1. Introduction
Effective treatments for a wide range of neurological disorders including epilepsy, neurodegenerative diseases (Dementia, Alzheimer’s, Parkinson’s), and neurodevelopmental disorders such as autism spectrum disorder (ASD), share a common feature: establishing early and accurate diagnoses [
1,
2,
3,
4,
5,
6]. However, this is particularly difficult in the presence of comorbidities, as neurological symptoms can result from many underlying causes [
7]. In addition, not only can many neurological disorders have their root in genetic factors, but exposure to environmental neurotoxicants can also impair cognitive health [
8,
9]. Effective monitoring prior to the symptomatic onset of disease is critical for combating the effects of neurotoxic exposures [
10,
11]. Brain tissue is sometimes obtainable for diagnostic purposes post-onset of symptomatic disease [
12], but brain biopsies are highly invasive and carry risk of potentially fatal complications [
13]. Non-invasive diagnostic biomarkers of brain-related disorders and neurotoxicity are highly sought [
14,
15].
Biofluids such as blood or cerebrospinal fluid (CSF) contain many potential biomarkers that hold promise as non-invasive diagnostics for early disease detection [
16,
17]. One such biomarker is cell-free DNA (cfDNA), which describes extracellular DNA in the bloodstream or fluid surrounding cells or tissues [
18]. CfDNA is believed to be largely generated through passive release by dying cells [
18], but cfDNA can also be actively extruded within extracellular vesicles [
19]. Characteristics of cfDNA including concentration, fragment size, sequence, and epigenetic modifications can all potentially be indicative of disease and tissue-level events [
16]. Currently, cfDNA already has several clinical applications in prenatal testing and cancer detection [
20,
21,
22,
23], but the full diagnostic potential of cfDNA has likely not yet been realized. Understanding what cfDNA profiles are characteristic of specific tissue types and which changes in cfDNA represent deleterious events are needed to expand the diagnostic spectrum of cfDNA. Some progress has already been made in exploring circulating nucleic acids as potential biomarkers of brain-related disorders. For example, alterations in cfDNA have been detected in patients with cancers of the central nervous system [
24], neurodegenerative diseases [
25], epilepsy [
26], and non-psychiatric neurological illness [
27]. Differences in nucleic acid content of extracellular vesicles have been observed in patients with Alzheimer’s Disease and proposed to have potential as biomarkers [
28]. In particular, levels of cfDNA from mitochondrial origin may be indicative of neurological and neurodegenerative disease [
29].
Yet, we still lack a complete picture of what cfDNA fragments are released specifically by brain tissue. Understanding the cfDNA profile of normal, healthy tissues is a key step towards identifying signatures of deleterious changes. Neuron-derived cfDNA has been identified in human plasma, further suggesting the potential of circulating nucleic acids to serve as biomarkers of neurological conditions [
30]. However, identification of novel cfDNA sequences originating from brain tissues in response to brain damage or disease can be challenging in vivo, due to the large proportion of cfDNA contributed from other cell and tissue sources such as white blood cells and erythrocyte progenitors [
31]. In addition, screening of toxicants to identify cfDNA signatures that are predictive, for example, of neurological damage is generally not feasible in human subjects.
Organoids provide a tractable means of examining disease states and screening toxic compounds in a human tissue background, with extensions in the study of individualized medicine [
32]. Organoids are more representative of organ tissues than 2D cell cultures or spheroids, but lack the full complexity of tissues in vivo, which contain cfDNA from numerous tissue sources [
31]. This permits a more isolated view of cell types and may aid identification of biomarkers specific to certain tissues and organs. For these reasons, we chose to examine cfDNA in organoid models. Successful isolation of cfDNA from pancreatic [
33], cardiac [
34], lung, and gastric organoids [
35] has been documented previously. However, outside of these few studies, whether cfDNA is released from additional organoid types remains unexplored.
Here, we isolated and characterized cfDNA from cerebral organoids. Our analyses revealed that cerebral organoids release cfDNA in quantities sufficient for whole-genome sequencing (WGS), droplet-digital PCR (ddPCR), and other downstream assays. Intriguingly, we observed that the recovered cfDNA contained many sequences that were aligned to genes associated with synaptic development and neurodevelopmental disorders. Overall, our study demonstrates the suitability of cerebral organoid systems as a tool for the study of cfDNA. Ultimately, this approach could be used to uncover potential novel cfDNA biomarkers associated with neurodevelopment, neurological disease, and neurotoxicity.
2. Results
2.1. Cerebral Organoids Generated for cfDNA Collection Express Markers of Neural Differentiation
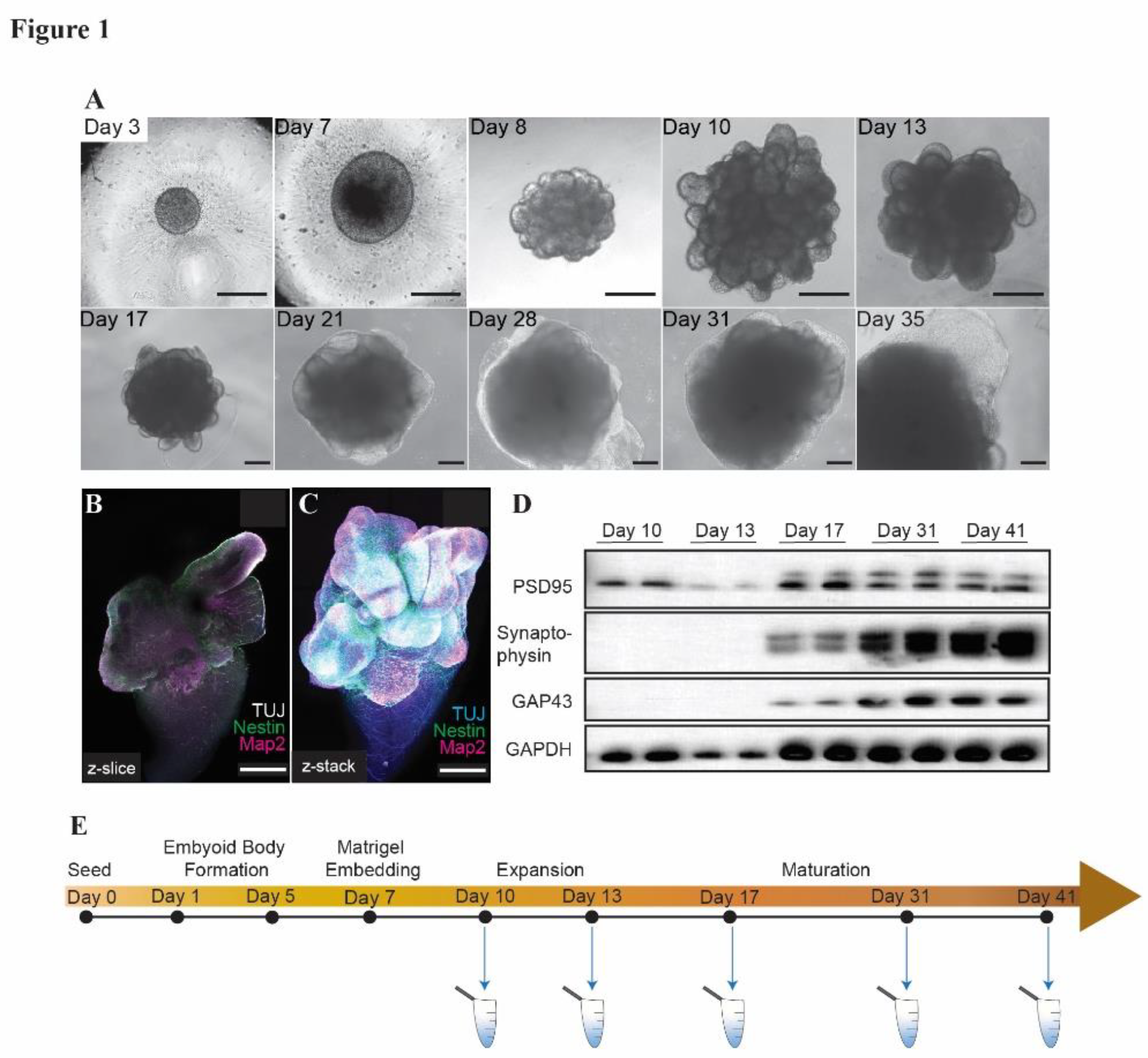
Cerebral organoids were derived from H9 human embryonic stem cells (hESCs) by seeding and differentiating into embryoid bodies, which were then embedded in Matrigel and guided towards neural lineage (
Figure 1A) using a commercially available kit (see Materials and Methods). Immunofluorescence staining of mature organoids revealed cells with colocalization of the neural markers β3-Tubulin and Map2, as well as populations of cells expressing the neural stem cell progenitor marker Nestin (Figure 1B, C). Consistently, we observed expression of proteins associated with neural differentiation as the organoids matured (
Figure 1D). Conditioned media was collected from the organoids at several time points during their growth for cfDNA extraction (
Figure 1E).
2.2. Cerebral Organoids Release cfDNA of Both Mitochondrial and Genomic Origin
We observed that total cfDNA concentration increased between growth day 31 and maturation (day 41) of the organoids (
Figure 2A). To account for changes in cell number during organoid growth, we normalized the concentrations of cfDNA to levels of genomic DNA (gDNA) collected on the same growth days. Normalized cfDNA concentration was highest on growth day 10 (
Figure 2B), indicating that cfDNA output per cell may be higher early in the organoid growth process, a finding we had previously observed in cardiac organoids [
34]. The recovered cfDNA from mature cerebral organoids was comprised primarily of small fragments (100-200 base pairs) (
Figure 2C).
Mitochondrial and nuclear DNA can both contribute to total cfDNA concentration, and the contribution of DNA from each of these sources can be used to further characterize cfDNA [
19]. To assess the abundance of each of these potential sources of cfDNA in our samples, we employed droplet digital PCR (ddPCR) using probes targeting regions within mitochondrial or nuclear-localized genes. We observed that sequences corresponding to cfDNA of mitochondrial origin (mt-cfDNA) were detectable in cfDNA taken from cerebral organoids at several time points during growth (
Figure 2D). Likewise, we were able to detect cfDNA sequences corresponding to gene regions within the nuclear-localized genes β-3 tubulin, nestin, NFATC1, and OCT 3/4 in cfDNA released from mature cerebral organoids (
Figure 2E). These data demonstrate that cerebral organoids release cfDNA in quantities sufficient for detection of specific sequences using ddPCR.
2.3. Whole-Genome Sequencing of Cerebral Organoid-Derived cfDNA Reveals Novel, Consistently Detectible Regions
To gain a more comprehensive understanding of the makeup of cfDNA derived from cerebral organoids, we performed whole-genome sequencing (WGS) of cfDNA isolated on growth days 10, 21, and 41. Surprisingly, this analysis revealed a high fraction of DNA of rodent origin in cfDNA taken from media conditioned by cerebral organoids on growth day 10. This non-human DNA fraction decreased on day 21, and nearly disappeared by day 41, leaving cfDNA of almost all human origin (Supplementary
Figure 1). We suspect that this contaminating rodent DNA in the early and mid- time points originated from the Matrigel in which the organoids are embedded on growth day 7. We therefore decided to focus our subsequent analyses on cfDNA recovered from mature organoids (growth day 41).
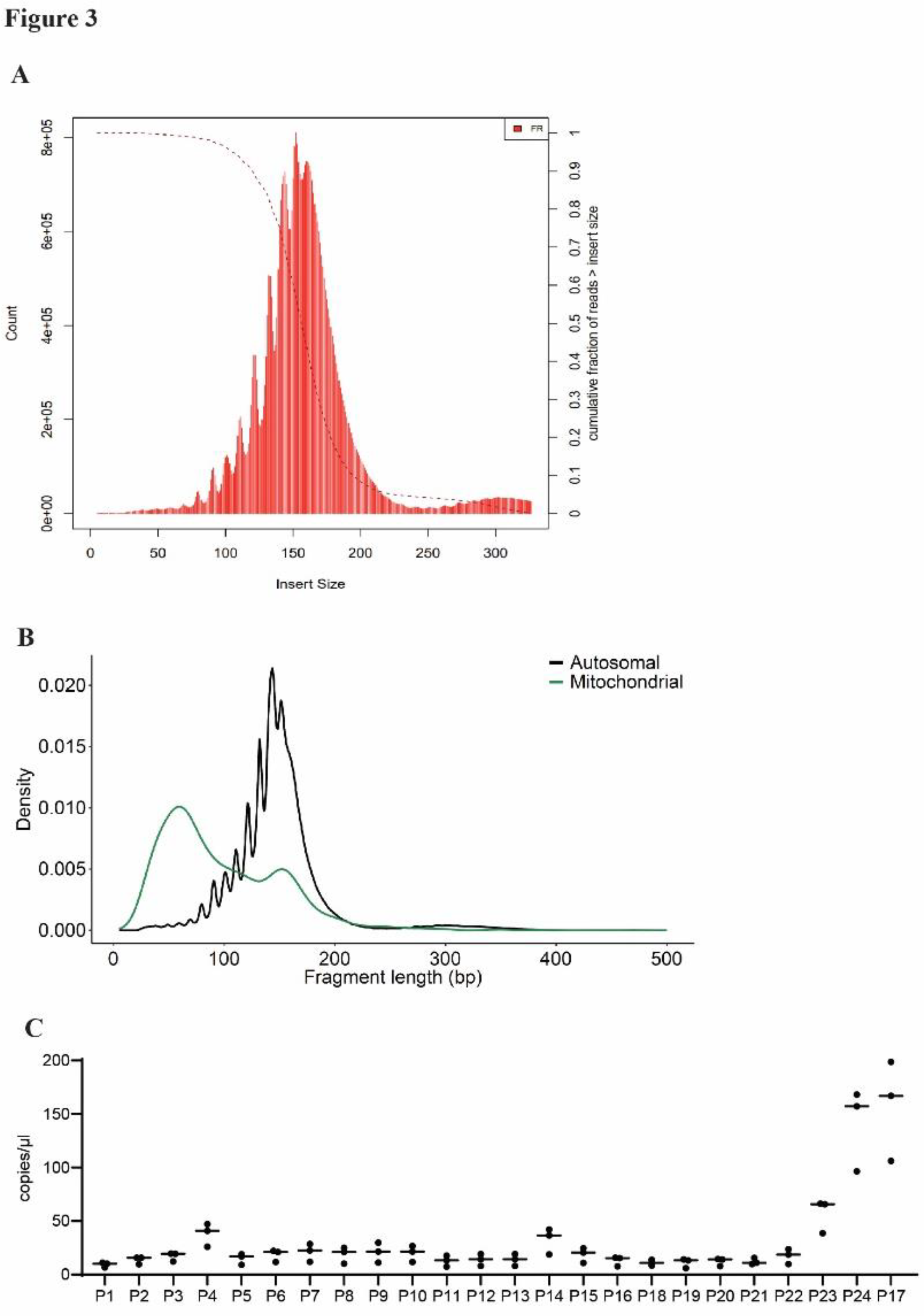
WGS showed the majority of cfDNA fragments isolated from day 41 cerebral organoids to be 100-200 bp long (
Figure 3A), consistent with our previous fragment analysis data. Sequences that aligned to the mitochondrial genome showed a higher density of shorter fragments (<100 bp) as compared to sequences aligning to autosomal regions (
Figure 3B). To confirm the presence of sequenced regions in our cfDNA samples, we designed custom ddPCR probes targeting a selection of cfDNA sequences from a range of FPKM values located across different genes. We were able to consistently detect these sequences in cfDNA from mature cerebral organoids (
Figure 3C). This indicates that novel sequenced reads are present and detectable in cerebral organoid-derived cfDNA using ddPCR.
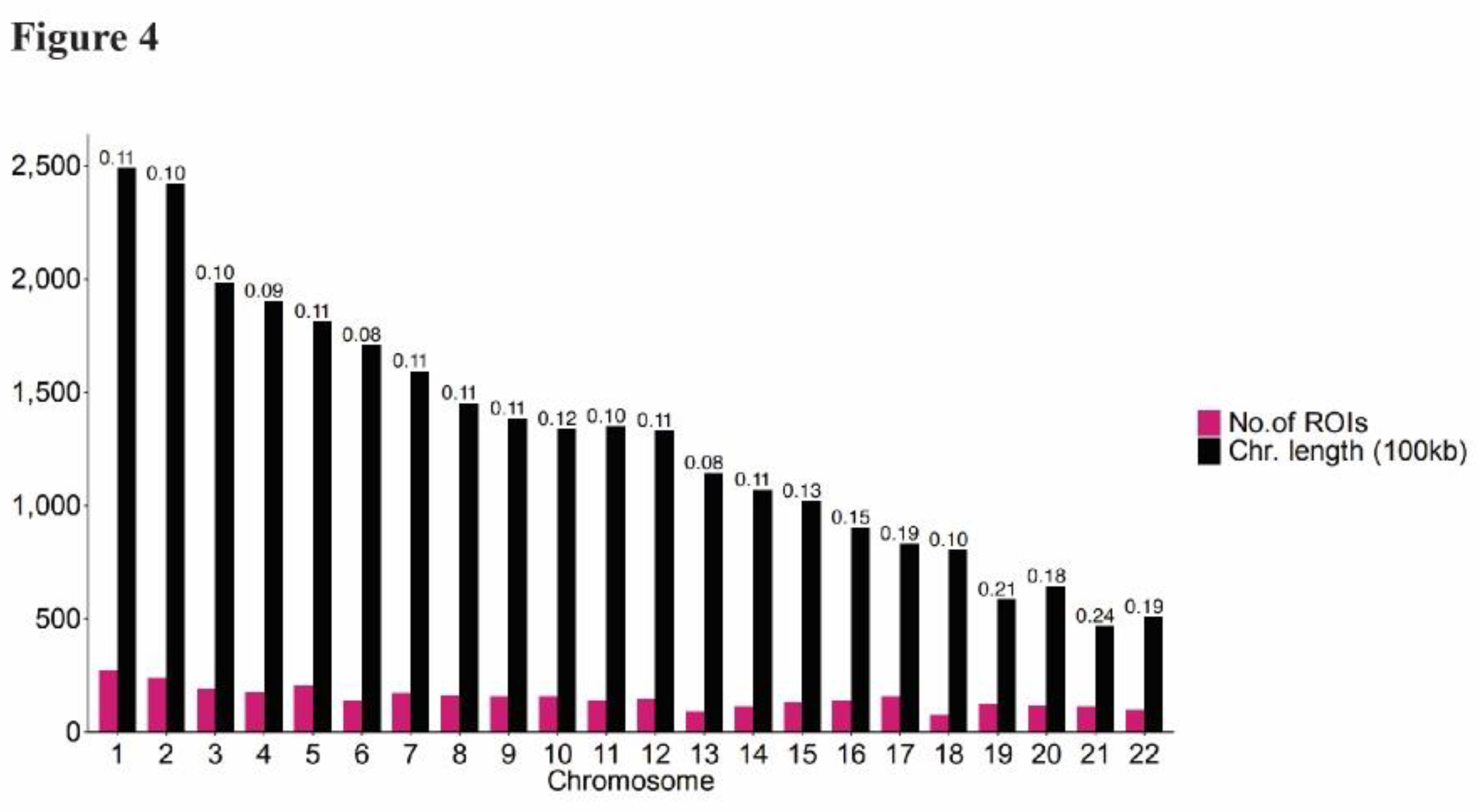
Genome-wide peak calling was performed to identify regions of increased signal in each of the biological replicates. The union of intersecting peaks was classified as regions of interest (ROIs) for downstream analyses (Supplementary
Figure 2). A total of 3,406 ROIs were identified, ranging from 169 to 6,314 bp in length. To examine the distribution of ROIs across the genome, the ratio of peak counts to chromosome length (e.g. number of peaks per 100 kb length) was calculated for each chromosome. (
Figure 4). The majority of peak ratios were <0.21 with a notable exception for chromosome 21, with a ratio of 0.24 (z-score = 2.55).
2.4. Cerebral Organoids Release cfDNA Corresponding to Gene Regions Associated with Brain Development and Neurological Disorders
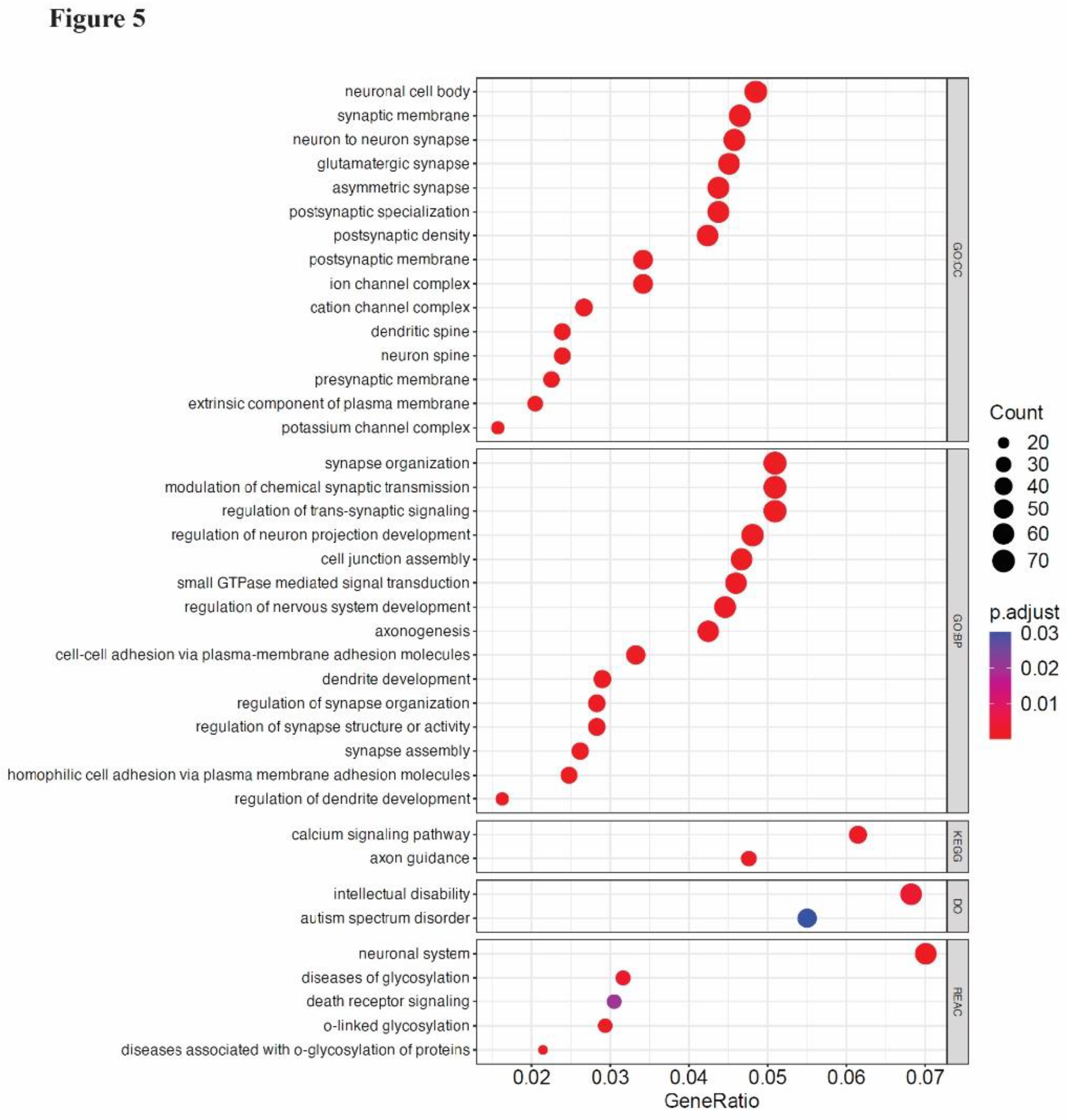
We found that 2,822 unique genes intersected the ROIs. Functional enrichment analysis of these genes including Gene Ontology (GO), Disease Ontology (DO), KEGG, and REACTOME revealed terms related to brain function and development (
Figure 5). Specifically, terms related to neuron and synapse formation were among the most prominent Cellular Component-associated GO terms. Synapse organization, regulation of nervous system development, and axonogenesis were highly represented in Biological Process-associated GO terms. Neuronal system was the most prominent pathway term identified using the REACTOME database [
36]. Additionally, calcium signaling and axon guidance pathways were identified using the KEGG database. Intriguingly, Disease Ontology terms associated with intellectual disability and autism spectrum disorder were also identified. Together, these data suggest that cfDNA released from cerebral organoids maps to gene regions associated with synaptic and neuronal development, and neurodevelopmental disorders. This observation supports our hypothesis that cerebral organoids can be used to study potential cfDNA biomarkers relevant to neurological development and disorders in a human tissue background.
2.5. Repetitive Elements May Influence Fragmentation of Cerebral Organoid-Derived cfDNA
Repetitive elements, particularly retrotransposons and satellite repeats [
37,
38,
39] have been previously reported to be implicated in neurological disease. We were therefore interested in exploring fragmentation patterns of cfDNA associated with repetitive elements in our cerebral organoid-derived dataset. Notably, we found that ROIs intersecting retrotransposons or acro satellite repeats showed a higher density of longer fragment lengths (
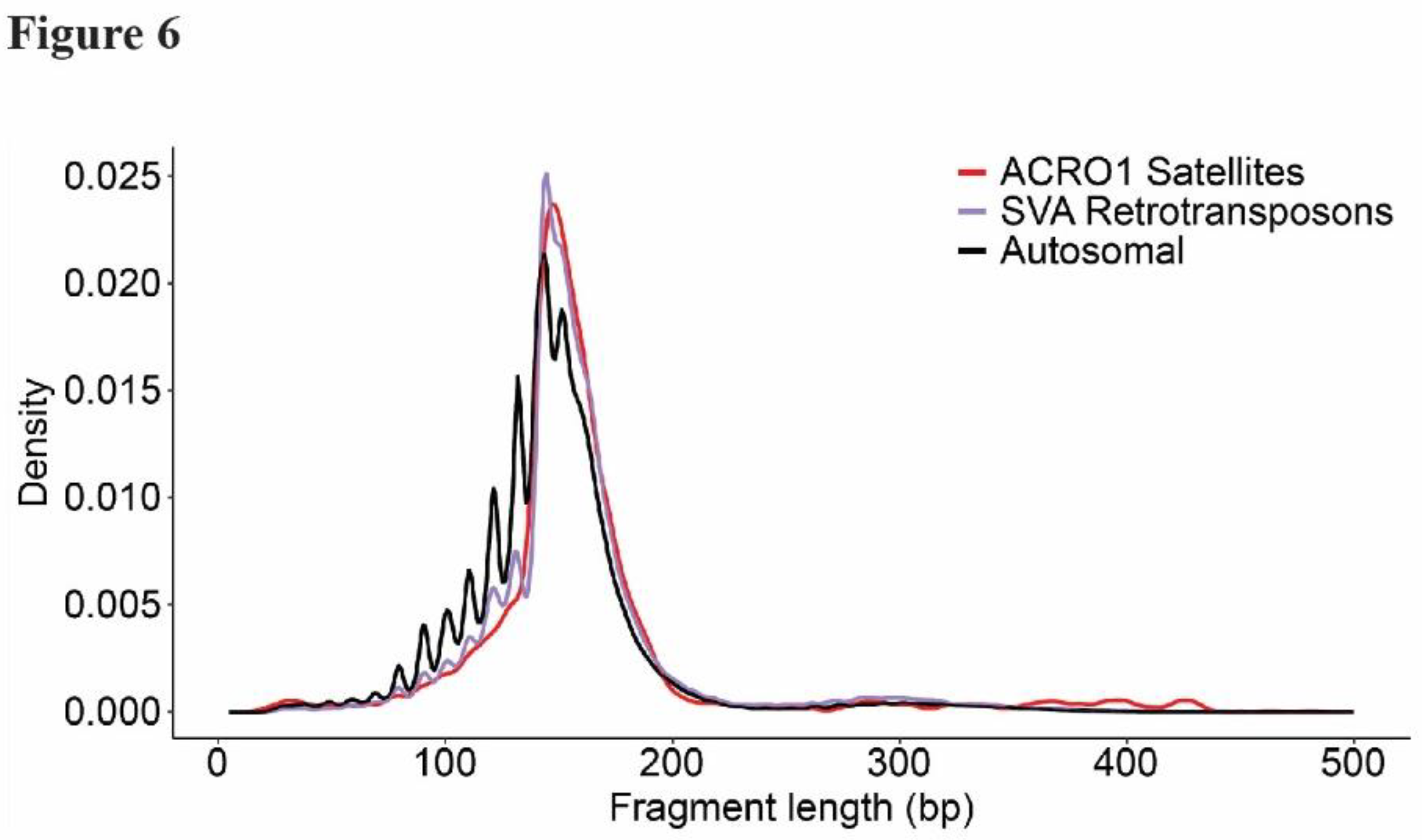
Figure 6). Together, from these data we conclude that cfDNA recovered from mature cerebral organoids shows non-random patterns of fragmentation and contains features of relevance to neurological development and disease.
3. Discussion
CfDNA has been previously isolated successfully from a few organoid systems [
33,
34,
35]. Yet, whether additional organoid models, such as brain organoids, release cfDNA in measurable quantities remains an open question. We have shown here that cfDNA can be consistently recovered from cerebral organoids in quantities sufficient for downstream analyses including WGS and ddPCR. Further, we have demonstrated that WGS is a tractable method to gain a broad understanding of the overall composition and patterns associated with cfDNA release from cerebral organoids. Our observation that cfDNA released from mature cerebral organoids was enriched in gene regions corresponding to GO terms such as “neuronal cell body”, “synaptic membrane”, “synapse organization”, “neuronal system”, and “axon guidance” was particularly intriguing, and suggested that cerebral organoids release cfDNA that is reflective of gene regions relevant to brain tissue. This methodology could be extended to additional organoid types, and potentially used to identify biomarkers associated with exposure to toxins or disease states in specific cell types or tissues.
Changes in gene expression during differentiation or upon neurotoxicant exposure may alter the abundance of sequences present in cfDNA through changes in histone wrapping, protein binding, or other epigenetic modifications that impact chromatin accessibility [
40]. Epigenetic modifications such as methylation are known to alter composition of cfDNA [
41,
42,
43]. Therefore, we wished to further explore cfDNA sequences in our dataset which might overlap with regions associated with epigenetic modifications. Previously, it has been observed that differentiation of hESCs to cerebral organoids changed patterns of methylation [
44]. These differentially methylated regions (DMRs) were defined by comparing mature cerebral organoids to undifferentiated hESCs of identical genetic background. We intersected our ROIs with the reported DMRs in this previously published dataset [
44]. We found that our ROIs overlapped hypomethylated regions significantly more than hypermethylated regions (
p-value = 4.514e-12). Increased cfDNA signal in these hypomethylated regions is suggestive of open, accessible chromatin which could be associated with altered transcriptional activity. Although speculative, this analysis suggests that cfDNA derived from cerebral organoids contains methylation signatures relevant to differentiated cerebral tissue. Future studies might aim to directly examine methylation marks in cerebral organoid-derived cfDNA.
The abundance of specific cfDNA sequences in a biofluid sample could potentially serve as biomarkers of tissue-level changes or disease. Recently, cfDNA of neuronal origin has been found to be elevated in blood plasma of patients with Alzheimer’s Disease [
30]. In addition, mt-cfDNA levels in CSF have been observed to fluctuate in persons with neurodegenerative disease. Specifically, decreased mt-cfDNA was observed in patients with Parkinson’s disease, while increased mt-cfDNA was correlated with Dementia [
25]. Notably, we were able to detect mt-cfDNA in media conditioned by cerebral organoids using ddPCR. This suggests that cerebral organoids indeed release cfDNA sequences that are relevant to human disease. Also, we found that specific sequences of cfDNA derived from cerebral organoids are consistently detectable with ddPCR. This demonstrates the potential of cerebral organoids to be used as a tool for the development of ddPCR assays to detect novel cfDNA sequences. Previously, ddPCR has been successfully used to identify the presence of brain somatic mutations in CSF-derived cfDNA, and proposed as an alternative to directly analyzing brain tissue, which is difficult to obtain [
26]. However, prediction of exactly which sequences will be present and detectable at the cfDNA level in human biofluids is difficult, as many mechanisms could contribute to cfDNA release [
45]. Further, cfDNA can originate from many cell types in vivo [
31], making it difficult to determine which cfDNA features or sequences may be especially relevant to a specific organ and reflective of tissue damage or disease. Organoid systems may provide a tractable means of identifying cfDNA sequences relevant to specific tissue types, toxicant exposures, or disease models that could then be evaluated in a more targeted manner in human biofluids.
In addition to the potential of cfDNA to serve as a diagnostic biomarker of neurodegenerative disease, cfDNA may have use in the identification of neurodevelopmental disorders. For example, early and accurate diagnosis of conditions such as ASD is essential for managing symptoms and improving function and quality of life in adulthood. However, the diagnosis of ASD is often difficult, especially in the presence of comorbidities, due to overlap of symptoms with other cognitive disorders [
46]. In addition to advancements in cognitive and behavioral testing, there is a growing need for identification of biomarkers of ASD [
47,
48]. We observed that cerebral organoids release cfDNA sequences aligning to gene regions associated with Disease Ontology terms related to ASD and intellectual disability. This finding was quite intriguing as it suggests that cerebral organoids might be used to identify features of cfDNA that could be investigated as potential biomarkers of ASD or other neurodevelopmental disorders.
In summary, here we evaluated the ability of cerebral organoids to release cfDNA in quantities sufficient for analyses including WGS and ddPCR. This strategy could be further applied to investigate potential cfDNA targets in cerebral organoid models of disease, neurodevelopmental disability, or neurotoxicant exposure. Future research might seek to investigate the presence of cerebral organoid-derived cfDNA sequences in human biofluids. Overall, this study demonstrates the utility of cerebral organoid models as a tool for identification of potential cfDNA targets relevant to brain tissue. Increasing our knowledge of non-invasive biomarkers such as cfDNA could ultimately help develop strategies for earlier detection and treatment of brain-related disease and disorders.
Study Limitations
Sequencing of cfDNA isolated from cerebral organoids on different growth days revealed a high degree of DNA from rodent origin early in development of the organoids. Therefore, care must be taken in interpreting differences in cfDNA concentration or sequence abundance between early and late time points during organoid maturation. For these reasons, we chose to focus our sequencing analyses on mature cerebral organoids (day 41), after deterioration of the Matrigel scaffold and disappearance of contamination by non-human DNA. Further research is needed to determine definitively whether quantitative differences exist between cfDNA sequences released between early and late time points of cerebral organoid growth. Although beyond the scope of this study, such efforts might explore Matrigel-free systems of culture, or methods of isolating human cfDNA sequences from potential contaminants present in cell culture systems.
4. Materials and Methods
4.1. hESC Culture
H9 human embryonic stem cells (hESCs) were maintained in mTeSR+ medium (Thermo Fisher) at 37oC and 5% CO 2. Cells were passaged between 60 and 80% confluence, using 0.5 mM EDTA/PBS (Gibco) to dissociate cells. Plates were pre-coated with a concentration of 1.2 µl/ml/cm2 Matrigel (Corning) for 30 min at 37oC in DMEM/F:12 (Thermo Fisher). Cultures were maintained for no more than 10 passages, provided no morphological changes indicative of differentiation were present.
4.2. Cerebral Organoid Generation
H9 ESCs were used to generate cerebral organoids with the StemDiff Cerebral Organoid Kit (StemCell Technologies) following the manufacturer’s instructions. Briefly, cells were trypsinized with 1x TrypLE (Gibco)/0.5mM EDTA and resuspended in EB Formation Medium (StemDiff Cerebral Differentiation Kit; StemCell Technologies) supplemented with 10 µM of Y27632 (Tocris). Cells were seeded at a density of 2x104 cells/mL, 50 µl per well in a 384 round bottom ultra-low attachment plate (Corning). The plate was centrifuged at 200g for 5 min to coalesce the cells. This was defined as day 0 of growth. On growth days 1 and 3, 40 µl of media was replaced with EB Formation Medium (StemDiff Cerebral Differentiation Kit; StemCell Technologies) and incubated for 48 hr at 37oC. On growth day 5, 40 µl of media was replaced with Induction Medium (StemDiff Cerebral Differentiation Kit; StemCell Technologies) and the plate was incubated again for 48 hr at 37oC. On growth day 7, the tissues were rinsed 1x with 30 µl Phosphate Buffered Saline (PBS; Thermo Fisher) and individually embedded in 15 µl of Matrigel (Corning) on an embedding sheet (StemCell Technologies). After 30 min incubation at 37oC to solidify the Matrigel, the tissues were transferred to a 24-well ultra-low attachment plate (Thermo Fisher) in 1 mL of Expansion Medium (StemDiff Cerebral Differentiation Kit; StemCell Technologies) at a density of four tissues/well and incubated at 37oC on an orbital shaker for 72 hr. On growth day 10, the media was changed to 1 ml of Maturation Medium (StemDiff Cerebral Differentiation Kit; StemCell Technologies), and the media changed every 3-4 days until maturity (day 41).
4.3. Nuclei Acid Extraction and Quantification of Gene Expression
Genomic DNA (gDNA) from organoids and cfDNA from conditioned media were extracted using the Promega Maxwell® RSC instrument with the Tissue DNA Kit or ccfDNA Plasma Kit, respectively. DNA concentrations were measured using the Promega QuantiFluor® ONE dsDNA System. For gDNA extraction, 3-4 organoids were homogenized according to the manufacturer’s protocol. CfDNA was isolated from 1 mL media conditioned by 4 organoids (2-3 technical replicates per batch, total of 3 independent batches). Prior to cfDNA extraction, all conditioned media was centrifuged for 10 min at 1,600g, the top 800 µl supernatant was centrifuged 10 min at 16,000g, and the top 400 µl collected and stored at -80oC until extraction. Prior to extraction, conditioned media was thawed at 4oC to prevent degradation. Gene expression and copy number were evaluated using droplet digital PCR (ddPCR; Bio-Rad) using either 0.5 or 1 ng of cfDNA per reaction.
4.4. Protein Detection and Quantification
In preparation for Western Blot analysis, organoid tissues were collected at several timepoints during growth (days 10, 13, 17, 31, 41) and rinsed 1x with PBS and homogenized in RIPA buffer (Thermo Fisher) supplemented with 25x Complete Protease Inhibitor (Thermo Fisher). Samples were centrifuged for 15 min at 14,000g at 4oC. The supernatant was removed and stored at -80o C until analysis. Protein concentration was calculated using the PierceTM BCA assay (Thermo Fisher). Protein (3.5 µg) was incubated for 10 min at 70oC with NuPage Reaction Buffer (Thermo Fisher) and NuPage Reducing Agent (Thermo Fisher) prior to loading onto NuPage gel (4 to 12%, Bis-Tris; Thermo Fisher). The gel was run for 35 min at 200V in NuPage MES Run Buffer (Thermo Fisher) to which was added NuPage Antioxidant (Thermo Fisher). After migrating through the gel, the proteins were transferred to nitrocellulose membranes using an iBlot™ Gel Transfer Device (Thermo Fisher). Ponceau S solution (Sigma) was used to confirm protein transfer to the membrane. Membranes were then blocked for 15 min in EveryBlot Blocking Reagent (Bio-Rad) and incubated in primary antibody solution overnight at 4oC. The following antibodies were diluted 1:1000 in Bio-Rad EveryBlot Blocking Reagent: rabbit anti-GAPDH (Bio-Rad), rabbit anti-PSD95 (Cell Signaling), rabbit anti-synaptophysin (Cell Signaling), rabbit anti-Gap43 (Cell Signaling). Membranes were washed 3x5 min in 1x tris-buffered saline (TBS; Bio-Rad) to which was added 0.1% Tween-20 (Sigma) (TBST). Membranes were then incubated for 45 min at room temperature in secondary antibody solution: anti-rabbit-HRP (Novus) diluted 1:5000 in EveryBlot Blocking Reagent (Bio-Rad). Membranes were washed 3x15 min in TBST. HRP was visualized using SuperSignal™ West Pico Plus Chemiluminescent Substrate (Thermo Fisher) according to the manufacturer’s protocol.
4.5. Immunofluorescent Staining
Mature cerebral organoids were fixed, cleared and immunostained following a published protocol [
49] as previously described [
34]. Primary antibodies were diluted 1:200 in blocking solution (3% Bovine Serum Albumin/PBS; Sigma): chick anti-β3 tubulin (Cell Signaling), mouse IgG1 anti-nestin (Cell Signaling), rabbit anti-map2 (Cell Signaling). Secondary antibodies were diluted 1:200 in blocking solution: Alexa 647 donkey-anti-rabbit (Invitrogen), Alexa 488 goat-anti-mouse (Invitrogen), Dylight 550 goat-anti-chicken (Thermo Fisher) and DAPI (1 µg/mL; Sigma). Tissues were imaged in 96-well optical plates (Thermo Fisher, M33089) on a Zeiss LSM 880 inverted confocal microscope with AiryScan (Jena, Germany).
4.6. Sequencing of cfDNA Fragments from Cerebral Organoids
CfDNA collected from media conditioned by cerebral organoids on growth days 10, 21, or 41 (three biological replicates each) was used to prepare sequencing libraries using the ThruPLEX Tag-seq Kit from Takara Bio following the manufacturer’s protocol. Libraries were sequenced on an Illumina NovaSeq 6000 platform to generate 150 base pair (bp) paired-end reads (Illumina, San Diego, CA, USA).
4.7. Bioinformatic Analyses
The Takara ThurPLEX Tag Seq HV processing pipeline for UMIs was applied for data processing (
https://www.takarabio.com/learning-centers/next-generation-sequencing/dna-seq-protocols/using-umis-with-thruplex-tag-seq-hv). In summary, adapter sequences were removed from raw fastq files with Trimmomatic v.0.39 [
50]. Unique molecular identifiers (UMIs) were appended to an unmapped BAM RX tag using the fgbio v.2.1.0 FastqToBam function and subsequently removed from the trimmed reads with picard v.2.26.9 SamToFastq (
https://fulcrumgenomics.github.io/fgbio/; https://broadinstitute.github.io/picard/). Processed reads were aligned to the Hg38 genome with Bowtie2 v.2.5.2 and converted to sorted BAM format with picard SortSam and samtools v.1.18 view functions [
51,
52]. The picard MergeBamAlignment functions was used to incorporate the UMI information from the unmapped BAM file to the sorted, aligned BAM. Filtered, deduplicated consensus reads were obtained in unmapped BAM format with the fgbio functions CorrectUmis, GroupReadsByUmis and CallMolecularConsensusReads. Consensus reads were extracted with SamToFastq and aligned with Bowtie2. The alignments were sorted and converted to BAM format with SortSam and the samtools view module. The UMIs present in the unmapped BAM files containing consensus reads were incorporated into the Bowtie2 alignments to generate the final BAM files used for all down-stream processes. Specific flag setting for all pipeline steps followed recommendations from the Takara pipeline (
https://www.takarabio.com/learning-centers/next-generation-sequencing/dna-seq-protocols/using-umis-with-thruplex-tag-seq-hv). Potential contamination of cfDNA with rodent DNA was assessed with the command line utility fastqscreen v.0.15.2 using the fastq files containing consensus reads [
53].
Fragment length analysis was performed to compare differences in fragment length density between autosomal, mitochondrial or genomic repeat classes. Day 41 cfDNA replicates were merged into a single BAM file using the samtools merge function. Estimated fragment lengths were extracted from the 5’ read of properly paired reads in the merged BAM file and plotted for relevant comparisons. The Repbase RepeatMasker library 20140231 was queried to determine reads mapping to Acro1 satellite or SVA retrotransposon regions in the genome [
54].
The macs2 v.2.2.9.1 peak caller with flags -f BAMPE --nomodel -p .01 -g 2652783500 --broad --broad-cutoff 0.01 was employed to identify regions of the signal enrichment in each of the Day 41 replicates [
55]. The union of broad peak intersections between the replicates comprised the final regions of interest (ROIs). Gencode v.32 Hg38 genes that intersected with ROIs were subject to pathway enrichment with clusterProfiler v.4.8.2 in the R v.4.3.1 programming environment [
36]. Genomic coordinates for hypo- and hypermethylated regions in day 40 organoids were retrieved from Luo et al. (2016) [
44]. The Hg19 methylation coordinates were lifted from Hg19 to Hg38 with the UCSC liftOver command line utility [
56]. Hypo- and hypermethylated regions were intersected with ROIs and Fisher’s exact test was applied to determine the significance of overlaps. All interval intersections and merges were performed with the bedtools v.2.25.0 intersect and merge functions unless otherwise stated [
57].
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: Sources of cfDNA in cerebral organoids during growth; Figure S2: Definition of regions of interest (ROI) in cerebral organoid-derived cfDNA sequencing data.
Author Contributions
B.B.S: conceptualization, methodology, investigation, formal analysis, writing—original draft preparation; A.B.: formal analysis, data curation, writing—review and editing; K.G.: conceptualization, resources, writing—review and editing, supervision, funding acquisition; E.J.T.: conceptualization, resources, writing—review and editing, supervision, funding acquisition.
Funding
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. This work was supported by NIEHS grant NTA-PMT ZIA: ES103378-02 to EJT.
Acknowledgments
We wish to acknowledge the NIEHS Fluorescence Microscopy and Imaging Center, NIEHS Molecular Genomics Core facility, and in particular Wesley Gladwell for generous assistance with the analysis and setup of Droplet Digital PCR experiments.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- National Academies of Sciences, E., and Medicine; Division of Behavioral and Social Sciences and Education; Board on Behavioral, Cognitive, and Sensory Sciences; Committee on the Decadal Survey of Behavioral and Social Science Research on Alzheimer's Disease and Alzheimer's Disease-Related Dementias. , Reducing the Impact of Dementia in America: A Decadal Survey of the Behavioral and Social Sciences. 2021, National Academies Press (US): Washington (DC). [CrossRef]
- Dubois, B., et al., Timely Diagnosis for Alzheimer's Disease: A Literature Review on Benefits and Challenges. J Alzheimers Dis, 2016. 49(3): p. 617-31. [CrossRef]
- Mirelman, A., A. Siderowf, and L. Chahine, Outcome Assessment in Parkinson Disease Prevention Trials: Utility of Clinical and Digital Measures. Neurology, 2022. 99(7 Suppl 1): p. 52-60. [CrossRef]
- Pagán, F.L., Improving Outcomes Through Early Diagnosis of Parkinson's Disease. American Journal of Managed Care, 2012. 18(7): p. S176-S182.
- Alessi, N., P. Perucca, and A.M. McIntosh, Missed, mistaken, stalled: Identifying components of delay to diagnosis in epilepsy. Epilepsia, 2021. 62(7): p. 1494-1504. [CrossRef]
- Zwaigenbaum, L., et al., Early Intervention for Children With Autism Spectrum Disorder Under 3 Years of Age: Recommendations for Practice and Research. Pediatrics, 2015. 136: p. S60-S81. [CrossRef]
- Moeller, J.J., et al., Diagnostic accuracy of neurological problems in the emergency department. Canadian Journal of Neurological Sciences, 2008. 35(3): p. 335-341. [CrossRef]
- Koger, S.M., T. Schettler, and B. Weiss, Environmental toxicants and developmental disabilities: a challenge for psychologists. Am Psychol, 2005. 60(3): p. 243-55. [CrossRef]
- Nabi, M. and N. Tabassum, Role of Environmental Toxicants on Neurodegenerative Disorders. Front Toxicol, 2022. 4: p. 837579. [CrossRef]
- Bocato, M.Z., J.P.B. Ximenez, C. Hoffmann, and F. Barbosa, An overview of the current progress, challenges, and prospects of human biomonitoring and exposome studies. Journal of Toxicology and Environmental Health-Part B-Critical Reviews, 2019. 22(5-6): p. 131-156. [CrossRef]
- Manzo, L., et al., Mechanisms of neurotoxicity: applications to human biomonitoring. Toxicol Lett, 1995. 77(1-3): p. 63-72. [CrossRef]
- Mathon, B., et al., Neurological diseases of unknown etiology: Brain-biopsy diagnostic yields and safety. Eur J Intern Med, 2020. 80: p. 78-85. [CrossRef]
- Jozsa, F., et al., Safety and efficacy of brain biopsy: Results from a single institution retrospective cohort study. Brain Spine, 2023. 3: p. 101763. [CrossRef]
- Roberts, R.A., et al., Translational Biomarkers of Neurotoxicity: A Health and Environmental Sciences Institute Perspective on the Way Forward. Toxicological Sciences, 2015. 148(2): p. 332-340. [CrossRef]
- Lleó, A., Biomarkers in Neurological Disorders: A Fast-Growing Market. Brain Communications, 2021. 3(2). [CrossRef]
- Zukowski, A., S. Rao, and S. Ramachandran, Phenotypes from cell-free DNA. Open Biol, 2020. 10(9): p. 200119. [CrossRef]
- Leal, A., D. Sidransky, and M. Brait, Tissue and Cell-Free DNA-Based Epigenomic Approaches for Cancer Detection. Clin Chem, 2020. 66(1): p. 105-116. [CrossRef]
- Grabuschnig, S., et al., Putative Origins of Cell-Free DNA in Humans: A Review of Active and Passive Nucleic Acid Release Mechanisms. Int J Mol Sci, 2020. 21(21). [CrossRef]
- Kustanovich, A., R. Schwartz, T. Peretz, and A. Grinshpun, Life and death of circulating cell-free DNA. Cancer Biol Ther, 2019. 20(8): p. 1057-1067. [CrossRef]
- Willems, P.J., et al., The first 3,000 Non-Invasive Prenatal Tests (NIPT) with the Harmony test in Belgium and the Netherlands. Facts Views Vis Obgyn, 2014. 6(1): p. 7-12.
- Jacobsen, D., G.R. Krog, and F.B. Clausen, Early and Accurate Sex Determination by qPCR of Y Chromosome Repetitive Sequence (YRS) In Cell-Free Fetal DNA from Maternal Plasma. J Appl Lab Med, 2018. 3(3): p. 346-356. [CrossRef]
- Cisneros-Villanueva, M., et al., Cell-free DNA analysis in current cancer clinical trials: a review. Br J Cancer, 2022. 126(3): p. 391-400. [CrossRef]
- Bronkhorst, A.J., V. Ungerer, and S. Holdenrieder, The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif, 2019. 17: p. 100087. [CrossRef]
- Wang, Y.X., et al., Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proceedings of the National Academy of Sciences of the United States of America, 2015. 112(31): p. 9704-9709. [CrossRef]
- Lowes, H., M. Kurzawa-Akanbi, A. Pyle, and G. Hudson, Post-mortem ventricular cerebrospinal fluid cell-free-mtDNA in neurodegenerative disease. Scientific Reports, 2020. 10(1). [CrossRef]
- Ye, Z.M., et al., Cerebrospinal fluid liquid biopsy for detecting somatic mosaicism in brain (vol 3, fcaa235, 2021). Brain Communications, 2021. 3(2). [CrossRef]
- Park, S.S., H. Jeong, and A.C. Andreazza, Circulating cell-free mitochondrial DNA in brain health and disease: A systematic review and meta-analysis. World Journal of Biological Psychiatry, 2022. 23(2): p. 87-102. [CrossRef]
- Aharon, A., et al., Extracellular Vesicles of Alzheimer's Disease Patients as a Biomarker for Disease Progression. Molecular Neurobiology, 2020. 57(10): p. 4156-4169. [CrossRef]
- Gambardella, S., et al., ccf-mtDNA as a Potential Link Between the Brain and Immune System in Neuro-Immunological Disorders. Frontiers in Immunology, 2019. 10. [CrossRef]
- Pollard, C., et al., Detection of neuron-derived cfDNA in blood plasma: a new diagnostic approach for neurodegenerative conditions. Frontiers in Neurology, 2023. 14. [CrossRef]
- Moss, J., et al., Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun, 2018. 9(1): p. 5068. [CrossRef]
- Kim, J., B.K. Koo, and J.A. Knoblich, Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol, 2020. 21(10): p. 571-584. [CrossRef]
- Dantes, Z., et al., Implementing cell-free DNA of pancreatic cancer patient-derived organoids for personalized oncology. JCI Insight, 2020. 5(15). [CrossRef]
- Silver, B., K. Gerrish, and E. Tokar, Cell-free DNA as a potential biomarker of differentiation and toxicity in cardiac organoids. eLife, 2023. 12: p. e83532. [CrossRef]
- Kim, J., et al., Multidimensional fragmentomic profiling of cell-free DNA released from patient-derived organoids. Hum Genomics, 2023. 17(1): p. 96. [CrossRef]
- Yu, G.C., L.G. Wang, Y.Y. Han, and Q.Y. He, clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. Omics-a Journal of Integrative Biology, 2012. 16(5): p. 284-287. [CrossRef]
- Billingsley, K.J., et al., Analysis of repetitive element expression in the blood and skin of patients with Parkinson's disease identifies differential expression of satellite elements. Scientific Reports, 2019. 9. [CrossRef]
- Copley, K.E. and J. Shorter, Repetitive elements in aging and neurodegeneration. Trends in Genetics, 2023. 39(5): p. 381-400. [CrossRef]
- Peze-Heidsieck, E., et al., Retrotransposons as a Source of DNA Damage in Neurodegeneration. Frontiers in Aging Neuroscience, 2022. 13. [CrossRef]
- Ulz, P., et al., Inference of transcription factor binding from cell-free DNA enables tumor subtype prediction and early detection. Nat Commun, 2019. 10(1): p. 4666. [CrossRef]
- Zhou, Q., et al., Epigenetic analysis of cell-free DNA by fragmentomic profiling. Proceedings of the National Academy of Sciences of the United States of America, 2022. 119(44). [CrossRef]
- Ivanov, M., et al., Non-random fragmentation patterns in circulating cell-free DNA reflect epigenetic regulation. Bmc Genomics, 2015. 16. [CrossRef]
- Lo, Y.M.D., D.S.C. Han, P.Y. Jiang, and R.W.K. Chiu, Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science, 2021. 372(6538): p. 144. [CrossRef]
- Luo, C.Y., et al., Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Reports, 2016. 17(12): p. 3369-3384.
- Aucamp, J., A.J. Bronkhorst, C.P.S. Badenhorst, and P.J. Pretorius, The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc, 2018. 93(3): p. 1649-1683. [CrossRef]
- Hus, Y. and O. Segal, Challenges Surrounding the Diagnosis of Autism in Children. Neuropsychiatr Dis Treat, 2021. 17: p. 3509-3529.
- McCarty, P. and R.E. Frye, Early Detection and Diagnosis of Autism Spectrum Disorder: Why Is It So Difficult? Semin Pediatr Neurol, 2020. 35: p. 100831. [CrossRef]
- Klin, A., Biomarkers in Autism Spectrum Disorder: Challenges, Advances, and the Need for Biomarkers of Relevance to Public Health. Focus (Am Psychiatr Publ), 2018. 16(2): p. 135-142. [CrossRef]
- Matsumoto, K., et al., Advanced CUBIC tissue clearing for whole-organ cell profiling. Nature protocols, 2019. 14(12): p. 3506-3537. [CrossRef]
- Bolger, A.M., M. Lohse, and B. Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 2014. 30(15): p. 2114-2120. [CrossRef]
- Langmead, B. and S.L. Salzberg, Fast gapped-read alignment with Bowtie 2. Nature Methods, 2012. 9(4): p. 357-U54. [CrossRef]
- Li, H., et al., The Sequence Alignment/Map format and SAMtools. Bioinformatics, 2009. 25(16): p. 2078-2079. [CrossRef]
- Wingett, S.W. and S. Andrews, FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res, 2018. 7: p. 1338. [CrossRef]
- Bao, W.D., K.K. Kojima, and O. Kohany, Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA, 2015. 6. [CrossRef]
- Zhang, Y., et al., Model-based Analysis of ChIP-Seq (MACS). Genome Biology, 2008. 9(9). [CrossRef]
- Hinrichs, A.S., et al., The UCSC Genome Browser Database: update 2006. Nucleic Acids Research, 2006. 34: p. D590-D598. [CrossRef]
- Quinlan, A.R. and I.M. Hall, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 2010. 26(6): p. 841-842. [CrossRef]
Figure 1.
Cerebral organoids derived from H9 ESCs express neural differentiation markers. (A) Brightfield images of cerebral organoids during growth and differentiation. Scale bars represent 100 µm. Immunofluorescence z slice (B) and z-stack (C) showing TUBB3, Nestin, and Map2 in mature (day 41) cerebral organoids. Scale bars represent 500 µm. (D) Western blot showing expression of PSD95, Synaptophysin, GAP43, and GAPDH (loading control) in cerebral organoids during maturation. (E) Schematic illustrating timepoints of cfDNA collection in cerebral organoids.
Figure 1.
Cerebral organoids derived from H9 ESCs express neural differentiation markers. (A) Brightfield images of cerebral organoids during growth and differentiation. Scale bars represent 100 µm. Immunofluorescence z slice (B) and z-stack (C) showing TUBB3, Nestin, and Map2 in mature (day 41) cerebral organoids. Scale bars represent 500 µm. (D) Western blot showing expression of PSD95, Synaptophysin, GAP43, and GAPDH (loading control) in cerebral organoids during maturation. (E) Schematic illustrating timepoints of cfDNA collection in cerebral organoids.
Figure 2.
CfDNA can be recovered from cerebral organoids in quantities sufficient for downstream analysis. (A) Shown are cfDNA concentrations taken from n=3 biological replicates during cerebral organoid differentiation. Data points represent the average of 2-3 technical replicates per batch. (B) Cerebral organoid-derived cfDNA concentrations from panel B, normalized to gDNA concentrations (multiplied by 100 to facilitate axis readability). Graphs show average concentrations of n=3 biological replicates +S.D. Samples were compared using an unpaired t-test with Welch’s correction. *, p<0.05; **, p<0.01; ***, p<0.001; ns, non-significant. (C) Representative electropherogram showing fragment sizes of cfDNA derived from mature cerebral organoids. (D) Abundance of cfDNA sequences detected using ddPCR with probes corresponding to mitochondrial genes: mtCO1, mtCO2, ND1, ND6. (E) Abundance of cfDNA sequences recovered from mature cerebral organoids detected using ddPCR with probes corresponding to sequences withing the genes β3-tubulin (TUBB3), nestin (NES), NFATC1, or Oct 3/4 (OCT). Graphs show individual data points and associated mean.
Figure 2.
CfDNA can be recovered from cerebral organoids in quantities sufficient for downstream analysis. (A) Shown are cfDNA concentrations taken from n=3 biological replicates during cerebral organoid differentiation. Data points represent the average of 2-3 technical replicates per batch. (B) Cerebral organoid-derived cfDNA concentrations from panel B, normalized to gDNA concentrations (multiplied by 100 to facilitate axis readability). Graphs show average concentrations of n=3 biological replicates +S.D. Samples were compared using an unpaired t-test with Welch’s correction. *, p<0.05; **, p<0.01; ***, p<0.001; ns, non-significant. (C) Representative electropherogram showing fragment sizes of cfDNA derived from mature cerebral organoids. (D) Abundance of cfDNA sequences detected using ddPCR with probes corresponding to mitochondrial genes: mtCO1, mtCO2, ND1, ND6. (E) Abundance of cfDNA sequences recovered from mature cerebral organoids detected using ddPCR with probes corresponding to sequences withing the genes β3-tubulin (TUBB3), nestin (NES), NFATC1, or Oct 3/4 (OCT). Graphs show individual data points and associated mean.
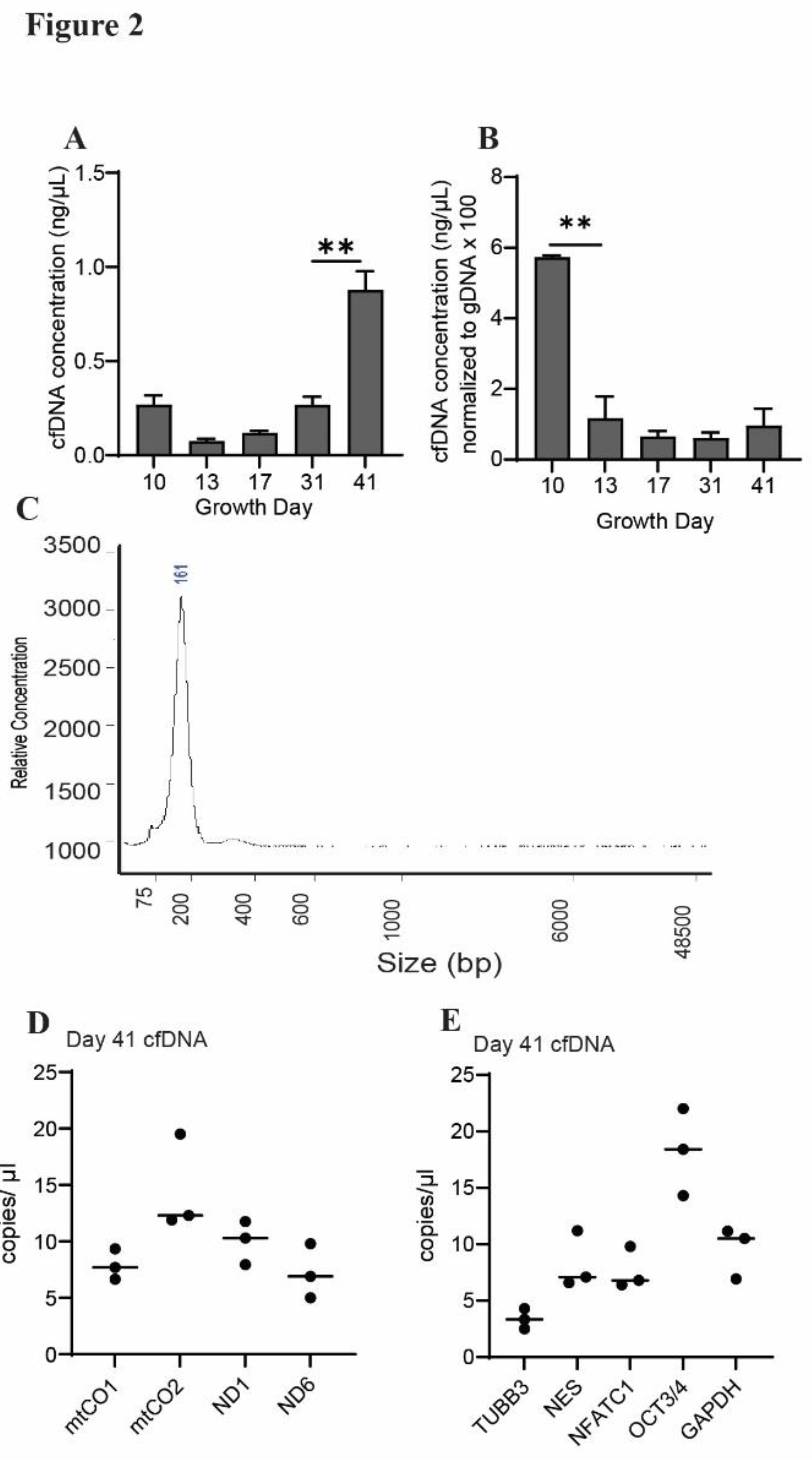
Figure 3.
WGS reveals novel and consistently detectable sequences in cfDNA recovered from cerebral organoids. (A) Histogram showing the length of sequenced cfDNA reads recovered from mature cerebral organoids. (B) Length distributions of sequenced cfDNA mapping to either mitochondrial or autosomal origin (C) Novel sequences detected in cfDNA from day 41 cerebral organoids using custom ddPCR probes (labeled P1-P24). Graphs show individual data points from n=3 biological replicates and associated mean.
Figure 3.
WGS reveals novel and consistently detectable sequences in cfDNA recovered from cerebral organoids. (A) Histogram showing the length of sequenced cfDNA reads recovered from mature cerebral organoids. (B) Length distributions of sequenced cfDNA mapping to either mitochondrial or autosomal origin (C) Novel sequences detected in cfDNA from day 41 cerebral organoids using custom ddPCR probes (labeled P1-P24). Graphs show individual data points from n=3 biological replicates and associated mean.
Figure 4.
Distribution of ROIs across chromosomes. Barplot displays the number of ROIs per chromosome (pink) and chromosome length in 100kb (black) on the y-axis for each chromosome, with the normalized ratio of ROI counts to chromosome length indicated above each bar.
Figure 4.
Distribution of ROIs across chromosomes. Barplot displays the number of ROIs per chromosome (pink) and chromosome length in 100kb (black) on the y-axis for each chromosome, with the normalized ratio of ROI counts to chromosome length indicated above each bar.
Figure 5.
Gene Ontology (GO) classification of genes aligning with sequenced cfDNA ROIs. Shown are the top 15 terms from GO: Cellular Component (CC), the top 15 terms from GO: Biological Pathway (BP), terms identified using KEGG and Disease Ontology (DO) databases, and the top 5 terms from the Reactome (REAC) database. GeneRatio is calculated as the number of input genes/total number of genes belonging to the source gene set while p.adjust indicates the Benjamini-Hochberg adjusted p-value for the over representation test, which is based on the hypergeometric distribution.
Figure 5.
Gene Ontology (GO) classification of genes aligning with sequenced cfDNA ROIs. Shown are the top 15 terms from GO: Cellular Component (CC), the top 15 terms from GO: Biological Pathway (BP), terms identified using KEGG and Disease Ontology (DO) databases, and the top 5 terms from the Reactome (REAC) database. GeneRatio is calculated as the number of input genes/total number of genes belonging to the source gene set while p.adjust indicates the Benjamini-Hochberg adjusted p-value for the over representation test, which is based on the hypergeometric distribution.
Figure 6.
CfDNA aligning with retrotransposons and acro satellite repeats shows distinct fragment length distributions. Shown are length distributions of sequenced cfDNA ROIs intersecting retrotransposons or acro satellite repeats.
Figure 6.
CfDNA aligning with retrotransposons and acro satellite repeats shows distinct fragment length distributions. Shown are length distributions of sequenced cfDNA ROIs intersecting retrotransposons or acro satellite repeats.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).