1. Introduction
After more than three years since the onset of the COVID-19 pandemic, attention has turned to the immunity of the population, either elicited by previous infections and/or by vaccination in light of emerging SARS-CoV-2 virus variants. Observational studies and clinical trials confirm the remarkable effectiveness of SARS-CoV-2 vaccines in preventing severe COVID-19 symptoms [
1,
2,
3,
4,
5,
6,
7]. Quite remarkably, the levels of IgG antibody against the SARS-CoV-2 Spike protein seems to correlate with protection against symptomatic or severe COVID-19 [
8,
9,
10].
In our previous studies, we demonstrated the excellent performance of the Brazilian COVID-19 vaccination program in inducing humoral immunity as measured by the levels of IgG reactive to SARS-CoV-2 Spike protein. We also reported a higher performance of the BNT162b2 vaccine (Pfizer-BioNTech) in inducing anti-Spike IgG antibodies after the initial vaccination and booster dose in comparison to other vaccines such as ChadoX and Coronavac [
11], findings that have been confirmed by other studies [
12,
13,
14].
It is important to note that the levels of IgG reactive to the SARS-CoV-2 Spike protein tend to decrease after vaccination, which may result in reduction in protection against novel SARS-CoV-2 infections and symptomatic COVID-19 [
15,
16,
17,
18]. Similarly, although prior SARS-CoV-2 infection provides protection against reinfection [
19,
20,
21], such protection also gradually diminishes [
15,
22].
SARS-CoV-2 Variants of Concern (VOCs), especially those carrying amino acid substitutions in the Spike protein, as observed in Omicron variants (B.1.1.529/BA, and sub-lineages BA.1, BA.2, BA.4, and BA.5), have the potential to overcome adaptive immunity elicited against the original SARS-CoV-2 epitopes [
23,
24]. Decreased vaccine-based protection has been reported for the Omicron variants, which became dominant on a global scale [
25,
26]. In Brazil, since the beginning of 2022, the Omicron variant and its sub-lineages have led to a significant increase in infection rates among fully vaccinated, as well as on boosted individuals, due to their high transmission capacity and potential for immune escape [
27,
28,
29].
Infection by Omicron variants usually results in asymptomatic and/or mildly symptomatic infections. The attenuation in the severity of symptoms observed in previously vaccinated individuals, compared to those not vaccinated, is presumably attributed to the partial protection provided by the residual repertoire of neutralizing antibodies (nAb) and the activation of the memory of B and T cells acquired during the vaccination process [
30,
31]. Omicron variants may cause less severe infections due to pre-mounted memory immunity and the predisposition of these variants to primarily infect the upper respiratory tract [
32,
33]. Subtle clinical symptoms caused by Omicron variants often result in underdiagnosis, contributing to continued global spread and evolution of SARS-CoV-2 [
34,
35].
Despite comprehensive research on the impact of SARS-CoV-2 viral mutations and its transmission capability [
36,
37,
38], more data is needed to understand how the previous humoral response, mounted against the original SARS-CoV-2 virus, correlates with infection and symptomatology to emerging variants, such as Omicron.
Here we analyzed the symptomatology and IgG levels before and after SARS-CoV-2 Omicron breakthrough infections in vaccinated individuals to understand the impact of previous immunity on the symptomatology of Omicron sub-lineage infections.
2. Materials and Methods
2.1. Study Design
This case study is a follow-up to a previous populational analysis of the COVID-19 antibody response [
11,
39]. Sampling campaigns took place between 2021 and 2022, at the Federal University of Paraná, Matinhos, Brazil. Individuals aged 18 and over were invited to participate in the study, with the flexibility to choose when and with what frequency they would volunteer for the sampling campaigns. Enrollments were done through the study’s website (
http://200.17.236.32/covid19/). Participants provided their consent, personal information, and details about positive COVID-19 tests (if any), vaccination history (including the vaccine manufacturer), and dates of administered doses.
From a cohort of 1,785 individuals in the current study, we selected 27 individuals who participated in at least three different sampling campaigns and underwent primary vaccination (1st and 2nd doses) and at least one booster dose (3rd dose). The selected participants contracted SARS-CoV-2 infection naturally after the primary vaccination and/or booster doses. The selected individuals were invited to fill out a form detailing the type and intensity of the symptoms, if any (
Table S1). The sum of the intensity level of all symptoms was used to understand the severity of each infection.
2.2. Serological Analysis
As detailed in our previous studies [
11,
39,
40,
41], serological analysis involved the collection of blood samples through capillary puncture. The obtained serum was used to detect IgG antibodies reactive to SARS-CoV-2 N and S antigens. The IgG titers were reported as normalized value in relation to a reference positive serum. The serology positivity cut off was set to a specificity of >99.5%.
2.3. SARS-CoV-2 Infection and Vaccination Events
The occurrence time of SARS-CoV-2 infections in volunteers was informed by the time of an official positive RT-qPCR or antigen test, if available. We considered “possible SARS-CoV-2 infection” had occurred on those individuals who seroconverted to IgG reactive to Nucleocapsid during this study. Individuals who seroconverted to anti-N IgG after receiving a shot of Coronavac were excluded as this vaccine can result in Nucleocapsid seroconversion [
42]. Although it is not possible to determine the precise date of these possible infections, they were reported to occur ten days before the detected anti-N IgG seroconversion. Vaccination of volunteers was reported on graphics according to the date of each dose and respective manufacturer as reported by each participant.
2.4. Variant Identification
For most of the individuals, the SARS-CoV-2 variant that caused the infection was presumed based on the month of the reported infection, along with the data indicating the dominant variant that circulated in the state of Paraná, Brazil, during the infection event. The data was retrieved from the Fiocruz Genomic Network - GISAID (
https://www.genomahcov.fiocruz.br/ - Accessed in October 2023).
A sample from one individual, as detailed in the text, was subject to SARS-CoV-2 genome sequencing using the Illumina platform. In brief, viral RNA was extracted from 0.2 ml of saliva samples using an automated magnetic extractor and the MagMAX™ Pathogen RNA/DNA Kit (Applied Biosystems™). Initial confirmation of SARS-CoV-2 in the sample was performed by RT-qPCR using Kit Biomol OneStep/Covid-19 PCR Master Mix Reverse Transcription Kit (IBMP) on a QuantStudio5 instrument (Thermo Fisher Scientific™). After confirming the presence of SARS-CoV-2 by RT-qPCR, the extracted RNA was subjected to cDNA synthesis and amplification of the viral genome by PCR in overlapping fragments of approximately 300 base pairs, with 2 primer pools provided in the Illumina COVIDSeq Test kit (Illumina, CA, USA). The viral genome was sequenced using reagents from the Illumina COVIDSeq Test kit and Nextseq 1000/2000 instrument (Illumina, CA, USA).
The raw data was processed using the COVID Lineage pipeline on the Dragen analysis platform. After quality control, the consensus sequence was assembly and mapped against the reference strain (NC_045512.2) to determine the identity of the mutations. The classification of lineages followed the dynamic classification system proposed by Rambaut and collaborators (2020) [
43], through the Phylogenetic Assignment of Named Global Outbreak LINages software, available at (
https://github.com/cov-lineages/pangolin) and also through NextClade. Definitive classification of lineages was made after confirmation by phylogenetic analysis containing representative sequences of the main circulating lineages.
2.5. Data Analysis
The statistical analyses were conducted using the software NCSS 11 and GraphPad Prism (version 8.4.3). Analysis such as Spearman correlation, Tukey ANOVA and Kruskal-Wallis ANOVA test was used as indicated in each figure.
3. Results
3.1. Cohort Description
Among the 1,785 participants from our previously populational studies we identified 27 participants who had been subjected to at least 3 rounds of serological analysis, had been fully vaccinated and had a SARS-CoV-2 infection after completing the primary and/or booster vaccination. This group had an average age of 40 years (SD 12; ranging from 19 to 74 years) with 70% being women. All participants completed the primary vaccination for COVID-19 during the study, 81% had taken the third and 19% up to the fourth booster dose. Vaccination schemes covered both homologous and heterologous combinations from different manufacturers.
Among the selected 27 individuals, 59% of the cohort underwent SARS-CoV-2 infections according to official qRT-PCR and/or antigen tests, with all these individuals having symptomatic infections. The remaining 41% of the individuals underwent asymptomatic SARS-CoV-2 infection, which were detected by anti-N IgG seroconversion during the study. All, except one participant (code B), seroconverted to anti-S IgG before the SARS-CoV-2 infection event. Relevant information from all individuals is provided in
Table S2.
3.2. Time Course IgG Titers, Variant and Disease Symptomatology
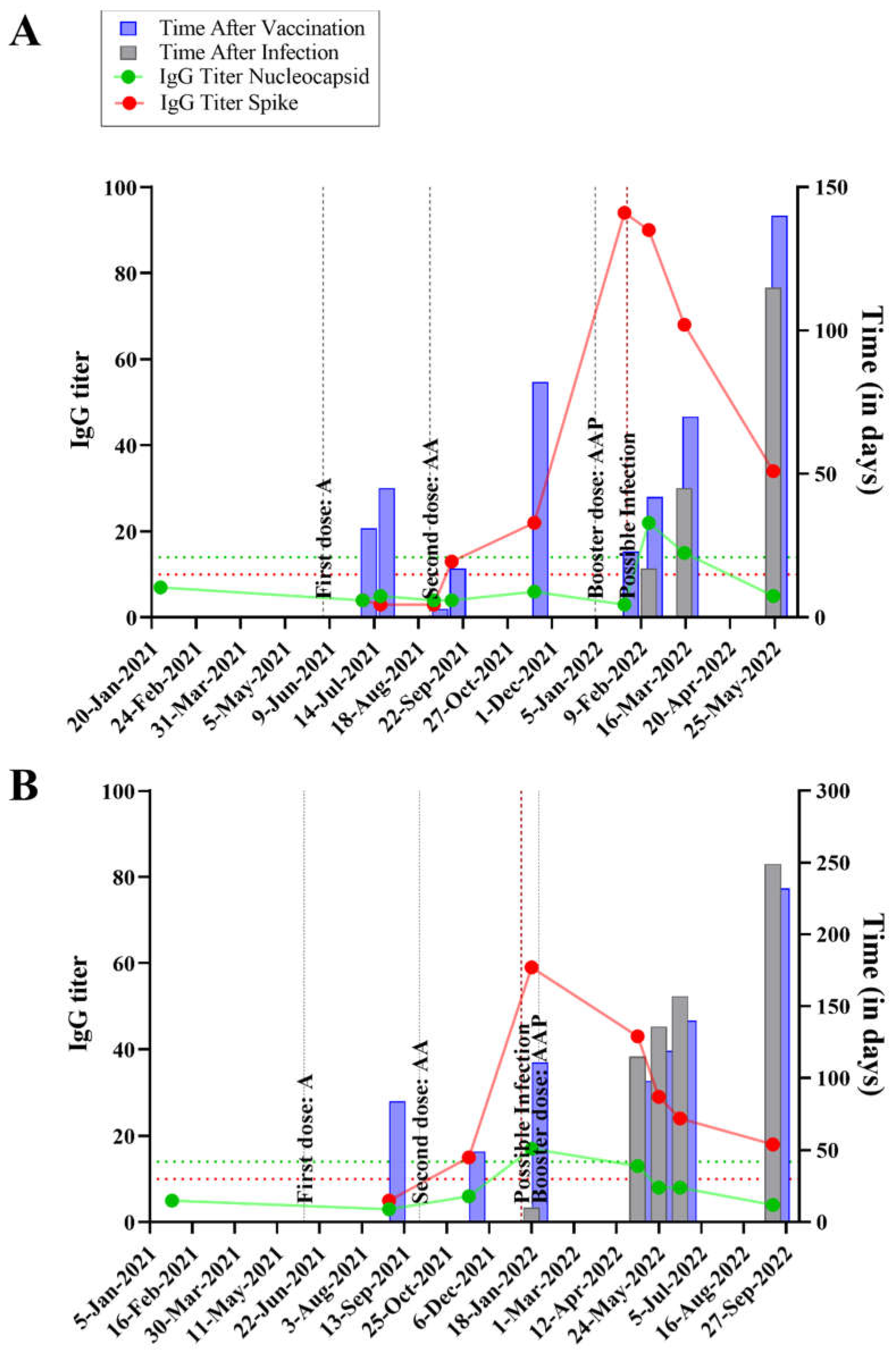
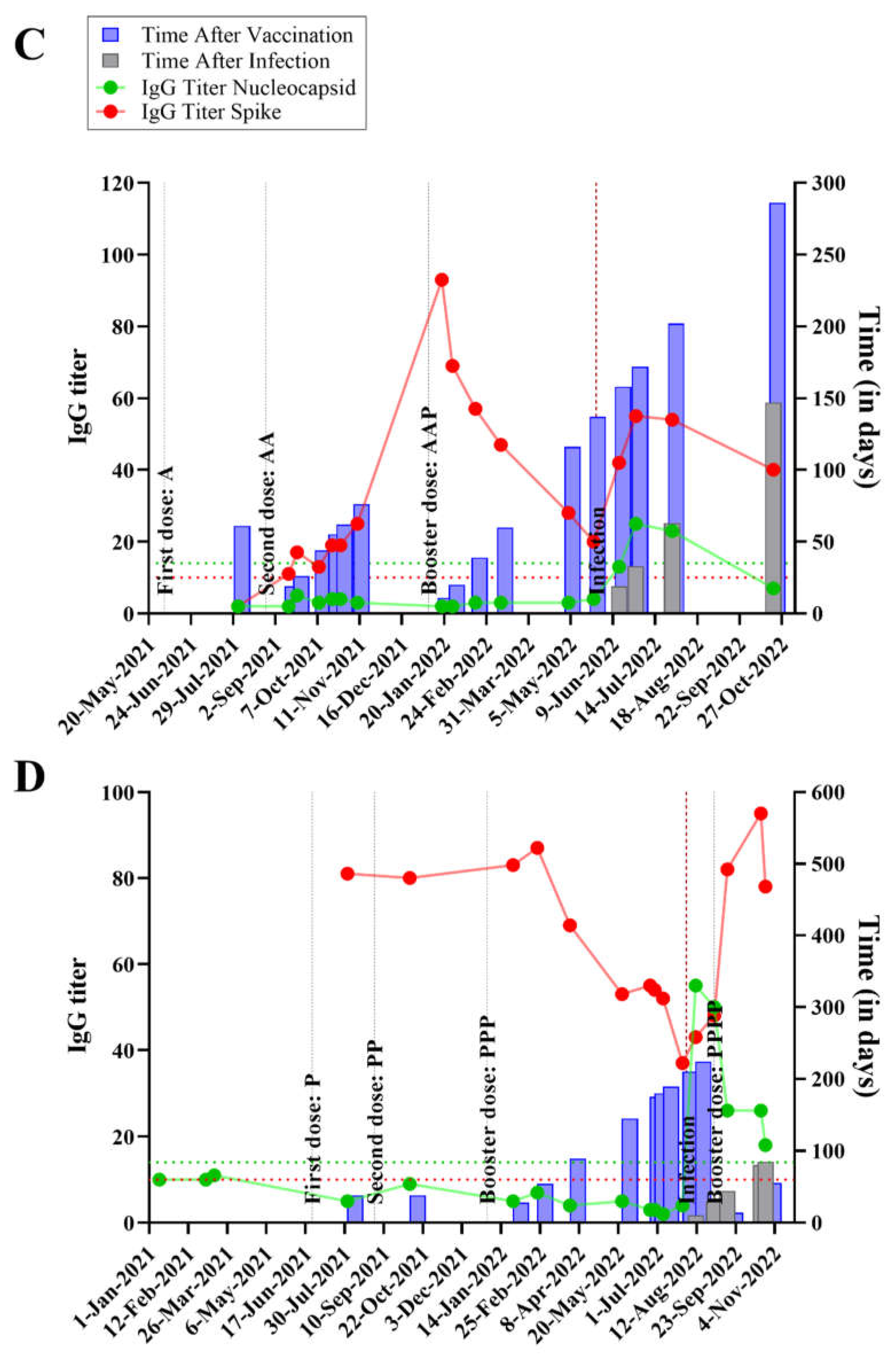
The time course, IgG titers and relevant events occurring in each individual are graphically expressed in
Figure S1. Specific examples, focusing on 4 individuals are shown, in
Figure 1, two of which were asymptomatic (panels A and B) and two were symptomatic for SARS-CoV-2 infection (panels C and D). The x-axis shows the temporal analysis of each individual with infection and vaccination events indicated by vertical lines. The anti-N and anti-S IgG titers followed the expected pattern, with anti-S titers raised after a vaccination event; whereas both anti-S and anti-N levels increased after an infection event. In the absence of further infection and/or vaccination both anti-S and anti-N titers waned over time, with anti-N showing a steeper decrease so that it became undetectable a few months after infection in most individuals (
Figure 1 and Figure S1).
Quite remarkably, the data depicted in
Figure 1 and Figure S1 shows that SARS-CoV-2 infections can occur even in individuals who had high anti-S IgG titers before the infection. During the course of the present study, all the vaccines in use in Brazil were produced based on the original Wuhan SARS-CoV-2 strain. Several studies showed that emerging SARS-CoV-2 variants, particularly Omicron, can escape the immune response which was elicited against the original version of the virus, considering either natural infections and/or vaccination.
To determine which SARS-CoV-2 variant was causing the infections, the date of each infection event was correlated with the predominant SARS-CoV-2 strain, with the predominancy data obtained from the Fiocruz Genomic surveillance Network - GISAID (
https://www.genomahcov.fiocruz.br/ - Accessed in October 2023) (
Figure S2). This analysis indicates that the Omicron variant caused all the cases reported in this study, with estimated frequencies of 10%, 40% and 50% for the BA.1, BA.2 and BA.5 variants, respectively (
Table S2).
A saliva sample collected during the active infection of one individual (
Table S2, code N) was subjected to SARS-CoV-2 genome sequencing which identified BA.2 (
supplementary information) as the infecting variant, corroborating the prediction based on strain predominance over time (
Table S2). The genome sequence-based strain identification can also apply to individuals L, P and Y (
Table S2), since these cases were related to individual N (
Figure S3).
Given that the cohort of this study had completed the vaccination scheme before being infected with different SARS-CoV-2 omicron variants, it would be interesting to understand the severity of the infection event. All the symptomatic participants answered a form where they rated nine different COVID-19 related symptoms (
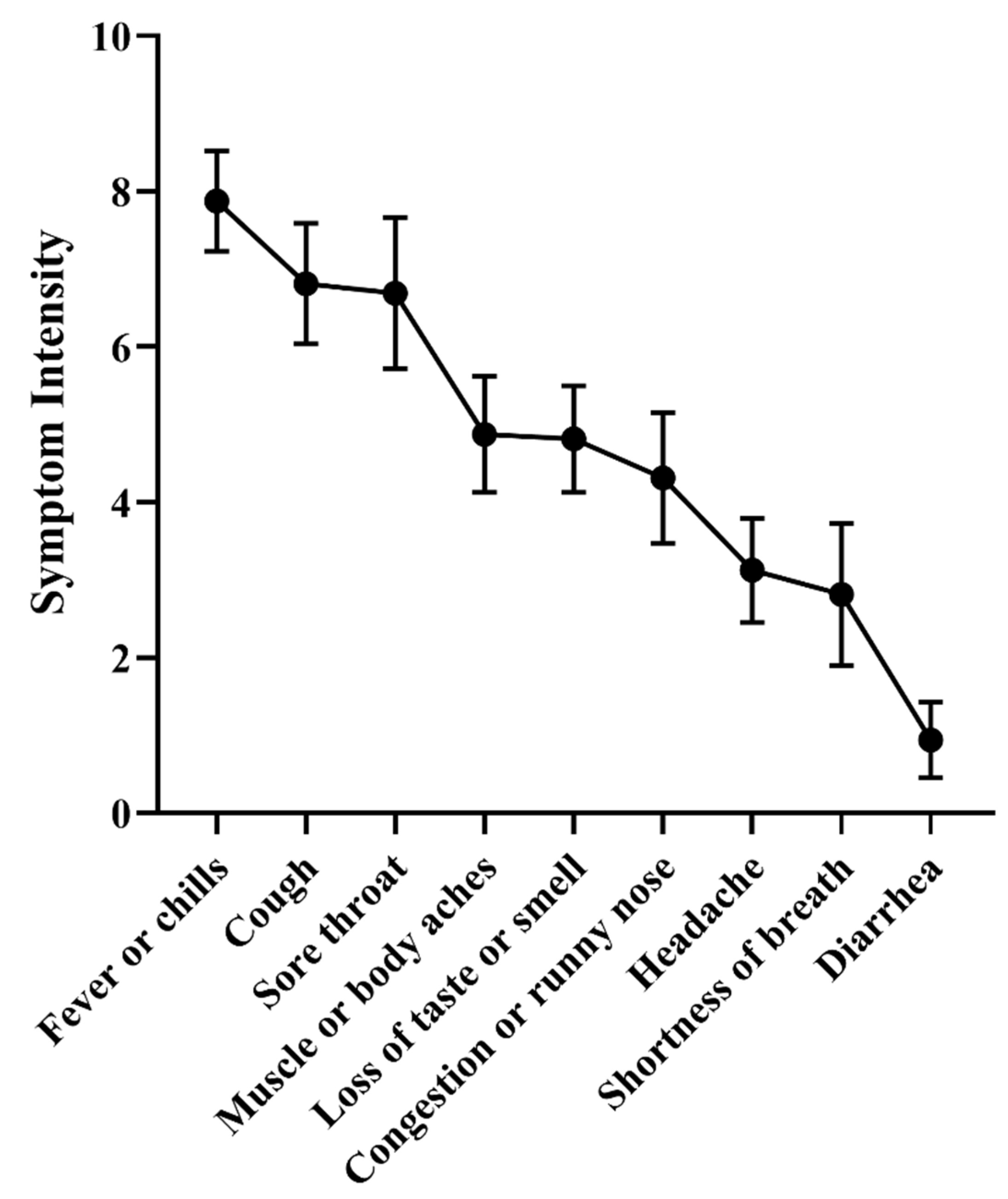
Table S1) on a scale from zero (no symptoms) to 10 (most severe symptom).
The three most common symptoms were: fever or chills; sore throat; cough; with average intensity of 7.9, 6.8 and 6.7, respectively (
Figure 2 and
Table S3). The sum of the intensity of all the symptoms was used as a proxy for the severity of each infection. The average was 42 (SD 13; ranging from 22 to 69).
3.3. Correlation between IgG Levels and COVID-19 Symptomatology
Several studies indicate that anti-S IgG titers elicited in response to vaccination and/or to previous infection correlate with protection against SARS-CoV-2 infections and symptoms. In this context, it is important to ascertain whether the IgG elicited against the Spike antigen of the original Wuhan isolate can provide protection against infections by emerging variants of concern, including Omicron.
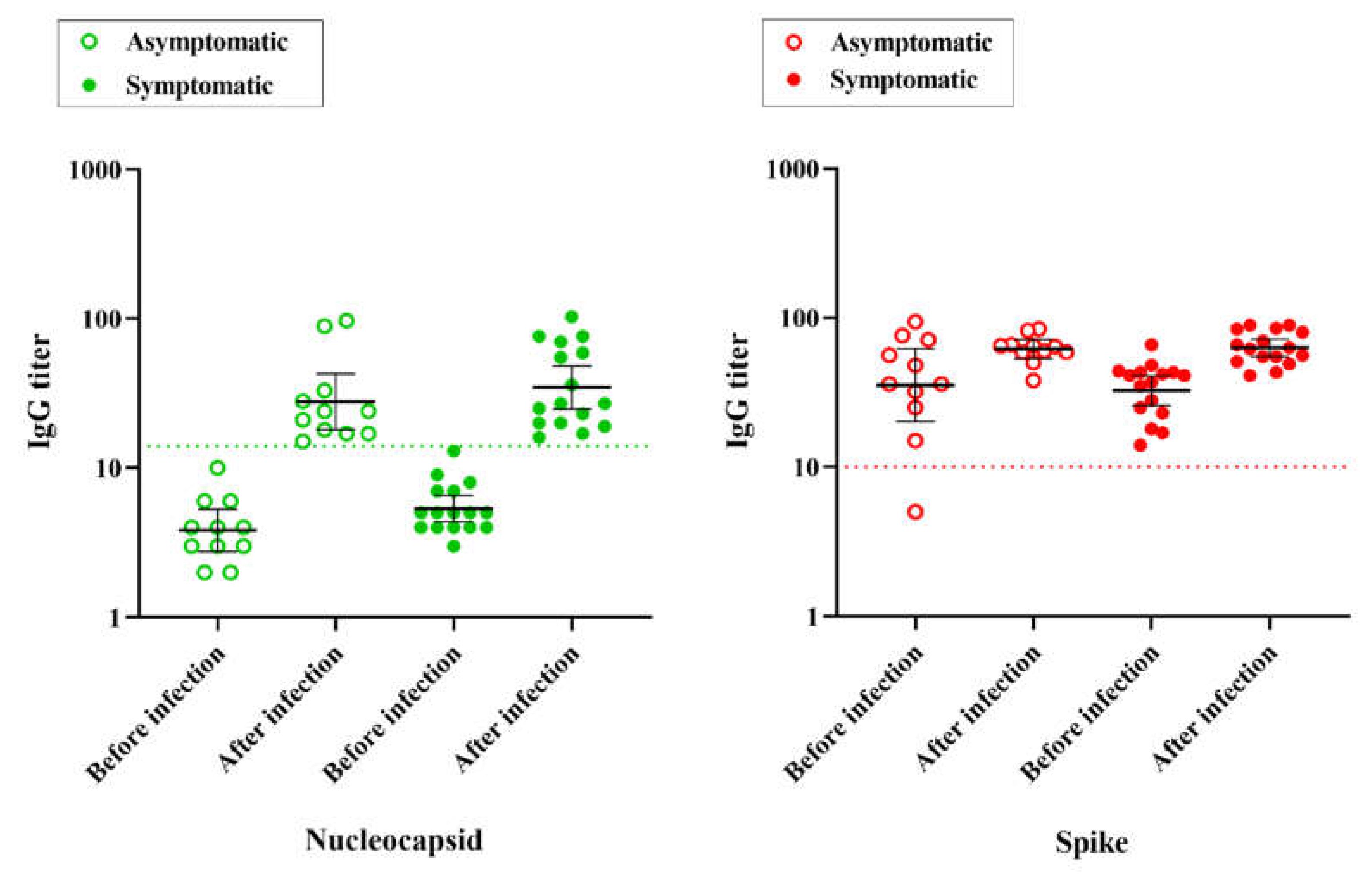
Within the cohort of this study, all except one participant (code B,
Table S2), seroconverted to anti-S IgG as a consequence of vaccination prior to SARS-CoV-2 infection (
Figure 3). The average anti-S IgG titer was 39.2 (SD 20), much higher than the assay cut-off which was 10. All 27 individuals were negative for anti-N IgG before infection, with an average anti-N signal of 5.1 (SD 2.5). As expected, all individuals seroconverted to anti-N IgG after infection, with an average anti-N signal of 39 (SD 28) and showed higher average anti-S IgG titers, with an average signal of 64 (SD 15) (
Figure 3).
Quite remarkably, anti-S IgG titers before the infection were similar among the symptomatic and asymptomatic groups (p = >0.99), with average anti-S IgG titers of 35 and 45, respectively. We noted that even participants having high pre-infection anti-S IgG titers underwent symptomatic infections (See
Figure S1), thereby confirming SARS-CoV-2 Omicron variants were able to escape the humoral response elicited against the previous version of the virus.
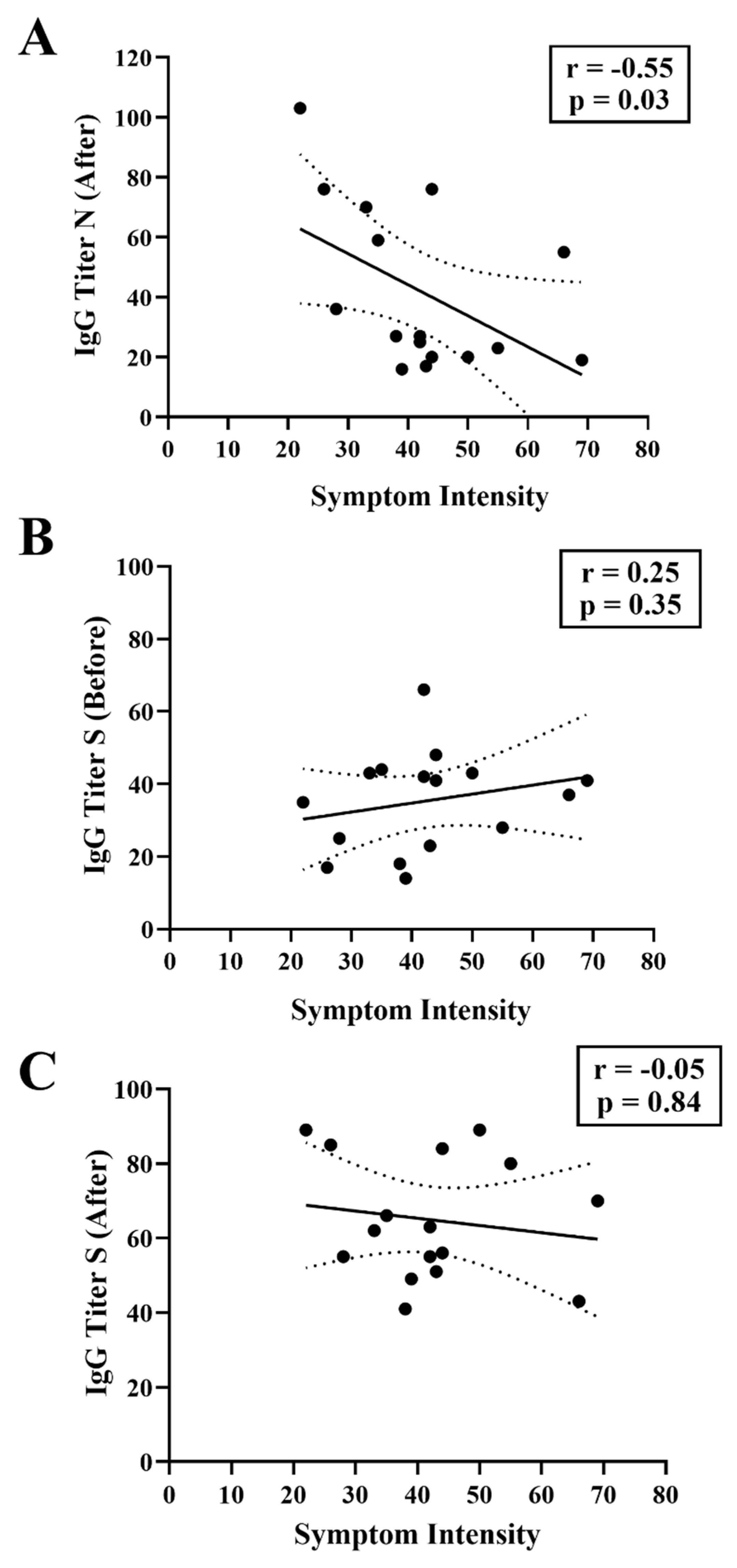
We sought to determine if there was a negative correlation between the anti-S IgG titer before the infection and the severity of the symptoms. However, this was not the case (
Figure 4B). This finding again supports the conclusion that preexisting anti-S IgG levels correlate poorly with symptomatology during infections caused by Omicron variants. There was also no correlation between the COVID-19 symptoms and anti-S IgG titers after the infections (
Figure 4C). Quite surprisingly, COVID-19 symptoms correlated with anti-N IgG levels detected after the infection (Spearman r -0.55, p=0.03). Thus individuals with lower anti-N IgG levels after infection were the ones who experienced the most intense COVID-19 symptoms (
Figure 4A). This observation suggests that human anti-N IgG antibodies may play an important role in resolution of the disease.
4. Discussion
Studies worldwide have indicated that the levels of human IgG antibodies, particularly those reactive to the SARS-CoV-2 Spike protein, correlate with protection against novel infections and to mild and severe COVID-19 [
44,
45,
46,
47]. However, other studies suggest that human anti-S IgG levels wane over time and this may negatively influence protection [
48,
49]. Furthermore, it remains to be determined to what extent the adaptive immunity response mounted against the original version of SARS-CoV-2 may confer protection against emerging viral variants, such as Omicron [
50]. This manuscript provides insights into the symptomatology and anti-N and anti-S IgG levels before and after SARS-CoV-2 Omicron breakthrough infections in vaccinated individuals.
Notably, all except one participant exhibited robust anti-S IgG titers post-vaccination, indicating the effective priming of the immune system against antigens based on the original Wuhan SARS-CoV-2 strain. However, the emergence of SARS-CoV-2 Omicron variants challenged the efficacy of pre-existing immunity, leading to breakthrough infections despite high anti-S IgG titers detected in the participants prior to infections (
Figure 1 and Figure S1). This data therefore implies that Omicron variants are well adapted to circumvent preexisting IgG reactive to the original virus.
Data obtained by others indicate that anti-S IgG levels correlate with protection against SARS-CoV-2 infections, with high IgG levels reducing the symptomatic infections and the possibility to develop severe disease [
44,
45,
51]. However, contrary to expectations, pre-existing anti-S IgG levels did not correlate with symptomatology in our study (
Figure 4B), nor prevent mild symptomatic infections by the Omicron variants (
Table S2), which has also been observed by others [
52,
53]. It is worth mentioning that our data is based on a limited number of individuals that were infected by a limited diversity of SARS-CoV-2 strains (
Table S2), which may explain the differences in the results observed in other studies. Furthermore, our study is limited by the fact that we did not encounter any critical COVID-19 case in our selected cohort of individuals. Nevertheless, our data highlight the importance of monitoring the spread of SARS-CoV-2 VOCs and the need to adapt vaccine formulations to address these emerging viral variants.
One striking observation was the inverse correlation between symptom severity and anti-N IgG levels (
Figure 4A). Symptom variables and post-infection anti-N IgG titers were highly correlated (
Figure 4A. Spearman r -0.55), suggesting that anti-N antibodies may play a role in resolution of the disease. Even though our study had a limited cohort, another study, using a larger cohort, also observed that lower anti-N IgG titers were associated with longer symptom duration [
54]. These observations, which need to be confirmed by further studies, provides novel insights into the relationship between the immune response and disease severity. Furthermore, such findings can help to guide novel vaccine formulations to include the Nucleocapsid along with the Spike antigen in the formulation.
In conclusion, this work provides an overview of Omicron breakthrough SARS-CoV-2 infections in vaccinated individuals, providing insights into immune response dynamics, variant susceptibility, and clinical outcomes which can guide future strategies to combat COVID-19.
Supplementary Materials
Supplementary information on SARS-CoV-2 genome sequencing of individual code N, Tables S1 to S3 and Figures S1 to S3 are available online.
Author Contributions
NMP and LFH, collected and analyzed the data. NMP and EJ, statistical analysis. AGG, supervision. LFH, conceptualization, formal analysis, and funding acquisition. VAB, EMS and FOP, genome sequence and analysis. NMP and LFH wrote the paper with inputs from all authors.
Funding
This research was funded by Federal University of Paraná.
Institutional Review Board Statement
Ethics approval was obtained from CEP/HEG (n# 54095221.0.0000.0098).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data supporting the findings of this study are available on request from the corresponding author (LFH).
Acknowledgments
We thank all the volunteers that contributed samples to this work. We acknowledge the financial support of the Alexander von Humboldt foundation, UFPR, CNPq, CAPES and Fundação Araucária. We acknowledge Profa Leda Castilho (LECC, UFRJ, Brazil) for providing the spike protein used in this study. We acknowledge the support of Ray Dixon (JIC, UK) for critical reading and language editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Iheanacho CO, Eze UIH, Adida EA. A systematic review of effectiveness of BNT162b2 mRNA and ChAdOx1 adenoviral vector COVID-19 vaccines in the general population. Bull Natl Res Cent. 2021; 45(1):150. [CrossRef]
- Cerqueira-Silva T, Andrews JR, Boaventura VS, et al. Effectiveness of CoronaVac, ChAdOx1 nCoV-19, BNT162b2, and Ad26.COV2.S among individuals with previous SARS-CoV-2 infection in Brazil: a test-negative, case-control study. Lancet Infect Dis. 2022; 22(6):791-801. [CrossRef]
- Cerqueira-Silva T, Katikireddi SV, de Araujo Oliveira V, et al. Vaccine effectiveness of heterologous CoronaVac plus BNT162b2 in Brazil. Nat Med. 2022; 28(4):838-843. [CrossRef]
- Fadlyana E, Setiabudi D, Kartasasmita CB, et al. Immunogenicity and safety in healthy adults of full dose versus half doses of COVID-19 vaccine (ChAdOx1-S or BNT162b2) or full-dose CoronaVac administered as a booster dose after priming with CoronaVac: a randomised, observer-masked, controlled trial in Indonesia. Lancet Infect Dis. 2023; 23(5):545-555. [CrossRef]
- Voysey M, Clemens SAC, Madhi SA, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK [published correction appears in Lancet. 2021 Jan 9; 397(10269):98]. Lancet. 2021; 397(10269):99-111. [CrossRef]
- Kahn R, Janusz CB, Castro MC, et al. The effectiveness of COVID-19 vaccines in Latin America, 2021: a multicenter regional case-control study. Lancet Reg Health Am. 2023; 20:100474. [CrossRef]
- Santos CVBD, Valiati NCM, et al. The effectiveness of COVID-19 vaccines against severe cases and deaths in Brazil from 2021 to 2022: a registry-based study. Lancet Reg Health Am. 2023 Apr; 20:100465. [CrossRef] [PubMed] [PubMed Central]
- Ravichandran S, Lee Y, Grubbs G, et al. Longitudinal antibody repertoire in “mild” versus “severe” COVID-19 patients reveals immune markers associated with disease severity and resolution. Sci Adv. 2021 Mar 5; 7(10): eabf2467. [CrossRef] [PubMed] [PubMed Central]
- Wu J, Liang BY, Fang YH, et al. Occurrence of COVID-19 Symptoms During SARS-CoV-2 Infection Defines Waning of Humoral Immunity. Front Immunol. 2021; 12:722027. Published 2021 Aug 16. [CrossRef]
- Hajilooi M, Keramat F, Moazenian A, et al. The quantity and quality of anti-SARS-CoV-2 antibodies show contrariwise association with COVID-19 severity: lessons learned from IgG avidity. Med Microbiol Immunol. 2023; 212(3):203-220. [CrossRef]
- Paula NM, Conzentino MS, Gonçalves ACA, et al. Population-Based Analysis of the Immunoglobulin G Response to Different COVID-19 Vaccines in Brazil. Vaccines. 2023; 11(1):21.
- Moreira ED Jr, Kitchin N, Xu X, et al. Safety and Efficacy of a Third Dose of BNT162b2 Covid-19 Vaccine. N Engl J Med. 2022;386(20):1910-1921. [CrossRef]
- Barin B, Kasap U, Selçuk F, Volkan E, Uluçkan Ö. Comparison of SARS-CoV-2 anti-spike receptor binding domain IgG antibody responses after CoronaVac, BNT162b2, ChAdOx1 COVID-19 vaccines, and a single booster dose: a prospective, longitudinal population-based study. Lancet Microbe. 2022;3(4):e274-e283. [CrossRef]
- Pratesi F, Caruso T, Testa D, et al. BNT162b2 mRNA SARS-CoV-2 Vaccine Elicits High Avidity and Neutralizing Antibodies in Healthcare Workers. Vaccines (Basel). 2021 Jun 18;9(6):672. [CrossRef] [PubMed] [PubMed Central]
- Feikin DR, Higdon MM, Abu-Raddad LJ, et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression [published correction appears in Lancet. 2022 Apr 4;:] [published correction appears in Lancet. 2023 Feb 25;401(10377):644]. Lancet. 2022;399(10328):924-944. [CrossRef]
- Ssentongo P, Ssentongo AE, Voleti N, et al. SARS-CoV-2 vaccine effectiveness against infection, symptomatic and severe COVID-19: a systematic review and meta-analysis. BMC Infect Dis. 2022;22(1):439. [CrossRef]
- Shrotri M, Navaratnam AMD, Nguyen V, et al. Spike-antibody waning after second dose of BNT162b2 or ChAdOx1. Lancet. 2021 Jul 31;398(10298):385-387. [CrossRef] [PubMed] [PubMed Central]
- Levin EG, Lustig Y, Cohen C, et al. Waning Immune Humoral Response to BNT162b2 Covid-19 Vaccine over 6 Months. N Engl J Med. 2021;385(24):e84. [CrossRef]
- Mao Y, Wang W, Ma J, Wu S, Sun F. Reinfection rates among patients previously infected by SARS-CoV-2: systematic review and meta-analysis. Chin Med J (Engl). 2022;135(2):145-152. Published 2022 Jan 20. [CrossRef]
- Hansen CH, Michlmayr D, Gubbels SM, Mølbak K, Ethelberg S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. 2021;397(10280):1204-1212. [CrossRef]
- Nordström P, Ballin M, Nordström A. Risk of SARS-CoV-2 reinfection and COVID-19 hospitalisation in individuals with natural and hybrid immunity: a retrospective, total population cohort study in Sweden [published correction appears in Lancet Infect Dis. 2022 Apr 8;:]. Lancet Infect Dis. 2022;22(6):781-790. [CrossRef]
- Chemaitelly H, Nagelkerke N, Ayoub HH, et al. Duration of immune protection of SARS-CoV-2 natural infection against reinfection. J Travel Med. 2022;29(8):taac109. [CrossRef]
- Schmidt, F., Weisblum, Y., Rutkowska, M. et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature 600, 512–516 (2021). [CrossRef]
- Wratil PR, Stern M, Priller A, et al. Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern. Nat Med. 2022;28(3):496-503. [CrossRef]
- Andrews N, Stowe J, Kirsebom F, et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N Engl J Med. 2022;386(16):1532-1546. [CrossRef]
- Altarawneh HN, Chemaitelly H, Ayoub HH, et al. Effects of Previous Infection and Vaccination on Symptomatic Omicron Infections. N Engl J Med. 2022;387(1):21-34. [CrossRef]
- Freire-Neto FP, Teixeira DG, da Cunha DCS, et al. SARS-CoV-2 reinfections with BA.1 (Omicron) variant among fully vaccinated individuals in northeastern Brazil. PLoS Negl Trop Dis. 2022;16(10):e0010337. Published 2022 Oct 3. [CrossRef]
- Oliveira JR, Ruiz CMR, Machado RRG, et al. Immunodominant antibody responses directed to SARS-CoV-2 hotspot mutation sites and risk of immune escape. Front Immunol. 2023;13:1010105. Published 2023 Jan 5. [CrossRef]
- Rhoden J, Hoffmann AT, Stein JF, et al. Diversity of Omicron sublineages and clinical characteristics in hospitalized patients in the southernmost state of Brazil. BMC Infect Dis. 2024;24(1):193. Published 2024 Feb 13. [CrossRef]
- Sette A, Crotty S. Immunological memory to SARS-CoV-2 infection and COVID-19 vaccines. Immunol Rev. 2022;310(1):27-46. [CrossRef]
- Goel RR, Apostolidis SA, Painter MM, et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci Immunol. 2021;6(58):eabi6950. [CrossRef]
- Otto SP, Day T, Arino J, et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr Biol. 2021;31(14):R918-R929. [CrossRef]
- Calistri P, Amato L, Puglia I, et al. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int J Infect Dis. 2021;105:753-755. [CrossRef]
- Zhang D, Zhong J, Xiong H, et al. Protective Effect of Inactivated COVID-19 Vaccines against Omicron BA.2 Infection in Guangzhou: A Test-Negative Case-Control Real-World Study. Vaccines (Basel). 2023;11(3):566. Published 2023 Mar 1. [CrossRef]
- Garrett N, Tapley A, Andriesen J, et al. High Rate of Asymptomatic Carriage Associated with Variant Strain Omicron. Preprint. medRxiv. 2022;2021.12.20.21268130. Published 2022 Jan 14. [CrossRef]
- Carabelli AM, Peacock TP, Thorne LG, et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. 2023;21(3):162-177. [CrossRef]
- Markov PV, Ghafari M, Beer M, et al. The evolution of SARS-CoV-2. Nat Rev Microbiol. 2023;21(6):361-379. [CrossRef]
- Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185(14):2422-2433.e13. [CrossRef]
- Huergo LF, or Paula NM et al. 2022. SARS-CoV-2 Seroconversion in Response to Infection and Vaccination: a Time Series Local Study in Brazil. Microbiol Spectr 10:e01026-22. [CrossRef]
- Conzentino MS, Gonçalves ACA, Paula NM, et al. A magnetic bead immunoassay to detect high affinity human IgG reactive to SARS-CoV-2 Spike S1 RBD produced in Escherichia coli. Braz J Microbiol. 2022;53(3):1263-1269. [CrossRef]
- Conzentino MS, Santos TPC, Selim KA, et al. Ultra-fast, high throughput and inexpensive detection of SARS-CoV-2 seroconversion using Ni2+ magnetic beads. Anal Biochem. 2021 Oct 15;631:114360. [CrossRef] [PubMed] [PubMed Central]
- Bochnia-Bueno L, De Almeida, S. M., Raboni, S. M. et al. Dynamic of humoral response to SARS-CoV-2 anti-Nucleocapsid and Spike proteins after CoronaVac vaccination. Diagnostic Microbiology and Infectious Disease, v. 102, n. 3, p. 115597, 2022.
- Rambaut, A., Holmes, E.C., O’Toole, Á. et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5, 1403–1407 (2020). [CrossRef]
- Wei, J., Pouwels, K.B., Stoesser, N. et al. Antibody responses and correlates of protection in the general population after two doses of the ChAdOx1 or BNT162b2 vaccines. Nat Med 28, 1072–1082 (2022). [CrossRef]
- Gilbert PB, Montefiori DC, McDermott AB, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375(6576):43-50. [CrossRef]
- Yamamoto S, Mizoue T, Ohmagari N. Analysis of Previous Infection, Vaccinations, and Anti-SARS-CoV-2 Antibody Titers and Protection Against Infection With the SARS-CoV-2 Omicron BA.5 Variant. JAMA Netw Open. 2023;6(3):e233370. Published 2023 Mar 1. [CrossRef]
- Murray, S.M., Ansari, A.M., Frater, J. et al. The impact of pre-existing cross-reactive immunity on SARS-CoV-2 infection and vaccine responses. Nat Rev Immunol 23, 304–316 (2023). [CrossRef]
- Yang ZR, Jiang YW, Li FX, et al. Efficacy of SARS-CoV-2 vaccines and the dose-response relationship with three major antibodies: a systematic review and meta-analysis of randomised controlled trials. Lancet Microbe. 2023;4(4):e236-e246. [CrossRef]
- Buhre JS et al. mRNA vaccines against SARS-CoV-2 induce comparably low long-term IgG Fc galactosylation and sialylation levels but increasing long-term IgG4 responses compared to an adenovirus-based vaccine. Frontiers in immunology, v. 13, p. 1020844, 2023.
- Murray SM, Ansari AM, Frater J, et al. The impact of pre-existing cross-reactive immunity on SARS-CoV-2 infection and vaccine responses. Nat Rev Immunol. 2023;23(5):304-316. [CrossRef]
- Goguet, Emilie et al. Immune and behavioral correlates of protection against symptomatic post-vaccination SARS-CoV-2 infection. Frontiers in Immunology, v. 15, p. 1287504, 2024.
- Spiteri G, D’Agostini M, Abedini M, et al. Protective role of SARS-CoV-2 anti-S IgG against breakthrough infections among European healthcare workers during pre and post-Omicron surge-ORCHESTRA project. Infection. Published online February 7, 2024. [CrossRef]
- Yang ZR, Jiang YW, Li FX, et al. Efficacy of SARS-CoV-2 vaccines and the dose-response relationship with three major antibodies: a systematic review and meta-analysis of randomised controlled trials. Lancet Microbe. 2023;4(4):e236-e246. [CrossRef]
- Jia X, Cao S, Lee AS, et al. Anti-nucleocapsid antibody levels and pulmonary comorbid conditions are linked to post-COVID-19 syndrome. JCI Insight. 2022;7(13):e156713. Published 2022 Jul 8. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).