1. Introduction
Oral leukoplakia (OL), as defined oral by the World Health Organization, is a condition where a white patch of oral mucosa cannot be erased or classified otherwise, either clinically or histopathologically. OL is the most common oral potentially malignant disorders (OPTMs), defined as any oral mucosal abnormality that is associated with a statistically increased risk of developing oral cancer.[
1] Oral cancer stands as a considerable global public health concern, constituting a substantial proportion of overall cancer incidence. Annually, millions of individuals receive diagnoses of oral cancer, contributing significantly to the global cancer burden. The American Cancer Society reports an anticipated 54,540 new cases of oral cavity or oropharyngeal cancer in 2023, with an estimated 11,580 fatalities attributed to these specific cancer types in the same year.[
2] The utilization of tobacco and the consumption of alcohol are recognized as established risk factors for oral cancer, with approximately 75% of all reported cancer cases being linked to these specific exposures.[
3,
4,
5,
6] Additional factors, including human papillomavirus (HPV) infection, dietary habits, nutritional considerations, gender, and exposure to radiation, contribute to an elevated likelihood of being diagnosed with oral cancer.[
7,
8,
9] Cancers also result in productivity losses, with oral cancer causing an economic impact of approximately
$0.74 billion in India and
$112,308 in South Africa.[
10]
Early detection of oral cancer is crucial for successful treatment.[
11,
12] The primary diagnostic methods for oral cancer involve clinical examination, biopsy, imaging studies (X-rays, CT scans, and MRI scans), and endoscopy.[
13,
14,
15] Clinical examination entails visual inspection by a healthcare professional to identify visible abnormalities.[
16] Biopsy is essential for confirming the presence of cancerous cells and determining cancer type and stage.[
17] Imaging studies, including X-rays and CT scans, provide detailed images aiding in tumor identification.[
18] These common diagnostic approaches facilitate early detection, crucial for effective treatment and improved outcomes in oral cancer cases. While there are variety of diagnostic methods, they have inherent limitations. Clinical examinations may be subjective and limited to surface observations, potentially missing deeper lesions. Biopsies, while definitive, are invasive and carry a risk of sampling errors due to the small tissue sample obtained.[
11,
19,
20] Imaging studies can produce false positives/negatives and expose patients to radiation.[
21] Acknowledging these limitations emphasizes the ongoing need for research to refine existing methods and explore complementary approaches for more accurate and comprehensive oral cancer diagnostics.
P90, alternatively identified as KIAA1524 and Cancerous Inhibitor of PP2A (CIP2A), exhibits elevated expression levels in both oral squamous cell carcinoma (OSCC) cell lines and tissues.[
22,
23,
24,
25] This protein is of considerable interest as a promising therapeutic target or a potential diagnostic marker given the relatively low levels of CIP2A expression in normal tissues.[
26] There are additional reports that implicate a positive role for protein phosphate 2A (PP2A) in inflammatory lung diseases like asthma and chronic obstructive pilmonary disease (COPD) and in heart function.[
27] These effects are attributed to PP2A‘s inhibitory effects on the mediators of inflammation. For the analysis of CIP2A in saliva, enzyme-linked immunoassay (ELISA) test kits are readily accessible, and the development of immunosensors utilizing carbon nanotubes has commenced.[
28] However, growing the nanotubes is time-consuming and also quite expensive. A recently introduced method for detecting CIP2A showcases high sensitivity through a transistor-based biosensor system. The P90 protein is treated with a PBS solution in this process, achieving a low limit of detection at 10
-15 g/mL. Notably, the sensitivity of this method surpasses that of commercially available ELISA test kits.[
24] While exhibiting high sensitivity and a low limit of detection, it is noteworthy that the standard calibration solution used in this method is PBS, rather than artificial saliva. Furthermore, human sample testing was not conducted. In contrast, our study established a calibration curve using P90 protein diluted in artificial saliva. A specially designed printed circuit board (PCB) was implemented for the detection of P90 protein concentrations. The accuracy of the newly developed method was demonstrated through testing human samples obtained from oral cancer patients.
2. Materials and Methods
Commercially available glucose test strips (Luvnshare Biomedical Inc. in Hsinchu, Taiwan) were used in this study, which is shown in
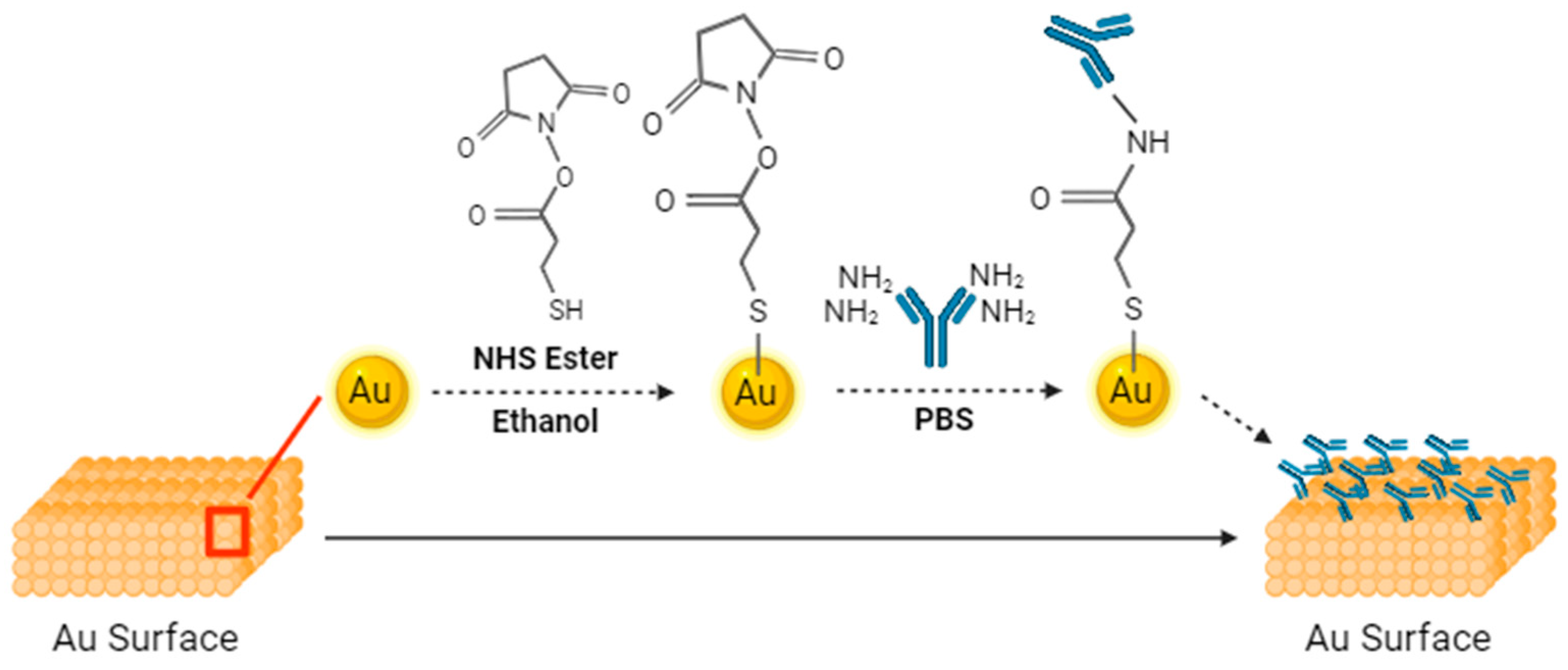
Figure 1. The tip of the strips features microfluidic channels for sample injection. A gold-plated electrode is present on the tip, undergoing a sequence of functionalization processes with the P90 antibody. The functionalization step is shown in
Figure 2. This functionalization enables the strips to discern variations between samples. The initial step in the functionalization process is the ozone treatment of the strips for a duration of 15 minutes, which effectively removes any carbon residues. Subsequently, a diluted ammonium hydroxide (NH
4OH) solution is used to eliminate gold oxide. Following this surface cleaning step, deionized (DI) water is employed to rinse the channels, and nitrogen is utilized for the drying process. The next phase involves preparing a 3-Mercaptopropanyl-N-hydroxysuccinimide ester (NHS ester) solution, which is dissolved in ethanol. NHS ester has a three-carbon chain ending in a thiol group, with an attached N-hydroxysuccinimide ester. This compound is utilized for bioconjugation, offering a reactive site for selective coupling with amine-containing molecules. The strips are submerged in this solution and left to react for 2 hours. The channels are then cleaned using DI water and nitrogen. Monoclonal CIP2A antibody 2G10-3B5 (Santa Cruz Biotechnology, Dallas, TX) at a concentration of 20 μg/mL was injected into the channel, and the strips were sealed and stored in a disk at 4°C for 18 hours. Lastly, ethanolamine was employed to deactivate un-functionalized groups, mitigating the risk of potential interference. The previous research provided confirmation of antibody functionalization through uniform methodologies, as indicated by current-voltage and capacitance measurements.[
29,
30,
31] CIP2A Protein (MyBioSource, San Diego, CA) were diluted into series of concentration with artificial saliva (Pickering Laboratories Inc., Mountain View, CA) to establish the calibration curve.
Seventeen human saliva samples and tissue samples were obtained from individuals, including both pre-oral cancer patients and healthy volunteers, through collaboration with the University of Florida Oral Pathology Clinic and Dental Clinical Research Unit. The age range of the healthy samples was from 20 to 80 years old, while samples from inflammatory oral lesions (clinically diagnosed as leukoplakia) ranged from 40 to 80 years old.
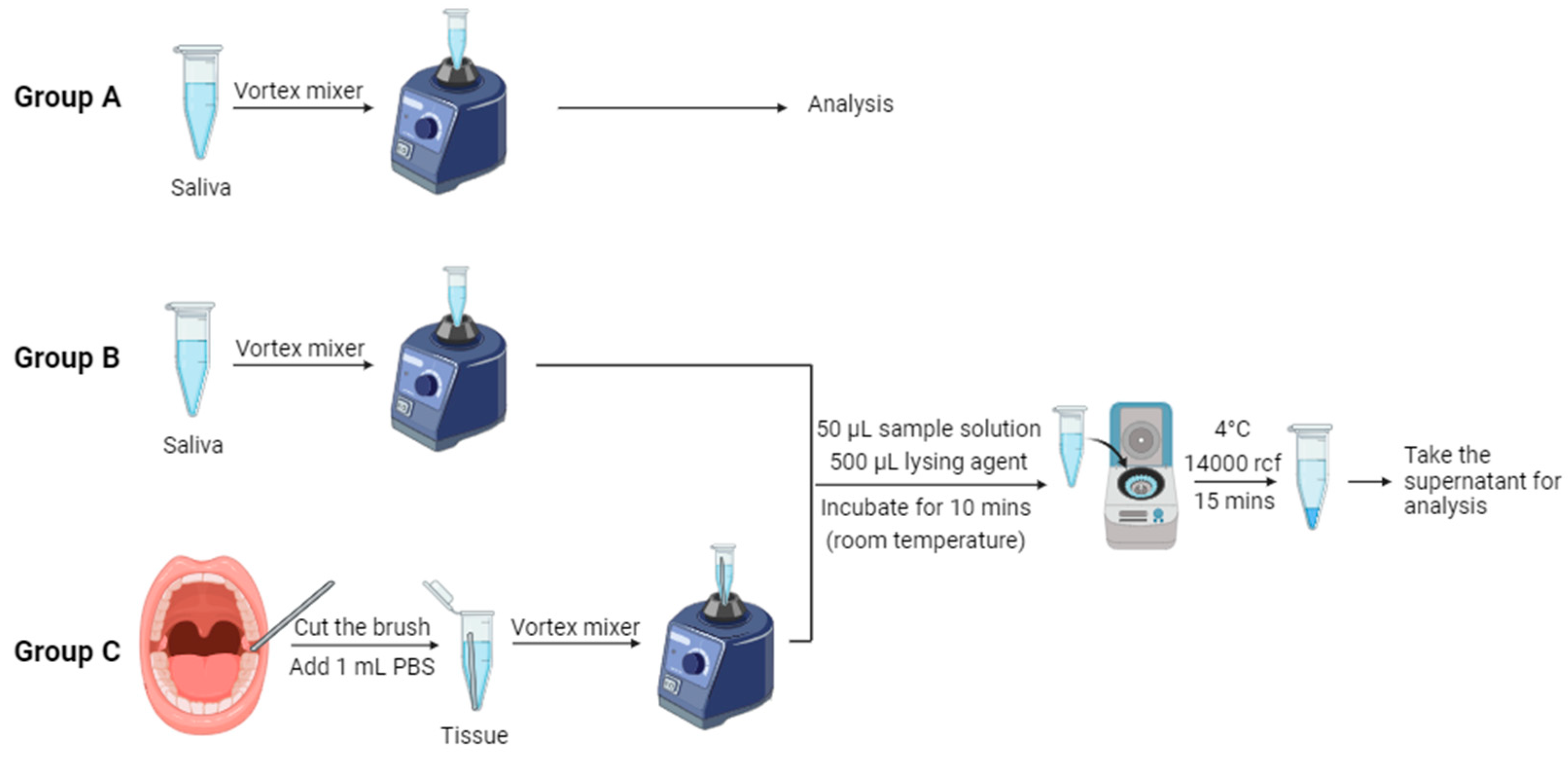
Table 1 displays comprehensive information regarding the patients examined in this study. We aim to investigate whether age influences the detection results. Brush kits (Andwin Scientific, Simi Valley, CA) were used in the collection of tissue samples. These specimens were carefully preserved in a deep-freeze storage unit at -78℃ . In this study, we tested three groups of samples: Group A consisted of saliva samples without cell lysis, Group B comprised lysed saliva samples, and Group C comprised of lysed tissue samples. The native lysis buffer used for saliva and tissue samples was purchased from Thermo Fisher Scientific (Massachusetts, U.S.). The sample collection and cell lysis procedure are shown in
Figure 3. Epithelium cells were obtained by turning the brushes against the mucosa inside the oral cavity. The head of the brushes was then cut and placed in a 1.5 mL microcentrifuge tube. 1 mL of 1x PBS buffer solution was added into the tube, and the tissue samples were suspended in the solution by vortex mixer. In the cell lysis procedure, the sample solution was initially combined with the native lysis buffer in a 1:10 ratio in a 1.5 mL microcentrifuge tube. Here, we took 50 μL (1 drop) sample solution with 50 μL lysing agent, followed by thorough mixing using a vortex mixer. Subsequently, the mixture was incubated at room temperature for 10 minutes. Finally, the mixture was centrifuged at 14000 rcf for 15 minutes at 4℃ to separate the resulting supernatant and pellet, which were then stored in the refrigerator for subsequent analysis.
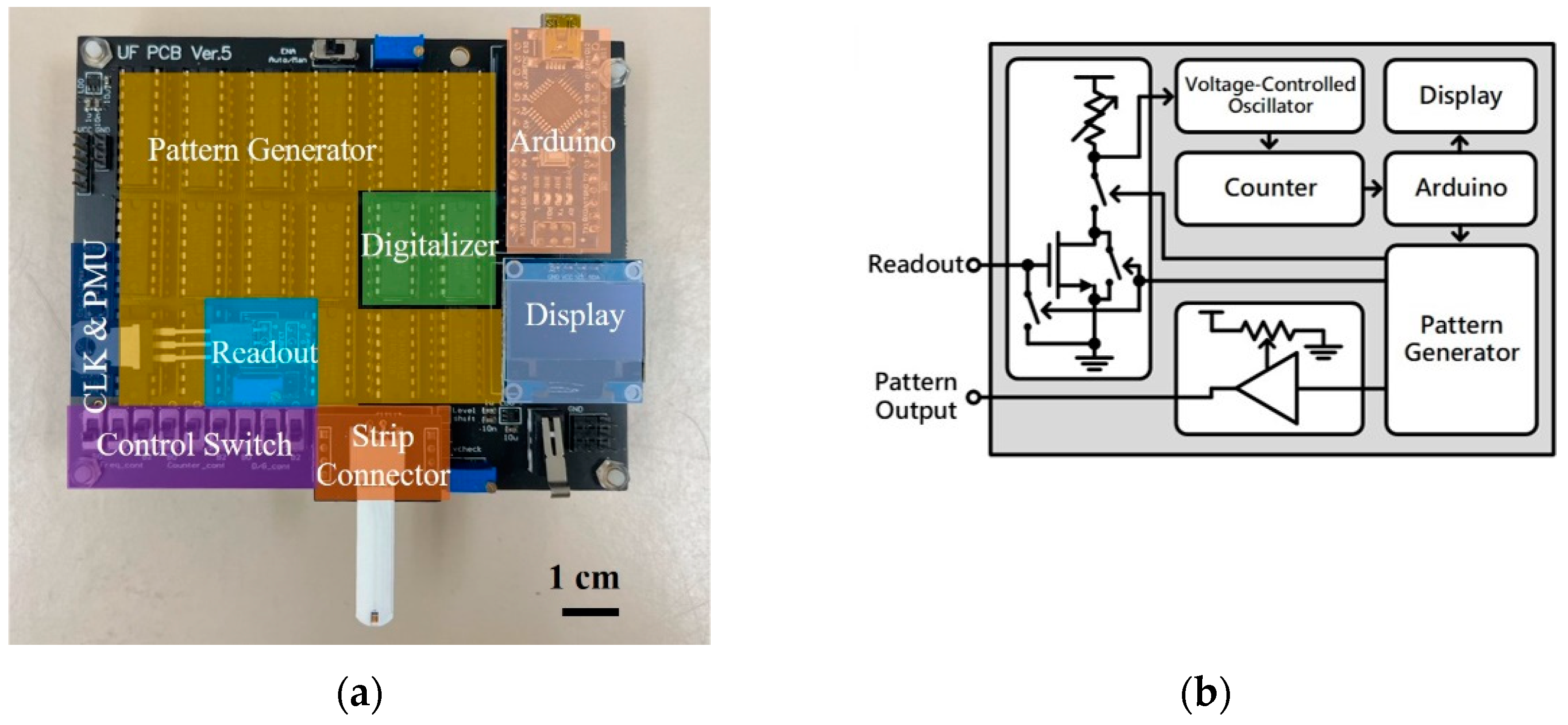
The printed circuit board (PCB) used in this study is shown in
Figure 4(a). This portable detection device consists of the readout block (Readout), pattern generator (Pattern Gen.), digitalizer, strip connector, Arduino, display, control switch, and system clock and power management unit (CLK & PMU).
Figure 4(b) demonstrates the simplified circuit diagram of the PCB. The following paragraphs will explain how this device operates and provide details about the device.
The process for the sensor readout is as follows. First, the strip is connected to the strip connector while the Arduino is activated. The Arduino generates signals triggering the pattern generator to generate a test pattern for measurement. The test pattern, passing through the strip, produces output signals of different magnitudes. This signal is then inputted into the gate terminal of the metal-oxide-semiconductor field-effect transistor (MOSFET) in the readout block, where amplification occurs using the MOSFET and its drain-side potentiometer. The amplified readout signal is converted into a frequency signal by the voltage-controlled oscillator (VCO) in the digitalizer. This frequency signal is then counted at fixed intervals by the counter. When the VCO reads a higher voltage, resulting in a higher output frequency, the counter outputs a larger value. Conversely, when the VCO reads a lower voltage, resulting in a lower output frequency, the counter outputs a smaller value. Because the counter's output varies with the MOSFET's output voltage, this value can be used as a digital representation of the readout voltage. Finally, the counter's output is processed by the Arduino and displayed on the mini-LCD screen, allowing users to directly read the numerical value to determine the concentration of the solution on the strip.
In each measurement, the device outputs multiple test patterns for repeated measurements, and the results are averaged to reduce measurement errors. To avoid the charge accumulation effect on the strip during the measurement process, the gate terminal of the MOSFET is grounded to release the accumulated charge on the strip and gate terminals after each test pattern measurement is completed. This ensures the accuracy of each measurement. Additionally, this device has high adjustability. For example, the control switch can adjust the length of the test pattern and control the timing of the digitalizer's voltage reading. The Arduino can control the time interval and frequency of test pattern generation. The voltage of the test pattern can be adjusted by the potentiometer on the PCB. Moreover, the MOSFET on the PCB uses an active socket, allowing for the replacement of MOSFET. Multiple adjustable parameters can be optimized according to the type of strip, ensuring that measurement parameters fall within the optimal range.
Several characteristic tests were performed on the PCB, including varying the load resistance, gate voltage, and also adding an external capacitor parallel to the MOSFET. The heightened concentration of the sample solution induces an elevation in capacitance, elucidated by the electric double layer theory.[
32] Consequently, commercial external capacitors were employed as the strip during the test to determine the optimal operational settings.
3. Results and Discussion
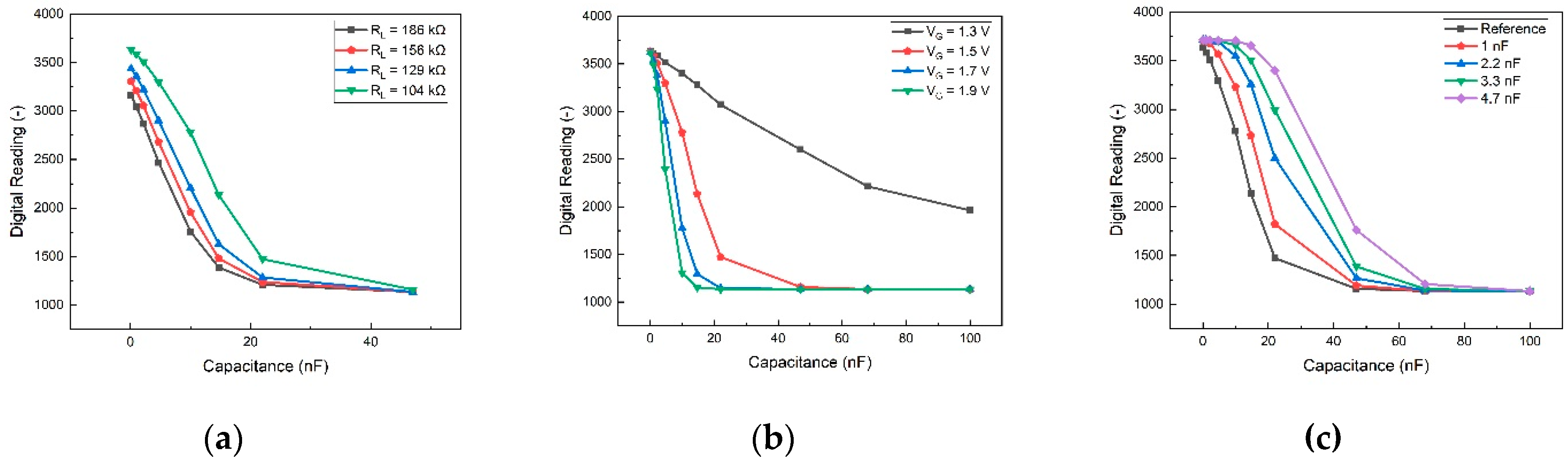
Tests were performed on the PCB, adjusting load resistance, gate voltage, and adding an external capacitor parallel to the MOSFET. The outcomes of these tests are illustrated in
Figure 3. These results serve as a guide for optimizing the board's conditions to enhance the sensitivity of our detection method. In
Figure 5(a), the impact of adjusting the load resistance connected to the supply voltage VDD on the PCB is demonstrated. A lower load resistance creates more space for voltage drop, expanding the operational range for testing and enhancing sensitivity. However, due to constraints imposed by other components on the PCB, the lowest permissible condition is set at 104 kΩ. Consequently, 104 kΩ was selected as the load resistance for subsequent tests.
Figure 5(b) presents the device's performance under varying gate voltages. Sensitivity increases with higher gate voltages, and although V
G=1.9 V exhibits the steepest slope for optimal sensitivity, it yields a smaller detection range compared to other gate voltages. Striking a balance between sensitivity and detection range, V
G=1.5 V was employed in subsequent biomarker tests. The outcomes of incorporating an external capacitor parallel to the MOSFET are depicted in
Figure 5(c). The total capacitance (
) of the strips and MOSFET can be expressed as:
Here, is the MOSFET capacitance, is the external capacitor capacitance, and is the strip capacitance. Notably, the digital reading exhibits significant differences only when the MOSFET capacitance approaches that of the strips. Consequently, when the sample's capacitance is substantial, the external capacitor proves beneficial in extending the detection range towards larger capacitance values.
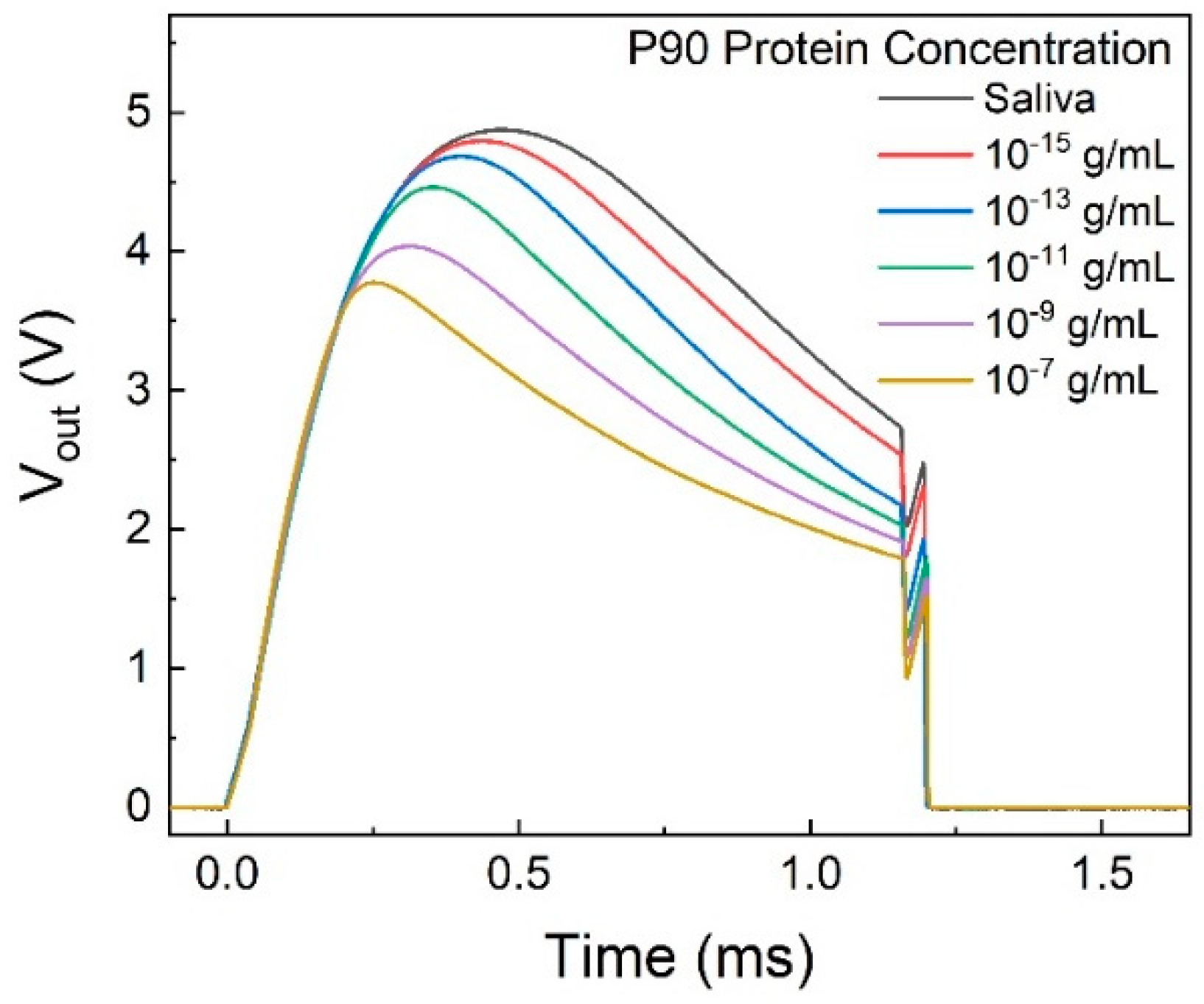
The output voltage from the printed circuit board (PCB) with a series of P90 concentration standard solutions is depicted in
Figure 6. The pattern of the voltage pulse can be elucidated by the double spring model.[
33]
The digital output reading is obtained by integrating the area under the curve of the output voltage.
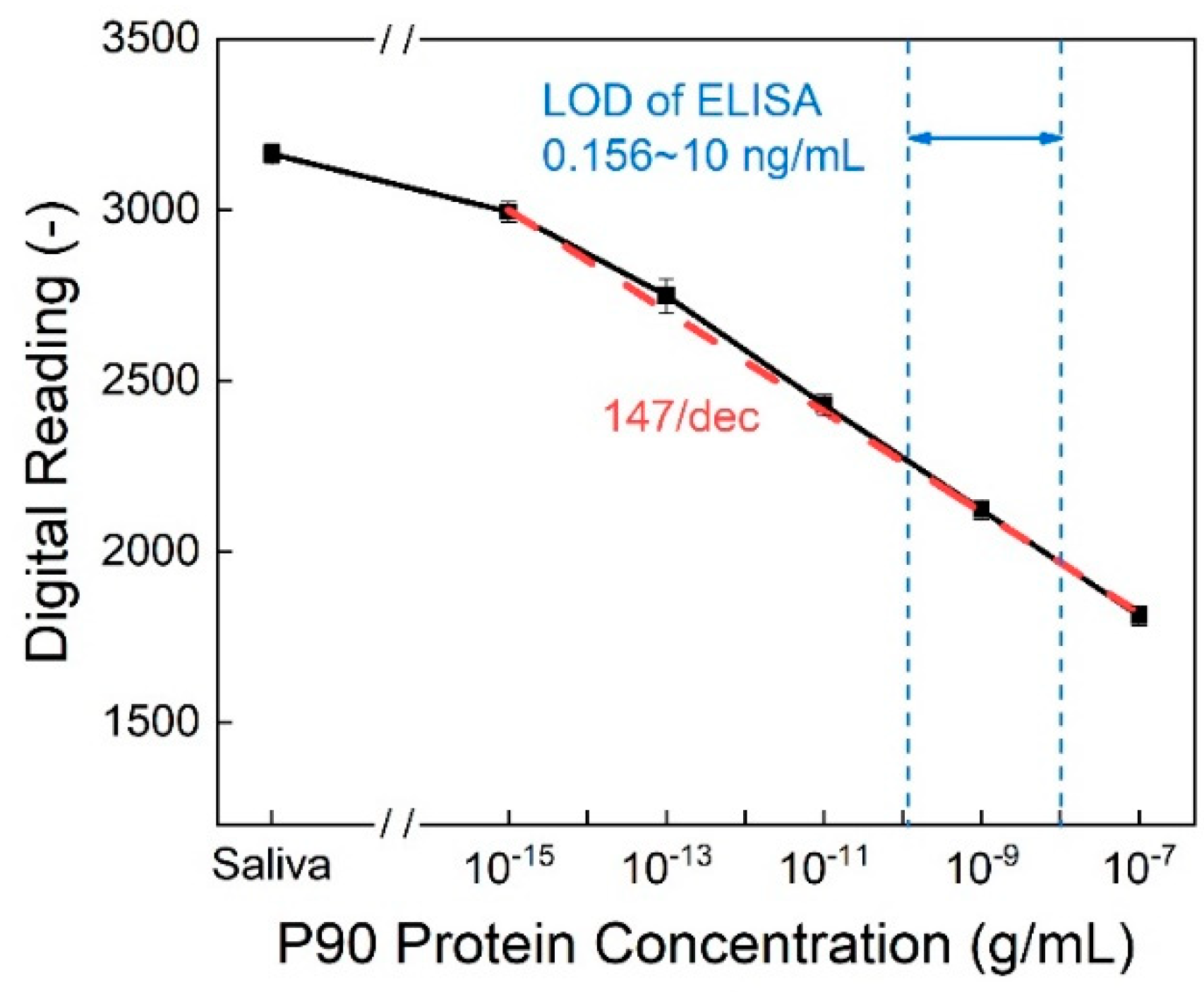
Figure 7 illustrates the calibration curve, showcasing a sensitivity of 147/dec. This implies that the digital reading decreases by approximately 147 when the protein concentration increases by one order of magnitude. In addition to its commendable sensitivity, the detection method achieves a limit of detection (LOD) as low as 10
-15 g/mL while the detection range of commercial ELISA kits is limited to 0.156 to 10 ng/mL.[
24]
In addition to evaluating the standard solution, human sample testing was conducted in this study.
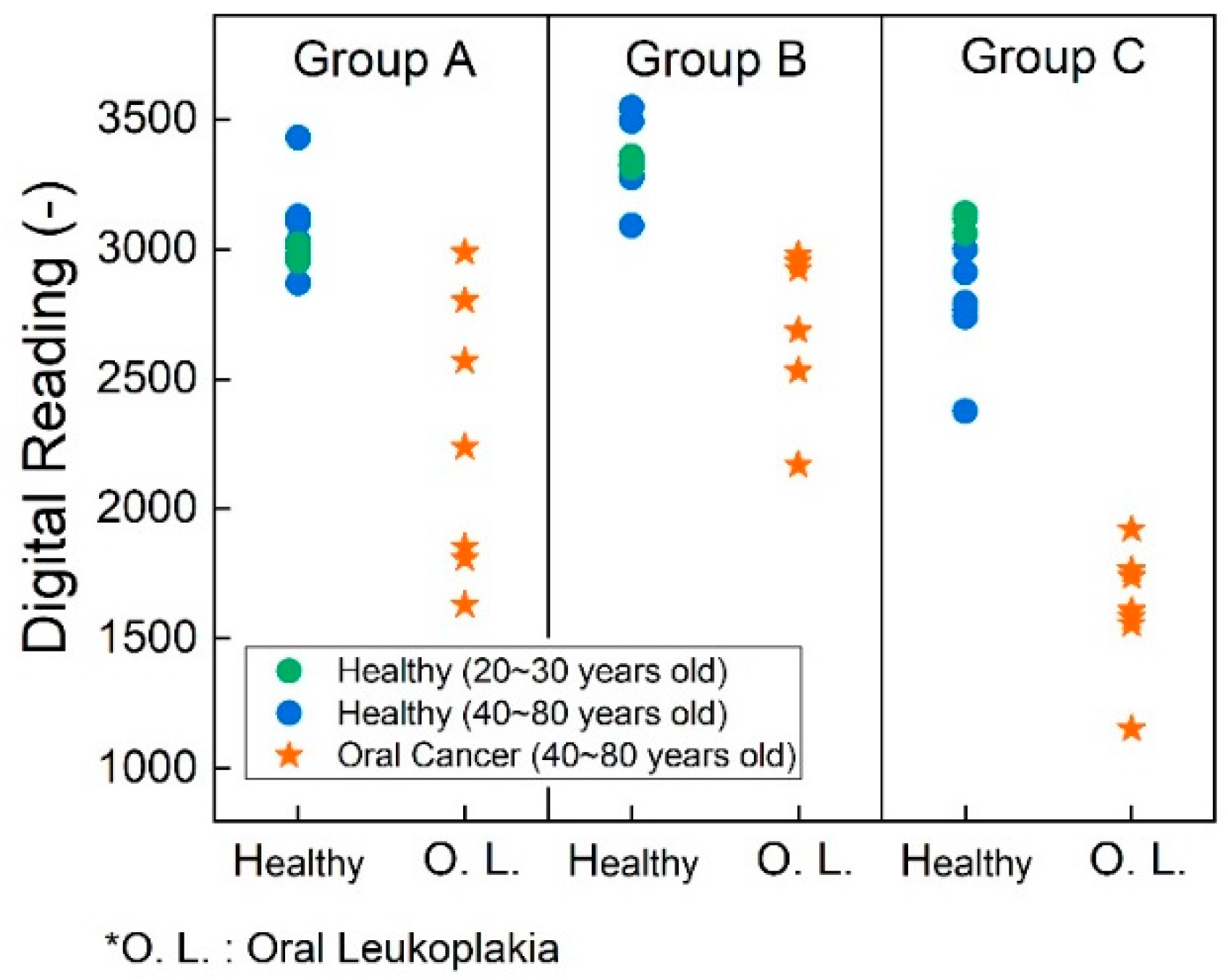
Figure 8 illustrates the test results of human sample test with saliva and tissue samples after the cell lysis process. We conducted experiments on three distinct sample groups. Group A comprises saliva samples without undergoing cell lysis processing. Group B underwent testing after saliva lysis, whereas Group C consisted of tissue samples subjected to lysis before testing. We employed different colors to distinguish age ranges in the figure: green dots represent data from healthy volunteers aged 20 to 30 years, while blue dots represent individuals aged 40 to 80 years. Across groups A, B, and C, there was no significant difference in P90 expression, consistent with Bockelman et al.’s findings.[
34] Notably, group A utilized distinct PCB settings compared to groups B and C, as determined by the earlier capacitance study, aimed at optimizing detection sensitivity.
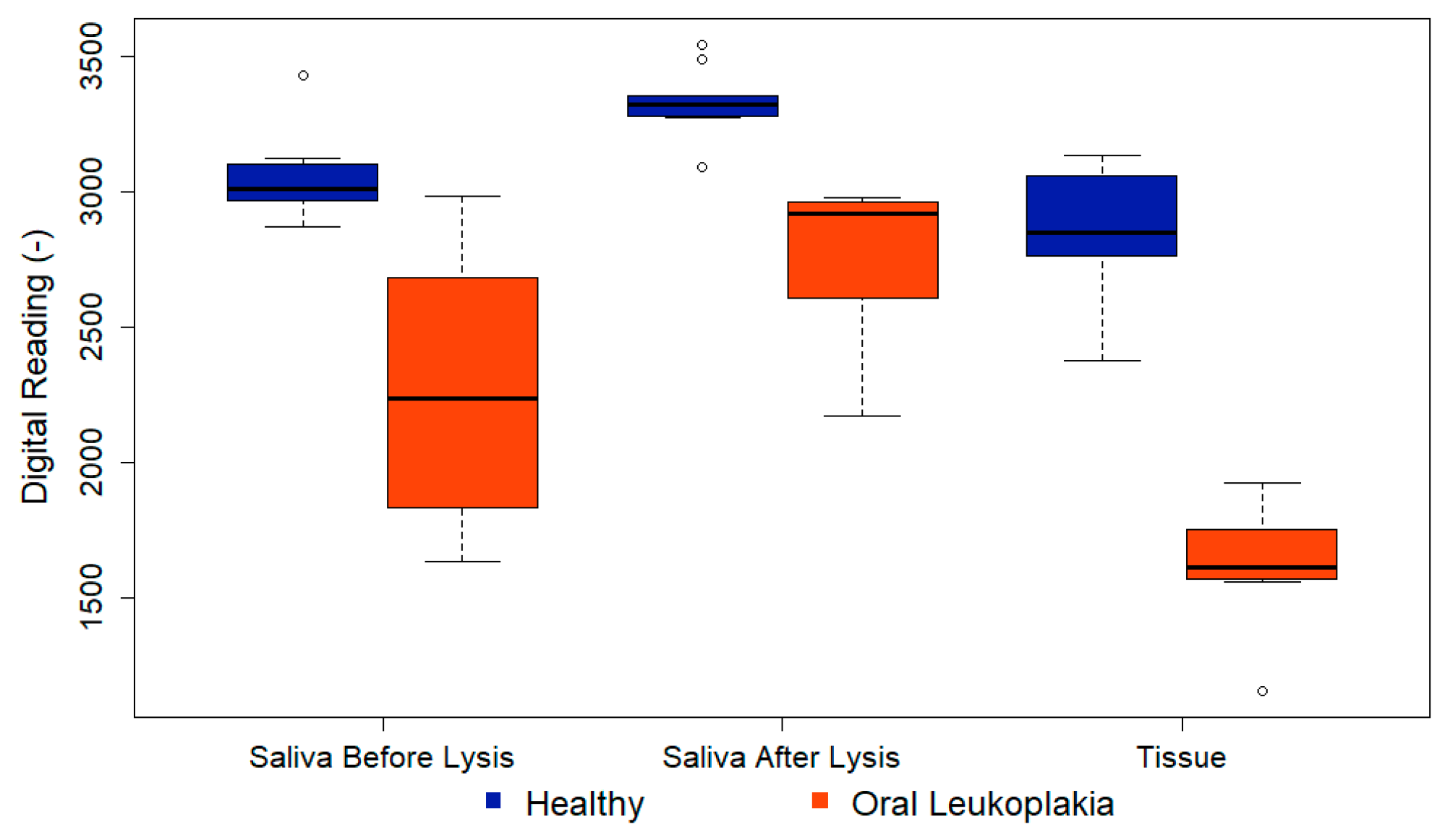
Figure 9 illustrates a boxplot representing the distribution of testing results. Within the boxplot, the bottom corresponds to the 25th percentile (Q1), the middle line indicates the median, and the top of the box signifies the 75th percentile (Q3). The upper and lower branches represent the maximum and minimum values of the data, respectively, excluding outliers. Outliers are depicted as individual data points. An outlier, which is represented by the hollow dot in the figure, is defined as a data point that falls significantly outside the expected variation around the median of the remaining dataset. For the interpretation of the first column, focusing on the healthy group, the saliva before lysis readings show a median just over 3000, with Q1 at approximately 2950 and Q3 at about 3150. The highest non-outlier observation was around 3200, while the lowest was approximately 2800. Notably, there was one observation at around 3450, which stands out as unusually high compared to the tightly clustered data around 3000. Thus, we defined it as the outlier. Similar interpretations apply to the second and third columns.
In group A, saliva samples were analyzed without cell lysis, revealing potential differences indicated by lower readings in pre-cancer samples and a significant p-value of 0.001. P-values were derived from Fisher’s exact tests for categorical variables and Mann-Whitney tests for continuous variables. Groups B and C had identical PCB settings to ensure comparable readings. As anticipated, all data points in group C were lower than those in group B, indicating a higher P90 concentration in tissue samples. The p-value for group B was 0.0001, demonstrating the efficacy of the technique in a non-invasive manner. Furthermore, for group C, the p-value decreased even further to 0.0008. The low p-value indicates a low probability of a false-positive result, underscoring the technique's high accuracy. Detailed data analysis findings are summarized in
Table 2.
Utilizing the calibration curve derived from the standard solution and the results of the human sample tests, we can integrate these findings to estimate the relative protein concentration in the human samples. Demonstrating a lower LOD compared to commercial ELISA kits, the sensor introduced in this study exhibits the capability to differentiate low-concentration samples and provides valuable insights for oral cancer detection.
4. Conclusions
In conclusion, this study marks a significant advancement in the realm of oral cancer diagnostics by introducing a transistor-based biosensor system for the detection of the P90 protein, this marker is involved in inflammatory processes and with oral squamous cell carcinoma. Our data indicated that P90 may be associated with leukoplakia that may have a potential for progression to oral malignant transformation. The specially designed printed circuit board (PCB) optimizes the functionality of the biosensor, ensuring accurate measurements and enhanced sensitivity. The superior sensitivity, low limit of detection, and successful human sample testing underscore the potential of this biosensor technique as an invaluable tool in the early detection of oral cancer, ultimately contributing to improved patient outcomes and a reduction in the global burden of this prevalent and impactful disease. The findings presented in this study mark a positive step forward in the ongoing efforts to enhance diagnostic accuracy and efficacy in oral cancer management.
Author Contributions
Conceptualization, F. R. Y. L., and J. E.; methodology, H. W., F. R., C. T., Y. L., and J. E..; software, H. W., C. T., and D. N.; validation, H. W, H. Z., C. C. J. L., and C. T.; formal analysis, H. W., C. T, D. N., and J. K.; investigation, H. W., H. Z., C. C., and C. T.; resources, F. R., Y. L., J. K., and J. E.; data curation, H. W., C. T., and D. N.; writing—original draft preparation, H. W.; writing—review and editing, F. R., Y. L., J. K., and J. E.; visualization, H. W. and C.T.; supervision, F. R., Y. L., J. K., and J. E.; project administration, F. R., Y. L., J. K., and J. E.; funding acquisition, F. R., Y. L., and J. E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by NIH-NIDCR Grant R56 DE025001.The authors at National Yang Ming Chiao Tung University, Hsinchu 30010, Taiwan would like to thank the National Science and Technology Council, Taiwan, for their financial support under the grants NSTC 112-2628-E-A49-015.
Institutional Review Board Statement
Human samples were through the University of Florida Oral Pathology Clinic and Dental Clinical Research Unit. Human subject research was approved by the UF IRB #202101643.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data that support the findings of this study are available within the article.
Acknowledgments
The authors extend their heartfelt gratitude to all the team members involved in this project.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Huang, Y.; Zhang, Q.; Guo, Z.; Deng, G.; Chen, R.; Zheng, Y. Potential noninvasive biomarkers for the malignant transformation of oral leukoplakia: A systematic review and meta-analysis. Cancer Medicine 2023, 12, 14718–14730. [Google Scholar] [CrossRef]
- Society, A.C. Key Statistics for Oral Cavity and Oropharyngeal Cancers. Availabe online: https://www.cancer.org/cancer/types/oral-cavity-and-oropharyngeal-cancer/about/key-statistics.html (accessed on ). 19 January.
- Blot, W.J.; McLaughlin, J.K.; Winn, D.M.; Austin, D.F.; Greenberg, R.S.; Preston-Martin, S.; Bernstein, L.; Schoenberg, J.B.; Stemhagen, A.; Fraumeni Jr, J.F. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer research 1988, 48, 3282–3287. [Google Scholar] [PubMed]
- Chaturvedi, P.; Singh, A.; Chien, C.-Y.; Warnakulasuriya, S. Tobacco related oral cancer. BMJ: British Medical Journal 2019, 365, I2142. [Google Scholar] [CrossRef] [PubMed]
- Heller, M.A.; Nyirjesy, S.C.; Balsiger, R.; Talbot, N.; VanKoevering, K.K.; Haring, C.T.; Old, M.O.; Kang, S.Y.; Seim, N.B. Modifiable risk factors for oral cavity cancer in non-smokers: A systematic review and meta-analysis. Oral Oncology 2023, 137, 106300. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli Pirzaman, A.; Ebrahimi, P.; Niknezhad, S.; Vahidi, T.; Hosseinzadeh, D.; Akrami, S.; Ashrafi, A.M.; Moeen Velayatimehr, M.; Hosseinzadeh, R.; Kazemi, S. Toxic mechanisms of cadmium and exposure as a risk factor for oral and gastrointestinal carcinomas. Human & Experimental Toxicology 2023, 42, 09603271231210262. [Google Scholar]
- Kumar, M.; Nanavati, R.; Modi, T.G.; Dobariya, C. Oral cancer: Etiology and risk factors: A review. Journal of cancer research and therapeutics 2016, 12, 458–463. [Google Scholar] [CrossRef]
- Chaturvedi, A.; Engels, E.; Anderson, W.; Gillison, M. Incidence trends for human papillomavirus-related and-unrelated oral squamous cell carcinomas in the United States. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology 2008, 26, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Goldoni, R.; Scolaro, A.; Boccalari, E.; Dolci, C.; Scarano, A.; Inchingolo, F.; Ravazzani, P.; Muti, P.; Tartaglia, G. Malignancies and biosensors: A focus on oral cancer detection through salivary biomarkers. Biosensors 2021, 11, 396. [Google Scholar] [CrossRef]
- Pearce, A.; Sharp, L.; Hanly, P.; Barchuk, A.; Bray, F.; de Camargo Cancela, M.; Gupta, P.; Meheus, F.; Qiao, Y.-L.; Sitas, F. Productivity losses due to premature mortality from cancer in Brazil, Russia, India, China, and South Africa (BRICS): A population-based comparison. Cancer epidemiology 2018, 53, 27–34. [Google Scholar] [CrossRef]
- Abati, S.; Bramati, C.; Bondi, S.; Lissoni, A.; Trimarchi, M. Oral cancer and precancer: a narrative review on the relevance of early diagnosis. International journal of environmental research and public health 2020, 17, 9160. [Google Scholar] [CrossRef]
- Silverman Jr, S. Early diagnosis of oral cancer. Cancer 1988, 62, 1796–1799. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Darvishi, S.; Preet, A.; Huang, T.-Y.; Lin, S.-H.; Girault, H.H.; Wang, L.; Lin, T.-E. A review: Electrochemical biosensors for oral cancer. Chemosensors 2020, 8, 54. [Google Scholar] [CrossRef]
- Megha, D.; Kiruthiga, S.; Lokesh, M.; Pavithra, P.; Indumathy, K.; Nirmala, E. Exploring the comprehensive review of diagnostic methods of oral cancer: A global scenario. 2023, 15 (01), 043-048.
- Bhosle, P.; Pathan, H.; Tapadiya, G.; Alam, M.I. Dusruptive Developments in Biomedical Applications; CRC Press: Boca Raton, FL, USA, 2022. [Google Scholar]
- Walsh, T.; Macey, R.; Kerr, A.R.; Lingen, M.W.; Ogden, G.R.; Warnakulasuriya, S. Diagnostic tests for oral cancer and potentially malignant disorders in patients presenting with clinically evident lesions. Cochrane Database of Systematic Reviews 2021, 2021(7), CD010276. [Google Scholar]
- Naito, Y.; Honda, K. Liquid Biopsy for Oral Cancer Diagnosis: Recent Advances and Challenges. Journal of Personalized Medicine 2023, 13, 303. [Google Scholar] [CrossRef]
- Pérez, M.G.S.; Bagán, J.V.; Jiménez, Y.; Margaix, M.; Marzal, C. Utility of imaging techniques in the diagnosis of oral cancer. Journal of Cranio-Maxillofacial Surgery 2015, 43, 1880–1894. [Google Scholar] [CrossRef]
- Chakraborty, D.; Natarajan, C.; Mukherjee, A. Advances in oral cancer detection. Advances in clinical chemistry 2019, 91, 181–200. [Google Scholar]
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of salivary biomarkers in oral cancer detection. Advances in clinical chemistry 2018, 86, 23–70. [Google Scholar] [PubMed]
- Huda, W. CT radiation exposure: an overview. Current Radiology Reports 2015, 3, 1–16. [Google Scholar] [CrossRef]
- Katz, J.; Jakymiw, A.; Ducksworth, M.K.; Stewart, C.M.; Bhattacharyya, I.; Cha, S.; Chan, E.K. CIP2A expression and localization in oral carcinoma and dysplasia. Cancer biology & therapy 2010, 10, 694–699. [Google Scholar]
- Velmurugan, B.; Wang, H.; Chung, C.; Lee, C.; Huang, L.; Yeh, K.; Lin, S. CIP2A overexpression in Taiwanese oral cancer patients. Cancer Management and Research 2019, 11, 2589–2594. [Google Scholar] [CrossRef]
- Xian, M.; Stephany, J.L.; Chiu, C.-W.; Chiang, C.-C.; Ren, F.; Tsai, C.-T.; Shan, S.-S.; Liao, Y.-T.; Esquivel-Upshaw, J.F.; Pearton, S.J. High sensitivity CIP2A detection for oral cancer using a rapid transistor-based biosensor module. Journal of Vacuum Science & Technology B 2023, 41, 013201. [Google Scholar]
- Soofiyani, S.R.; Hejazi, M.S.; Baradaran, B. The role of CIP2A in cancer: a review and update. Biomedicine & Pharmacotherapy 2017, 96, 626–633. [Google Scholar]
- Sangodkar, J.; Farrington, C.C.; McClinch, K.; Galsky, M.D.; Kastrinsky, D.B.; Narla, G. All roads lead to PP 2A: exploiting the therapeutic potential of this phosphatase. The FEBS journal 2016, 283, 1004–1024. [Google Scholar] [CrossRef] [PubMed]
- Nader, C.P.; Cidem, A.; Verrills, N.M.; Ammit, A.J. Protein phosphatase 2A (PP2A): a key phosphatase in the progression of chronic obstructive pulmonary disease (COPD) to lung cancer. Respiratory Research 2019, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Das, S.R.; Brownlee, B.J.; Parate, K.; Davis, T.M.; Stromberg, L.R.; Chan, E.K.; Katz, J.; Iverson, B.D.; Claussen, J.C. CIP2A immunosensor comprised of vertically-aligned carbon nanotube interdigitated electrodes towards point-of-care oral cancer screening. Biosensors and Bioelectronics 2018, 117, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Xian, M.; Chiu, C.-W.; Carey, P.H.; Fares, C.; Chen, L.; Wu, R.; Ren, F.; Tsai, C.-T.; Shan, S.-S.; Liao, Y.-T. Digital biosensor for human cerebrospinal fluid detection with single-use sensing strips. Journal of Vacuum Science & Technology B 2022, 40, 023202. [Google Scholar]
- Xian, M.; Luo, H.; Xia, X.; Fares, C.; Carey, P.H.; Chiu, C.-W.; Ren, F.; Shan, S.-S.; Liao, Y.-T.; Hsu, S.-M. Fast SARS-CoV-2 virus detection using disposable cartridge strips and a semiconductor-based biosensor platform. Journal of Vacuum Science & Technology B 2021, 39, 033202. [Google Scholar]
- Wan, H.-H.; Zhu, H.; Chiang, C.-C.; Li, J.-S.; Ren, F.; Tsai, C.-T.; Liao, Y.-T.; Neal, D.; Esquivel-Upshaw, J.F.; Pearton, S.J. High sensitivity saliva-based biosensor in detection of breast cancer biomarkers: HER2 and CA15-3. Journal of Vacuum Science & Technology B 2024, 42, 023202. [Google Scholar]
- Yang, J.; Carey, P.; Ren, F.; Wang, Y.-L.; Good, M.L.; Jang, S.; Mastro, M.A.; Pearton, S. Rapid detection of cardiac troponin I using antibody-immobilized gate-pulsed AlGaN/GaN high electron mobility transistor structures. Appl. Phys. Lett. 2017, 111, 202104. [Google Scholar] [CrossRef]
- Yang, J.; Carey, P.; Ren, F.; Mastro, M.A.; Beers, K.; Pearton, S.; Kravchenko, I.I. Zika virus detection using antibody-immobilized disposable cover glass and AlGaN/GaN high electron mobility transistors. Appl. Phys. Lett. 2018, 113, 032101. [Google Scholar] [CrossRef]
- Böckelman, C.; Hagström, J.; Mäkinen, L.K.; Keski-Säntti, H.; Häyry, V.; Lundin, J.; Atula, T.; Ristimäki, A.; Haglund, C. High CIP2A immunoreactivity is an independent prognostic indicator in early-stage tongue cancer. British journal of cancer 2011, 104, 1890–1895. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).