1. Introduction
There has been a great emphasis on the data mining fields of computer science that can be applied in different fields. Specifically, text and graph mining have been extensively used to extract hidden interesting findings, insights, and features from large amounts of data in various domains (Alamro et al., 2023; Dong et al., 2017; Thafar, Albaradie, et al., 2020; Thafar et al., 2022; Thafar, Olayan, et al., 2020). The exponential growth of academic publications in recent years has made it increasingly difficult for researchers to identify significant literature for their research. Citation numbers, such as those obtained from databases like Google Scholar, SCOPUS, ORCID, and Web of Science (WOS), are often used to measure a research paper's popularity, quality, and significance (Ding et al., 2014). However, citation numbers cannot be a reliable indicator of the importance of a recent paper. As a result, more attention has been given to the problem of predicting future citation levels for publications across different domains (Butun & Kaya, 2020). Machine learning (ML) and deep learning (DL) have been utilized by incorporating text and graph features to solve such problems (Ali, Kefalas, et al., 2020).
Previous studies have emphasized the importance of text-based features using text mining and text embedding techniques in different domains (Alshahrani et al., 2022; Gupta et al., 2020; Thafar et al., 2023), including our problem of citation level prediction (Akujuobi et al., 2018; Castano et al., 2018). Those text-based features have been obtained through feature extraction, a critical aspect of ML, as it involves transforming raw data into a set of features used to train the ML model. For instance, West and coauthors (West et al., 2016) designed a citation recommendation system based on scientific papers' content and citation relationships. They employed text-based features such as titles, abstracts, and keywords to capture the scope and topics of publications and enhance the citation recommendation's accuracy.
Although the above and other studies focused on employing text-based attributes to predict the future literature impact and citation counts, they should have considered crucial features derived from complex networks formed with citations, articles, and authors. Therefore, recent studies utilized graph-based features to predict the importance of the publications (Ali, Qi, et al., 2020; Weis & Jacobson, 2021; Xia et al., 2023). For example, CiteRivers (Heimerl et al., 2016) depends on visualization capabilities, including graph visualization analysis. Thus, they extended the capability of the previous methods, which were dependent only on visualization, and represented a new visual interactive analysis system for investigating and analyzing the citation patterns as well as contents of scientific documents and then spotting the trends they obtained. Another method (Lu et al., 2018) developed an enhanced recommendation system based on the Author Community Topic Time Model (ACTTM) and bilayer citation network. They created an author–article citation network model consisting of author citation layers and paper citation layers. After that, they proposed a topic modeling method for recommending authors/papers to scientists and researchers. The third study utilized graph-based attributes, formulating the problem as a supervised link prediction in directed, weighted, and temporal networks (Pobiedina & Ichise, 2016). They developed a method to predict the links and their weights and evaluated it by conducting two experiments that demonstrated the effectiveness of their approach. The last study to mention is a graph centrality-based (Samad et al., 2019) that applied graph mining techniques to find papers' importance by computing the centrality measures such as PageRank, Degree, and Betweenness Closeness. They demonstrated their finding that topological-based similarity using Jaccard and Cosine similarity algorithms outperforms when using those similarity algorithms applied to textual data. Moreover, a few recent studies have taken advantage of graph-based and text-based features to predict future research paper citations and proved their efficiency (Kanellos et al., 2019). One of these studies, ExCiteSearch (Sterling & Montemore, 2022), is a framework developed to enable researchers to choose relevant and important research papers. ExCiteSearch implemented a novel research paper recommendation system that utilizes both abstract textual similarity and citation network information by conducting unsupervised clustering on sets of scientific papers.
Recent studies take advantage of graph embedding techniques to encode nodes or edges into low-dimensional space and generate feature representation through unsupervised learning methods, successfully enhancing the performance of different downstream tasks such as node classification and link prediction (Jiang et al., 2021; Thafar et al., 2021). For instance, another work that utilized text and graph data has been developed to generate a scientific paper representation called Paper2Vec (Ganguly & Pudi, 2017). Unlike methods that rely on traditional graph mining for feature extraction, Paper2Vec employs a graph embedding technique to generate a paper representation that can be used for different tasks, including node classification and link prediction related to predicting the paper's significance. Paper2Vec leveraged the power of unsupervised feature learning from graphs and text documents by utilizing a neural network to generate paper embedding. This method demonstrated the robustness of paper embeddings using three real-world academic datasets. The significance of the research paper embeddings generated by the Paper2Vec method has been validated via different experiments on three real-world academic datasets, indicating Paper2Vec's capability to generate strong and rich representations.
Even though several approaches have been proposed to predict future citation levels using different feature models, there are limitations and more room to improve the prediction performance. Moreover, the problem of citation level prediction remains challenging due to the imbalance in citation distribution and the need for effective feature selection and extraction techniques. In this regard, we contribute to the existing literature by presenting an effective method called FutureCite for predicting citation levels using text and graph mining techniques. FutureCite is designed to provide researchers with a tool to identify promising research papers that are influential in their field and better understand research impact. We utilized various feature extraction techniques and a dataset of thousands of publications from the data mining field to develop our method. This study contributes to the field of citation analysis by addressing the challenge of predicting future citation levels.
The overall structure of this paper is as follows.
Section 1 introduces the motivation for predicting future citation problems, conducts a literature review of recent research that has been published contributing to this topic, and finally presents the fundamentals of some important graph and text mining concepts used in this work.
Section 2 describes the datasets that have been utilized in this project.
Section 3 presents the FutureCite model's methodology phases and introduces the problem formulation.
Section 4 discusses the results and performance of our proposed approach. Finally, we conclude our work, highlight some limitations, and outline potential future directions for research in this area.
2. Materials
To develop the FutureCite model, We utilized a popular scholar dataset (Akujuobi & Zhang, 2017), which has been collected from scientific research articles that have been published in different venues (i.e., conferences and journals) such as ICDE, KDD, TKDE, VLDB, CIKM, NIPS, ICML, ICDM, PKDD, SDM, WSDM, AAAI, IJCAI, DMKD, WWW, KAIS and TKDD. This dataset was extended by adding the references for all documents (i.e., research articles).
In our work, we utilized part of this scholarly dataset that focuses on data mining publications provided by the Delve system's authors (Akujuobi & Zhang, 2017). Thus, the dataset we used to develop the FutureCite model is the data mining publications dataset. Each research article in the dataset has several attributes: paper ID, venue name, authors' names, publication year, paper title, index keys (i.e., keywords), the paper abstract, and references. We divided our input dataset into four types based on the feature categories we will extract.
First, the research paper data we utilized to extract text and metadata features in our dataset consists of 11,941 papers. However, after applying different filters to the dataset (described in the cleaning and preprocessing phase), we obtained 6,560 research papers. Thus, the size of our dataset is moderate.
Second, the citation graph data, where all papers and their corresponding references are provided, formulate citation edge lists consisting of 133,482 papers and 335,531 relationships (i.e., citations). This citation graph is utilized to define the class labels using different thresholds for each label. Those thresholds have been selected based on the distribution of citation counts in the dataset, as will be explained later.
The third type is the Author-Coauthor Graph, comprising 10,270 authors and 26,963 author-coauthor relationships.
The fourth and last data type, the Author-Research graph, consists of 10,270 authors and 6,560 publications and the relations between them.
The details of each type, with its descriptions, are discussed later in the feature extraction section.
3. Methods
3.1. Problem Formulation
This study describes the objective of predicting the research paper’s future citation level as supervised learning, specifically multilabel classification. Thus, given a dataset of data mining publications, we are trying to predict each paper citation level based on predefined class labels. As mentioned in the material section, all data samples (i.e., published research papers) in our dataset have their features and can be represented as vector X = {x1, x2, ..., xn} where n is the number of all data samples. In addition to the data samples, we also provided all data samples with their class labels Y = {y1, y2, ..., yn} since our problem is supervised learning. We prepared the class labels based on the citation numbers of the paper in our dataset, and the process of creating and preparing the class labels will be explained in
Section 3.3.
We extracted different features from different perspectives for each data sample (research paper), explained later. The classification model aims to find the hidden patterns and associations between research papers and their class labels based on the features we have extracted and then predict the true class labels.
3.2. FutureCite Model Workflow
Figure 1 provides the workflow to develop the FutureCite model, which involves five main steps. These steps are summarized as follows
Data preparation and preprocessing,
Class label preparation,
Feature extraction and integration. These features are grouped based on three categories: graph-based, text-based, and metadata-based.
Data normalization and sampling,
-
ML classification, where several classifiers are built for class prediction,
A detailed explanation of each step is provided in the following subsections.
3.3. Class Label Creation
Since our model is supervised learning, as explained earlier, we need to provide the class labels along with the data samples. Therefore, each research paper in the dataset must have a class label. All class labels have been created using the citation graph based on the in-degree measurement. To do so, first, we utilized a specific part of the dataset, including the research papers and their references, to build the citation graph. A graph G = (V, E) consists of a set of vertices (i.e., nodes) V and a set of edges E. In our study, each node in the citation graph represents the research paper, and each edge between two nodes represents the citation relationship (i.e., paper 1 is a reference for paper 2, or paper 2 cites paper 1). Therefore, we obtained a directed unweighted graph consisting of 133,482 nodes (including papers and references) and 335,531 edges (links). Next, we applied a graph mining technique to calculate each node in-degree feature that reflects the citation number of each paper. An example of a top-5 paper in degree is shown in
Table 1.
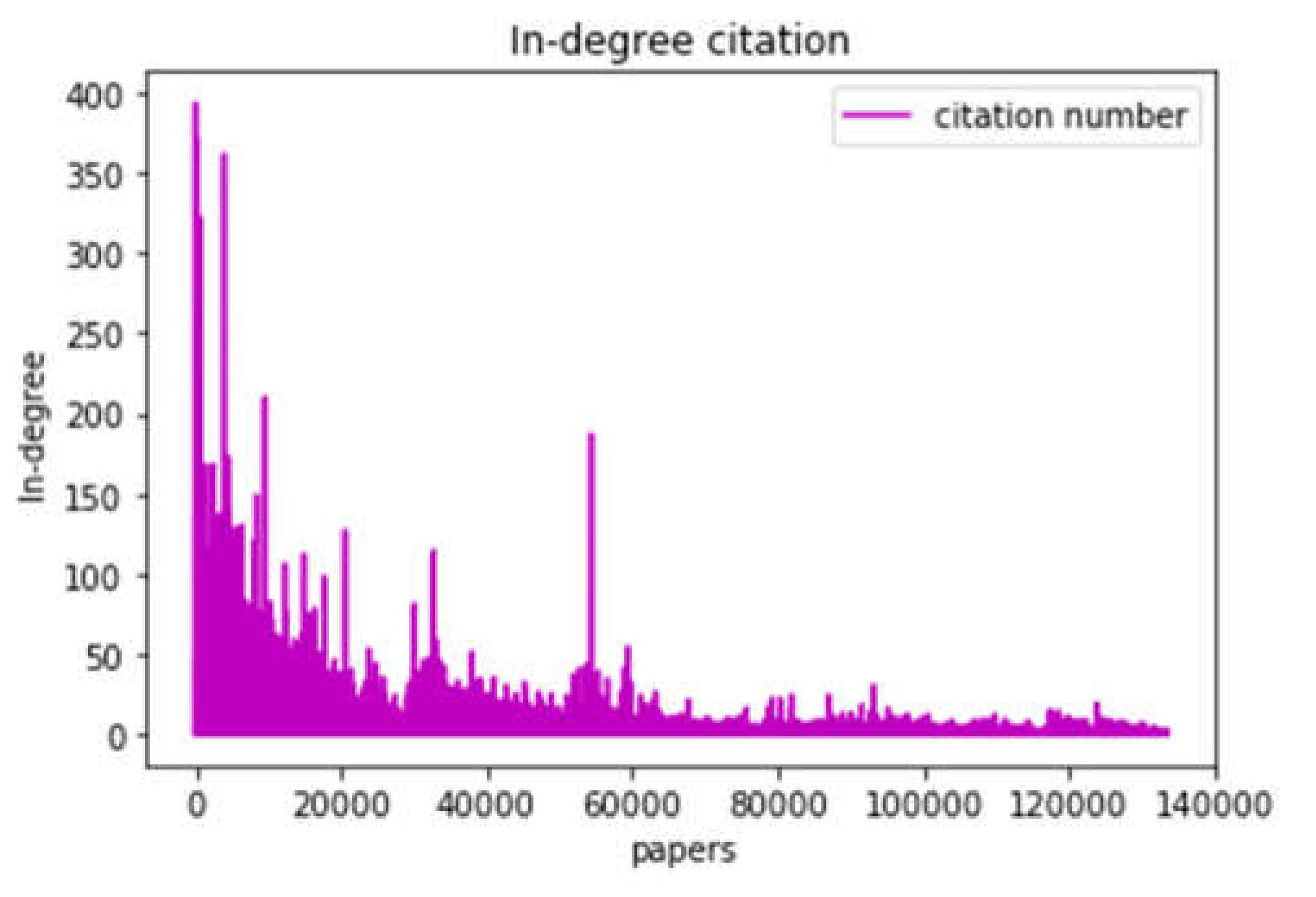
Next, we applied a graph mining technique to calculate each node in-degree feature that reflects the citation number of each research paper. After that, we obtained the in-degree distribution of all research papers, as shown in
Figure 2. Also, we provide an illustrative example of a top-5 research paper in degree measurement is shown in
Table 1. As illustrated in
Figure 2, we observed extremely low values for in-degree features across the dataset. We calculated the in-degree average to investigate this metric further, which equals 2.2. This average in-degree provides a baseline for acquiring the class labels in our data.
Although the average in-degree is 2.2, we modified the threshold average to 5. This procedure was crucial because many references (i.e., research articles) within our dataset have no citations (i.e., in-degree = 0). This choice benefits to account for the skew in our data caused by research articles with no citations and allows for additional meaningful classification. After this adjustment, we conducted an empirical analysis of the distribution of the in-degree values concerning the new threshold average. Based on this analysis, we determined four class labels for research papers in our dataset. These classes correspond to different ranges of citation counts, as described in
Table 2. Each paper in the dataset is assigned to one of these classes according to its in-degree value, which measures its citation count.
3.4. Data Preprocessing and Feature Extraction
In the FutureCite study, since we performed the data preprocessing differently considering the feature category, we will explain each feature group's data preprocessing and extraction steps together. As a result, the data preprocessing step was applied three times, removing certain data samples on each occasion.
The feature extraction techniques we employed, based on text and graph mining, are specifically designed to capture various aspects of the data mining publications datasets. Three categories of features have been utilized: text-based, graph-based, and metadata-based. The features for each research paper cover various perspectives, including the quality and significance of the publication venue, the collaboration between authors, the individual authors' expertise and influences, the content of the publications themselves, and other features. These features were then used to develop and train a classification model using machine ML algorithms to predict paper citation levels based on predefined class labels, ranging from low to highly cited. Each feature category comprises multiple characteristics that have been created based on specific attributes of the dataset. Each category is explained in more detail in the following subsections and illustrated in
Figure 1.
3.4.1. Metadata-Based Features
By metadata, we refer to the extra information that we can add to each research paper, which is, in our context, the publication venues and years. Venue-related features are generated based on the venue's name, known as the impact factor, and the h-index of each publication venue. The h-index is a metric used to evaluate the conference or journal's quality (Mingers et al., 2012). To ensure the quality of our publication’s datasets and the relevance of the research papers to the same domain, we implemented a preprocessing procedure on all research papers in our datasets, consisting of 11,941 papers. During this process, we filtered out any research paper unrelated to the data mining conferences or journals and those with a count of less than 50 in our datasets. In this step, it is important to have all publications datasets related to the same field to find hidden patterns and associations in the data. Consequently, the number of research papers we have utilized has been reduced, resulting in obtaining 7,069 research papers published in 23 venues. Also, we filtered out all papers published before 2001, resulting in 6,560 research papers. The data mining venues within our limited datasets and the corresponding number of research papers published in each venue from our datasets are shown in
Figure 3. Finally, the h-indices are collected from the Google Scholar website and incorporated as a new feature (venue-h-index) for each research paper. For instance, the Knowledge Discovery and Data Mining (KDD) conference has a good reputation and exhibited an h-index of 42.
Another example is the IEEE Transactions on Knowledge and Data Engineering journal, one of the most popular and strong data mining conferences, with an h-index of 99. Also, we collected the impact factors of all venues as another feature. Although only one feature is related to the venue, it indicates the research paper's significance considering the conference or journal reputation, which, in turn, affects the predictions of paper citation futures.
3.4.2. Text-Based Features
Text-related features have a crucial role in capturing the topics and contents of the research papers. These features include the research paper title and abstract. We started by combining and then preprocessing all titles and abstracts for the 6,560 documents (i.e., research papers). The data preprocessing steps we applied involved text tokenization, punctuation elimination, stop and common word removal, and text stemming. After that, a bag-of-words (BOW) feature vector is constructed using the Term Frequency-Inverse Document Frequency (TF-IDF) vectorizer technique (W. Zhang et al., 2011), which is a popular natural language processing (NLP) technique that is a commonly used technique for determining the importance of a term in a document. TF-IDF reflects the relevance of a term to a document by measuring the frequency of the term in the document and inversely scaling it by the frequency of the term across all documents in the corpus. It is used to build a bag of word feature vectors that have been used extensively for several methods that rely on text-based features for prediction. We optimized several hyperparameters for TF-IDF vectorizers such as max_df, min_df, ngram_range, and max features to obtain the best and most effective bag-of-words FV. For example, if max_df = 0.4, it means, when building the vocabulary, ignore terms that appear in more than 40% of the documents (i.e., have a document frequency strictly higher than the given threshold). Likewise, if min_df = 0.01, it means "ignore terms that appear in less than 1% of the documents.
As a result of this process, we obtained 802 features from the text-based data. However, we considered this number of features a high dimensionality of the feature space. Therefore, dimensionality reduction approaches are employed to mitigate the issue of having many correlated features. This means we need to reduce the number of features into fewer uncorrelated variables by taking advantage of the existing correlations between the input variables in the dataset. We have utilized two approaches: Principal component analysis (PCA) and singular value decomposition (SVD). However, after comparing the performance of those two approaches and PCA demonstrating that it provides superior results to SVD, we have selected the PCA approach. PCA effectively reduces the number of features to 420, which captures around 85% of the variance. As a consequence, the resulting FV from the text-based category is 420.
3.4.3. Graph-Based Features
In the graph-based features, we focus on extracting the information relevant to the authors of each research paper. Two graphs, the author-coauthor graph, and the author-paper graph, are constructed to extract author-based features. Since we used all author and co-author names in both graphs, we began with the preprocessing step on all authors' and co-authors' names before constructing any graph. This preprocessing step comprises converting the names to uppercase, removing all dots ('.'), eliminating middle names, and matching some first names with corresponding initials.
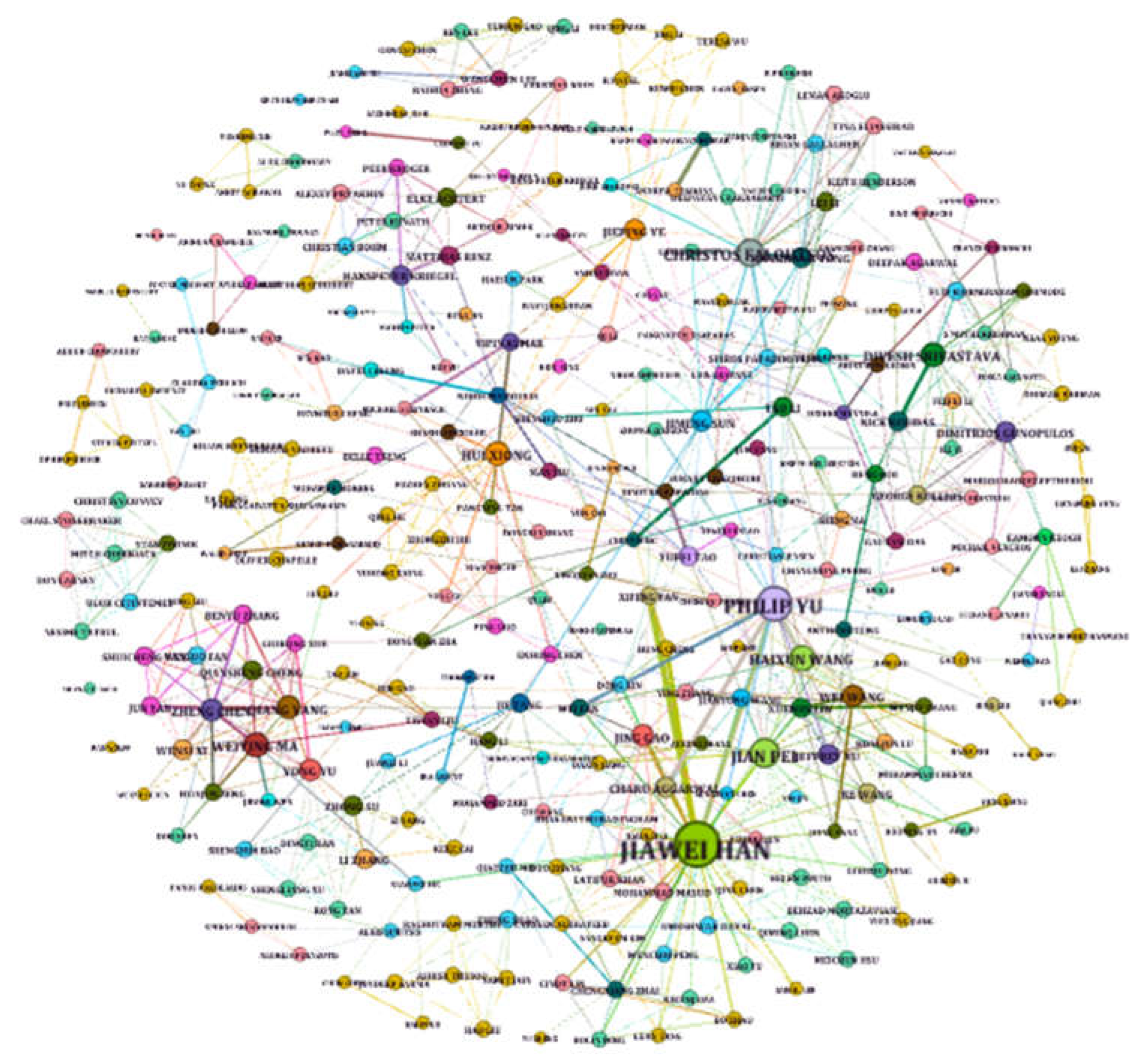
The first graph (G1(V, E))) represents the author-coauthor relationship. This undirected, unweighted graph is built by converting all author-coauthor relationships into an edge list, resulting in 10,270 nodes V (authors) and 26,963 edges E (relation). We visualize the author-coauthor graph to represent the relationships among authors since data visualization (in our case graph visualization) is an essential tool used excessively in different domains, allowing researchers to explore large and complex datasets more effectively and gain useful insights from the data (Aljehane et al., 2015; Shakeel et al., 2022). However, to achieve the best visualization, we applied a filtering procedure by removing all authors that have no connection with other authors or the authors with connections below a specific threshold. This process helps to reduce less important authors and enhance the visualization for more impactful authors.
Figure 4 visualizes the author-coauthor graph where the node size indicates the importance of the corresponding authors that appear the most in our dataset, as we can see, the largest node represents Jiawei Han, one of the pioneering scientists in data mining, has current citation records of more than 250,000. Another insight we can obtain from the visualized graph in
Figure 4, is the edge width that represents the connection strength between authors based on their author-coauthor relationship in several publications. Furthermore, the color indicates the community of authors.
After formulating the graph, graph mining techniques then employed three global ranking algorithms to extract three graph properties for each author, which are degree centrality (DC), betweenness centrality (BC), and PageRank (Newman, 2010). Those three features derived by graph mining prove their significance and efficiency in several ML problems across various domains (Albaradei et al., 2023, 2021; De et al., 2020; Salavati et al., 2019) and predict future citation problems. The definition and equation for each measurement are as follows:
Page Rank (P. Zhang et al., 2022), equivalent to the eigenvector centrality CE of node v, measures the node's importance based on its neighboring nodes' importance.
where σst is the total number of shortest paths between nodes vs and vt, and σst(vi) is the number of shortest paths between nodes vs and vt that pass along node vi.
After employing the equations above, we obtained three properties of each author, as shown in
Table 3, where we list several examples of the Top-5 prominent authors in our datasets. Furthermore, three features for each data sample (i.e., research paper) were calculated. Those features are the average degree, the average page rank, and the average Betweenness Centrality of all authors associated with each paper.
Additionally, we constructed a second graph known as the author-paper graph. We created an edge list that connects the 10,270 authors to the 6,560 publications, resulting in 16,830 nodes. An edge is created if an author's name appears in the publication authors list. The degree of each author node is computed as a feature for each author. Consequently, the average of all authors' degrees is calculated as a fourth graph-based feature and assigned to each paper. The resulting FV from the graph-based category is 4, but they are important in citation-level prediction.
3.4.4. Features Integration
Upon completing the feature extraction process, we obtained one venue-related feature, 420 text-related features, and four graph-related features. The order of the documents remained consistent across all feature category extraction processes, facilitating the concatenation process to integrate all features into a single large comprehensive FV. After that, a min-max normalization process is applied to all features based on each column, ensuring that all features have the same scale.
3.4. FutureCite Predictive Model
3.4.1. Sampling Techniques for Imbalanced Data
Figure 3 shows that the number of data samples in each class is different. We can see clearly that the numbers of data samples in classes 3 and 4 (i.e., classes with average and above average labels) are much larger than in classes 1 and 2. This issue is called imbalanced data and must be addressed since the ML classifiers face a problem in prediction based on imbalanced data. The imbalanced data affects the ML models' ability to classify most test samples into the majority class when the minority class lacks information. We addressed this issue by applying random oversampling (Liu et al., 2007) in the training data to balance the data. This technique is implemented using an imblearn Python package (Lemaître et al., 2017).
3.4.2. Multilabel Classification Model
In the FutureCite model, after completing the feature extraction process and obtaining a feature vector (FV) for all research papers, we fed the FVs with their class labels into several supervised ML classifiers, which are support vector machine (SVM) (Suthaharan, 2016), Naïve Bayes (NB) (Ting et al., 2011), and random forests (RF) (Ho, 1995).
SVM classifier works by finding the hyperplane that maximally separates the classes in the feature space. SVM uses a kernel function to transform the input features into a higher dimensional space where the classes can be more easily separated. The optimal hyperplane is found by maximizing the margin, which is the distance between the hyperplane and the closest data points from each class. Naïve Bayes is a probabilistic algorithm that is based on Bayes' theorem. It works by calculating the probability of each class given the input features and selecting the class with the highest probability as the output. Both SVM and NB perform well in many applications, particularly when dealing with high-dimensional data such as text classification, and this is the reason we have picked them in our prediction. The third classifier we applied is RF, which demonstrates its effectiveness in prediction since it operates efficiently on extensive datasets and is less prone to the overfitting issue.
In our study, for each classifier, we optimized several critical parameters using the training datasets to improve the classifier's performance. For Example, for the SVM classifier, we optimized the Regularization parameter C, the kernel function where we set it to Radial Basis Function (RBF), and the class weight to be balanced. For the NB classifier, the parameters include fit_priorbool, specified to learn the class's prior probabilities or not, and class_prior, which is the prior probabilities of the classes. Finally, for the RF classifier, the parameters include the number of trees in the forest (i.e., n_estimators), the maximum depth of the trees, and the function to measure the quality of a split (criterion).
To implement our FutureCite method, we utilized the following tools:
Python 3.3 (Van Rossum, 2007) is used for implementing all project phases, including preprocessing, feature extraction, training and validation, and classification. We utilized several supported packages, including scikit-learn for ML algorithms, NetworkX for graph mining (Platt, 2019), imblearn package (He & Ma, 2013) to handle imbalanced class labels, Matplotlib to plot different figures, and pandas data frame to deal with the data preparation and preprocessing (Nelli, 2015).
Gephi (Yang et al., 2017): Gephi is used to visualize and analyze graph data, including the author-coauthor and citation graphs. The data is preprocessed and prepared to be in a suitable shape for this tool. Gephi allows us to explore the structure of the citation network and gain valuable insights into the relationships between different research papers, authors, and venues. These tools are essential in developing a comprehensive method for predicting the future citation level of research papers.
4. Results and Discussion
This section describes the evaluation protocols, the experiments conducted, and the results of our FutureCite prediction performance based on several evaluation metrics. We further highlighted several possible characteristics that could boost the FutureCite method prediction performance.
4.1. Evaluation Metrics and Protocols
Several performance metrics can be used to evaluate the prediction performance of the multilabel classification models. However, since our datasets are highly imbalanced, typical accuracy is not accurate or meaningful. Therefore, we employed three evaluation metrics derived from the confusion matrix (Maria Navin & Pankaja, 2016), which are precision (also called positive predictive value), recall (also called sensitivity or true positive rate (TPR)), and F1 score (a combination of precision and recall metrics) (Powers, 2011). The calculation of these evaluation metrics is based on true positive (TP), and false positive (FP), which represent correctly and incorrectly predicted samples, and the true negative (TN) and false negative (FN), which represent the samples that are correctly and incorrectly predicted, respectively. Equations (4), (5), and (6) illustrate the precision, recall, and F1-score calculations.
For the evaluation setting and protocol, we applied a hold-out validation approach. We split the dataset into 80% for training and 20% for testing in a stratified manner where we ensure the same proportion of each class label in both training and test splits. However, we repeated this process five times for sufficient test samples to generate reliable evaluation metrics. This setup is similar to 5-fold cross-validation. In particular, we initially shuffled the data and then divided it into 20% and 80% for training and testing, respectively. Subsequently, we selected another 20% for testing and allocated the remaining 80% for training, and so on. In each cycle, we tested each classifier to predict the 20% of hold-out samples (i.e., test set) and calculated the three evaluation metrics identified above. Finally, we averaged the results over the five test split for each evaluation metric.
4.2. FutureCite Prediction Performance Evaluation
The results of the SVM, NB, and RF classifiers, based on precision, recall, and F1-score evaluation matric, are presented in
Table 4 and reveal interesting findings.
From the results reported in
Table 3, it is evident that the RF classifier demonstrates better performance than both Naïve Bayes and SVM. It achieves a precision of 83.8%, a recall of 84.1%, and an F1-score of 83.9%. The reason behind the RF classifier’s efficiency can be attributed to its ensemble learning approach, which proves its effectiveness in dealing with imbalanced datasets. Another reason can related to the extensive optimization of several hyperparameters of RF resulting in improved performance. The second-best performance is achieved by the SVM classifier in all evaluation metrics, obtaining results lower than the RF classifier by 2.1%, 2.6%, and 2.3% in terms of precision, recall, and F1 score, respectively. SVM demonstrates its robustness in dealing with document data. On the other hand, the Naïve Bayes obtained the lowest results, although it approves its efficiency in dealing with document data as reported in the literature. This might be because of the feature combinations we derived, consisting of text and graph data.
Furthermore, we conducted the last evaluation process to demonstrate the practicality of the FutureCite model’s predictive power by analyzing and validating some results using the literature review. In particular, we selected some published research papers from our test data and analyzed the prediction results. For example, we picked a research paper titled “Simrank: a measure of structural-context similarity” (Jeh & Widom, 2002), which is predicted to be a high-quality paper (i.e., the predicted class label is 1) by our model using the three classifiers, SVM, NB, and RF. We verified this result by examining the research paper details, such as the authors, venues of the paper, and the current citation count. Our examination shows that the current citation for this paper is 2700 and was published in SIG KDD, one of the most important conferences in the Data Mining and Machine learning fields. Furthermore, the authors are popular, have a strong reputation, and have scholarly profiles with impressive citation records. All these factors confirm that this research paper is indeed significant, as predicted by our model.
Overall, the results indicate that the classification model, based on citation graph mining and machine learning, effectively classifies research papers into different classes based on their citation counts. This model can be utilized to identify papers with high impact and potential significance, as well as papers that fall below average. Such insights can be valuable and useful for researchers and decision-makers across various fields interested in identifying and analyzing research papers with different levels of impact.
5. Conclusions Remarks and Recommendation
In this study, we developed the FutureCite Model, which mainly predicts the future citation of research papers to reveal their importance and significance. We formulated the problem as a multi-label classification task, where each paper was predicted to belong to one of four classes that we have derived: 'Highly cited,' 'Good cited,' 'Above/in average cited,' and 'Below average cited.' Three classifiers were used for prediction; two demonstrated their robustness and efficiency in such a problem. The results highlight the effectiveness of our approach. We conducted results analysis and validation to identify highly cited papers and show evidence for those papers to be predicted as significant. The FutureCite model leverages a combination of graph-based and text-based features, which capture both the structure and content of the data mining publications dataset, improving the model's performance in predicting the citation level and demonstrating its effectiveness. The outcomes of our work have implications for diverse stakeholders such as researchers, funding agencies, and academic institutions, as they can make informed decisions based on our models' predictive capability.
Although our method proves its efficiency and robustness, it also faces some limitations and challenges that must be addressed to ensure its effectiveness and reliability in practice. One of the major limitations of this study is the size of the used dataset, which may not accurately represent the full range of research papers. It also can lead to another challenge of data availability, where our method relies on access to a large dataset of published research papers and their citation information. This information may not be available or may require considerable effort and resources to collect. Another limitation is the prediction performance, which can be greatly improved using massive data, especially the deep learning models we omit due to our dataset size.
Despite the above limitations, the FutureCite model contributes to the research paper citation prediction field as a powerful and useful application that demonstrates its effectiveness using graph and test mining and machine learning techniques. The approach can be applied to various fields, provide insights into the impact and significance of research papers, and aid researchers in their research and literature.
Future work can involve various areas of exploration and improvement. One aspect is to utilize larger datasets, as working with massive amounts of data can allow us to employ deep learning techniques for feature generation and prediction. Deep learning models usually perform better when trained on extensive datasets. Another aspect of future work is refining our model, FutureCite, and improving its performance. This can be achieved by exploring and incorporating additional features into the model. One potential approach to integrating more essential features is to leverage graph embedding techniques such as node2vec, which can automatically generate crucial features based on the underlying graph structure and topological information. Furthermore, incorporating recent text embedding techniques like Bidirectional Encoder Representations from Transformers (BERT) embedding can enhance the model's ability to capture semantic information and relationships. Combining text and graph features from these embeddings can comprehensively represent the research papers.
Authors’ Contributions
MAT conceptualized and designed the study. MA implemented the code. MAT, MA, and SA wrote the manuscript and designed the figures. MAT, MA, and SA validated and analyzed the results. All authors revised/edited the manuscript and approved the final version.
Data Availability of Data and Materials
Competing Interests
The authors have declared that no conflict of interest exists.
References
- Akujuobi, U., Sun, K., & Zhang, X. (2018). Mining top-k Popular Datasets via a Deep Generative Model. 2018 IEEE International Conference on Big Data (Big Data), 584–593.
- Akujuobi, U., & Zhang, X. (2017). Delve: A Dataset-Driven Scholarly Search and Analysis System. SIGKDD Explor. Newsl., 19(2), 36–46.
- Alamro, H., Thafar, M. A., Albaradei, S., Gojobori, T., Essack, M., & Gao, X. (2023). Exploiting machine learning models to identify novel Alzheimer’s disease biomarkers and potential targets. Scientific Reports, 13(1), 4979. [CrossRef]
- Albaradei, S., Alganmi, N., Albaradie, A., Alharbi, E., Motwalli, O., Thafar, M. A., Gojobori, T., Essack, M., & Gao, X. (2023). A deep learning model predicts the presence of diverse cancer types using circulating tumor cells. Scientific Reports, 13(1), 21114. [CrossRef]
- Albaradei, S., Uludag, M., Thafar, M. A., Gojobori, T., Essack, M., & Gao, X. (2021). Predicting bone metastasis using gene expression-based machine learning models. Frontiers in Genetics, 12. [CrossRef]
- Ali, Z., Kefalas, P., Muhammad, K., Ali, B., & Imran, M. (2020). Deep learning in citation recommendation models survey. Expert Systems with Applications, 162, 113790. [CrossRef]
- Ali, Z., Qi, G., Kefalas, P., Abro, W. A., & Ali, B. (2020). A graph-based taxonomy of citation recommendation models. Artificial Intelligence Review, 53(7), 5217–5260. [CrossRef]
- Aljehane, S., Alshahrani, R., & Thafar, M. (2015). Visualizing the Top 400 Universities. Proceedings of The. https://www.researchgate.net/profile/Maha-Thafar/publication/285927843_Visualizing_the_Top_400_Universities/links/5664c6cd08ae192bbf90aa9c/Visualizing-the-Top-400-Universities.pdf.
- Alshahrani, M., Almansour, A., Alkhaldi, A., Thafar, M. A., Uludag, M., Essack, M., & Hoehndorf, R. (2022). Combining biomedical knowledge graphs and text to improve predictions for drug-target interactions and drug-indications. PeerJ, 10, e13061. [CrossRef]
- Butun, E., & Kaya, M. (2020). Predicting Citation Count of Scientists as a Link Prediction Problem. IEEE Transactions on Cybernetics, 50(10), 4518–4529. [CrossRef]
- Castano, S., Ferrara, A., & Montanelli, S. (2018). Topic summary views for exploration of large scholarly datasets. Journal on Data Semantics, 7(3), 155–170. [CrossRef]
- De, S. S., Dehuri, S., & Cho, S.-B. (2020). Research contributions published on betweenness centrality algorithm: modelling to analysis in the context of social networking. International Journal of Social Network Mining, 3(1), 1–34.
- Ding, Y., Rousseau, R., & Wolfram, D. (2014). Measuring Scholarly Impact: Methods and Practice. Springer.
- Dong, Y., Chawla, N. V., & Swami, A. (2017). Metapath2Vec: Scalable Representation Learning for Heterogeneous Networks. Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 135–144.
- Evans, T. S., & Chen, B. (2022). Linking the network centrality measures closeness and degree. Communications Physics, 5(1), 1–11. [CrossRef]
- Ganguly, S., & Pudi, V. (2017). Paper2vec: Combining Graph and Text Information for Scientific Paper Representation. Advances in Information Retrieval, 383–395.
- Gupta, A., Dengre, V., Kheruwala, H. A., & Shah, M. (2020). Comprehensive review of text-mining applications in finance. Financial Innovation, 6(1), 1–25. [CrossRef]
- He, H., & Ma, Y. (2013). Imbalanced Learning: Foundations, Algorithms, and Applications. John Wiley & Sons.
- Heimerl, F., Han, Q., Koch, S., & Ertl, T. (2016). CiteRivers: Visual Analytics of Citation Patterns. IEEE Transactions on Visualization and Computer Graphics, 22(1), 190–199.
- Ho, T. K. (1995). Random decision forests. Proceedings of 3rd International Conference on Document Analysis and Recognition, 1, 278–282 vol.1.
- Jeh, G. , & Widom, J. (2002). SimRank: a measure of structural-context similarity. Proceedings of the Eighth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 538–543.
- Jiang, S. , Koch, B., & Sun, Y. (2021). HINTS: Citation Time Series Prediction for New Publications via Dynamic Heterogeneous Information Network Embedding. Proceedings of the Web Conference 2021, 3158–3167. [Google Scholar]
- Kanellos, I., Vergoulis, T., Sacharidis, D., Dalamagas, T., & Vassiliou, Y. (2019). Impact-based ranking of scientific publications: A survey and experimental evaluation. IEEE Transactions on Knowledge and Data Engineering, 33(4), 1567–1584. [CrossRef]
- Lemaître, G., Nogueira, F., & Aridas, C. K. (2017). Imbalanced-learn: A python toolbox to tackle the curse of imbalanced datasets in machine learning. Journal of Machine Learning Research: JMLR, 18(1), 559–563.
- Liu, A., Ghosh, J., & Martin, C. E. (2007). Generative Oversampling for Mining Imbalanced Datasets. DMIN, 66–72.
- Lu, M., Qu, Z., Wang, M., & Qin, Z. (2018). Recommending authors and papers based on ACTTM community and bilayer citation network. China Communications, 15(7), 111–130. [CrossRef]
- Maria Navin, J. R., & Pankaja, R. (2016). Performance analysis of text classification algorithms using confusion matrix. International Journal of Engineering and Technical Research (IJETR), 6(4), 75–78.
- Mingers, J., Macri, F., & Petrovici, D. (2012). Using the h-index to measure the quality of journals in the field of business and management. Information Processing & Management, 48(2), 234–241. [CrossRef]
- Nelli, F. (2015). Python Data Analytics: Data Analysis and Science using pandas, matplotlib and the Python Programming Language. Apress.
- Newman, M. (2010). Networks: An Introduction. OUP Oxford.
- Platt, E. L. (2019). Network Science with Python and NetworkX Quick Start Guide: Explore and visualize network data effectively. Packt Publishing Ltd.
- Pobiedina, N., & Ichise, R. (2016). Citation count prediction as a link prediction problem. Applied Intelligence, 44(2), 252–268. [CrossRef]
- Powers, D. M. (2011). Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. https://dspace2.flinders.edu.au/xmlui/handle/2328/27165.
- Prountzos, D., & Pingali, K. (2013). Betweenness centrality: algorithms and implementations. Proceedings of the 18th ACM SIGPLAN Symposium on Principles and Practice of Parallel Programming, 35–46.
- Salavati, C., Abdollahpouri, A., & Manbari, Z. (2019). Ranking nodes in complex networks based on local structure and improving closeness centrality. Neurocomputing, 336, 36–45. [CrossRef]
- Samad, A., Islam, M. A., Iqbal, M. A., & Aleem, M. (2019). Centrality-Based Paper Citation Recommender System. EAI Endorsed Transactions on Industrial Networks and Intelligent Systems, 6(19), e2–e2.
- Shakeel, H. M., Iram, S., Al-Aqrabi, H., Alsboui, T., & Hill, R. (2022). A Comprehensive State-of-the-Art Survey on Data Visualization Tools: Research Developments, Challenges and Future Domain Specific Visualization Framework. IEEE Access, 10, 96581–96601. [CrossRef]
- Sterling, J. A., & Montemore, M. M. (2022). Combining Citation Network Information and Text Similarity for Research Article Recommender Systems. IEEE Access, 10, 16–23. [CrossRef]
- Suthaharan, S. (2016). Support Vector Machine. In S. Suthaharan (Ed.), Machine Learning Models and Algorithms for Big Data Classification: Thinking with Examples for Effective Learning (pp. 207–235). Springer US.
- Thafar, M. A., Albaradei, S., Uludag, M., Alshahrani, M., Gojobori, T., Essack, M., & Gao, X. (2023). OncoRTT: Predicting novel oncology-related therapeutic targets using BERT embeddings and omics features. Frontiers in Genetics, 14, 1139626. [CrossRef]
- Thafar, M. A., Albaradie, S., Olayan, R. S., Ashoor, H., Essack, M., & Bajic, V. B. (2020). Computational Drug-target Interaction Prediction based on Graph Embedding and Graph Mining. Proceedings of the 2020 10th International Conference on Bioscience, Biochemistry and Bioinformatics, 14–21.
- Thafar, M. A., Alshahrani, M., Albaradei, S., Gojobori, T., Essack, M., & Gao, X. (2022). Affinity2Vec: drug-target binding affinity prediction through representation learning, graph mining, and machine learning. Scientific Reports, 12(1), 4751. [CrossRef]
- Thafar, M. A., Olayan, R. S., Albaradei, S., Bajic, V. B., Gojobori, T., Essack, M., & Gao, X. (2021). DTi2Vec: Drug-target interaction prediction using network embedding and ensemble learning. Journal of Cheminformatics, 13(1), 71. [CrossRef]
- Thafar, M. A., Olayan, R. S., Ashoor, H., Albaradei, S., Bajic, V. B., Gao, X., Gojobori, T., & Essack, M. (2020). DTiGEMS+: drug–target interaction prediction using graph embedding, graph mining, and similarity-based techniques. Journal of Cheminformatics, 12(1), 44.
- Ting, S. L., Ip, W. H., Tsang, A. H. C., & Others. (2011). Is Naive Bayes a good classifier for document classification. International Journal of Software Engineering and Its Applications, 5(3), 37–46.
- Van Rossum, G. (2007). Python Programming Language. USENIX Annual Technical. http://kelas-karyawan-bali.kurikulum.org/IT/en/2420-2301/Python_3721_kelas-karyawan-bali-kurikulumngetesumum.html.
- Weis, J. W., & Jacobson, J. M. (2021). Learning on knowledge graph dynamics provides an early warning of impactful research. Nature Biotechnology, 39(10), 1300–1307. [CrossRef]
- West, J. D., Wesley-Smith, I., & Bergstrom, C. T. (2016). A Recommendation System Based on Hierarchical Clustering of an Article-Level Citation Network. IEEE Transactions on Big Data, 2(2), 113–123. [CrossRef]
- Xia, W., Li, T., & Li, C. (2023). A review of scientific impact prediction: tasks, features and methods. Scientometrics, 128(1), 543–585. [CrossRef]
- Yang, J., Cheng, C., Shen, S., & Yang, S. (2017). Comparison of complex network analysis software: Citespace, SCI 2 and Gephi. 2017 IEEE 2nd International Conference on Big Data Analysis (ICBDA), 169–172.
- Zhang, P., Wang, T., & Yan, J. (2022). PageRank centrality and algorithms for weighted, directed networks. Physica A: Statistical Mechanics and Its Applications, 586, 126438.
- Zhang, W., Yoshida, T., & Tang, X. (2011). A comparative study of TF*IDF, LSI and multi-words for text classification. Expert Systems with Applications, 38(3), 2758–2765. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).