Submitted:
29 April 2024
Posted:
30 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
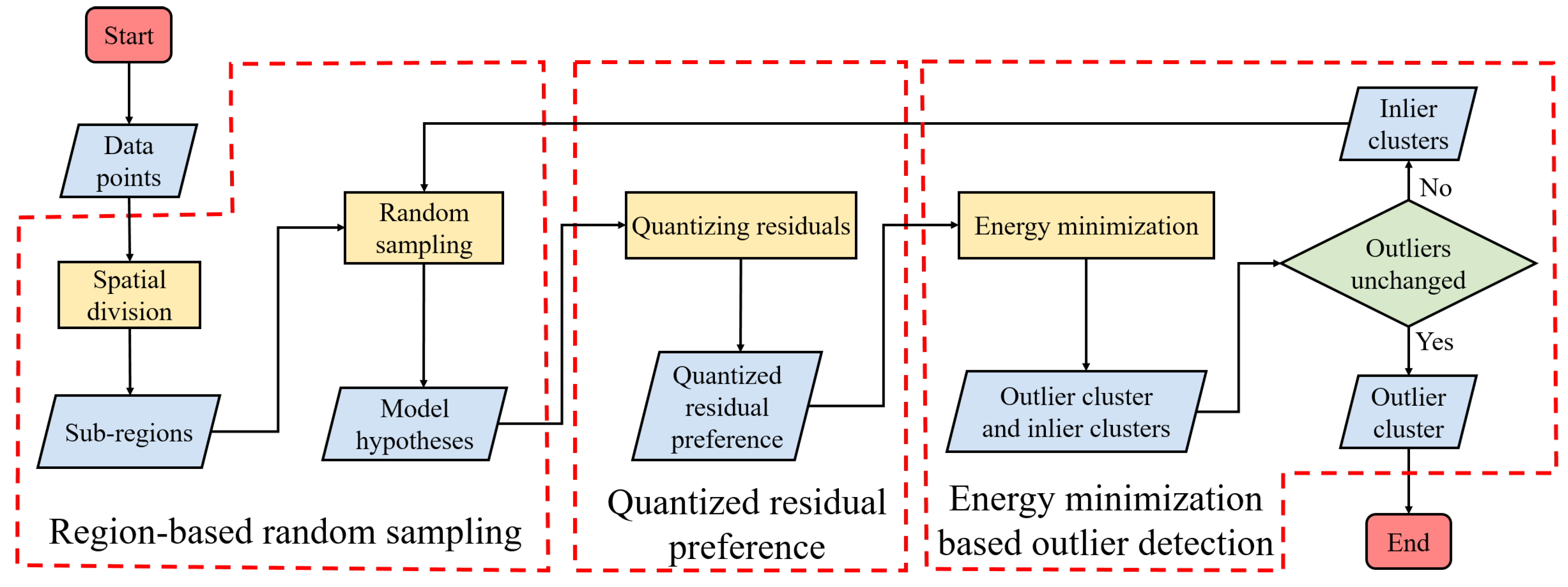
2. Materials and Methods
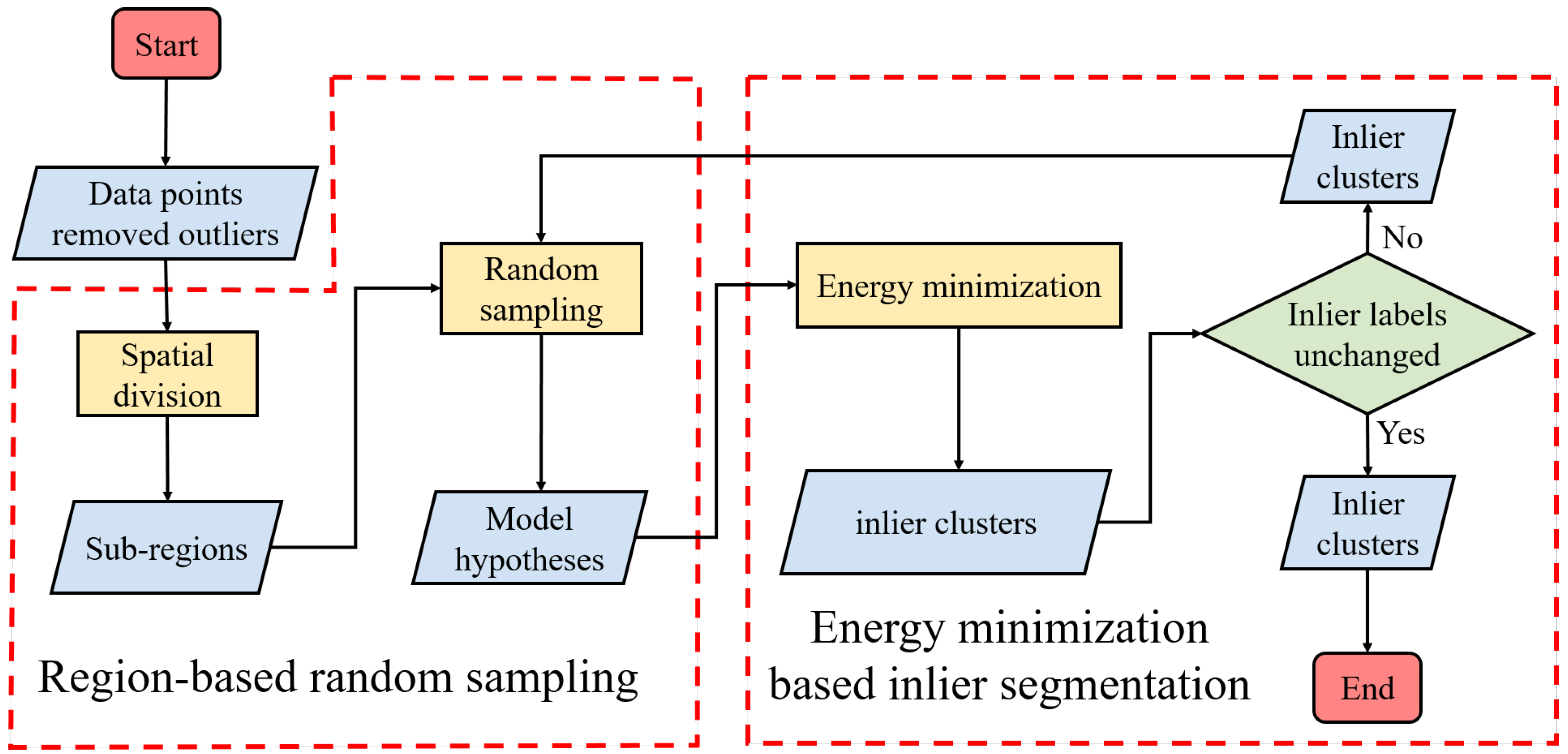
2.1. Outlier Detection
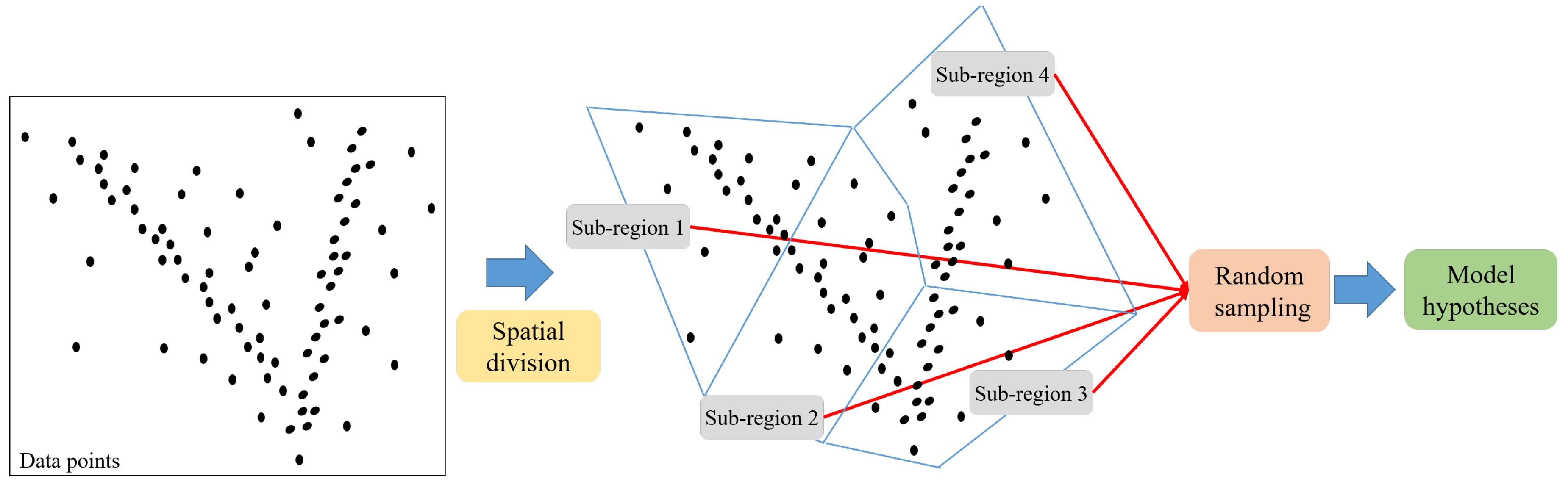
2.1.1. Region-based Random Sampling
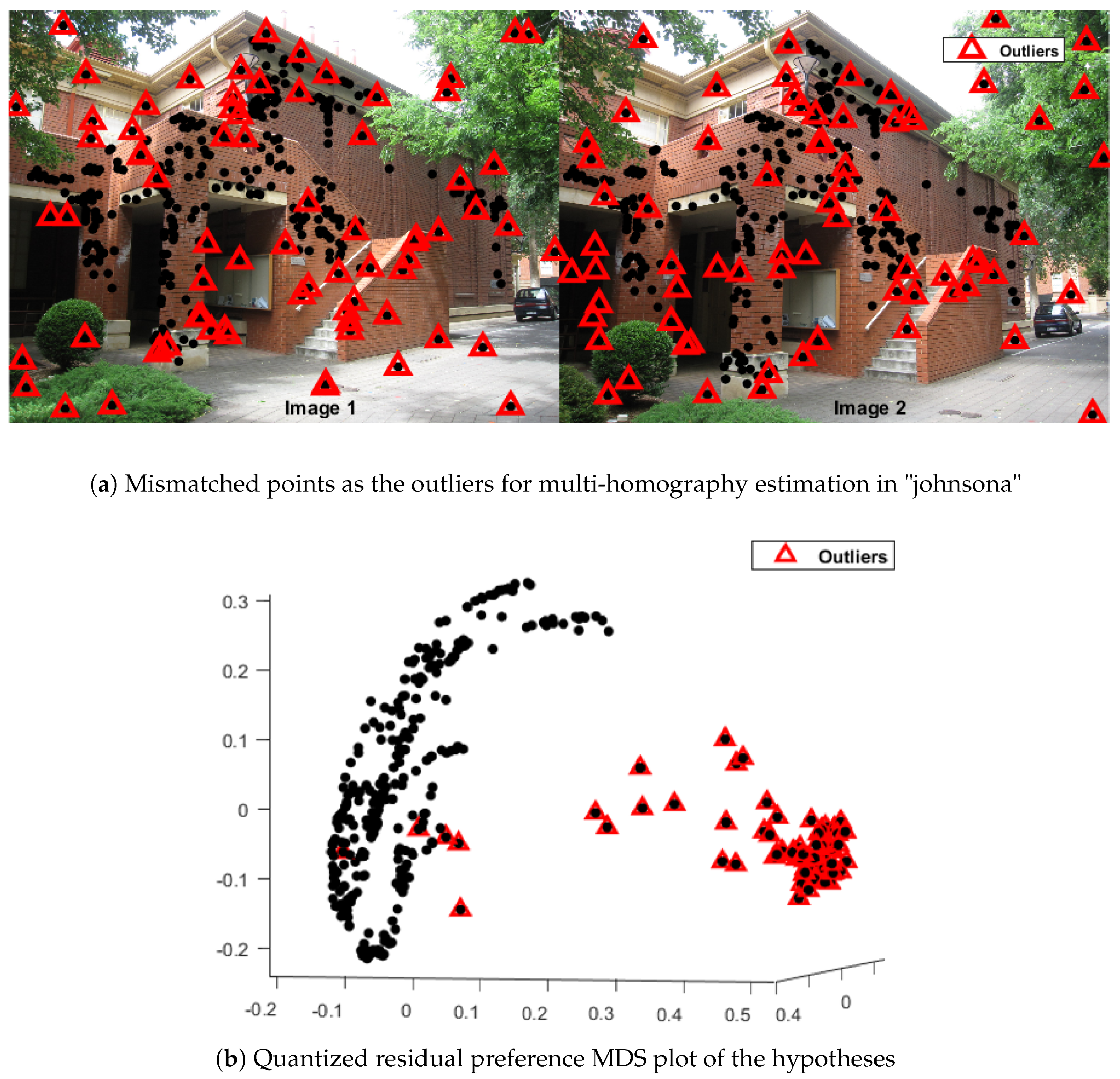
2.1.2. Quantized Residual Preference
2.1.3. Energy Minimization Based Outlier Detection
| Algorithm 1 Energy Minimization Based Outlier Detection |
|
2.2. Inlier Segmentation
| Algorithm 2 Inlier Segmentation |
|
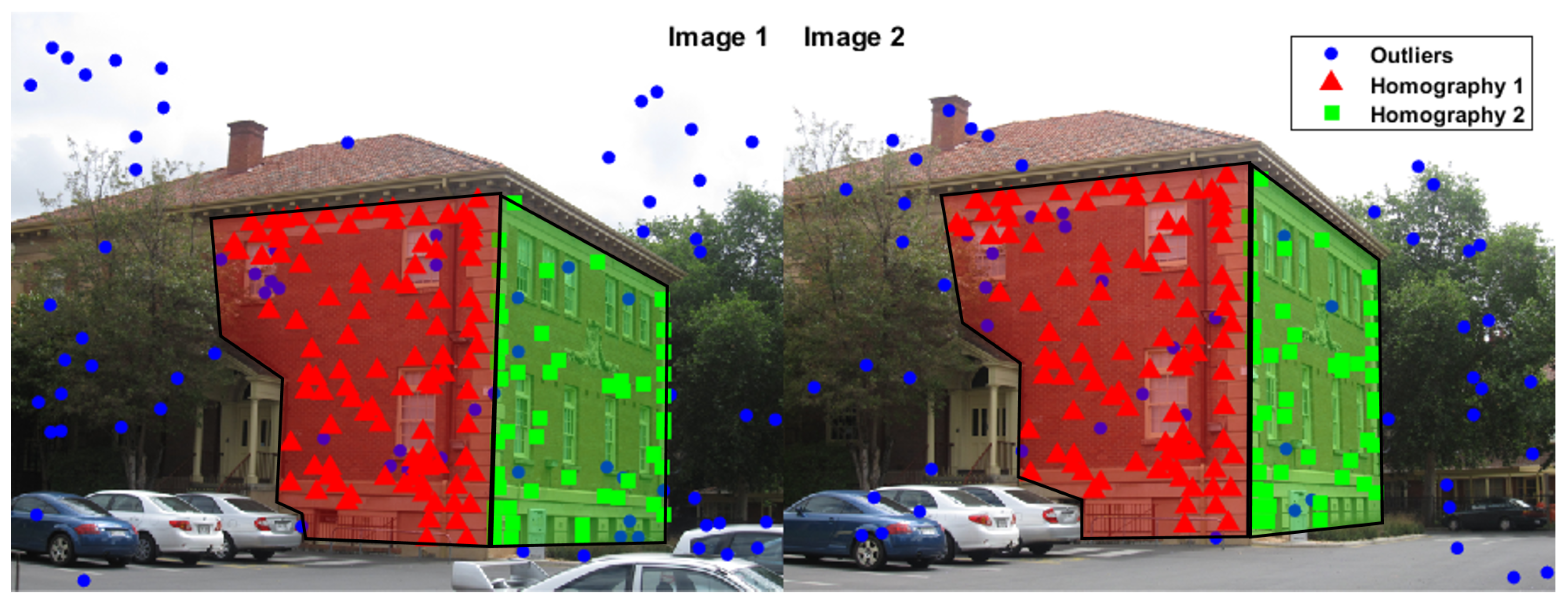
3. Results
4. Conclusions
Author Contributions
References
- Wong, H.S.; Chin, T.J.; Yu, J.; Suter, D. Dynamic and hierarchical multi-structure geometric model fitting. International Conference on Computer Vision, 2011, pp. 1044–1051.
- Fischler, M.A.; Bolles, R.C. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Communications of the ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Choi, S.; Kim, T.; Yu, W.; Choi, S.; Kim, T.; Yu, W.; Choi, S.; Kim, T.; Yu, W. Performance evaluation of RANSAC family. British Machine Vision Conference, BMVC 2009, London, UK, September 7-10, 2009. Proceedings, 2009.
- Vincent, E.; Laganiére, R. Detecting planar homographies in an image pair. Proceedings of the 2nd International Symposium on Image and Signal Processing and Analysis, 2001, pp. 182–187.
- Kanazawa, Y.; Kawakami, H. Detection of Planar Regions with Uncalibrated Stereo using Distributions of Feature Points. BMVC, 2004, pp. 1–10.
- Zuliani, M.; Kenney, C.S.; Manjunath, B. The multiransac algorithm and its application to detect planar homographies. IEEE International Conference on Image Processing 2005. IEEE, 2005, Vol. 3, pp. III–153.
- Stewart, C.V. Bias in robust estimation caused by discontinuities and multiple structures. IEEE Transactions on Pattern Analysis and Machine Intelligence 1997, 19, 818–833. [Google Scholar] [CrossRef]
- Amayo, P.; Pinies, P.; Paz, L.M.; Newman, P. Geometric Multi-Model Fitting with a Convex Relaxation Algorithm 2017.
- Wong, H.S.; Chin, T.; Yu, J.; Suter, D. Mode seeking over permutations for rapid geometric model fitting. Pattern Recognition 2013, 46, 257–271. [Google Scholar] [CrossRef]
- Toldo, R.; Fusiello, A. Robust Multiple Structures Estimation with J-Linkage. European Conference on Computer Vision, 2008, pp. 537–547.
- Toldo, R.; Fusiello, A. Real-time incremental j-linkage for robust multiple structures estimation. International Symposium on 3D Data Processing, Visualization and Transmission (3DPVT), 2010, Vol. 1, p. 6.
- Magri, L.; Fusiello, A. T-linkage: A continuous relaxation of j-linkage for multi-model fitting. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2014, pp. 3954–3961.
- Magri, L.; Fusiello, A. Multiple Model Fitting as a Set Coverage Problem. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 3318–3326.
- Magri, L.; Fusiello, A. Fitting Multiple Models via Density Analysis in Tanimoto Space; Springer International Publishing, 2015; pp. 73–84.
- Magri, L.; Fusiello, A. Multiple structure recovery via robust preference analysis. Image and Vision Computing 2017, 67, 1–15. [Google Scholar] [CrossRef]
- Magri, L.; Fusiello, A. Robust Multiple Model Fitting with Preference Analysis and Low-rank Approximation. British Machine Vision Conference, 2015.
- Chin, T.J.; Wang, H.; Suter, D. Robust fitting of multiple structures: The statistical learning approach. IEEE International Conference on Computer Vision, 2009, pp. 413–420.
- Chin, T.J.; Wang, H.; Suter, D. The ordered residual kernel for robust motion subspace clustering. International Conference on Neural Information Processing Systems, 2009, pp. 333–341.
- Xiao, G.; Wang, H.; Yan, Y.; Zhang, L. Mode seeking on graphs for geometric model fitting via preference analysis. Pattern Recognition Letters 2016, 83, 294–302. [Google Scholar] [CrossRef]
- Wong, H.S.; Chin, T.J.; Yu, J.; Suter, D. A simultaneous sample-and-filter strategy for robust multi-structure model fitting. Computer Vision and Image Understanding 2013, 117, 1755–1769. [Google Scholar] [CrossRef]
- Pham, T.T.; Chin, T.J.; Yu, J.; Suter, D. The Random Cluster Model for robust geometric fitting. IEEE Conference on Computer Vision and Pattern Recognition, 2012, pp. 710–717.
- Chin, T.J.; Yu, J.; Suter, D. Accelerated Hypothesis Generation for Multistructure Data via Preference Analysis. IEEE Transactions on Pattern Analysis & Machine Intelligence 2012, 34, 625. [Google Scholar]
- Torr, P.H.S.; Zisserman, A. MLESAC: A New Robust Estimator with Application to Estimating Image Geometry. Computer Vision and Image Understanding 2000, 78, 138–156. [Google Scholar] [CrossRef]
- Lai, T.; Wang, W.; Liu, Y.; Li, Z.; Lin, S. Robust model estimation by using preference analysis and information theory principles. Applied Intelligence 2023, 53, 22363–22373. [Google Scholar] [CrossRef]
- Isack, H.; Boykov, Y. Energy-Based Geometric Multi-model Fitting. International Journal of Computer Vision 2012, 97, 123–147. [Google Scholar] [CrossRef]
- Delong, A.; Osokin, A.; Isack, H.N.; Boykov, Y. Fast Approximate Energy Minimization with Label Costs. International Journal of Computer Vision 2012, 96, 1–27. [Google Scholar] [CrossRef]
- Pham, T.T.; Chin, T.J.; Schindler, K.; Suter, D. Interacting geometric priors for robust multimodel fitting. IEEE Transactions on Image Processing A Publication of the IEEE Signal Processing Society 2014, 23, 4601–10. [Google Scholar] [CrossRef] [PubMed]
- Isack, H.N.; Boykov, Y. Energy Based Multi-model Fitting & Matching for 3D Reconstruction. CVPR, 2014, pp. 1–4.
- Barath, D.; Matas, J. Multi-class model fitting by energy minimization and mode-seeking. Proceedings of the European Conference on Computer Vision (ECCV), 2018, pp. 221–236.
- Barath, D.; Matas, J. Progressive-x: Efficient, anytime, multi-model fitting algorithm. Proceedings of the IEEE/CVF international conference on computer vision, 2019, pp. 3780–3788.
- Boykov, Y.; Veksler, O.; Zabih, R. Fast approximate energy minimization via graph cuts. IEEE Transactions on Pattern Analysis and Machine Intelligence 2001, 23, 1222–1239. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, X.; Hotta, K.; Yang, X. G2MF-WA: Geometric multi-model fitting with weakly annotated data. Computational Visual Media 2020, 6, 135–145. [Google Scholar] [CrossRef]
- Barath, D.; Matas, J. Graph-cut RANSAC: Local optimization on spatially coherent structures. IEEE transactions on pattern analysis and machine intelligence 2021, 44, 4961–4974. [Google Scholar] [CrossRef]
- Purkait, P.; Chin, T.J.; Ackermann, H.; Suter, D. Clustering with Hypergraphs: The Case for Large Hyperedges. European Conference on Computer Vision, 2014, pp. 672–687.
- Wang, H.; Xiao, G.; Yan, Y.; Suter, D. Mode-Seeking on Hypergraphs for Robust Geometric Model Fitting. IEEE International Conference on Computer Vision, 2015, pp. 2902–2910.
- Xiao, G.; Wang, H.; Lai, T.; Suter, D. Hypergraph modelling for geometric model fitting. Pattern Recognition 2016, 60, 748–760. [Google Scholar] [CrossRef]
- Lee, K.; Meer, P.; Park, R. Robust adaptive segmentation of range images. IEEE Transactions on Pattern Analysis and Machine Intelligence 1998, 20, 200–205. [Google Scholar]
- Babhadiashar, A.; Suter, D. Robust segmentation of visual data using ranked unbiased scale estimate. Robotica 1999, 17, 649–660. [Google Scholar] [CrossRef]
- Wang, H.; Suter, D. Robust adaptive-scale parametric model estimation for computer vision. IEEE Transactions on Pattern Analysis and Machine Intelligence 2004, 26, 1459–1474. [Google Scholar] [CrossRef]
- Fan, L. Robust Scale Estimation from Ensemble Inlier Sets for Random Sample Consensus Methods. European Conference on Computer Vision, 2008, pp. 182–195.
- Toldo, R.; Fusiello, A. Automatic Estimation of the Inlier Threshold in Robust Multiple Structures Fitting. International Conference on Image Analysis and Processing, 2009, pp. 123–131.
- Raguram, R.; Frahm, J.M. RECON: Scale-adaptive robust estimation via Residual Consensus. International Conference on Computer Vision, 2011, pp. 1299–1306.
- Wang, H.; Chin, T.; Suter, D. Simultaneously Fitting and Segmenting Multiple-Structure Data with Outliers. IEEE Transactions on Pattern Analysis and Machine Intelligence 2012, 34, 1177–1192. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, Y.; Qin, Q.; Luo, B. Quantized residual preference based linkage clustering for model selection and inlier segmentation in geometric multi-model fitting. Sensors 2020, 20, 3806. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, Y.; Xie, S.; Qin, Q.; Wu, S.; Luo, B. Outlier detection based on residual histogram preference for geometric multi-model fitting. Sensors 2020, 20, 3037. [Google Scholar] [CrossRef] [PubMed]
- Nasuto, D.; Craddock, J.B.R. Napsac: High noise, high dimensional robust estimation-it’s in the bag. Proc. Brit. Mach. Vision Conf., 2002, pp. 458–467.
- Ni, K.; Jin, H.; Dellaert, F. GroupSAC: Efficient consensus in the presence of groupings. 2009 IEEE 12th International Conference on Computer Vision. IEEE, 2009, pp. 2193–2200.
- Sattler, T.; Leibe, B.; Kobbelt, L. SCRAMSAC: Improving RANSAC’s efficiency with a spatial consistency filter. 2009 IEEE 12th International Conference on Computer Vision. IEEE, 2009, pp. 2090–2097.
- Hartley, R.; Zisserman, A. Multiple view geometry in computer vision; Cambridge university press, 2003.
- Hartley, R.I. In defense of the eight-point algorithm. IEEE Transactions on Pattern Analysis and Machine Intelligence 1997, 19, 580–593. [Google Scholar] [CrossRef]
- Tron, R.; Vidal, R. A Benchmark for the Comparison of 3-D Motion Segmentation Algorithms. Computer Vision and Pattern Recognition, 2007. CVPR ’07. IEEE Conference on, 2007, pp. 1–8.
- Xiao, G.; Wang, H.; Ma, J.; Suter, D. Segmentation by continuous latent semantic analysis for multi-structure model fitting. International Journal of Computer Vision 2021, 129, 2034–2056. [Google Scholar] [CrossRef]
- Mittal, S.; Anand, S.; Meer, P. Generalized Projection-Based M-Estimator. IEEE Transactions on Pattern Analysis and Machine Intelligence 2012, 34, 2351–2364. [Google Scholar] [CrossRef]
| 1 | |
| 2 | |
| 3 |

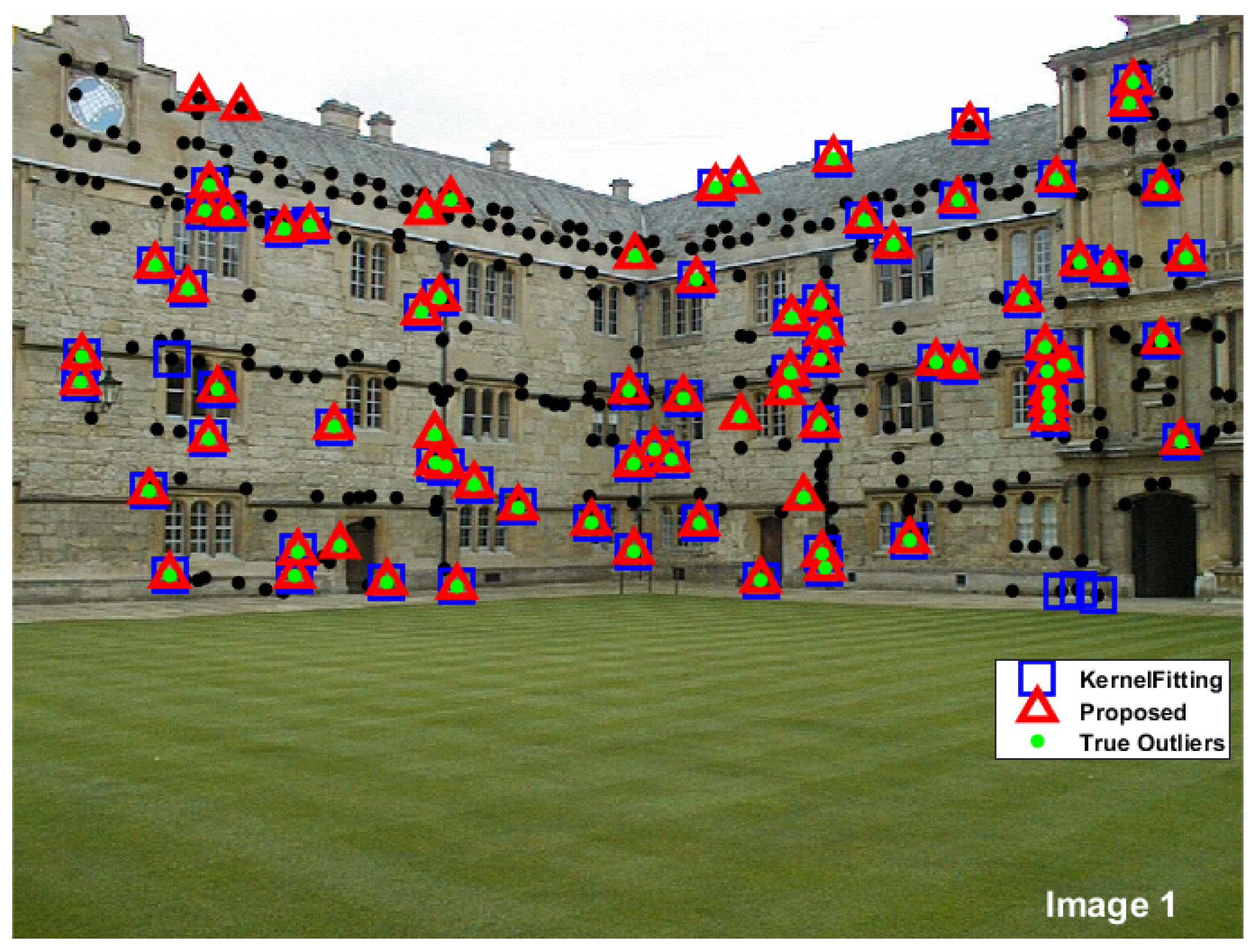

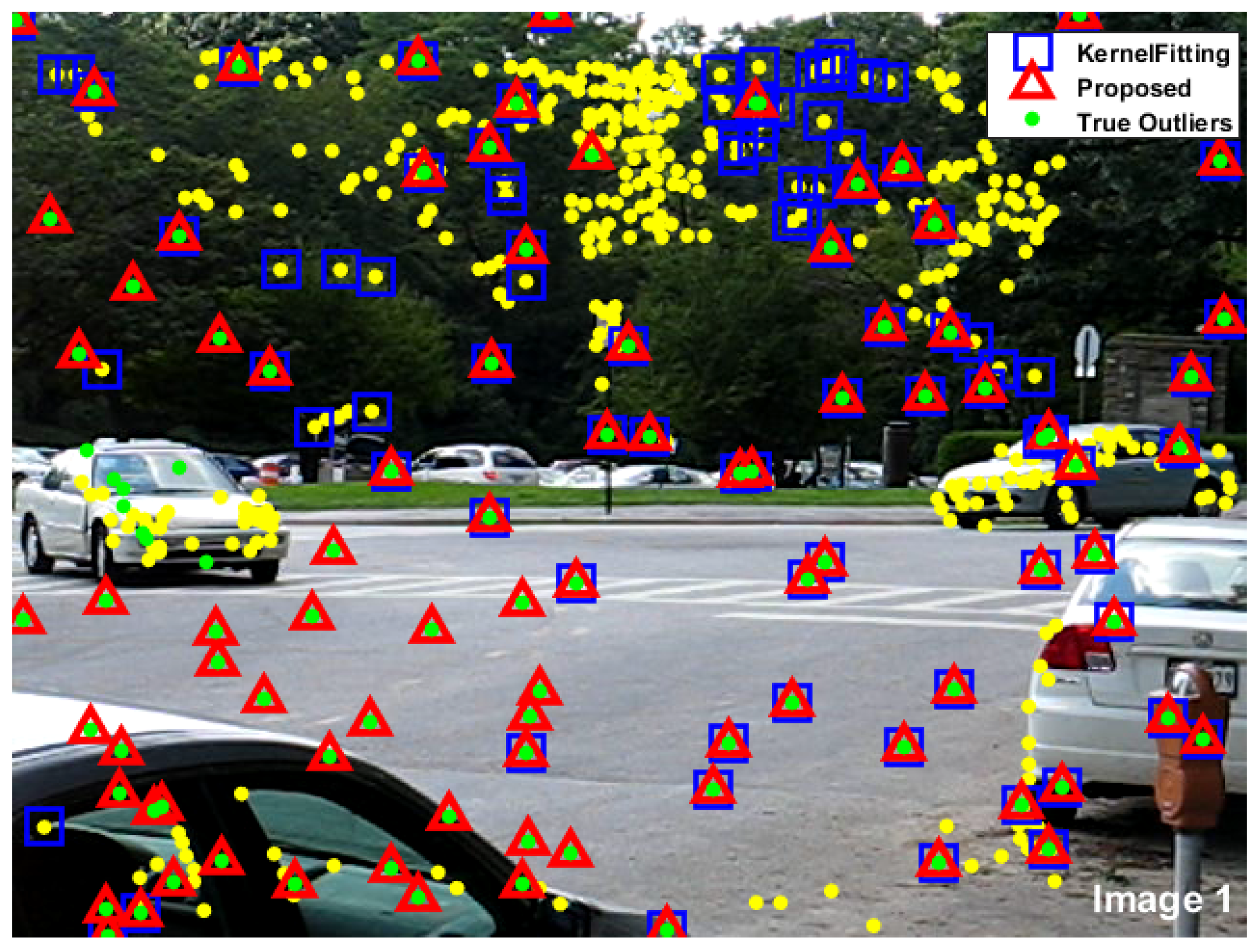

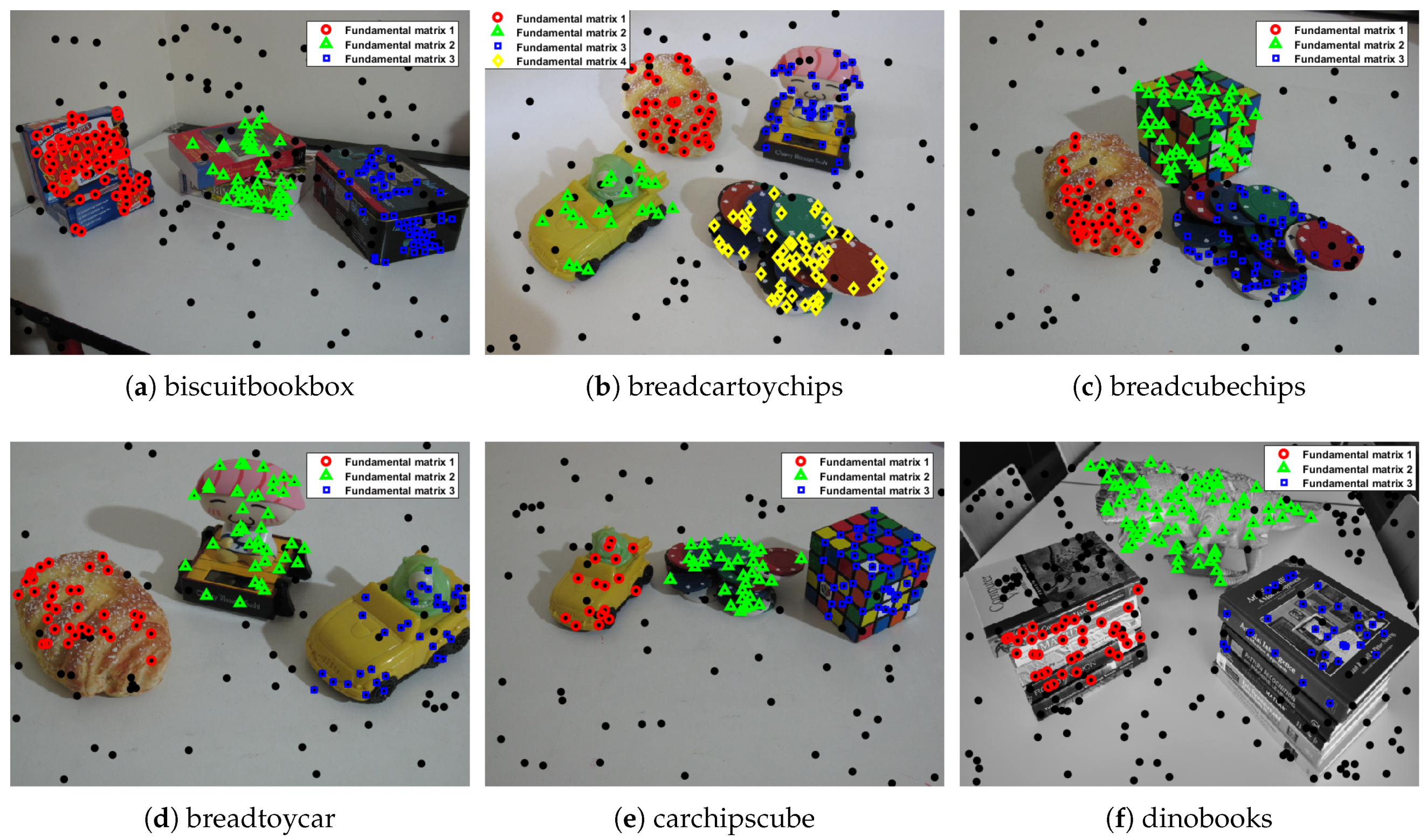
| Total | Total | Kernel fitting | Proposed | |||||
| points | outliers | Correct | Missing | False | Correct | Missing | False | |
| two-view plane segmentation | ||||||||
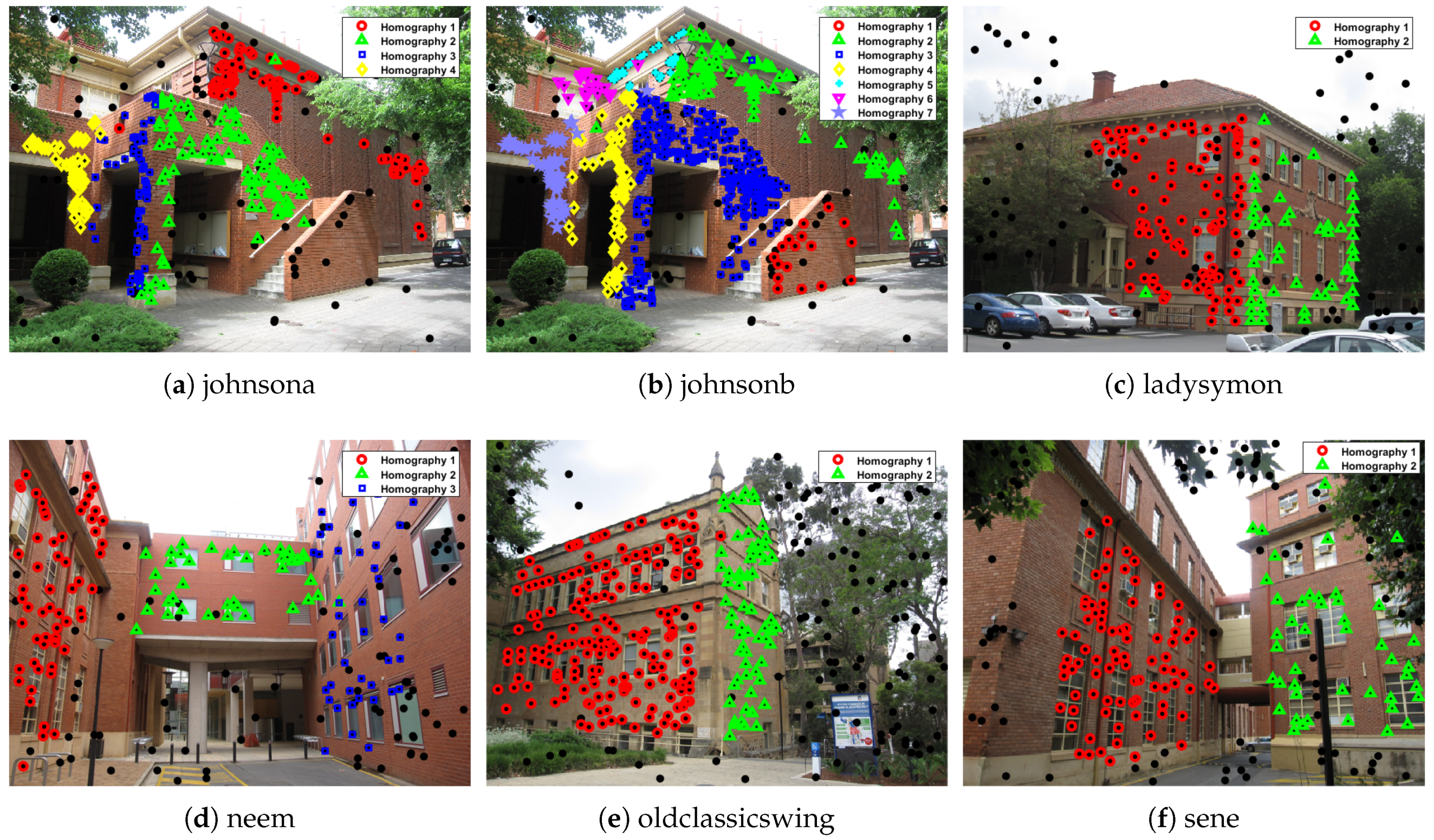
| johnsona | 373 | 78 | 70 | 8 | 0 | 75 | 3 | 0 |
| johnsonb | 649 | 78 | 63 | 15 | 33 | 71 | 7 | 0 |
| ladysymon | 273 | 77 | 70 | 7 | 0 | 76 | 1 | 0 |
| neem | 241 | 88 | 88 | 0 | 4 | 88 | 0 | 0 |
| oldclassicswing | 379 | 123 | 123 | 0 | 0 | 123 | 0 | 0 |
| sene | 250 | 118 | 106 | 12 | 3 | 117 | 1 | 0 |
| two-view motion segmentation | ||||||||
| biscuitbookbox | 259 | 97 | 90 | 7 | 3 | 97 | 0 | 0 |
| breadcartoychips | 237 | 82 | 76 | 6 | 0 | 81 | 1 | 0 |
| breadcubechips | 230 | 81 | 69 | 12 | 4 | 80 | 1 | 0 |
| breadtoycar | 166 | 56 | 43 | 13 | 1 | 53 | 3 | 0 |
| carchipscube | 165 | 60 | 52 | 8 | 0 | 60 | 0 | 0 |
| dinobooks | 360 | 155 | 128 | 27 | 25 | 151 | 4 | 41 |
| Methods | PEARL | J-linkage | T-linkage | SA-RCM | Prog-X | CLSA | Proposed |
| johnsona | 4.02 | 5.07 | 4.02 | 5.90 | 5.07 | 6.00 | 1.61 |
| johnsonb | 18.18 | 18.33 | 18.33 | 17.95 | 6.12 | 20.0 | 3.39 |
| ladysymon | 5.49 | 9.25 | 5.06 | 7.17 | 3.92 | 1.00 | 2.11 |
| neem | 5.39 | 3.73 | 3.73 | 5.81 | 6.75 | 1.00 | 0.83 |
| oldclassicswing | 1.58 | 0.27 | 0.26 | 2.11 | 0.52 | 0.00 | 0.26 |
| sene | 0.80 | 0.84 | 0.40 | 0.80 | 0.40 | 0.00 | 0.4 |
| Methods | PEARL | J-linkage | T-linkage | SA-RCM | Prog-X | CLSA | Proposed |
| biscuitbookbox | 4.25 | 1.55 | 1.54 | 7.04 | 3.11 | 1.00 | 0 |
| breadcartoychips | 5.91 | 11.26 | 3.37 | 4.81 | 2.87 | 5.00 | 0.42 |
| breadcubechips | 4.78 | 3.04 | 0.86 | 7.85 | 1.33 | 1.00 | 0.43 |
| breadtoycar | 6.63 | 5.49 | 4.21 | 3.82 | 3.06 | 0.00 | 1.81 |
| carchipscube | 11.82 | 4.27 | 1.81 | 11.75 | 13.90 | 3.00 | 0 |
| dinobooks | 14.72 | 17.11 | 9.44 | 8.03 | 7.66 | 10.00 | 12.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





