1. Introduction
Maize is considered one of the most important cereal crops worldwide. It serves as a staple food for billions of people and is also a crucial feed for livestock. Environmental factors like drought-stress have detrimental effects on the growth and productivity of maize, reducing yield and impairing quality [
1]. This has necessitated the development of effective strategies for early detection and mitigation of drought-stress in maize.
Traditional methods for assessing drought-stress in maize, such as visual inspection and manual measurements, are labor-intensive, time-consuming, and often subjective. In recent years, there has been a growing interest in leveraging computer vision and deep learning techniques to automate the process of drought-stress analysis in maize.[
2,
3,
4,
5,
6]. These technologies offer the potential to provide rapid, non-destructive, and objective assessments of drought-stress, enabling farmers and researchers to make informed decisions in a timely manner.
High-throughput imaging systems have been pivotal in advancing maize plant phenotyping, enabling researchers to efficiently analyze large numbers of plants for various traits, including those related to drought-stress. [
7,
8,
9,
10]. These systems typically involve the use of cameras and sensors to capture detailed images of plants at various growth stages, allowing for the extraction of quantitative data on plant morphology, physiology, and response to environmental stressors.
In the case of visual phenotyping, the near-infrared (NIR) reflectance is known to be particularly responsive to drought-stress [
11,
12]. Well-watered healthy plants are known to reflect more NIR wavelengths to reduce net heat gain as a result of photosynthesis. Similarly, drought-stressed plants show lesser NIR reflectance due to improper photosynthesis and nutrient deficiency. Studies have shown that the growth of the youngest expanding leaf (YEL) is most sensitive to water stress [
13,
14,
15]. We utilize these visual cues observed during plant phenotyping to distinguish between well-watered and drought-stressed maize plants.
In this paper, we extend the work by Rafael et al [
16] on developing a high throughput low-cost system for water stress detection in maize plants. This platform is utilized to collect a dataset of images of maize plants placed in a controlled chamber. A group of four maize plants placed side-by-side were imaged with alternating plants treated to two different watering protocols. One set of maize plants were subjected to induced drought-stress over several days whereas the other set remained well-watered. The protocol for each trial lasted for 6 to 11 days. We primarily used NIR reflectance captured using Raspberry Pi cameras in our analysis [
17,
18,
19]. Similar to the approach adopted in [
16], we used fasterRCNN [
20] to detect the region of interest on the maize plants and a NIR workflow implemented by plantCV [
21] to segment out the background. In addition to generating histograms and computing the Earth Mover’s Distance (EMD) [
22] to study the effect of drought-stress on NIR reflectance, we also made use of a Vision Transformer (ViT) [
23] to perform a simple binary classification between drought-stress and well-watered plants. We studied the impact of inclusion of leaves in our analysis (with respect to YEL and stems) by considering larger regions around detection boxes. We also explored experimental settings for training the ViT model to determine its generalization.
The rest of the paper is organized as follows. In
Section 2, we give a brief description of the automated imaging platform introduced by the authors in [
16], capable of monitoring plants placed inside a controlled chamber. In
Section 3, we introduce our model pipeline used for the detection and analysis of drought-stress in the plants.
Section 4 discusses the experimental setup used in our analysis and presents results on NIR reflectance distributions and classification performance by our VIT model. We also present our findings corresponding to our experiments on locating an ideal region on the plant, other than the YEL, that is most sensitive to drought-stress.
2. Dataset
2.1. Data Collection Platform
The Plant Data Collection (PDC) system, as described in [
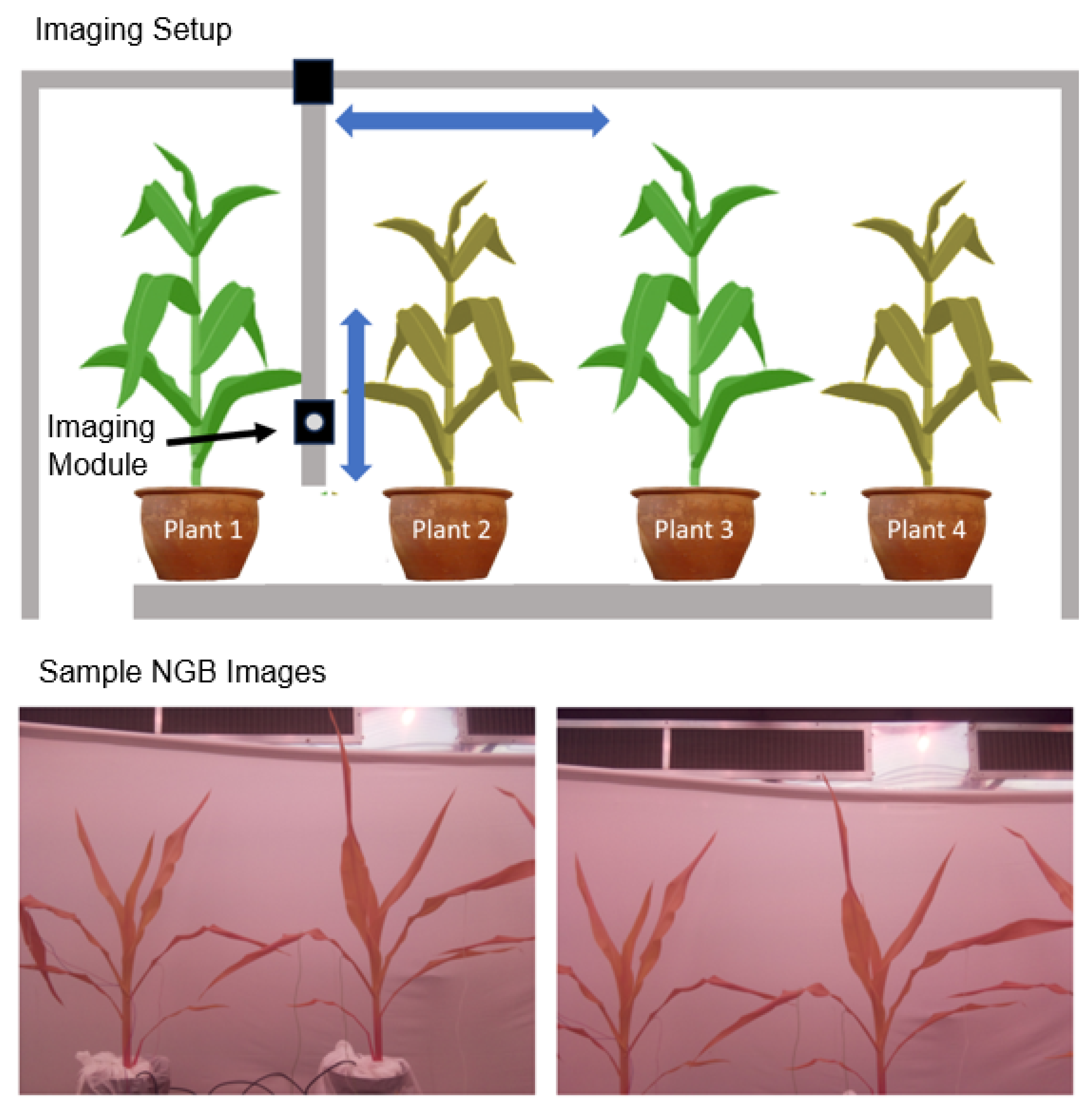
16], was installed in a controlled chamber with regulated temperature, humidity, luminosity, and watering conditions. The imaging system consisted of a gantry with two carts controlled by stepper motors, offering two degrees of freedom: vertical and horizontal movement.
Figure 1 illustrates the setup. A NoIR Pi Camera V2 without an infrared (IR) filter was mounted on the cart for vertical imaging. To improve the camera’s performance, a Roscolux Cinegel R2007 Storaro Blue film filter was placed over the Pi Camera aperture. This filter primarily allows blue and IR light to pass through while blocking much of the red and green spectrum, enhancing the camera’s ability to capture color changes related to photosynthetic activity. The resulting images are NIR (Near IR), Green and Blue (NGB) having a high resolution of 3280×2464 pixels.
The PDC system was remotely controlled by a Raspberry Pi 3 [
24] running Node-RED software [
25], which provided real-time diagnostics and ensured regular monitoring of the system status. The data collected was temporarily stored in the Pi and automatically backed up daily to a local storage drive. Images were saved into their respective folders corresponding to the date of data collection. The date, time and spot location was appended to the image file name. They were then normalized using the approach mentioned in [
16].
During data collection, each vertical scan involved capturing 1 image per step. A number of vertical steps (which differed based on the trial) were followed by a horizontal step. This cycle was repeated 21 times, resulting in 800 images per scan within approximately 30 minutes, constituting a single session.
2.2. Water Stress Protocol and Image Dataset
The drought-stress protocol was conducted on three imaging trials from October to December of 2020. Data collection was performed on four Syngenta Agrisure Viptera maize plants, each placed side by side in 6.1-liter pots containing a 50%/50% peat lite/sand mix. Alternate pots received regular watering while the remaining were unwatered throughout the experiment.
Table 1 shows the details of the three trials. The growth stage indicates the stage of the maize plants at the start of the trial. V and H steps are the number of vertical and horizontal scans undertaken by the Raspberry Pi camera per session of the trial. For each trial, 2 sessions were conducted daily, at 6:10 am and 3:10 pm respectively. Many days of the imaging trials have session 2 absent, due to interruption in data collection. Hence, our experiments were conducted only on Session 1 of each trial. For details on the availability of our imaging trials see the corresponding section at the end of this paper.
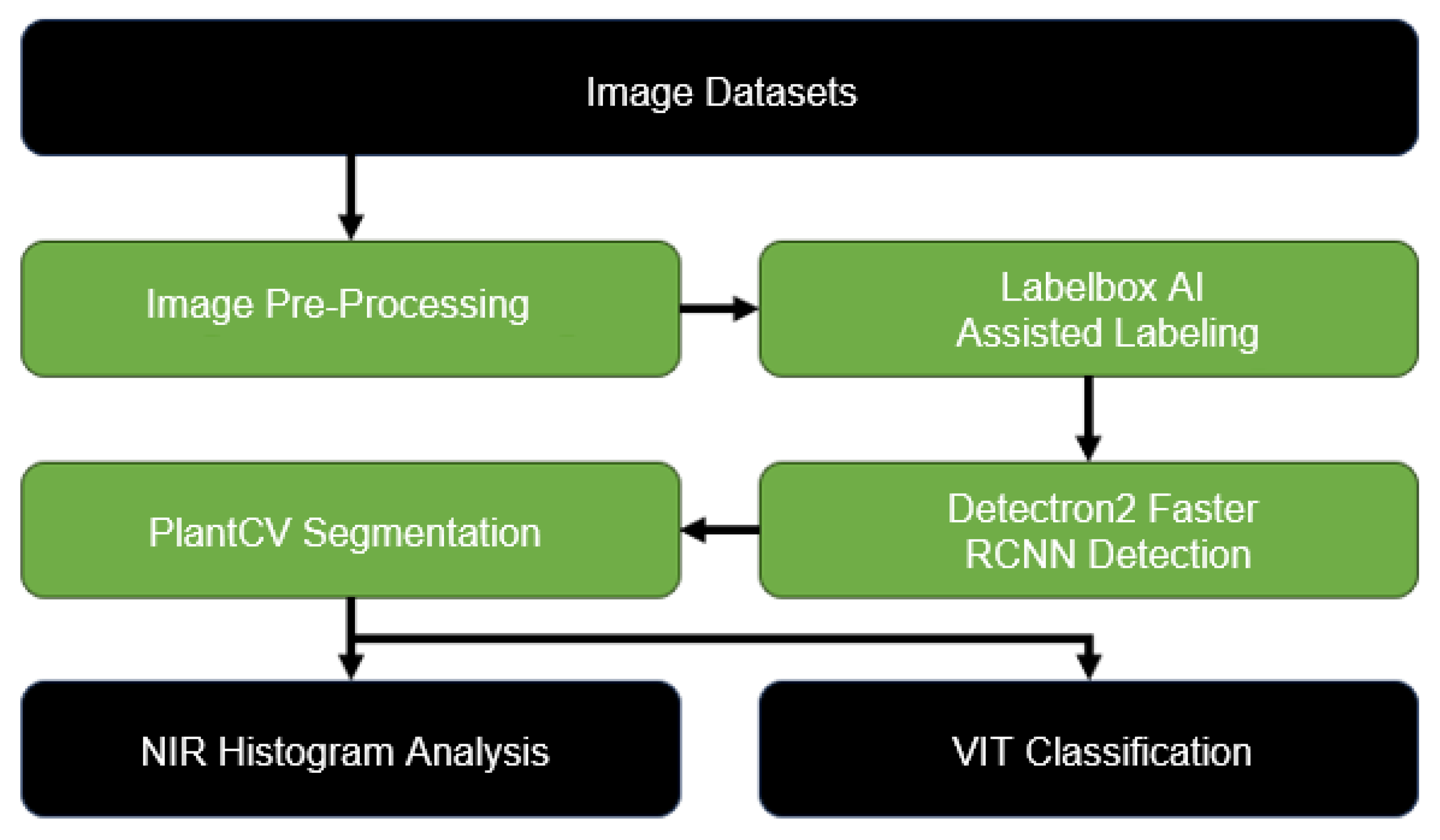
3. Methodology
The overall processing pipeline is shown in
Figure 2.
3.1. Image Pre-Processing
First, image normalization was done per session, using the equation norm=
.
stands for pixel value,minimum pixel value and maximum pixel value respectively.The next step of data pre-processing included creating patches to separate the four plants and sorting them into well-watered and drought-stressed categories. This was done using a semi-automatic script that took advantage of the repetitive motion of the camera with the locations indexed by the image ID. The script assigned a patch with height same as that of the image and allowed the user to decide where to cut the image vertically to crop out the plant. This patch size was allowed to automatically propagate across the images for two consecutive horizontal scans after which the user was prompted to choose a new vertical cut. This process was repeated for each day in a trial. Out of the four plants imaged, we conducted our analysis on Plants 2 and 3 since they were the most visible throughout each scan sessions. Plant 1 and 4 being located towards either end of the row, appeared inconsistently in between image frames due to the horizontal motion and edge distortions of the camera (see
Figure 1). As a result only few patches of Plants 1 and 4 were extracted and hence were left out of the analysis. Additionally views that either had no plant part visible or just the top of some leaves visible were also removed. This was again done semi-automatically utilizing the repetitive movement of the camera.
In our attempt to scale up the data collection and conduct drought-stress analysis on multiple trials, we faced additional challenges not addressed in [
16]. As mentioned in
Section 1, the YEL of maize plants has been shown to be significantly impacted by water stress. While we still wanted to rely on analysis of NIR pixels extracted from the YEL, accurately and consistently detecting the smallest leaf posed a significant challenge in our pre-processing steps. Being small and usually hidden between surrounding leaves, the YEL is usually difficult to detect in images. Also occasional shifting of pots in the chamber at the time of watering and manual measurements as well as movement of the camera in each session led to images with changing views of the plants. Another major drawback present in the dataset, was the significant overlap between leaves of adjacent plants, particularly in the later days of a trial when leaves were overgrown or at their maximum length. This led to occlusions of parts of the plant which negatively impacted our analysis.
In order to overcome the above challenges, we decided to adopt a different approach in our analysis than the one undertaken in [
16]. Instead of training an object detection model to detect just the YEL, we chose a region that approximately surrounded the YEL and the stem for each plant. Whenever the YEL wasn’t visible we annotated what we considered to be the next youngest expanding leaf. Annotations were done by drawing tight bounding boxes enclosing the tip of the youngest leaf to the base of the stem.
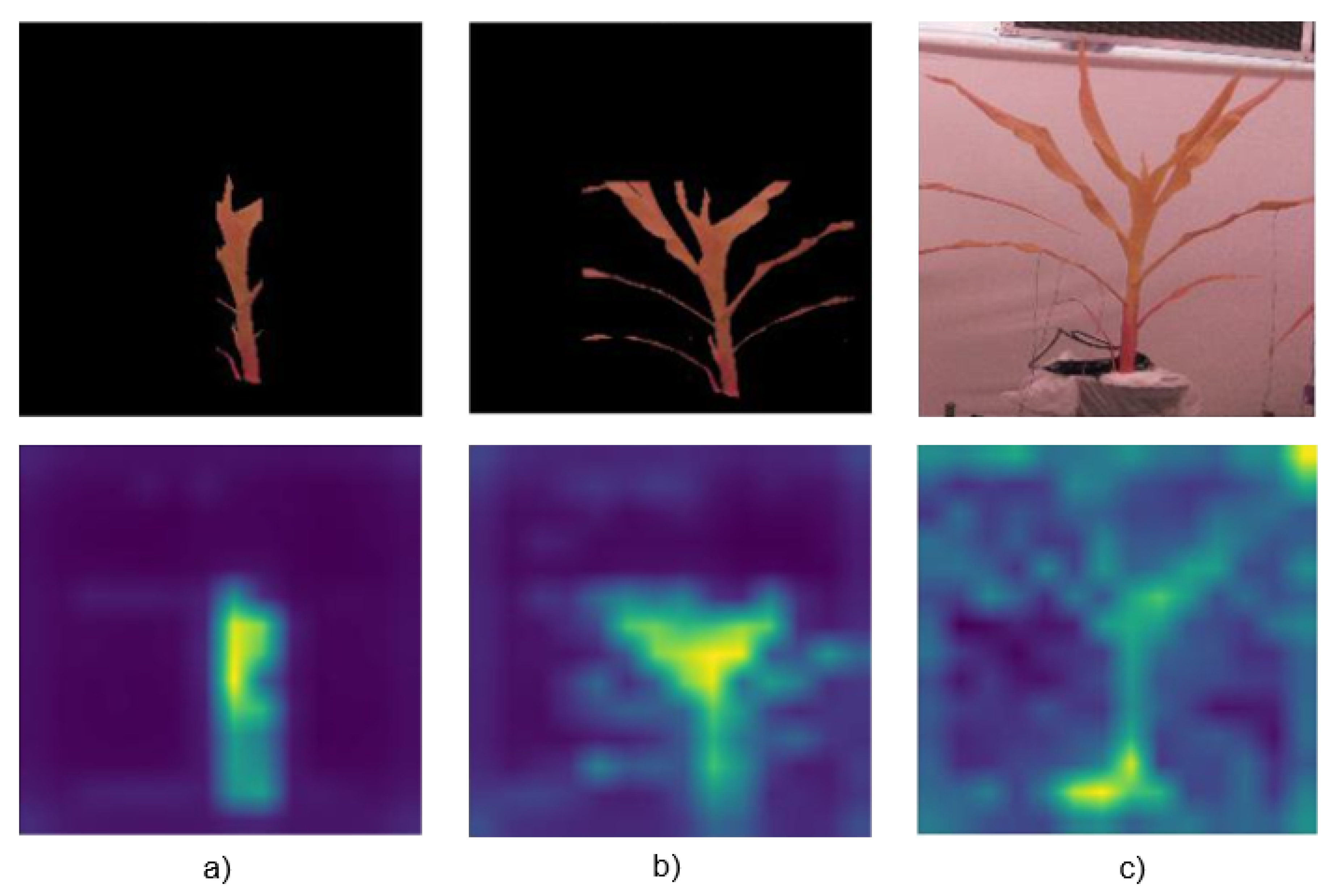
Figure 3 provides an example of the detections using our Deep Learning (DL) model. The online labeling platform LabelBox [
26] was used for generating the ground truth bounding boxes. Labelbox is a powerful labeling tool that allows efficient and accurate annotations of datasets for various machine learning projects. Through its online platform and easy to use interface, users can import data, define labels and annotation types, collaborate with team members to assign, annotate and review labels as well as export the labeled data in the desired format. Additionally, LabelBox’s Model Assisted Labeling (MAL) feature can significantly reduce annotation time by allowing computer generated predictions to be uploaded as pre-labels. This feature especially proved useful in our analysis as it helped us annotate our dataset in a short amount of time as well as improve on the detection accuracy.
3.2. Detection and Segmentation
We utilized Facebook’s Detectron2 framework [
27] with a FasterRCNN backbone [
20] for the purpose of detection of the YEL and stem regions within each plant patch.For training our Detectron2 model, we used a curated dataset combining our three imaging trials.
Two views from each vertical scan from the first sessions for each day were selected from all three trials. The two views were selected such that one showed the plant in full view while another showed the plant with the camera shifted up by a few vertical steps.
Table 2 shows details of our annotation process using Labelbox and it’s MAL feature. For each trial we select a small subset of images and label them manually. We train a detectron2 model on this subset with a train-validation split of 90%-10% and report the annotation time and model evaluation in terms of average precision (AP and AP75 values). We use the predictions as pre-labels on our second set of images (using MAL) and compare the annotation time with our manually labeled subset. We observed that with MAL we were able to annotate twice the number of images in less than half the time compared to manual labeling. We also report the evaluation of our final 3 trained models with the full dataset (combining manually labeled and MAL images). Due to more number of images in this case, we see a significant improvement in the AP values.The 3 trained models were then used to run inference on all images in their respective trials. The predictions were then used to extract the YEL and Stem region from each plant.
For segmenting the plant part within these regions, we adopted a method similar to the NIR workflow in PlantCV [
21]. NIR images comprise grayscale pixels representing NIR light reflected from plants, requiring adequate lighting and sufficient contrast between plants and the background. To conduct water stress analysis, we extracted the NIR channel from the NGB image, as depicted in
Figure 3. Initially, we converted the NGB image to the Hue-Saturation-Value (HSV) scale using the OpenCV library [
28]. Next, we created a binary mask using the saturation channel from the HSV image and the OpenCV thresholding techniques to eliminate the background. Finally, we applied this binary mask to the NIR image to obtain a masked NIR image with the background removed.
3.3. Drought Stress Analysis
We performed an analysis on the NIR values over time by tracking changes in their distributions for the control and drought-stressed plants and used the NIR images as inputs to a DL classifier quantifying its performance over time as a way to monitor differences in the plants. For the first analysis, to study the distributions of the NIR pixels extracted from the detected regions, we computed their histograms and EMDs. The shape of the histograms as well as their mean helps us to visualize the pixel distributions of each plant as we progress through the days of the trial. Additionally, the EMDs capture how reflectance evolved for each plant over time by comparing each day with the first day of the trial.
The segmented plants were used to train a DL classifier to separate drought-stress and well-watered plants. We chose a Vision Transformer (ViT) [
23] to perform the binary classification. The transformer architecture is considered to be the state-of-the-art for natural language processing and computer vision tasks. A ViT is a model based on the transformer architecture that performs image classification based on patches of images. It divides an image into fixed-size patches, adds positional embedding to these patches, which are then used as an input to the transformer encoder. ViTs have achieved the best results in various computer vision tasks like image classification, object detection and semantic segmentation. They are known to outperform CNNs. Like the original transformer, ViTs are also equipped with the self-attention mechanism that allows to capture long range dependencies and contextual information present in the input data. The self learnt attention weights allows the model to focus on relevant areas in the image and provides more interpretability in the form of attention heatmaps. In
Figure 6, we show some attention plots indicating what our trained ViT has learnt.
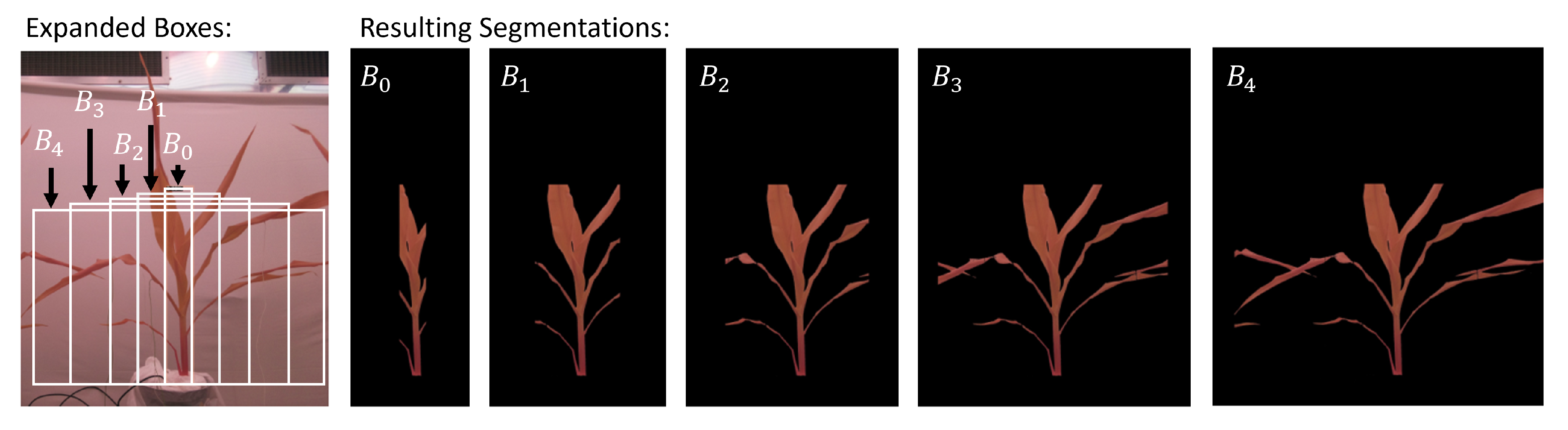
In order to quantify the impact of having the adjacent leaves as part of both analysis pipelines, we increased the width of our bounding boxes. They were expanded on both sides by
and 1000 pixels. This generates bounding boxes
through
.
Figure 4 illustrates the original detected bounding box and the subsequent gradual expansion. The motivation behind conducting these experiments was to find a suitable bounding box size encompassing the YEL and stem region of the plants, that would yield the best results for drought-stress detection. It was also done to verify the negative effect of leaf occlusion by adjacent plants, as bounding boxes with larger widths were expected to perform worse.
4. Results
4.1. Experimental Setup
To select our training views for our ViT classifier,we separated some views for each trial based on the protocol adopted for the Detectron2 training, discussed in
Section 3.2.This gave us a total of 1278 images in the training set. A train-validation split of 85%-15% was used. We used the remaining images as the test set. A total of six experiments were considered using different combinations of trials for training and testing.
Table 3 shows the details.Note that we did not include Trial 1 as train set for Experiments D and E, for reasons discussed later in the next subsection.
Transfer learning was used to fine-tune the ViT model pre-trained on ImageNet-21k at a resolution of 224x224. Training was done over 100 epochs with a batch size of 32. Data augmentation on the train set included horizontal flip, rotate, zoom and brightness adjustment. An ADAM optimizer [
29] with a learning rate of 5e-5 was used for training. All experiments were run on an Ubuntu workstation with an NVIDIA GeForce GTX 1080 Ti GPU.
4.2. Results and Discussion
In this section we present the results of our detailed analysis on our three imaging trials. First, we show results for our extracted NIR pixels in the form of histogram distributions and EMD, and then of our ViT experiments with classification accuracy plots.
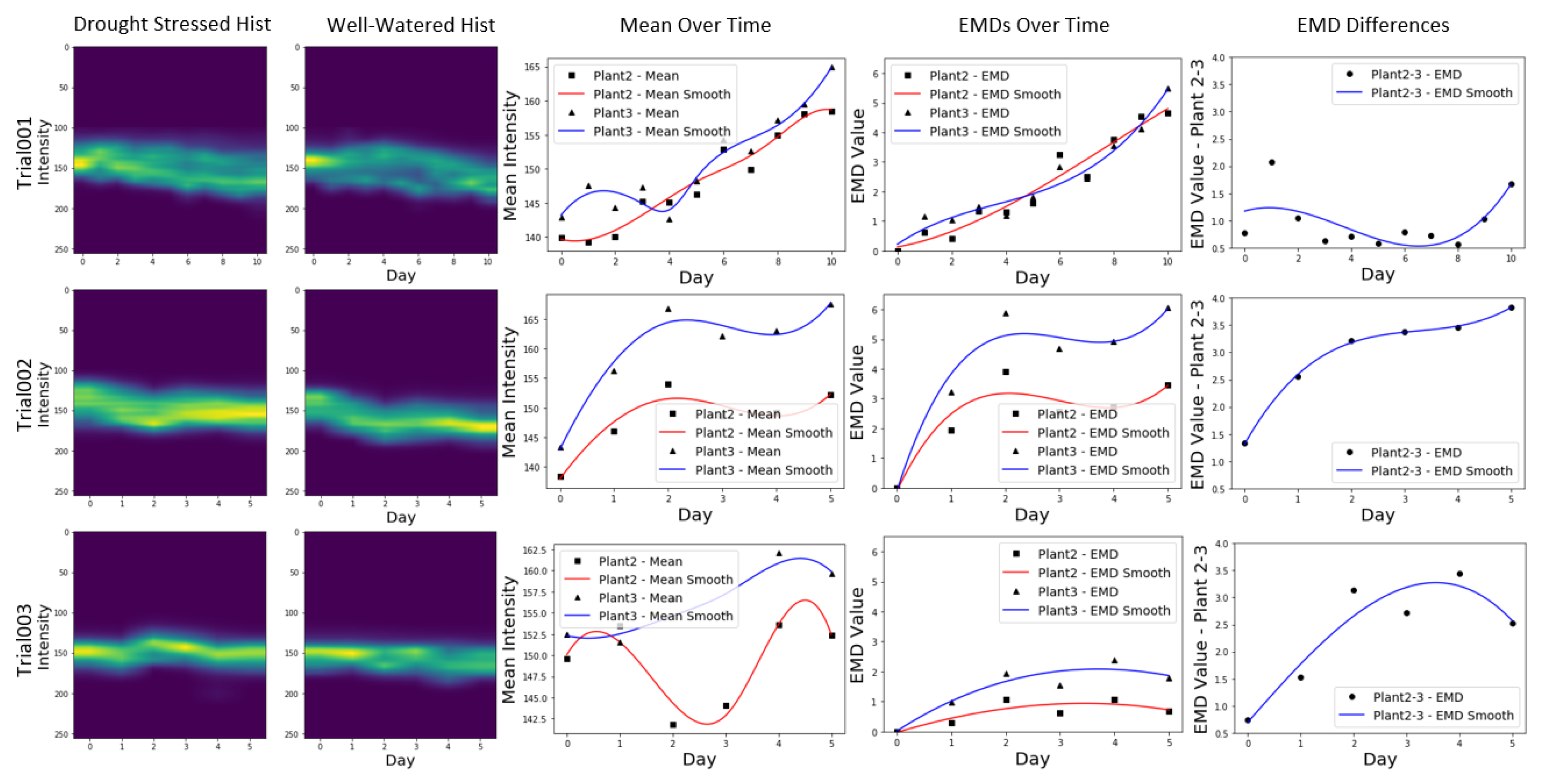
Figure 5 shows histograms and the mean of the distributions for the three trials as well as the EMDs between the well-watered and drought-stressed group. For the histogram plots we have NIR intensity and time along the y and x axis respectively and the frequency is represented by the colors. The mean of the histograms over time are also plotted. The EMDs over time compare EMD each day with the first day of the experiment for both groups of plants. This gives us an estimate of how the NIR reflectance evolves for each plant over time. The EMD differences give us a cross comparison between the well-watered and the drought-stressed plant per day. To smooth the data values we use spline representation and B spline basis elements utilizing scipy’s interpolation package [
30]. We observe no separation between the groups for trial 1. One of the possible explanations for this is that trial 1 had almost twice the number of days compared to the other trials. This prolonged period of drought-stress could be a reason why expected trends were not observed. More number of days also meant plants were overgrown towards the end of the trial which led to more occlusion and in turn more noisy data.Furthermore, evaporation and stomatal conductance are very dynamic and can change not only with plant dehydration, but also with light, temperature, vapor pressure deficit (VPD) etc. In a controlled chamber setting, even though it is possible to control most of these parameters, some like VPD, are prone to significant fluctuations, which in turn can affect plant transpiration rate and photosynthesis. [
31,
32,
33]
Trial 2 showed the overall best separation between the two distributions. We observe a clear separation between the means of the NIR pixel distributions as well as the EMD over time plot. The EMD difference between the two sets of plants also show a continuous increase as we progress through the trial. For trial 3, even though a separation can be observed in all of the plots, this is not as clear as in the case of trial 2. We observe that the separation between the plants decreases towards the end of the trial as seen in the mean over time plot.Unlike trial 2, the EMD values for each plant do not show a sharp increase compared to the first day of the trial.
The EMD cross comparison between the two plants also show a slight dip in the otherwise increasing curve, towards the end of the trial. These results could be attributed to more leaf occlusions present in trial 3 in comparison to trial 2.
For the classification task,
Figure 6 shows attention masks extracted from the self-attention layers of our ViT. We observe maximum brightness around the YEL region for both sets of images, indicating that this region of the plants is most sensitive to changes in the watering regime. We also show an example of what the ViT is learning from images that do not have the background segmented out. We observe that the attention in this scenario is no longer near the YEL region but on other parts of the background. To prevent the model from learning to identify objects on the background, our ViT classifier was trained on images with the background removed.
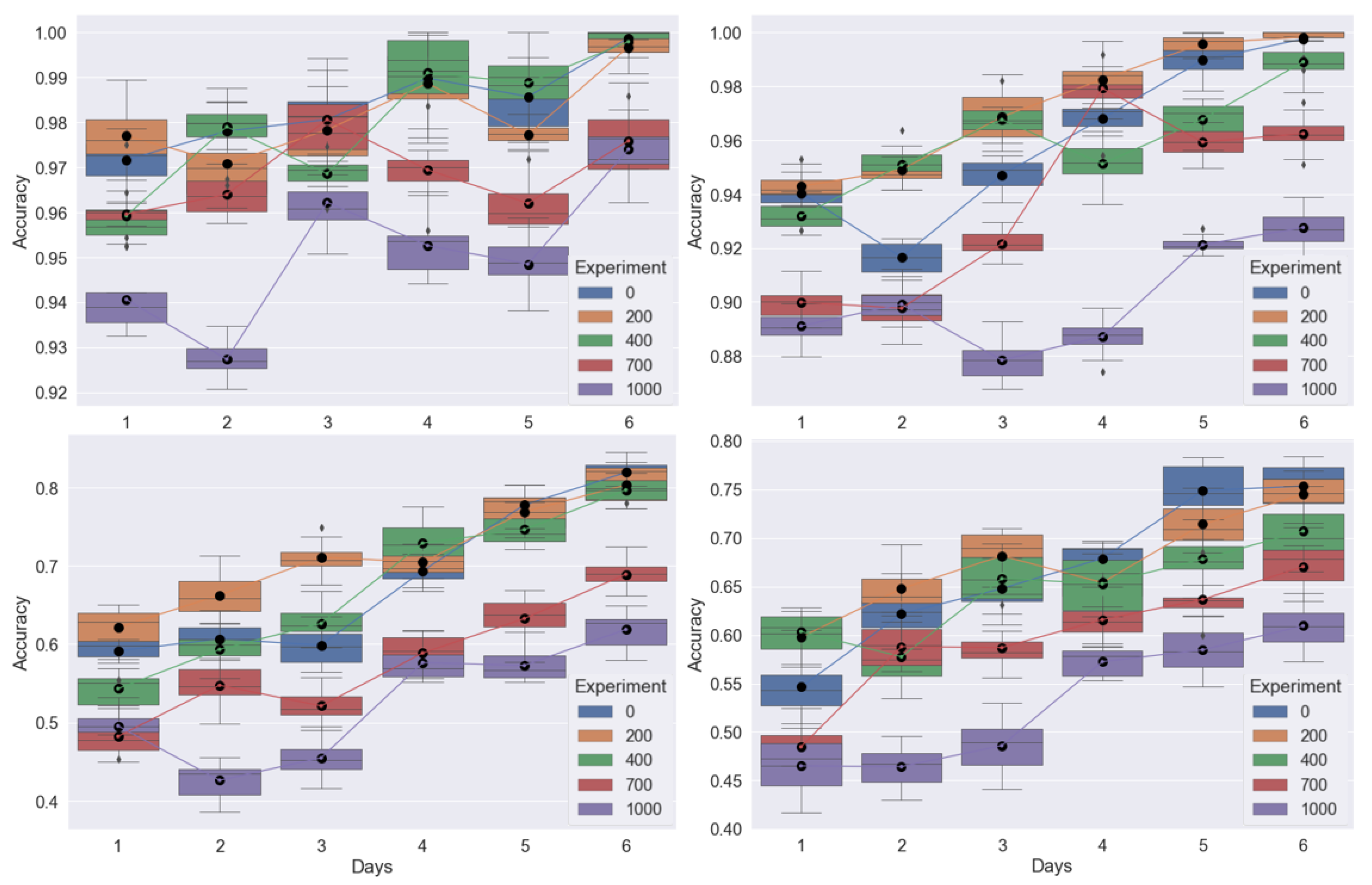
Figure 7 shows classification accuracy for the different combinations of train and test images introduced in
Table 3. In order to obtain smoother trends we performed multiple model runs of our ViT classifier corresponding to each bounding box. Each model was run 10 times and boxplots of the classification accuracy over time were generated for each trial.
Table 3 show mean and standard deviation values for bounding box
. Mean1 and STD1 are values averaged over all runs and all days of the test trial. Mean2 and STD2 show the values averaged over all runs for the last day of each test trial. For experiments A,B and C, selected train views from all trials are combined and tested on each trial separately. As a result an overall high classification accuracy is achieved for these experiments.
Similar to the NIR pixel analysis results, we observe a much lower mean accuracy for Experiment D (tested on trial 1).Overall for trial 1, we did not observe a clear separation between the two sets of plants and hence chose not to show boxplots for A and D. In
Figure 7 B and C, we combine all trials for training and test on trials 2 and 3 respectively. E and F show results on trials 2 and 3 respectively when trained on a different trial. Compared to B and C there is an overall drop in accuracy for these experiments as we introduced hold out trials for testing. For E and F, we observe a gradual increase in accuracy as we move from the first day to the last day of the trial. Such a trend is expected, as we progress through the trial the drought-stress gets prolonged, thus creating a larger separation between the two groups. This presumably makes it easier for the model to classify the two sets of plants as the trial progresses. This increasing trend across the trial is also slightly observable in B and C.
A second trend observed in B, C, E and F is a drop in accuracy for bounding boxes and when compared to , and . Particularly for bounding box , which includes the entire plant for classification, we see a significant drop in accuracy for most days in the trial. This is also expected as a full view of the plant includes more occlusions from the leaves of adjacent plants which are treated with the opposite watering protocol. These occlusions are absent or quite small for bounding boxes , and which is why overall accuracy is higher for these sets of images. Overall trial 2 performs better than trial 3 with the highest classification accuracy of 85 % observed at the last day of the trial for bounding box . From these experiments we believe that in controlled chamber settings and through a multi-analysis approach, early drought stress detection from NIR images of maize plants is possible before it becomes evident through visual inspection.
5. Conclusions
In this paper, we used an existing high throughput low-cost system developed by [
16] for water stress detection from NIR images of maize plant. In our effort to scale up the analysis on more imaging trials, we faced additional challenges not addressed by the authors in [
16]. These challenges involved difficulties in annotating and detecting the YEL of the plants as well as occlusion of leaves from adjacent plants. To overcome these, we introduced a pipeline for automatic detection of drought-stress that did not solely rely on the YEL but instead focused on the region surrounding the YEL and the stem. Our analysis on three imaging trials revealed several key findings. The Vision Transformer implemented for classifying drought-stressed and well-watered plants showed an increase in accuracy across the days of the trial which could be correlated to an increase in drought-stress from the first to last day of the trial. Our bounding box experiments also showed a significant drop in accuracy in case of analysis on the entire plant vs. areas surrounding the YEL and the stem region. We showed that this region can act as a more reliable alternative to the YEL. Finally, we were able to show that out of the three trials performed, early drought-stress detection was clearly observable in two of them based on our classification metric and NIR distribution analysis. We intend to use our pipeline for remote monitoring of plants in controlled chamber settings which could aid future research in the field of crop science.
Author Contributions
Conceptualization: Michael Daniele, Alper Bozkurt, Edgar Lobaton; Methodology: Sanjana Banerjee, Edgar Lobaton; Software: Sanjana Banerjee; Experiments: James Reynolds, Matthew Taggart; Data curation: James Reynolds, Matthew Taggart, Sanjana Banerjee; Writing—original draft preparation: Sanjana Banerjee; Writing—review and editing: James Reynolds, Matthew Taggart, Michael Daniele, Alper Bozkurt, Edgar Lobaton; Supervision: Michael Daniele, Alper Bozkurt, Edgar Lobaton. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the United States Department of Agriculture—National Institute of Food and Agriculture under the grant number 1015796 and the United States National Science Foundation under grant numbers ECCS-2231012, EF-2319389, ITE-2344423, IIS-2037328 and EEC-1160483 for the Nanosystems Engineering Research Center for Advanced Self-Powered Systems of Integrated Sensors and Technologies (ASSIST)
Data Availability Statement
The original data presented in the study are openly available on the data sharing platform Zenodo
https://zenodo.org/records/10991581 with DOI 10.5281/zenodo.10991581.The repository contains raw images before any of the pre-processing steps mentioned in
Section 3. Images in the repository were down-sampled from their original resolution (see
Section 2 by a factor of 2 due to a size restriction on Zenodo. The resized dataset is 16.8 GB.For any other questions about the dataset, or data availability please reach out to the corresponding author.
Acknowledgments
The authors would like to thank Dr. Carole Saravitz, Joe Chiera, and other staff members of the NC State University Phytotron for their assistance with plant care and chamber maintenance.
Conflicts of Interest
The authors declare no conflicts of interest.The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Sheoran, S.; Kaur, Y.; Kumar, S.; Shukla, S.; Rakshit, S.; Kumar, R. Recent advances for drought stress tolerance in maize (Zea mays l.): Present status and future prospects. Frontiers In Plant Science 2022, 13, 872566. [Google Scholar] [CrossRef]
- Goyal, P.; Sharda, R.; Saini, M.; Siag, M. A deep learning approach for early detection of drought stress in maize using proximal scale digital images. Neural Computing And Applications 2024, 36, 1899–1913. [Google Scholar] [CrossRef]
- Gao, Y.; Qiu, J.; Miao, Y.; Qiu, R.; Li, H.; Zhang, M. Prediction of leaf water content in maize seedlings based on hyperspectral information. IFAC-PapersOnLine 2019, 52, 263–269. [Google Scholar] [CrossRef]
- An, J.; Li, W.; Li, M.; Cui, S.; Yue, H. Identification and classification of maize drought stress using deep convolutional neural network. Symmetry 2019, 11, 256. [Google Scholar] [CrossRef]
- Zhuang, S.; Wang, P.; Jiang, B.; Li, M.; Gong, Z. Early detection of water stress in maize based on digital images. Computers And Electronics In Agriculture 2017, 140, 461–468. [Google Scholar] [CrossRef]
- Jiang, B.; Wang, P.; Zhuang, S.; Li, M.; Li, Z.; Gong, Z. Detection of maize drought based on texture and morphological features. Computers And Electronics In Agriculture 2018, 151, 50–60. [Google Scholar] [CrossRef]
- Asaari, M.; Mertens, S.; Dhondt, S.; Inzé, D.; Wuyts, N.; Scheunders, P. Analysis of hyperspectral images for detection of drought stress and recovery in maize plants in a high-throughput phenotyping platform. Computers And Electronics In Agriculture 2019, 162, 749–758. [Google Scholar] [CrossRef]
- Dar, Z.; Dar, S.; Khan, J.; Lone, A.; Langyan, S.; Lone, B.; Kanth, R.; Iqbal, A.; Rane, J.; Wani, S. Others Identification for surrogate drought tolerance in maize inbred lines utilizing high-throughput phenomics approach. Plos One 2021, 16, e0254318. [Google Scholar] [CrossRef]
- Souza, A.; Yang, Y. High-throughput corn image segmentation and trait extraction using chlorophyll fluorescence images. Plant Phenomics 2021. [Google Scholar] [CrossRef]
- Romano, G.; Zia, S.; Spreer, W.; Sanchez, C.; Cairns, J.; Araus, J.; Müller, J. Use of thermography for high throughput phenotyping of tropical maize adaptation in water stress. Computers And Electronics In Agriculture 2011, 79, 67–74. [Google Scholar] [CrossRef]
- Gausman, H.; Allen, W. Optical parameters of leaves of 30 plant species. Plant Physiology 1973, 52, 57–62. [Google Scholar] [CrossRef]
- Humplík, J.; Lazár, D.; Husičková, A.; Spíchal, L. Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses–a review. Plant Methods 2015, 11, 1–10. [Google Scholar] [CrossRef]
- Hsiao, T.; Acevedo, E.; Henderson, D. Maize leaf elongation: continuous measurements and close dependence on plant water status. Science 1970, 168, 590–591. [Google Scholar] [CrossRef]
- Acevedo, E.; Hsiao, T.; Henderson, D. Immediate and subsequent growth responses of maize leaves to changes in water status. Plant Physiology 1971, 48, 631–636. [Google Scholar] [CrossRef]
- Tardieu, F.; Reymond, M.; Hamard, P.; Granier, C.; Muller, B. Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: a synthesis of the effects of soil water status, evaporative demand and temperature. Journal Of Experimental Botany 2000, 51, 1505–1514. [Google Scholar] [CrossRef]
- Silva, R.; Starliper, N.; Bhosale, D.; Taggart, M.; Ranganath, R.; Sarje, T.; Daniele, M.; Bozkurt, A.; Rufty, T.; Lobaton, E. Feasibility study of water stress detection in plants using a high-throughput low-cost system. 2020 IEEE SENSORS, 2020; 1–4. [Google Scholar]
- Valle, B.; Simonneau, T.; Boulord, R.; Sourd, F.; Frisson, T.; Ryckewaert, M.; Hamard, P.; Brichet, N.; Dauzat, M.; Christophe, A. PYM: a new, affordable, image-based method using a Raspberry Pi to phenotype plant leaf area in a wide diversity of environments. Plant Methods 2017, 13, 1–17. [Google Scholar] [CrossRef]
- Lee, U.; Chang, S.; Putra, G.; Kim, H.; Kim, D. An automated, high-throughput plant phenotyping system using machine learning-based plant segmentation and image analysis. PloS One 2018, 13, e0196615. [Google Scholar] [CrossRef]
- Brichet, N.; Fournier, C.; Turc, O.; Strauss, O.; Artzet, S.; Pradal, C.; Welcker, C.; Tardieu, F.; Cabrera-Bosquet, L. A robot-assisted imaging pipeline for tracking the growths of maize ear and silks in a high-throughput phenotyping platform. Plant Methods 2017, 13, 1–12. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. Advances In Neural Information Processing Systems 2015, 28. [Google Scholar] [CrossRef]
- Gehan, M.; Fahlgren, N.; Abbasi, A.; Berry, J.; Callen, S.; Chavez, L.; Doust, A.; Feldman, M.; Gilbert, K.; Hodge, J. Others PlantCV v2: Image analysis software for high-throughput plant phenotyping. PeerJ 2017, 5, e4088. [Google Scholar] [CrossRef]
- Rubner, Y.; Tomasi, C.; Guibas, L. A metric for distributions with applications to image databases. Sixth International Conference On Computer Vision (IEEE Cat. No. 98CH36271).1998, pp. 59-66.
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. Others An image is worth 16x16 words: Transformers for image recognition at scale. ArXiv 2020, arXiv:2010.11929. [Google Scholar]
- Raspberry pi Hardware Documnetation.. Available online: https://www.raspberrypi.com/documentation/computers/raspberry-pi.html.
- Node-RED [Online]. Available online: https://nodered.org/.
- Labelbox, "Labelbox," Online, 2024. [Online]. Available online: https://labelbox.com.
- Wu, Y.; Kirillov, A.; Massa, F.; Lo, W.Y.; Girshick, R. Detectron2. Available online: https://github.com/facebookresearch/detectron2, 2019.
- Bradski, G. The OpenCV Library. Dr. Dobb’s Journal Of Software Tools. 2000.
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. ArXiv 2014, arXiv:1412.6980. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.; Haberl, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0 Contributors SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Hsiao, J.; Swann, A.; Kim, S. Maize yield under a changing climate: The hidden role of vapor pressure deficit. Agricultural And Forest Meteorology 2019, 279, 107692. [Google Scholar] [CrossRef]
- Devi, M.; Reddy, V.; Timlin, D. Drought-induced responses in maize under different vapor pressure deficit conditions. Plants 2022, 11, 2771. [Google Scholar] [CrossRef]
- Inoue, T.; Sunaga, M.; Ito, M.; Yuchen, Q.; Matsushima, Y.; Sakoda, K.; Yamori, W. Minimizing VPD fluctuations maintains higher stomatal conductance and photosynthesis, resulting in improvement of plant growth in lettuce. Frontiers In Plant Science 2021, 12, 646144. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).