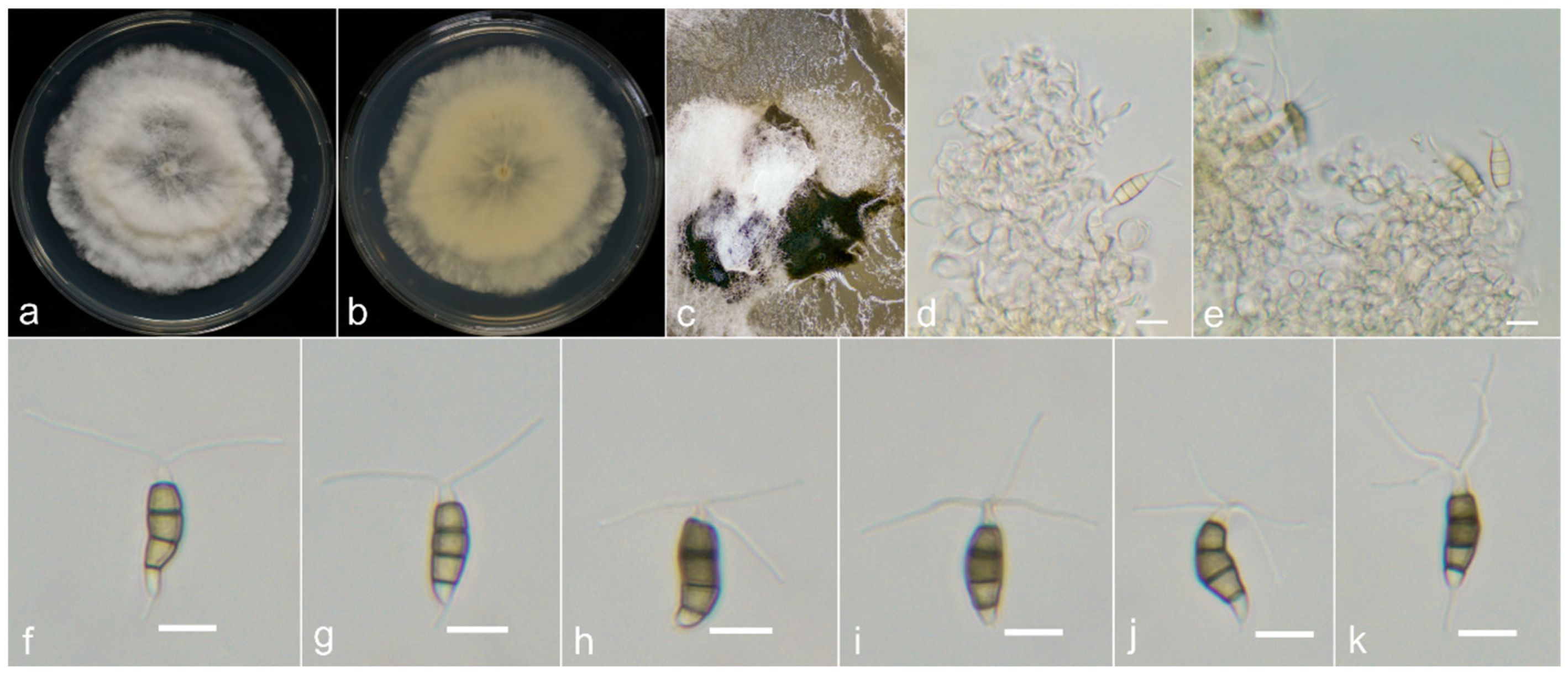
2.1. Sample Collection, Fungi Isolation and Morphological Examination
2.2. DNA Extraction, Gene Sequencing, and Phylogenetic Analyses
2.3. Pathogenicity Test
3. Results
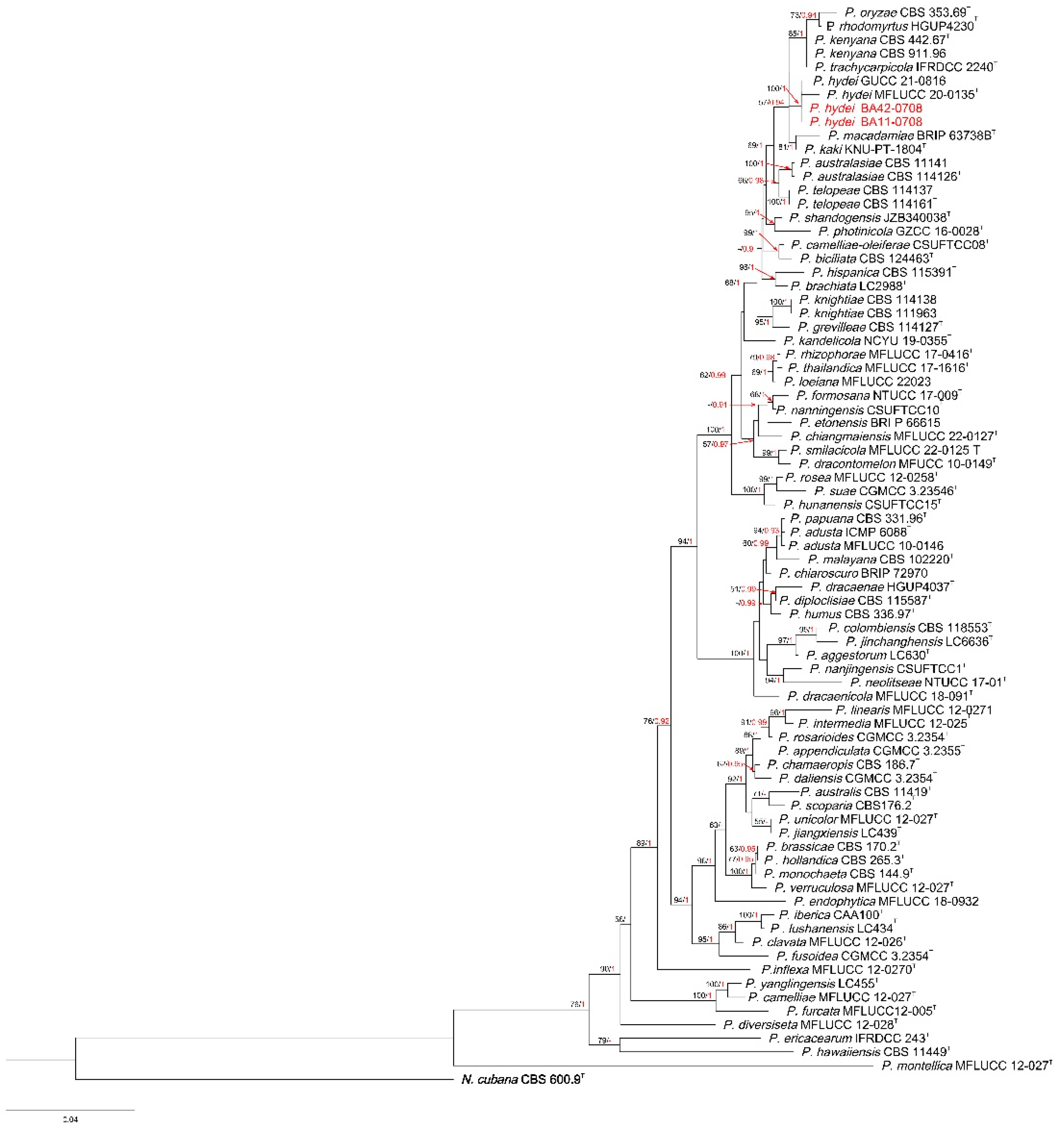
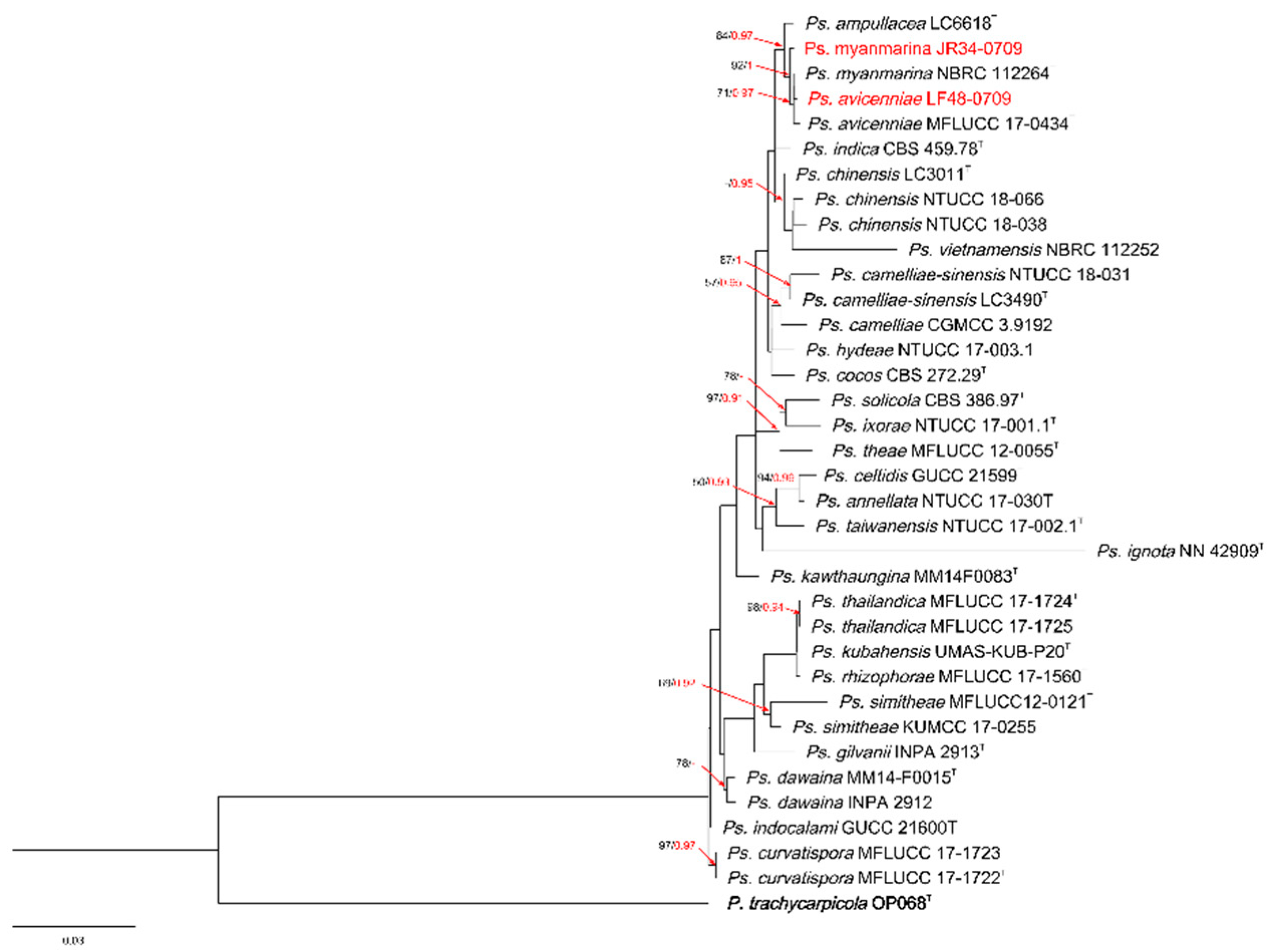
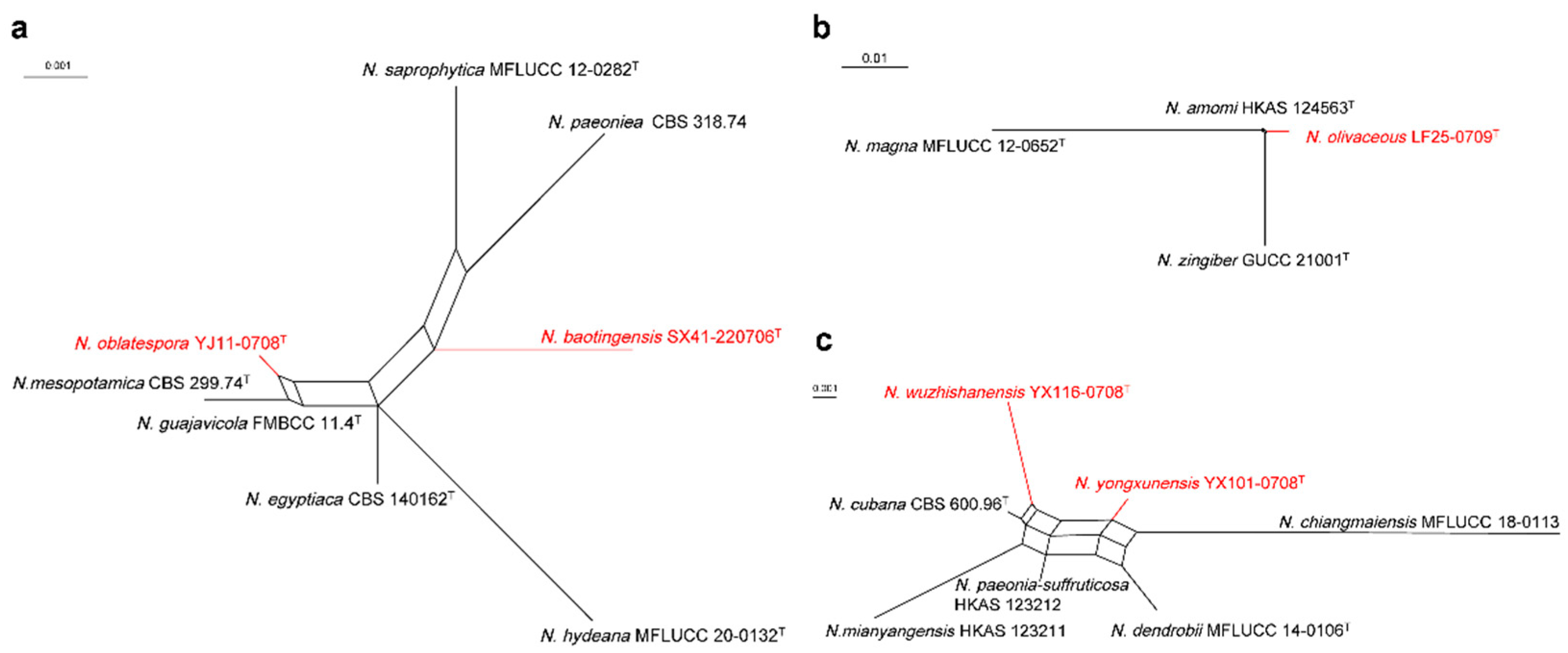
3.1. Phylogenetic Analyses
3.2. PHI Analyses
3.3. Taxonomy
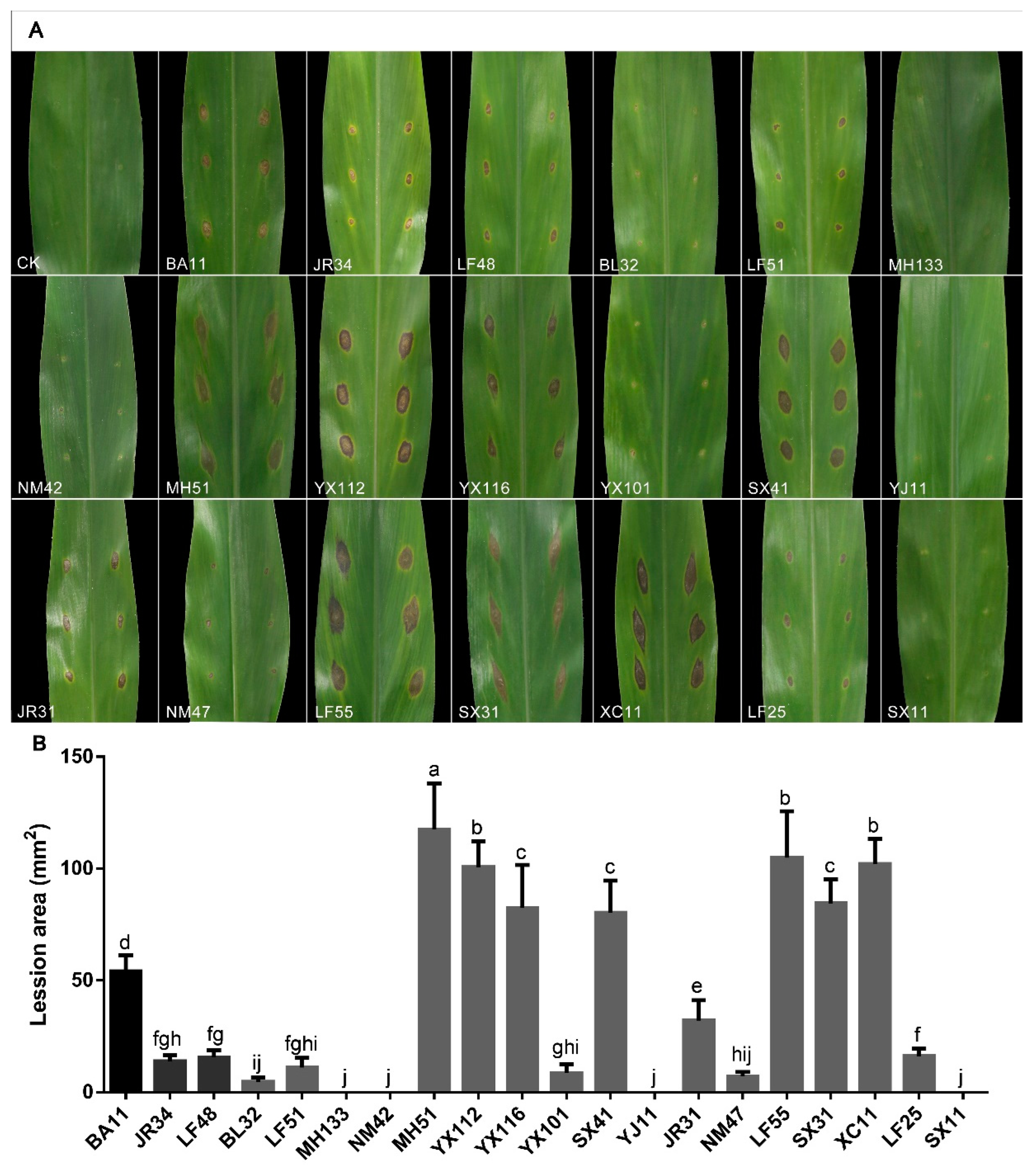
3.4. Pathogenicity Assay
4. Discussion
Author Contributions
Data Availability Statement
Acknowledgements
Conflict of Interest
References
- Qiu, L.Y. Investigation of germplasm resources of Alpinia oxyphylla Miq. in Hainan Island. Master's thesis,Hainan University, Hainan, China, 2015.
- Pharmacopoeia of the People's Republic of China. Alpiniae oxyphyllae fructus; The Medicine Science and Technology Press of China, China, 2020, 1, 303.
- Zhang, Q.; Zheng, Y.L.; Hu, X.J.; Hu, X.L.; Lv, W.W.; Lv, D.; Chen, J.J.; Wu, M.L.; Song, Q.C.; Shentu, J.Z. Ethnopharmacological uses, phytochemistry, biological activities, and therapeutic applications of Alpinia oxyphylla Miguel: A review. Journal of Ethnopharmacology 2018, 224, 149-168. [CrossRef]
- Zhang, J.Q.; Wang, S.; Li, Y.H.; Xu, P.; Chen, F.; Tan, Y.F.; Duan, J.A. Anti-diarrheal constituents of Alpinia oxyphylla. Fitoterapia 2013, 89, 149-156. [CrossRef]
- Zhou, J.Z.; Zhang, K.; Zhang, L.Y.; Xian, Q.Q.; Sun, P.Z.; Du, Z.Y. Geographical distribution and prediction of potential region of Alpinia oxyphylla Miq. Anhui Agric. Sci.2018, 46, 1-3+8. [CrossRef]
- Wang, Y.F. Exploration of metabolites and related genes of volatile oil from wild Alpinia oxyphylla Miq. in Hainan Province. Central University for Nationalities, Beijing, China, 2022.
- Chen, L.Z.; Liufu, Y.Q.; Lin, S.C.; Xian, S.Q. Analysis of the basic biological characteristics and high yield cultivation techniques of Alpinia oxyphylla Miq. South China Agriculture 2018, 12, 27+29. [CrossRef]
- Zhang, C.; Wu, Y.G.; Yang, H.G.; Yao, G.L.; Zhang, J.F.; Yu, J.; Yang, D.M.; Chen, P. The current planting situation and industrial development countermeasures of Alpinia oxyphylla Miq. in Hainan Province. Modern Horticulture 2021, 44, 63-67. [CrossRef]
- He, H.M. Analysis of biological characteristics and environment for high production of Alpinia Oxyphylla. Chinese Journal of Mordern Applied Pharmacy 1992, 205-207.
- Yan, X.X.; Ren, B.L.; Wang, M.Y.; Wang, Q.L.; Yang, Q.; Tang, H.; Wang, Z.N. Present situation and development strategy of Alpinia oxyphylla. China Journal of Chinese Materia Medica 2019, 44, 1960-1964. [CrossRef]
- Hong, X.Q.; Chen, J.J. Three important new diseases of Alpinia oxyphylla Miq. Chinese Journal of Tropical Agriculture 1986, 50-53.
- Xu, X.R.; Zhou, J.; Wu, X.P.; Lei, X.T. The main diseases and pests of Alpinia oxyphylla and their prevention and control. China Tropical Agriculture 2012, 48, 56-58.
- Fu, M.Y.; Zeng, X.P.; Rui, K.; Wang, H.F.; Chen, M.C. The occurrence and control of the leaf blight disease of Alpinia oxyphylla in Hainan Island. Modern Agricultural Science and Technology 2015, 655, 170-171.
- Maharachchikumbura, S.S.N.; Guo, L.D.; Cai, L.; Chukeatirote, E.; Wu, W.P.; Sun, X.; Crous, P.W.; Bhat, D.J.; McKenzie, E.H.C.; Bahkali, A.H.; et al. A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Diversity 2012, 56, 95-129. [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Groenewald, J.Z.; Xu, J.; Crous, P.W. Pestalotiopsis revisited. Studies in Mycology 2014, 121-186. [CrossRef]
- Sun, Y.R.; Jayawardena, R.S.; Sun, J.E.; Wang, Y. Pestalotioid species associated with medicinal plants in southwest China and Thailand. Microbiology Spectrum 2023, 11, e0398722. [CrossRef]
- Li, L.L.; Yang, Q.; Li, H. Morphology, phylogeny, and pathogenicity of pestalotioid species on Camellia oleifera in China. Journal of Fungi 2021, 7, 1080. [CrossRef]
- Diogo, E.; Goncalves, C.I.; Silva, A.C.; Valente, C.; Braganca, H.; Phillips, A.J.L. Five new species of Neopestalotiopsis associated with diseased Eucalyptus spp. in Portugal. Mycological Progress 2021, 20, 1441-1456. [CrossRef]
- Lin, H.F.; Chen, T.H.; Liu, S.D. Bioactivity of antifungal substance iturin a produced by Bacillus subtilis strain BS-99-H against Pestalotiopsis eugeniae, a causal pathogen of wax apple fruit rot. Plant Pathology Bulletin 2010, 19, 225-233.
- Prasannath, K.; Shivas, R.G.; Galea, V.J.; Akinsanmi, O.A. Neopestalotiopsis species associated with flower diseases of Macadamia integrifolia in Australia. Journal of Fungi 2021, 7, 771. [CrossRef]
- Tsai, I.; Chung, C.L.; Lin, S.R.; Hung, T.H.; Shen, T.L.; Hu, C.Y.; Hozzein, W.N.; Ariyawansa, H.A. Cryptic diversity, molecular systematics, and pathogenicity of genus Pestalotiopsis and allied genera causing gray blight disease of tea in Taiwan, With a description of a new Pseudopestalotiopsis species. Plant Disease 2021, 105, 425-443. [CrossRef]
- Maharachchikumbura, S.S.N.; Larignon, P.; Hyde, K.D.; Al-Sadi, A.M.; Liu, Z.Y. Characterization of Neopestalotiopsis, Pestalotiopsis and Truncatella species associated with grapevine trunk diseases in France. Phytopathologia Mediterranea 2016, 55, 380-390. [CrossRef]
- Ayoubi, N.; Soleimani, M.J. Strawberry fruit rot caused by Neopestalotiopsis iranensis sp nov., and N. mesopotamica. Current Microbiology 2016, 72, 329-336. [CrossRef]
- Chamorro, M.; Aguado, A.; De los Santos, B. First report of root and crown rot caused by Pestalotiopsis clavispora (Neopestalotiopsis clavispora) on strawberry in Spain. Plant Disease 2016, 100, 1495-1495. [CrossRef]
- Maharachchikumbura, S.S.N.; Guo, L.D.; Chukeatirote, E.; Bahkali, A.H.; Hyde, K.D. Pestalotiopsis-morphology, phylogeny, biochemistry and diversity. Fungal Diversity 2011, 50, 167-187. [CrossRef]
- Qi, P.K. Fungal diseases of cultivated medicinal plants in Guangdong Province; Guangdong Science and Technology Press: Guangdong, China, 1994.
- Jeewon, R.; Liew, E.C.Y.; Hyde, K.D. Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Molecular Phylogenetics and Evolution 2002, 25, 378-392. [CrossRef]
- Jeewon, R.; Liew, E.C.Y.; Simpson, J.A.; Hodgkiss, I.J.; Hyde, K.D. Phylogenetic significance of morphological characters in the taxonomy of Pestalotiopsis species. Molecular Phylogenetics and Evolution 2003, 27, 372-383. [CrossRef]
- Gualberto, G.F.; Catarino, A.D.; Sousa, T.F.; Cruz, J.C.; Hanada, R.E.; Caniato, F.F.; Silva, G.F. Pseudopestalotiopsis gilvanii sp. nov. and Neopestalotiopsis formicarum leaves spot pathogens from guarana plant: a new threat to global tropical hosts. Phytotaxa 2021, 489, 121-139. [CrossRef]
- He, Y.K.; Yang, Q.; Sun, Y.R.; Zeng, X.Y.; Jayawardena, R.S.; Hyde, K.D.; Wang, Y. Additions to Neopestalotiopsis (Amphisphaeriales, Sporocadaceae) fungi: two new species and one new host record from China. Biodiversity Data Journal 2022, 10, e90709. [CrossRef]
- Li, W.L.; Dissanayake, A.J.; Zhang, T.; Maharachchikumbura, S.S.N.; Liu, J.K. Identification and pathogenicity of pestalotioid fungi associated with woody oil plants in Sichuan Province, China. Journal of Fungi 2022, 8. [CrossRef]
- Prematunga, C.J.Y., L.Q.; Gomdola, D.; Balasuriya, A.; Yang, Y.H.; Jayawardena, R.S.; Luo, M. An addition to pestalotioid fungi in China: Neopestalotiopsis fragariae sp. nov. causing leaf spots on Fragaria × ananassa. Asian Journal of Mycology 2022, 5, 220-238, https://asianjournalofmycology.org/pdf/AJOM_5_2_10-1.
- Yang, Q.; He, Y.K.; Yuan, J.; Wang, Y. Two new Pseudopestalotiopsis species isolated from Celtis sinensis and Indocalamus tessellatus plants in southern China. Phytotaxa 2022, 543, 274-282. [CrossRef]
- Yang, Q.; Zeng, X.Y.; Yuan, J.; Zhang, Q.; He, Y.K.; Wang, Y. Two new species of Neopestalotiopsis from southern China. Biodiversity Data Journal 2021, 9. [CrossRef]
- White, T.J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pcr Protocols, Academic Press, Inc, 1990, 315-322.
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553-556. [CrossRef]
- O'Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 1998, 95, 2044-2049. [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 1995, 61, 1323-1330. [CrossRef]
- Odonnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 1997, 7, 103-116. [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171-180. [CrossRef]
- Norphanphoun, C.; Jayawardena, R.S.; Chen, Y.; Wen, T.C.; Meepol, W.; Hyde, K.D. Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. Mycosphere 2019, 10, 531-578. [CrossRef]
- Kumar, V.; Cheewangkoon, R.; Gentekaki, E.; Maharachchikumbura, S.S.N.; Brahmanage, R.S.; Hyde, K.D. Neopestalotiopsis alpapicalis sp. nov. a new endophyte from tropical mangrove trees in Krabi Province (Thailand). Phytotaxa 2019, 393, 251-262. [CrossRef]
- Bezerra, J.D.P.; Machado, A.R.; Firmino, A.L.; Rosado, A.W.C.; de Souza, C.A.F.; de Souza-Motta, C.M.; Freire, K.; Paiva, L.M.; Magalhaes, O.M.C.; Pereira, O.L.; et al. Mycological diversity description I. Acta Botanica Brasilica 2018, 32, 656-666. [CrossRef]
- Jiang, N.; Fan, X.; Tian, C. Identification and characterization of leaf-inhabiting fungi from Castanea Plantations in China. Journal of Fungi (Basel, Switzerland) 2021, 7. [CrossRef]
- Liu, X.F.; Tibpromma, S.; Zhang, F.; Xu, J.C.; Chethana, K.W.T.; Karunarathna, S.C.; Mortimer, P.E. Neopestalotiopsis cavernicola sp. nov. from Gem Cave in Yunnan Province, China. Phytotaxa 2021, 512, 1-27. [CrossRef]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.C.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 2018, 93, 1-160. [CrossRef]
- Song, Y.; Geng, K.; Zhang, B.; Hyde, K.D.; Zhao, W.S.; Wei, J.G.; Kang, J.C.; Wang, Y. Two new species of Pestalotiopsis from Southern China. Phytotaxa 2013, 126, 22-30. [CrossRef]
- Peng, C.; Crous, P.W.; Jiang, N.; Fan, X.; Liang, Y.; Tian, C. Diversity of Sporocadaceae (pestalotioid fungi) from rosa in China. Persoonia 2022, 49, 201-260. [CrossRef]
- Ma, X.Y.; Maharachchikumbura, S.S.N.; Chen, B.W.; Hyde, K.D.; McKenzie, E.H.C.; Chomnunti, P.; Kang, J.C. Endophytic pestalotioid taxa in Dendrobium orchids. Phytotaxa 2019, 419, 268-286. [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Le Roux, J.J.; Richardson, D.M.; Strasberg, D.; Shivas, R.G.; Alvarado, P.; Edwards, J.; Moreno, G.; Sharma, R.; et al. Fungal Planet description sheets: 371-399. Persoonia 2015, 35, 264-327. [CrossRef]
- Konta, S.; Tibpromma, S.; Karunarathna, S.C.; Samarakoon, M.C.; Steven, L.S.; Mapook, A.; Boonmee, S.; Senwanna, C.; Balasuriya, A.; Eungwanichayapant, P.D.; et al. Morphology and multigene phylogeny reveal ten novel taxa in Ascomycota from terrestrial palm substrates (Arecaceae) in Thailand. Mycosphere 2023, 14, 107-152. [CrossRef]
- Ul Haq, I.; Ijaz, S.; Khan, N.A. Genealogical concordance of phylogenetic species recognition-based delimitation of Neopestalotiopsis species associated with leaf spots and fruit canker disease affected guava plants. Pakistan Journal of Agricultural Sciences 2021, 58, 1301-1313. [CrossRef]
- Zhang, Z.; Liu, R.; Liu, S.; Mu, T.; Zhang, X.; Xia, J. Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys 2022, 88, 171-192. [CrossRef]
- Freitas, E.F.S.; Da Silva, M.; Barros, M.V.P.; Kasuya, M.C.M. Neopestalotiopsis hadrolaeliae sp. nov., a new endophytic species from the roots of the endangered orchid Hadrolaelia jongheana in Brazil. Phytotaxa 2019, 416, 211-220. [CrossRef]
- Huanaluek, N.; Jayawardena, R.S.; Maharachchikum-Bura, S.S.N.; Harishchandra, D.L. Additions to pestalotioid fungi in Thailand: Neopestalotiopsis hydeana sp. nov. and Pestalotiopsis hydei sp. nov. Phytotaxa 2021, 479, 23-43. [CrossRef]
- Akinsanmi, O.A.; Nisa, S.; Jeff-Ego, O.S.; Shivas, R.G.; Drenth, A. Dry flower disease of macadamia in Australia caused by Neopestalotiopsis macadamiae sp. nov. and Pestalotiopsis macadamiae sp. nov. Plant Disease 2017, 101, 45-53. [CrossRef]
- Maharachchikumbura, S.S.; Guo, L.D.; Chukeatirote, E.; Hyde, K.D. Improving the backbone tree for the genus Pestalotiopsis; addition of P. steyaertii and P. magna sp. nov. Mycological Progress 2014, 13, 617-624. [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet description sheets: 1042-1111. Persoonia 2020, 44, 301-459. [CrossRef]
- Liu, F.; Bonthond, G.; Groenewald, J.Z.; Cai, L.; Crous, P.W. Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Studies in Mycology 2019, 92, 287-415. [CrossRef]
- Silverio, M.L.; Calvacanti, M.A.d.Q.; Silva, G.A.d.; Oliveira, R.J.V.d.; Bezerra, J.L. A new epifoliar species of Neopestalotiopsis from Brazil. Agrotropica 2016, 28, 151-158.
- Crous, P.W.; Summerell, B.A.; Swart, L.; Denman, S.; Taylor, J.E.; Bezuidenhout, C.M.; Palm, M.E.; Marincowitz, S.; Groenewald, J.Z. Fungal pathogens of Proteaceae. Persoonia 2011, 27, 20-45. [CrossRef]
- Chaiwan, N.; Jeewon, R.; Pem, D.; Jayawardena, R.S.; Nazurally, N.; Mapook, A.; Promputtha, I.; Hyde, K.D. Fungal species from Rhododendron sp.: Discosia rhododendricola sp. nov, Neopestalotiopsis rhododendricola sp. nov. and Diaporthe nobilis as a new host record. Journal of Fungi (Basel, Switzerland) 2022, 8. [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D. Additions to Pestalotiopsis in Taiwan. Mycosphere 2018, 9, 999-1013. [CrossRef]
- Santos, J.; Hilario, S.; Pinto, G.; Alves, A. Diversity and pathogenicity of pestalotioid fungi associated with blueberry plants in Portugal, with description of three novel species of Neopestalotiopsis. European Journal of Plant Pathology 2022, 162, 539-555. [CrossRef]
- Fiorenza, A.; Gusella, G.; Aiello, D.; Polizzi, G.; Voglmayr, H. Neopestalotiopsis siciliana sp. nov. and N. rosae causing stem lesion and dieback on avocado plants in Italy. Journal of Fungi 2022, 8. [CrossRef]
- Jayawardena, R.S.; Liu, M.; Maharachchikumbura, S.S.N.; Zhang, W.; Xing, Q.; Hyde, K.D.; Nilthong, S.; Li, X.; Yan, J. Neopestalotiopsis vitis sp. nov. causing grapevine leaf spot in China. Phytotaxa 2016, 258, 63-74. [CrossRef]
- Liu, X.F.; Tibpromma, S.; Hughes, A.C.; Chethana, K.W.T.; Wijayawardene, N.N.; Dai, D.Q.; Du, T.Y.; Elgorban, A.M.; Stephenson, S.L.; Suwannarach, N.; et al. Culturable mycota on bats in central and southern Yunnan Province, China. Mycosphere 2023, 14, 497-662. [CrossRef]
- Liu, F.; Hou, L.w.; Raza, M.; Cai, L. Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Scientific Reports 2017, 7. [CrossRef]
- Gu, R.; Bao, D.F.; Shen, H.W.; Su, X.J.; Li, Y.X.; Luo, Z.L. Endophytic Pestalotiopsis species associated with Rhododendron in Cangshan Mountain, Yunnan Province, China. Frontiers in Microbiology 2022, 13. [CrossRef]
- Crous, P.W.; Boers, J.; Holdom, D.; Osieck, E.R.; Steinrucken, T.V.; Tan, Y.P.; Vitelli, J.S.; Shivas, R.G.; Barrett, M.; Boxshall, A.G.; et al. Fungal Planet description sheets: 1383–1435. Persoonia 2022, 48, 261-371. [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.W.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lucking, R.; et al. Fungal diversity notes 111-252-taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 2015, 75, 27-274. [CrossRef]
- Chaiwan, N.; Wanasinghe, D.N.; Mapook, A.; Jayawardena, R.S.; Norphanphoun, C.; Hyde, K.D. Novel species of Pestalotiopsis fungi on Dracaena from Thailand. Mycology 2020, 11, 306-315. [CrossRef]
- Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Ariyawansa, H.A.; Bhat, D.J.; Boonmee, S.; Maharachchikumbura, S.S.N.; McKenzie, E.H.C.; Phookamsak, R.; Phukhamsakda, C.; et al. Fungal diversity notes 1-110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 2015, 72, 1-197. [CrossRef]
- Silva, N.; Maharachchikumbura, S.; Thambugala, K.; Bhat, D.J.; Hyde, K.D. Morpho-molecular taxonomic studies reveal a high number of endophytic fungi from Magnolia candolli and M. garrettii in China and Thailand. Mycosphere 2021, 12, 163–237.
- Yan, M.Z.; Maharachchikumbura, S.; Tian, Q.; Hyde, K.D. Pestalotiopsis species on ornamental plants in Yunnan Province, China. Sydowia -Horn 2013, 65, 113–128.
- Maharachchikumbura, S.S.N.; Chukeatirote, E.; Guo, L.D.; Crous, P.W.; Mckenzie, E.H.C.; Hyde, K.D. Pestalotiopsis species associated with Camellia sinensis (tea). Mycotaxon 2013, 123, 47-61(15). [CrossRef]
- Monteiro, P.; Gonçalves, M.F.M.; Pinto, G.; Silva, B.; Martín-García, J.; Diez, J.J.; Alves, A. Three novel species of fungi associated with pine species showing needle blight-like disease symptoms. European Journal of Plant Pathology 2022, 162, 183-202. [CrossRef]
- Hyde, K.D.; Jeewon, R.; Chen, Y.-J.; Bhunjun, C.S.; Calabon, M.S.; Jiang, H.-B.; Lin, C.-G.; Norphanphoun, C.; Sysouphanthong, P.; Pem, D.; et al. The numbers of fungi: is the descriptive curve flattening? Fungal Diversity 2020, 103, 219-271. [CrossRef]
- Das, K.; Lee, S.Y.; Jung, H.Y. Pestalotiopsis kaki sp. nov., a Novel species isolated from persimmon tree (Diospyros kaki) Bark in Korea. Mycobiology 2020, 49, 54-60. [CrossRef]
- Chen, Y.Y.; Maharachchikumbura, S.S.N.; Liu, J.K.; Hyde, K.D.; Nanayakkara, R.R.; Zhu, G.S.; Liu, Z.Y. Fungi from Asian Karst formations I. Pestalotiopsis photinicola sp nov., causing leaf spots of Photinia serrulata. Mycosphere 2017, 8, 103-110. [CrossRef]
- Harishchandra, D.L.; Aluthmuhandiram, J.V.S.; Yan, J.; Hyde, K.D. Molecular and morpho-cultural characterisation of Neopestalotiopsis and Pestalotiopsis species associated with ornamental and forest plants in China. Lett Appl Microbiol 2020, 73(3), 352-362.
- Zhang, Y.; Maharachchikumbura, S.S.N.; McKenzie, E.H.C.; Hyde, K.D. A novel species of Pestalotiopsis causing leaf spots of Trachycarpus fortunei. Cryptogamie Mycologie 2012, 33, 311-318. [CrossRef]
- Wei, J.G.; Phan, C.K.; Wang, L.; Xu, T.; Luo, J.T.; Sun, X.; Guo, L.D. Pestalotiopsis yunnanensis sp. nov., an endophyte from Podocarpus macrophyllus (Podocarpaceae) based on morphology and ITS sequence data. Mycological Progress 2013, 12, 563-568. [CrossRef]
- Maharachchikumbura, S.S.N.; Guo, L.D.; Liu, Z.Y.; Hyde, K.D. Pseudopestalotiopsis ignota and Ps. camelliae spp. nov associated with grey blight disease of tea in China. Mycological Progress 2016, 15. [CrossRef]
- Catarino, A.d.M.; Hanada, R.E.; de Queiroz, C.A.; Sousa, T.F.; Lima, Í.N.; Gasparotto, L.; da Silva, G.F. First report of Pseudopestalotiopsis dawaina causing spots in Caryota mitis in Brazil. Plant Disease 2020, 104, 989. [CrossRef]
- Nozawa, S.; Ando, K.; Phay, N.; Watanabe, K. Pseudopestalotiopsis dawaina sp. nov. and Ps. kawthaungina sp. nov.: two new species from Myanmar. Mycological Progress 2018, 17, 865-870. [CrossRef]
- Tsai, I.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Ariyawansa, H.A. Molecular phylogeny, morphology and pathogenicity of Pseudopestalotiopsis species on Ixora in Taiwan. Mycological Progress 2018, 17, 941-952. [CrossRef]
- Lateef, A.A.; Sepiah, M.; Bolhassan, M.h. Description of Pseudopestalotiopsis kubahensis sp. nov., a new species of microfungi from Kubah National Park, Sarawak, Malaysia. Current Research in Environmental & Applied Mycology 2015. 5, 376–381.
- Nozawa, S.; Yamaguchi, K.; Yen, L.T.H.; Hop, D.V.; Phay, N.; Ando, K.; Watanabe, K. Identification of two new species and a sexual morph from the genus Pseudopestalotiopsis. Mycoscience 2017, 58, 328-337. [CrossRef]
- Song, Y.; Tangthirasunun, N.; Maharachchikumbura, S.S.N.; Jiang, Y.L.; Xu, J.J.; Hyde, K.D.; Wang, Y. Novel Pestalotiopsis species from Thailand point to the rich undiscovered diversity of this chemically creative genus. Cryptogamie Mycologie 2014, 35, 139-149. [CrossRef]
- Miller, M.A.; Pfeiffer, W.T.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 2010, 1-8. [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572-1574. [CrossRef]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1-40. [CrossRef]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665-2681. [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 2006, 23, 254-267. [CrossRef]
- Pornsuriya, C.; Chairin, T.; Thaochan, N.; Sunpapao, A. Identification and characterization of Neopestalotiopsis fungi associated with a novel leaf fall disease of rubber trees (Hevea brasiliensis) in Thailand. Journal of Phytopathology 2020, 168, 416-427. [CrossRef]
- Khoo, Y.W.; Tan, H.T.; Khaw, Y.S.; Li, S.F.; Chong, K.P. First report of Neopestalotiopsis cubana causing leaf blight on Ixora chinensis in Malaysia. Plant Disease 2022, 106, 2749-2749. [CrossRef]


| Locus | Primes Name | Sequence (5’to 3’) | PCR procedures | Reference |
|---|---|---|---|---|
| ITS | ITS5 | GGAAGTAAAAGTCGTAACAAGG | 95°C 5 min; 94°C 25 s; 52°C 25 s; 72°C 10 s; repeat 2 to 4 for 35 cycles; 72°C 5 min; 4°C on hold | [35] |
| ITS4 | TCCTCCGCTTATTGATATGC | |||
| tef-1α | EF1-728F | CATCGAGAAGTTCGAGAAGG | 95°C 5 min; 94°C 25 s; 52°C 25 s; 72 °C 10 s (15s); repeat 2 to 4 for 35 cycles; 72°C 5 min; 4°C on hold | [36,37] |
| EF1-526F | GTCGTYGTYATYGGHCAYGT | |||
| EF2 | GGARGTACCAGTSATCATGTT | |||
| tub2 | T1 | AACATGCGTGAGATTGTAAGT | 95°C 5 min; 94°C 25 s; 55°C 25 s; 72°C 15 s; repeat 2 to 4 for 35 cycles; 72°C 5 min; 4°C on hold | [38,39] |
| Bt2b | ACCCTCAGTGTAGTGACCCTTGGC |
| Taxonomic status | Strain No. | Host/Substrate | Origin | Gen Bank Accessions Numbers | References | ||
|---|---|---|---|---|---|---|---|
| ITS | tub 2 | tef-lα | |||||
| Neopestalotiopsis acrostichi | MFLUCC 17-1754T | Acrostichum aureum | Thailand | MK764272 | MK764338 | MK764316 | [41] |
| N. acrostichi | MFLUCC 17-1755 | Acrostichum aureum | Thailand | MK764273 | MK764339 | MK764317 | [41] |
| N. alpapicalis | MFLUCC 17-2544T | Rhizophora mucronata | Thailand | MK357772 | MK463545 | MK463547 | [42] |
| N. alpapicalis | MFLUCC 17-2545 | Rhizophora mucronata | Thailand | MK357773 | MK463546 | MK463548 | [42] |
| N. amomi | HKAS 124563T | Amomum villosum | China | OP498012 | OP752133 | OP653489 | [16] |
| N. amomi | HKAS 124564 | Amomum villosum | China | OP498013 | OP765913 | OP753382 | [16] |
| N. aotearoa | CBS 367.54T | Canvas | New Zealand | KM199369 | KM199454 | KM199526 | [14] |
| N. asiatica | MFLUCC 12-0286T | Prunus dulcis | China | JX398983 | JX399018 | JX399049 | [15] |
| N. australis | CBS 114159T | Telopea sp. | Australia | KM199348 | KM199432 | KM199537 | [15] |
| N. brachiata | MFLUCC 17-1555 | Rhizophora apiculata | Thailand | MK764274 | MK764340 | MK764318 | [41] |
| N. brasiliensis | COAD 2166T | Psidium guajava | Brazil | MG686469 | MG692400 | MG692402 | [43] |
| N. brasiliensis | CFCC 54341 | Castanea mollissima | China | MW166229 | MW218522 | MW199748 | [44] |
| N. camelliae-oleiferae | CSUFTCC81T | Camellia oleifera | China | OK493585 | OK562360 | OK507955 | [17] |
| N. camelliae-oleiferae | CSUFTCC82 | Camellia oleifera | China | OK493586 | OK562361 | OK507956 | [17] |
| N. cavernicola | KUMCC 20-0269T | Cave | China | MW545802 | MW557596 | MW550735 | [45] |
| N. chiangmaiensis | MFLUCC 18–0113 | Pandanus sp. | Thailand | NA | MH412725 | MH388404 | [46] |
| N. chrysea | MFLUCC 12-0261T | Dead leaves | China | JX398985 | JX399020 | JX399051 | [14] |
| N. chrysea | MFLUCC 12-0262 | Dead leaves | China | JX398986 | JX399021 | JX399052 | [14] |
| N. clavispora | MFLUCC 12-0281T | Magnolia sp. | China | JX398979 | JX399014 | JX399045 | [14] |
| N. clavispora | MFLUCC 12-0280 | Magnolia sp. | China | JX398978 | JX399013 | JX399044 | [14] |
| N. cocoes | MFLUCC 15-0152T | Cocos nucifera | Thailand | KX789687 | NA | KX789689 | [41] |
| N. coffeae-arabicae | HGUP4015 | Coffea arabica | China | KF412647 | KF412641 | KF412644 | [47] |
| N. coffeae-arabicae | HGUP4019T | Coffea arabica | China | KF412649 | KF412643 | KF412646 | [47] |
| N. concentrica | CFCC 55162T | Rosa chinensis | China | OK560707 | OM117698 | OM622433 | [48] |
| N. cubana | CBS 600.96T | Leaf litter | Cuba | KM199347 | KM199438 | KM199521 | [15] |
| N. dendrobii | MFLUCC 14-0106T | Dendrobium cariniferum | Thailand | MK993571 | MK975835 | MK975829 | [49] |
| N. dendrobii | MFLUCC 14-0099 | Dendrobium cariniferum | Thailand | MK993570 | MK975834 | MK975828 | [49] |
| N. drenthii | BRIP 72263a | Macadamia integrifolia | Australia | MZ303786 | MZ312679 | MZ344171 | [20] |
| N. drenthii | BRIP 72264aT | Macadamia integrifolia | Australia | MZ303787 | MZ312680 | MZ344172 | [20] |
| N. egyptiaca | CBS 140162T | Mangifera indica | Egypt | KP943747 | KP943746 | KP943748 | [50] |
| N. elaeagni | HGUP10002T | Elaeagnus pungens | China | MW930716 | MZ683391 | MZ203452 | [30] |
| N. elaeidis | MFLUCC 15-0735 | Elaeis guineensis | Thailand | ON650689 | NA | ON734012 | [51] |
| N. ellipsospora | MFLUCC 12-0283T | Dead plant | China | JX398980 | JX399016 | JX399047 | [14] |
| N. eucalyptorum | CBS 147684T | Eucalyptus globulus | Portugal | MW794108 | MW802841 | MW805397 | [18] |
| N. eucalypticola | CBS 264.37T | Eucalyptus globulus | NA | KM199376 | KM199431 | KM199551 | [15] |
| N. fragariae | ZHKUCC 22-0115 | Fragaria x ananassa | China | ON651146 | ON685199 | ON685197 | [32] |
| N. foedans | CGMCC 3.9123T | Mangrove plant | China | JX398987 | JX399022 | JX399053 | [14] |
| N. foedans | CGMCC 3.9178 | Neodypsis decaryi | China | JX398989 | JX399024 | JX399055 | [14] |
| N. formicarum | CBS 362.72T | Dead ant | Cuba | KM199358 | KM199455 | KM199517 | [15] |
| N. formicarum | CBS 115.83 | Plant debris | Cuba | KM199344 | KM199444 | KM199519 | [15] |
| N. guajavae | FMBCC 11.1T | Psidium guajava | Pakistan | MF783085 | MH460871 | MH460868 | [52] |
| N. guajavicola | FMBCC 11.4T | Psidium guajava | Pakistan | MH209245 | MH460873 | MH460870 | [52] |
| N. haikouensis | SAUCC212271T | Ilexchinensis sp. | China | OK087294 | OK104870 | OK104877 | [53] |
| N. hadrolaeliae | COAD 2637T | Hadrolaelia jongheana | Brazil | MK454709 | MK465120 | MK465122 | [54] |
| N. hispanica | CBS 147686T | Eucalyptus globulus | Portugal | MW794107 | MW802840 | MW805399 | [18] |
| N. honoluluana | CBS 114495T | Telopea sp. | USA | KM199364 | KM199457 | KM199548 | [15] |
| N. hydeana | MFLUCC 20-0132T | Artocarpus heterophyllus | Thailand | MW266069 | MW251119 | MW251129 | [55] |
| N. hyperici | HKAS 124561 | Hypericum monogynum | China | OP498010 | OP765908 | OP713768 | [16] |
| N. iberica | CBS 147688T | Eucalyptus globulus | Portugal | MW794111 | MW802844 | MW805402 | [18] |
| N. javaensis | CBS 257.31T | Cocos nucifera | Indonesia | KM199357 | KM199437 | KM199543 | [15] |
| N. lusitanica | CBS 147690T | Eucalyptus globulus | Portugal | MW794110 | MW802843 | MW805406 | [18] |
| N. longiappendiculata | CBS 147692T | Eucalyptus globulus | Portugal | MW794112 | MW802845 | MW805404 | [18] |
| N. macadamiae | BRIP 63737cT | Macadamia integrifolia | Australia | KX186604 | KX186654 | KX186627 | [56] |
| N. macadamiae | BRIP 63742a | Macadamia integrifolia | Australia | KX186599 | KX186657 | KX186629 | [56] |
| N. maddoxii | BRIP 72266aT | Macadamia integrifolia | Australia | MZ303782 | MZ312675 | MZ344167 | [56] |
| N. magna | MFLUCC 12-0652T | Pteridium sp. | France | KF582795 | KF582793 | KF582791 | [57] |
| N. mesopotamica | CBS 336.86T | Pinus brutia | Iraq | KM199362 | KM199441 | KM199555 | [15] |
| N. mesopotamica | CBS 299.74 | Eucalyptus sp. | Turkey | KM199361 | KM199435 | KM199541 | [15] |
| N. mianyangensis | HKAS 123211 | Paeonia suffruticosa | China | OP546681 | OP672161 | OP723490 | [31] |
| N. musae | MFLUCC 15-0776T | Musa sp. | Thailand | KX789683 | KX789686 | KX789685 | [41] |
| N. natalensis | CBS 138.41T | Acacia mollissima | South Africa | KM199377 | KM199466 | KM199552 | [15] |
| N. nebuloides | BRIP 66617T | Sporobolus elongatus | Australia | MK966338 | MK977632 | MK977633 | [58] |
| N. olumideae | BRIP 72273aT | Macadamia integrifolia | Australia | MZ303790 | MZ312683 | MZ344175 | [20] |
| N. paeoniea | CBS 318.74 | Anacardium occidentale | Nigeria | MH554031 | MH554707 | NA | [59] |
| N. paeonia-suffruticosa | HKAS 123212T | Paeonia suffruticosa | China | OP082292 | OP235980 | OP204794 | [31] |
| N. pernambucana | URM 7148-01T | Vismia guianensis | Brazil | KJ792466 | NA | KU306739 | [60] |
| N. perukae | FMBCC11.3T | Guava | Pakistan | MH209077 | MH460876 | MH523647 | [52] |
| N. petila | MFLUCC 17-1737 | Rhizophora mucronata | Thailand | MK764275 | MK764341 | MK764319 | [41] |
| N. petila | MFLUCC 17-1738T | Rhizophora mucronata | Thailand | MK764276 | MK764342 | MK764320 | [41] |
| N. phangngaensis | MFLUCC 18-0119T | Pandanus sp. | Thailand | MH388354 | MH412721 | MH388390 | [46] |
| N. piceana | CBS 254.32 | Cocos nucifera | Indonesia | KM199372 | KM199452 | KM199529 | [15] |
| N. piceana | CBS 394.48T | Picea sp. | UK | KM199368 | KM199453 | KM199527 | [15] |
| N. photiniae | MFLUCC 22-0129T | Photinia serratifolia | China | OP498008 | OP752131 | OP753368 | [16] |
| N. protearum | CBS 114178T | Leucospermum cuneiforme | Zimbabwe | JN712498 | KM199463 | LT853201 | [61] |
| N. psidii | FMBCC 11.2T | Psidium guajava | Pakistan | MF783082 | MH477870 | MH460874 | [52] |
| N. rhapidis | GUCC 21501T | Rhododendron simsii | China | MW931620 | MW980441 | MW980442 | [34] |
| N. rhizophorae | MFLUCC 17-1551T | Rhizophora mucronata | Thailand | MK764277 | MK764343 | MK764321 | [41] |
| N. rhizophorae | MFLUCC 17 1550 | Rhizophora mucronata | Thailand | MK764278 | MK764344 | MK764322 | [41] |
| N. rhododendri | GUCC 21504T | Rhododendron simsii | China | MW979577 | MW980443 | MW980444 | [34] |
| N. rhododendricola | KUN-HKAS-123204T | Rhododendron sp. | China | OK283069 | OK274147 | OK274148 | [62] |
| N. rosae | CBS 101057T | Rosa sp. | New Zealand | KM199359 | KM199429 | KM199523 | [15] |
| N. rosicola | CFCC 51992T | Rosa chinensis | China | KY885239 | KY885245 | KY885243 | [63] |
| N. rosicola | CFCC 51993 | Rosa chinensis | China | KY885240 | KY885246 | KY885244 | [63] |
| N. samarangensis | CBS 115451 | Unidentified Tree | China | KM199365 | KM199447 | KM199556 | [15] |
| N. saprophytica | MFLUCC 12-0282T | Magnolia sp. | China | JX398982 | JX399017 | JX399048 | [15] |
| N. scalabiensis | CAA 1029T | Vaccinium corymbosum | Portugal | MW969748 | MW934611 | MW959100 | [64] |
| N. sichuanensis | CFCC 54338T | Castanea mollissima | China | MW166231 | MW218524 | MW199750 | [44] |
| N. siciliana | AC46 | Persea americana | Italy | ON117813 | ON209162 | ON107273 | [65] |
| N. sonneratae | MFLUCC 17-1744 | Sonneronata alba | Thailand | MK764279 | MK764345 | MK764323 | [41] |
| N. sonneratae | MFLUCC 17-1745T | Sonneronata alba | Thailand | MK764280 | MK764346 | MK764324 | [41] |
| N. steyaertii | IMI 192475T | Eucalyptus viminalis | Australia | KF582796 | KF582794 | KF582792 | [15] |
| N. surinamensis | CBS 450.74T | Soil under Elaeis guineensis | Suriname | KM199351 | KM199465 | KM199518 | [15] |
| N. subepidermalis | CFCC 55160 | Rosa chinensis | China | OK560699 | OM117690 | OM622425 | [48] |
| N. suphanburiensis | MFLUCC 22-0126T | Unknown | Thailand | OP497994 | OP752135 | OP753372 | [16] |
| N. terricola | HKAS 123213 | Paeonia suffruticosa | China | OP082294 | OP235982 | OP204796 | [31] |
| N. thailandica | MFLUCC 17-1730T | Rhizophora mucronata | Thailand | MK764281 | MK764347 | MK764325 | [41] |
| N. thailandica | MFLUCC 17-1731 | Rhizophora mucronata | Thailand | MK764282 | MK764348 | MK764326 | [41] |
| N. umbrinospora | MFLUCC 12-0285T | Unidentified plant | China | JX398984 | JX399019 | JX399050 | [14] |
| N. vaccinii | CAA 1059T | Vaccinium corymbosum | Portugal | MW969747 | MW934610 | MW959099 | [64] |
| N. vacciniicola | CAA 1055T | Vaccinium corymbosum | Portugal | MW969751 | MW934614 | MW959103 | [64] |
| N. vheenae | BRIP 72293aT | Macadamia integrifolia | Australia | MZ303792 | MZ312685 | MZ344177 | [20] |
| N. vitis | MFLUCC 15-1265T | Vitis vinifera cv. “Summer black” | China | KU140694 | KU140685 | KU140676 | [66] |
| N. vitis | MFLUCC 15-1270 | Vitis vinifera cv. “Kyoho” | China | KU140699 | KU140690 | KU140681 | [66] |
| N. xishuangbannaensis | KUMCC 21-0424T | Kerivoula hardwickii (Bat) | China | ON426865 | OR025934 | OR025973 | [67] |
| N. xishuangbannaensis | KUMCC 21-0425 | Kerivoula hardwickii (Bat) | China | ON426866 | OR025935 | OR025974 | [67] |
| N. zakeelii | BRIP 72282aT | Macadamia integrifolia | Australia | MZ303789 | MZ312682 | MZ344174 | [20] |
| N. zimbabwana | CBS 111495T | Leucospermum cunciforme | Zimbabwe | NA | KM199456 | KM199545 | [15] |
| N. zingiber | GUCC 21001T | Zingiber officinale | China | MW930715 | MZ683390 | MZ683389 | [34] |
| Neopestalotiopsis sp.2 | CFCC 54340 | Castanea mollissima | China | MW166235 | MW218528 | MW199754 | [44] |
| Neopestalotiopsis sp.2 | ZX22B | Castanea mollissima | China | MW166236 | MW218529 | MW199755 | [44] |
| Neopestalotiopsis sp. nov. | GUCC 210003 | Unknown | China | MW930717 | MZ683392 | MZ540914 | [34] |
| Neopestalotiopsis sp.1 | CFCC 54337 | Castanea mollissima | China | MW166233 | MW218526 | MW199752 | [44] |
| Neopestalotiopsis sp.1 | ZX121 | Castanea mollissima | China | MW166234 | MW218527 | MW199753 | [44] |
| Pestalotiopsis adusta | ICMP 6088T | Refrigerator door | Fiji | JX399006 | JX399037 | JX399070 | [14] |
| P. adusta | MFLUCC 10-0146 | Syzygium sp. | Thailand | JX399007 | JX399038 | JX399071 | [14] |
| P. aggestorum | LC6301T | Camellia sinensis | China | KX895015 | KX895348 | KX895234 | [68] |
| P. appendiculata | CGMCC 3.23550T | Rhododendron sp. | China | OP082431 | OP185516 | OP185509 | [69] |
| P. australasiae | CBS 114126T | Knightia sp. | New Zealand | KM199297 | KM199409 | KM199499 | [15] |
| P. australasiae | CBS 11141 | Protea sp. | New Sout Wales | KM199298 | KM199410 | KM199501 | [15] |
| P. australis | CBS 114193T | Grevillea sp. | New South Wales | KM199332 | KM199383 | KM199475 | [15] |
| P. biciliata | CBS 124463T | Platanus×hispanica | Slovakia | KM199308 | KM199399 | KM199505 | [15] |
| P. brachiata | LC2988T | Camellia sp. | China | KX894933 | KX895265 | KX895150 | [14] |
| P. brassicae | CBS 170.26T | Brassica napus | New Zealand | KM199379 | NA | KM199558 | [15] |
| P. camelliae | MFLUCC 12-0277T | Camellia japonica | China | JX399010 | JX399041 | JX399074 | [14] |
| P. camelliae-oleiferae | CSUFTCC08T | Camellia oleifera | China | OK493593 | OK562368 | OK507963 | [17] |
| P. chamaeropis | CBS 186.71T | Chamaerops humilis | Italy | KM199326 | KM199391 | KM199473 | [14] |
| P. chiangmaiensis | MFLUCC 22-0127T | Phyllostachys edulis | Thailand | OP497990 | OP752137 | OP753374 | [16] |
| P. chiaroscuro | BRIP 72970 | Sporobolus natalensis | Australia | OK422510 | OK423752 | OK423753 | [70] |
| P. clavata | MFLUCC 12-0268T | Buxus sp. | China | JX398990 | JX399025 | JX399056 | [14] |
| P. colombiensis | CBS 118553T | Eucalyptus eurograndis | Colombia | KM199307 | KM199421 | KM199488 | [15] |
| P. daliensis | CGMCC 3.23548T | Rhododendron sp. | China | OP082429 | OP185518 | OP185511 | [69] |
| P. diploclisiae | CBS 115587T | Diploclisia glaucescens | China | KM199320 | KM199419 | KM199486 | [15] |
| P. diversiseta | MFLUCC 12-0287T | Rhododendron sp. | China | NR 120187 | JX399040 | JX399073 | [14] |
| P. dracaenae | HGUP4037T | Dracaena fragrans | China | NA | MT598645 | MT598644 | [71] |
| P. dracaenicola | MFLUCC 18-0913T | Dracaena sp. | Thailand | MN962731 | MN962733 | MN962732 | [72] |
| P. dracontomelon | MFUCC 10-0149T | Dracontomelon dao | Thailand | KP781877 | NA | KP781880 | [73] |
| P. endophytica | MFLUCC 18-0932 | Endophytic on healthy leaves of Magnolia candoll | Thailand | NR 172439 | NA | MW417119 | [74] |
| P. ericacearum | IFRDCC 2439T | Rhododendron delavayi | China | KC537807 | KC537821 | KC537814 | [75] |
| P. etonensis | BRI P 66615 | Sporobolus jacquemontii | Australia | MK966339 | MK977634 | MK977635 | [58] |
| P. formosana | NTUCC 17-009T | dead grass | China | MH809381 | MH809385 | MH809389 | [63] |
| P. furcata | MFLUCC 12-0054T | Camellia sinensis | Thailand | JQ683724 | JQ683708 | JQ683740 | [76] |
| P. fusoidea | CGMCC 3.23545T | Endophytic in fresh Rhododendron delavayi leaves. | China | OP082427 | OP185519 | OP185512 | [69] |
| P. grevilleae | CBS 114127T | Grevillea sp. | Australia | KM199300 | KM199407 | KM199504 | [15] |
| P. hawaiiensis | CBS 114491T | Leucospermum sp. | Hawaii | KM199339 | KM199428 | KM199514 | [15] |
| P. hispanica | CBS 115391T | Protea ‘Susara’ | Spain | MH553981 | MH554640 | MH554399 | [59] |
| P. hydei | MFLUCC 20-0135T | Litsea Petiolata | Thailand | MW266063 | MW251112 | MW251113 | [55] |
| P. hydei | GUCC 21-0816 | dead twigs | China | OP753660 | OP765909 | OP753383 | [16] |
| P. hollandica | CBS 265.33T | Sciadopitys verticillata | Netherlands | KM199328 | KM199388 | KM199481 | [15] |
| P. humus | CBS 336.97T | Soil | Papua New Guinea | KM199317 | KM199420 | KM199484 | [15] |
| P. hunanensis | CSUFTCC15T | Camellia oleifera | China | OK493599 | OK562374 | OK507969 | [17] |
| P. iberica | CAA1006T | Pinus radiata | Spain | MW732249 | MW759036 | MW759039 | [77] |
| P. inflexa | MFLUCC 12-0270T | Unidentified tree | China | JX399008 | JX399039 | JX399072 | [14] |
| P. intermedia | MFLUCC 12-0259T | Unidentified tree | China | JX398993 | JX399028 | JX399059 | [14] |
| P. jiangxiensis | LC4399T | Eurya sp. | China | KX895009 | KX895341 | KX895227 | [68] |
| P. jinchanghensis | LC6636T | Camellia sinensis | China | KX895028 | KX895361 | KX895247 | [68] |
| P. kandelicola | NCYU 19-0355T | Kandelia candel | China | MT560722 | MT563099 | MT563101 | [78] |
| P. kaki | KNU-PT-1804T | Diospyros kaki | Korea | LC552953 | LC552954 | LC553555 | [79] |
| P. kenyana | CBS 442.67T | Coffea sp. | Kenya | KM199302 | KM199395 | KM199502 | [15] |
| P. kenyana | CBS 911.96 | raw material from agar-agar | NA | KM199303 | KM199396 | KM199503 | [15] |
| P. knightiae | CBS 114138T | Knightia sp. | New Zealand | KM199310 | KM199408 | KM199497 | [15] |
| P. knightiae | CBS 111963 | Knightia sp. | New Zealand | KM199311 | KM199406 | KM199495 | [15] |
| P. linearis | MFLUCC 12-0271 | Trachelospermum sp. | China | JX398992 | JX399027 | JX399058 | [14] |
| P. loeiana | MFLUCC 22-0123 | dead leaves | Thailand | OP497988 | OP713769 | OP737881 | [16] |
| P. lushanensis | LC4344T | Camellia sp. | China | KX895005 | KX895337 | KX895223 | [68] |
| P. macadamiae | BRIP 63738BT | Macadamia integrifolia | Australia | KX186588 | KX186680 | KX186621 | [56] |
| P. malayana | CBS 102220T | Macaranga triloba | Malaysia | KM199306 | KM199411 | KM199482 | [15] |
| P. monochaeta | CBS 144.97T | Quercus robur | Netherlands | KM199327 | KM199386 | KM199479 | [15] |
| P. jesteri | MFLUCC 12-0279T | Fagraea bodenii | China | JX399012 | JX399043 | JX399076 | [14] |
| P. nanjingensis | CSUFTCC16T | Camellia oleifera | China | OK493602 | OK562377 | OK507972 | [17] |
| P. nanningensis | CSUFTCC10T | Camellia oleifera | China | OK493596 | OK562371 | OK507966 | [17] |
| P. neolitseae | NTUCC 17-011T | Neolitsea villosa | Taiwan | MH809383 | MH809387 | MH809391 | [63] |
| P. oryzae | CBS 353.69T | Oryza sativa | Denmark | KM199299 | KM199398 | KM199496 | [15] |
| P. papuana | CBS 331.96T | Coastal soil | Papua New Guinea | KM199321 | KM199413 | KM199491 | [15] |
| P. photinicola | GZCC 16-0028T | Photinia serrulata | China | KY092404 | KY047663 | KY047662 | [80] |
| P. rhizophorae | MFLUCC 17-0416T | Rhizophora apiculata | Thailand | MK764283 | MK764349 | MK764327 | [41] |
| P. rhodomyrtus | HGUP4230T | Rhodomyrtus tomentosa | China | KF412648 | KF412642 | KF412645 | [47] |
| P. rosarioides | CGMCC 3.23549T | Rhododendron decorum | China | OP082430 | OP185520 | OP185513 | [69] |
| P. rosea | MFLUCC 12-0258T | Pinus sp. | China | JX399005 | JX399036 | JX399069 | [14] |
| P. scoparia | CBS176.25T | Chamaecyparis sp. | NA | KM199330 | KM199393 | KM199478 | [15] |
| P. shandogensis | JZB340038T | unknow | China | MN625275 | MN626729 | MN626740 | [81] |
| P. smilacicola | MFLUCC 22-0125T | Smilax sp. | Thailand | OP497991 | OP762673 | OP753376 | [16] |
| P. suae | CGMCC 3.23546T | Rhododendron delavayi | China | OP082428 | OP185521 | OP185514 | [69] |
| P. telopeae | CBS 114161T | Telopea sp. | Australia | KM199296 | KM199403 | KM199500 | [15] |
| P. telopeae | CBS 114137 | Protea sp. | Australia | KM199301 | KM199469 | KM199559 | [15] |
| P. thailandica | MFLUCC 17-1616T | Rhizophora apiculata | Thailand | MK764285 | MK764351 | MK764329 | [41] |
| P. trachycarpicola | OP068T | Trachycarpus fortunei | China | JQ845947 | JQ845945 | JQ845946 | [82] |
| P. unicolor | MFLUCC 12-0276T | Rhododendron sp. | China | JX398999 | JX399030 | NA | [14] |
| P. verruculosa | MFLUCC 12-0274T | Rhododendron sp. | China | JX398996 | NA | JX399061 | [14] |
| P. yanglingensis | LC4553T | Camellia sinensis | China | KX895012 | KX895345 | KX895231 | [83] |
| Pseudopestalotiopsis ampullacea | LC6618T | Camellia sinensis | China | KX895025 | KX895358 | KX895244 | [68] |
| Ps. annellata | NTUCC 17-030T | Camellia sinensis | China, Taiwan | MT322087 | MT321889 | MT321988 | [21] |
| Ps. avicenniae | MFLUCC 17-0434T | Avicennia marina | Thailand | MK764287 | MK764353 | MK764331 | [41] |
| Ps. camelliae | CGMCC 3.9192 | Camellia sinensis | China | NA | KU562851 | KU562850 | [84] |
| Ps. camelliae-sinensis | NTUCC 18-031 | Camellia sinensis | China, Taiwan | MT322047 | MT321849 | MT321948 | [21] |
| Ps. camelliae-sinensis | LC3490T | Camellia sinensis | China | KX894985 | KX895316 | KX895202 | [68] |
| Ps. chinensis | NTUCC 18-066 | Camellia sinensis | China, Taiwan | MT322083 | MT321885 | MT321984 | [21] |
| Ps. chinensis | LC3011T | Camellia sinensis | China | KX894937 | KX895269 | KX895154 | [68] |
| Ps. chinensis | NTUCC 18-038 | Camellia sinensis | China, Taiwan | MT322055 | MT321857 | MT321956 | [21] |
| Ps. cocos | CBS 272.29T | Cocos nucifera | Java | MH855069 | KM199467 | KM199553 | [15] |
| Ps. celtidis | GUCC 21599T | Celtis sinensis | China | OL423535 | OL439010 | OL439012 | [33] |
| Ps. curvatispora | MFLUCC 17-1723 | Rhizophora mucronata | Thailand | MK764290 | MK764356 | MK764334 | [41] |
| Ps. curvatispora | MFLUCC 17-1722T | Rhizophora mucronata | Thailand | MK764289 | MK764355 | MK764333 | [41] |
| Ps. dawaina | INPA 2912 | Caryota mitis | Brazil | MN096659 | MN151310 | MN151308 | [85] |
| Ps. dawaina | MM14-F0015T | unknown | Dawei, Myanmar | LC324750 | LC324751 | LC324752 | [86] |
| Ps. gilvanii | INPA 2913T | Paullinia cupana | Brazil | MN385951 | MN385954 | MN385957 | [29] |
| Ps. hydeae | NTUCC 17-003.1 | Diospyros sp. | China, Taiwan | MG816313 | MG816323 | MG816333 | [87] |
| Ps. ignota | NN 42909T | Camellia sinensis | China | KU500020 | NA | KU500016 | [84] |
| Ps. indica | CBS 459.78T | Hibiscus rosa-sinensis | India | KM199381 | KM199470 | KM199560 | [15] |
| Ps. indocalami | GUCC 21600T | Indocalamus tessellatus | China | OL423536 | OL439011 | OL439013 | [33] |
| Ps. ixorae | NTUCC 17-001.1T | Lxora sp. | NA | MG816316 | MG816326 | MG816336 | [87] |
| Ps. kawthaungina | MM14F0083T | unknown | Kawthaung, Myanmar | LC324753 | LC324754 | LC324755 | [86] |
| Ps. kubahensis | UMAS-KUB-P20T | Macaranga sp. | Sarawak, Malaysia | MG818971 | NA | NA | [88] |
| Ps. myanmarina | NBRC 112264T | Averrhoa carambola | Dawei, Myanmar | LC114025 | LC114045 | LC114065 | [89] |
| Ps. rhizophorae | MFLUCC 17-1560T | Rhizophora apiculata | Thailand | MK764291 | MK764357 | MK764335 | [41] |
| Ps. simitheae | KUMCC 17-0255 | Magnolia candolli | China | MW244023 | MW602387 | MW273930 | [74] |
| Ps. simitheae | MFLUCC12-0121T | Pandanus odoratissimus | Thailand | KJ503812 | KJ503815 | KJ503818 | [90] |
| Ps. solicola | CBS 386.97T | soil in tropical forest | Papua New Guinea | MH554039 | MH554715 | MH554474 | [59] |
| Ps. Taiwanensis | NTUCC 17-002.1T | Ixora sp. | China, Taiwan | MG816319 | MG816329 | MG816339 | [87] |
| Ps. Thailandica | MFLUCC 17-1724T | Rhizophora mucronata | Thailand | MK764292 | MK764358 | MK764336 | [41] |
| Ps. Thailandica | MFLUCC 17-1725 | Rhizophora mucronata | Thailand | MK764293 | MK764359 | MK764337 | [41] |
| Ps. Theae | MFLUCC 12-0055T | Camellia sinensis | Thailand | JQ683727 | JQ683711 | JQ683743 | [14] |
| Ps. vietnamensis | NBRC 112252 | Fragaria sp. | Hue, Vietnam | LC114034 | LC114054 | LC114074 | [89] |
| Species | Isolate Number | Conidial Size (μm) | Apical Appendages (μm) | Basal Appendage | |
|---|---|---|---|---|---|
| Number | Length | ||||
| N. baotingensis | SX41-0706 | 18-26×5-7.2 | 2-4 | 3-30.5 | 2.5-10 |
| N. Saprophytica | MFLUCC 12-0282 | 22–30×5–6 | 2-4 | 23–35 | 4-7 |
| N. brachiata | SX31-0706 | 18.5-25.3×5.5-7.5 | 1-3 | 3.7-38.7 | 2.5-8 |
| N. brachiata | MFLUCC 17-1555 | 18.5–25×5.5–6 | 1-3 | 9.5–33 | 4–9 |
| N. coffeae-arabicae | BL32-0708 | 19.2-25.3×5.3-7 | 2-4 | 10.9-22.6 | 1.4-5.4 |
| N. coffeae-arabicae | LF51-0709 | 17.8-24.2×5-7 | 2-4 | 6.6-21.6 | 2.5-6.8 |
| N. coffeae-arabicae | NM42-0706 | 17.5-23.8×5.8-7.8 | 2-4 | 12.7-31 | 2.7-9.2 |
| N. coffeae-arabicae | HGUP4019 | 16-20×5-7 | 2-4 | 11-16 | 3-5 |
| N. cubana | MH51-0708 | 19.7-30×5-6.8 | 2-4 | 15.5-32.2 | 4-7.5 |
| N. cubana | YX112-0708 | 21-29×5.6-7.3 | 2-4 | 18.7-36.5 | 3.3-10.3 |
| N. cubana | CBS 600.96 | 20-25×8-9.5 | 2-4 | 21-27 | 4-7 |
| N. oblatespora | YJ11-0708 | 18-23.2×5.5-7 | 2-4 | 10-26.5 | 2-9 |
| N. guajavicola | FMBCC 11.4 | 23.3×6.5 | 2-3 | 21.8 | 4.4 |
| N. olivaceous | LF25-0709 | 21.5-33.8×5.5-7.7 | 2-5 | 9.5-22.5 | (0)1.2-4.8 |
| N. amomi | HKAS 124563 | 18-30×4-7 | 2-3 | 7-17 | 2-5 |
| N. zingiberis | GUCC 21001 | 21‒31×6‒9.5 | 1-3 | 12-15 | 0-6 |
| N. oxyphylla | LF55-0709 | 18.8-23.5×5.3-7.0 | 2-4 | 10-25.3 | 2.5-8 |
| N. aotearoa | CBS 367.54 | 21–28×6.5–8.5 | 2-3 | 5-12 | 1.5-4 |
| N. elaeidis | MFLUCC 15-0735 | 10-20×3-7 | 2-3 | 10-20 | (0)2-6 |
| N. petila | MFLUCC 17-1738 | 21–26.5×6–7 | 2-3 | 22-29 | 3-8 |
| N. piceana | CBS 394.48 | 19.5–25×7.5–9 | 3 | 21-31 | 6-23 |
| N. samarangensis | MFLUCC 12-0233 | 18–21×6.5–7.5 | 3 | 12–18 | 3.5–5.2 |
| N. rosicola | NM47-0708 | 16.9-24.6×5.5-7.2 | 2-4 | 10-25 | 1.7-7 |
| N. rosicola | CFCC 51992 | 20.2-25.5×5.5-8 | 2-4 | 17-22.8 | 2-9.5 |
| N. vaccinii | JR31-0709 | 14.5-20.6×5.5-7.4 | 2-3 | 10-22.5 | 1.3-5.1 |
| N. vaccinii | CAA1059 | 20.9×6.4 | 2-3 | 8.9–25.3 | 1.7–6.6 |
| N. hispanica | CBS 147686 | 24.4–25.3×7.2–7.8 | 3-4 | 19.5–22.6 | 5.1–15.5 |
| N. wuzhishanensis | YX116-0708 | 19.5-26.5×4.5-6.3 | 1-3 | 9-20.8 | (0)0.8-3.8 |
| N. mianyangensis | UESTCC 22.0006 | 19–23×5.5–7 | 3 | 5.5–11 | 3–4 |
| N. yongxunensis | YX101-0708 | 18.2-25.5×5.8-7.5 | 2-4 | 10.5-24.7 | 1.7-7 |
| N. dendrobii | MFLUCC 14-0106 | 20.5–23×6.5–7.5 | 2–3 | 5–6.5 | NA |
| N. paeonia-suffruticosa | CGMCC3.23554 | 20–23×9–11 | 3–4 | 22.5–34 | 3.5–7.5 |
| P. hydei | BA11-0708 | 20.3-27.8×4.5-6.6 | 1-3 | 3.4-17.2 | 1-9.5 |
| P. hydei | MFLUCC 20-0135 | 18-35×3-6 | 1-3 | 3-12 | 2-8 |
| Ps. avicenniae | LF48-0709 | 20.8-30.7×5.8-7.9 | 1-3 | 17.2-33.3 | 2-7.8 |
| Ps. avicenniae | MFLUCC 17-0434 | 22.5-26.5×5.5-6 | 1-3 | 15.5–28.5 | 3-4 |
| Ps. myanmarina | JR34-0709 | 25.4-34.8×5.8-7.4 | 2-3 | 18.1-36.9 | 2.7-7 |
| Ps. myanmarina | NBRC 11226 | 31-38.5×6.5-9 | 2-3 | 22.5-38.5 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





