Submitted:
30 April 2024
Posted:
01 May 2024
You are already at the latest version
Abstract
Keywords:
MSC: 60E05; 62H05; 62E10; 62F10; 62F15; 62P05
1. Introduction
1.1. Sample Size in SEM and BSEM
1.2. Objectives
- How do the Bayes and ML goodness of fit indices compare across varying sample sizes?
- How does the Bayes and ML model parameter estimation precision compare across varying sample sizes?
2. Methods
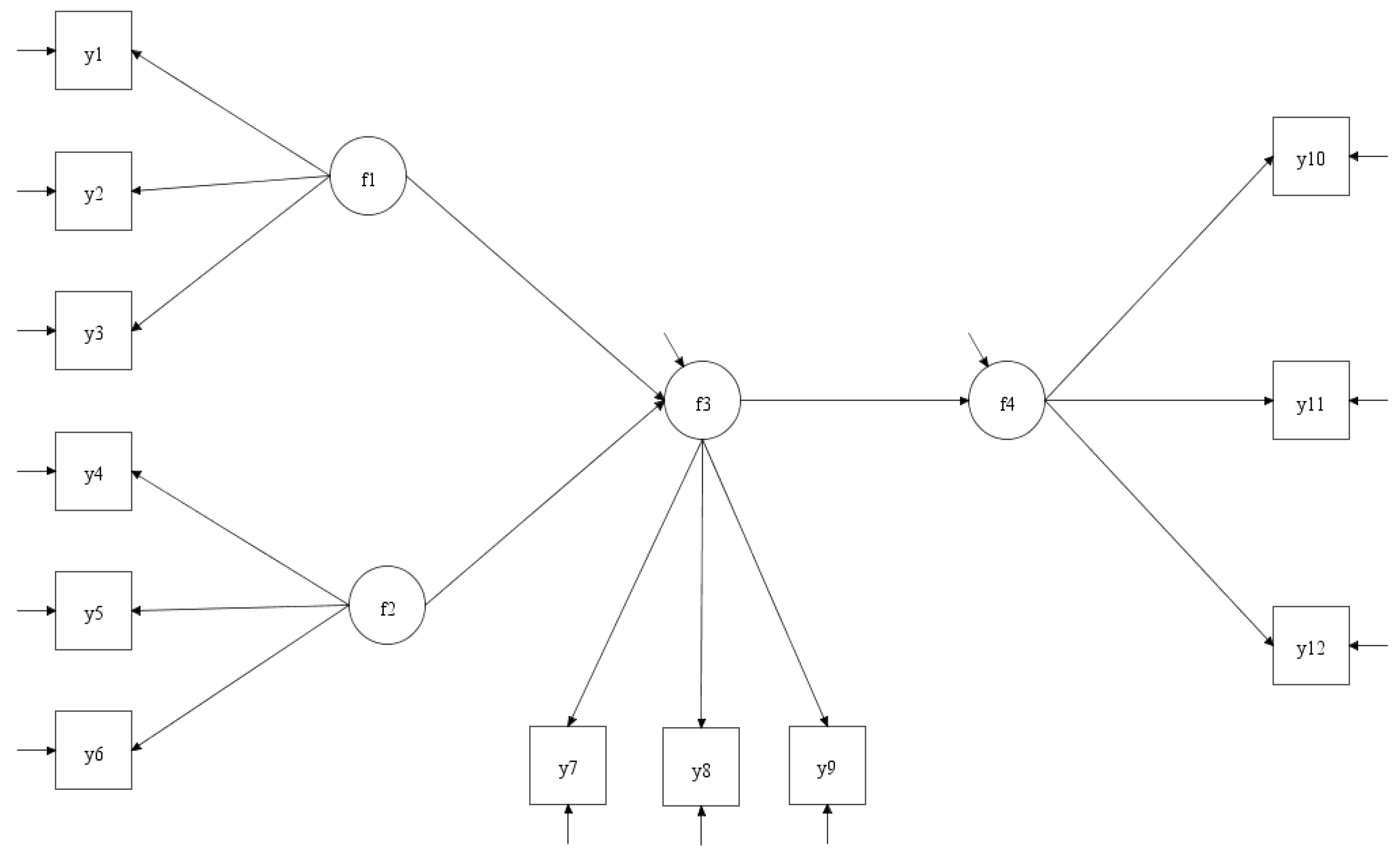
2.1. Structural Equation Modeling
- Y is a vector of observed variables.
- η is a vector of latent variables.
- ξ is a vector of exogenous latent variables (if any).
- Λ and Γ are matrices of factor loadings representing the relationships between latent and observed variables and between latent variables, respectively.
- ε and ζ are vectors of error terms.
2.2. Bayesian Structural Equation Modeling
- Structural Equations: Like traditional SEM, BSEM describes relationships between latent and observed variables These equations are typically written in the form:where Y is a vector of observed variables, Λ is a matrix of factor loadings representing the relationships between latent variables (η) and observed variables, and ϵ is a vector of error terms.Y=Λη+ϵ
- Priors for Parameters: In Bayesian analysis, prior distributions are specified for model parameters. These priors reflect prior beliefs or knowledge about the parameters before observing the data. The choice of priors can vary depending on the specific model and research question. Commonly used priors include normal, uniform, or informative priors based on previous studies or expert knowledge.
-
Likelihood Function: The likelihood function specifies the probability of observing the data given the model parameters. In BSEM, this typically involves assuming a distribution for the observed variables conditional on the latent variables and error terms. Common distributions include the normal distribution for continuous variables or the categorical distribution for categorical variables. The joint posterior distribution of the parameters given the data is then obtained using Bayes' theorem:where:p(θ∣data)∝p(data∣θ)×p(θ)
2.3. Goodness of Fit
2.3.1. The χ2 Test of Model Fit
- N is the sample size,
- ∣Smodel∣ is the determinant of the model-implied covariance matrix, and
- ∣S∣ is the determinant of the observed covariance matrices.
- N is the sample size,
- k is the number of estimated parameters in the model, and
- df represents the degrees of freedom.
2.3.2. The Bayes Information Criteria
- L is the likelihood of the model,
- k is the number of estimated parameters in the model, and
- N is the sample size.
2.3.3. The Tucker-Lewis Index
- df(H0) is the degrees of freedom of the baseline (null) model,
- df(M) is the degrees of freedom of the hypothesized model,
- TL(M) is the TLI value of the hypothesized model, and
- TL(H0) is the TLI value of the baseline model.
2.3.4. The Comparative Fit Index
- TLP is the Tucker-Lewis Index of the proposed model,
- TLO is the Tucker-Lewis Index of the baseline (null) model, and
- df represents the degrees of freedom of the model.
2.3.5. The Root Mean Square Error of Approximation
- 𝜒2 is the chi-square statistic,
- 𝑑𝑓 is the degrees of freedom, and
- 𝑁 is the sample size.
2.3.6. The Posterior Predictive P-Value (PPV)
- 𝑀 is the number of simulated datasets,
- 𝑇(𝑖) is the test statistic computed from the 𝑖th simulated dataset,
- 𝑇obs is the test statistic computed from the observed dataset, and
- 1(⋅) is the indicator function, which equals 1 if the condition is true and 0 otherwise.
2.4. Data Analysis
Simulation Study

3. Results
4. Discussion
5. Conclusions
6. Limitations and Suggestions for Further Research
References
- Kline, R.B. (2015). Principles and Practice of Structural Equation Modeling (4th ed.). Guilford Press.
- Bollen, K.A.; Pearl, J. (2013). Eight Myths About Causality and Structural Equation Models. In Handbook of Causal Analysis for Social Research (pp. 301-328). Springer. [CrossRef]
- Hair, J.F.; Black, W.C.; Babin, B.J.; Anderson, R.E. (2019). Multivariate Data Analysis, (8th ed.). Cengage Learning.
- Muthén, B.; Asparouhov, T. Bayesian structural equation modeling: A more flexible representation of substantive theory. Psychological Methods 2012, 17, 313–335. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Song, X.Y. Evaluation of the Bayesian and maximum likelihood approaches in analyzing structural equation models with small sample sizes. Multivariate Behavioral Research 2004, 39, 653–686. [Google Scholar] [CrossRef]
- Asparouhov, T.; Muthén, B. Exploratory structural equation modeling. Structural Equation Modeling: A Multidisciplinary Journal 2009, 16, 397–438. [Google Scholar] [CrossRef]
- Hair, J.F.; Black, W.C.; Babin, B.J.; Anderson, R.E. (2019). Multivariate Data Analysis, (8th ed.). Cengage Learning.
- Wolf, E.J.; Harrington, K.M.; Clark, S.L.; Miller, M.W. Sample Size Requirements for Structural Equation Models: An Evaluation of Power, Bias, and Solution Propriety. Educational and Psychological Measurement 2013, 73, 913–934. [Google Scholar] [CrossRef]
- MacCallum, R.C.; Browne, M.W.; Sugawara, H.M. Power analysis and determination of sample size for covariance structure modeling. Psychological Methods 1996, 1, 130–149. [Google Scholar] [CrossRef]
- Cohen, J. (1988). Statistical Power Analysis for the Behavioral Sciences, (2nd ed.). Lawrence Erlbaum Associates.
- Maxwell, S.E. The Persistence of Underpowered Studies in Psychological Research: Causes, Consequences, and Remedies. Psychological Methods 2004, 9, 147–163. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.F. (2013). Introduction to Mediation, Moderation, and Conditional Process Analysis: A Regression-Based Approach. Guilford Press.
- Hox, J.J.; Boeije, H.R. (2005). Data Collection, Primary versus Secondary. In International Encyclopedia of Social & Behavioral Sciences (pp. 3247-3252). Elsevier.
- Schreiber, J.B.; Nora, A.; Stage, F.K.; Barlow, E.A.; King, J. Reporting Structural Equation Modeling and Confirmatory Factor Analysis Results: A Review. The Journal of Educational Research 2006, 99, 323–338. [Google Scholar] [CrossRef]
- Muthén, B.; Asparouhov, T. Bayesian structural equation modeling: A more flexible representation of substantive theory. Psychological methods 2012, 17, 313–335. [Google Scholar] [CrossRef] [PubMed]
- van de Schoot, R.; Kaplan, D.; Denissen, J.; Asendorpf, J.B.; Neyer, F.J.; van Aken, M.A. G. A gentle introduction to Bayesian analysis: Applications to developmental research. Child Development 2014, 85, 842–860. [Google Scholar] [CrossRef] [PubMed]
- Muthén, L.K.; Muthén, B.O. (2012). Mplus User’s Guide (7th ed.). Muthén & Muthén.
- Byrne, B.M. (2016). Structural Equation Modeling with AMOS: Basic Concepts, Applications, and Programming (3rd ed.). Routledge.
- Kline, R.B. (2016). Principles and Practice of Structural Equation Modeling (4th ed.). Guilford Press.
- Hu, L.T.; Bentler, P.M. Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives. Structural Equation Modeling: A Multidisciplinary Journal 1999, 6, 1–55. [Google Scholar] [CrossRef]
- Brown, T.A. (2015). Confirmatory Factor Analysis for Applied Research (2nd ed.). Guilford Press.
- Schwarz, G. Estimating the dimension of a model. The Annals of Statistics 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Tucker, L.R.; Lewis, C. A reliability coefficient for maximum likelihood factor analysis. Psychometrika 1973, 38, 1–10. [Google Scholar] [CrossRef]
- Bentler, P.M. Comparative fit indexes in structural models. Psychological Bulletin 1990, 107, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Steiger, J.H. Structural model evaluation and modification: An interval estimation approach. Multivariate Behavioral Research 1990, 25, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Browne, M.W.; Cudeck, R. Alternative ways of assessing model fit. Sociological Methods & Research 1992, 21, 230–258. [Google Scholar]
- Gelman, A.; Meng, X.L.; Stern, H. Posterior predictive assessment of model fitness via realized discrepancies. Statistica Sinica 1996, 6, 733–760. [Google Scholar]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Dunson, D.B.; Vehtari, A.; Rubin, D.B. (2013). Bayesian Data Analysis (3rd ed.). Chapman and Hall/CRC.
- Asparouhov, T.; Muthén, B. (2010). Bayesian Analysis using Mplus: Technical Implementation, Mplus Web Notes, No. 14.
- Davison, A.C.; Hinkley, D.V. (1997). Bootstrap methods and their application, Cambridge University Press.
- Wilcox, R.R. (2017). Introduction to robust estimation and hypothesis testing (4th ed.). Academic Press.
- Rubinstein, R.Y.; Kroese, D.P. (2016). Simulation and the Monte Carlo Method, (3rd ed.). Wiley.
- Gilks, W.R.; Richardson, S.; Spiegelhalter, D.J. (1995). Markov Chain Monte Carlo in Practice, Chapman and Hall/CRC.
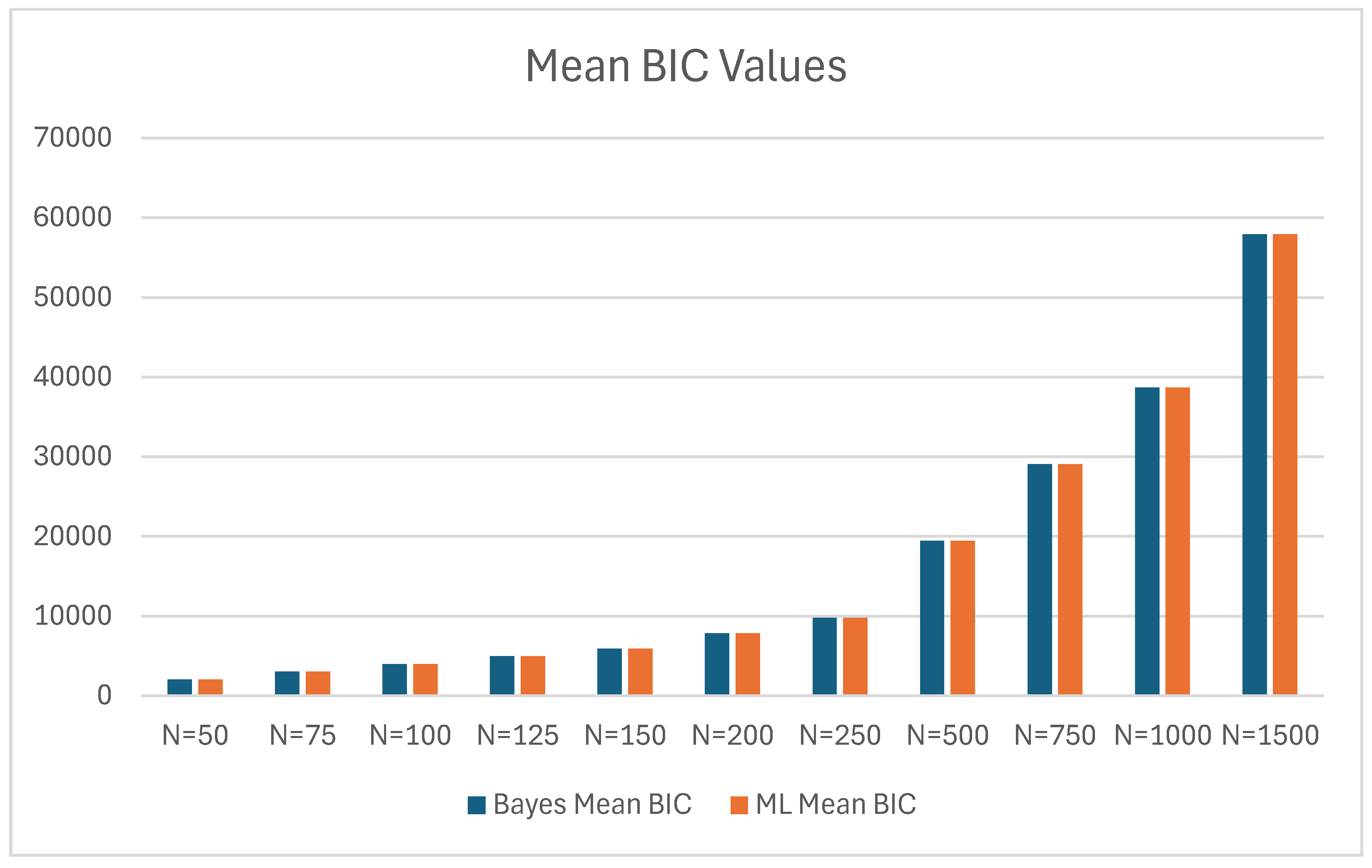
| Bayes | ML | |||||||
|---|---|---|---|---|---|---|---|---|
| Posterior Predictive Value (PPV) | Chi-Square Test of Model Fit (df=50) | |||||||
| N | Mean PPV | Mean PPV – 0.5 | SD | N Successful Computations | Mean ꭓ2 | ꭓ2/df | SD | N Successful Computations |
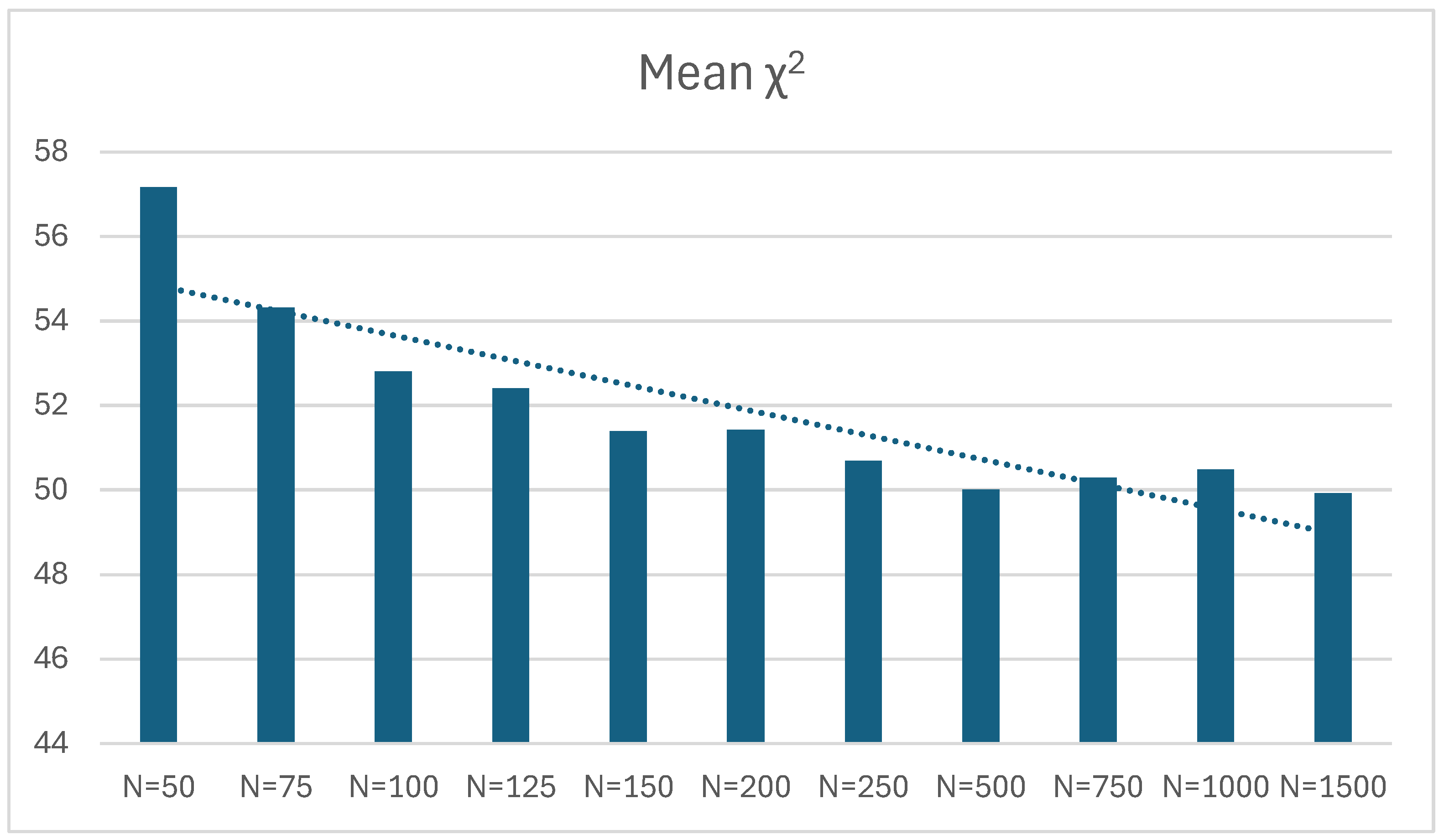
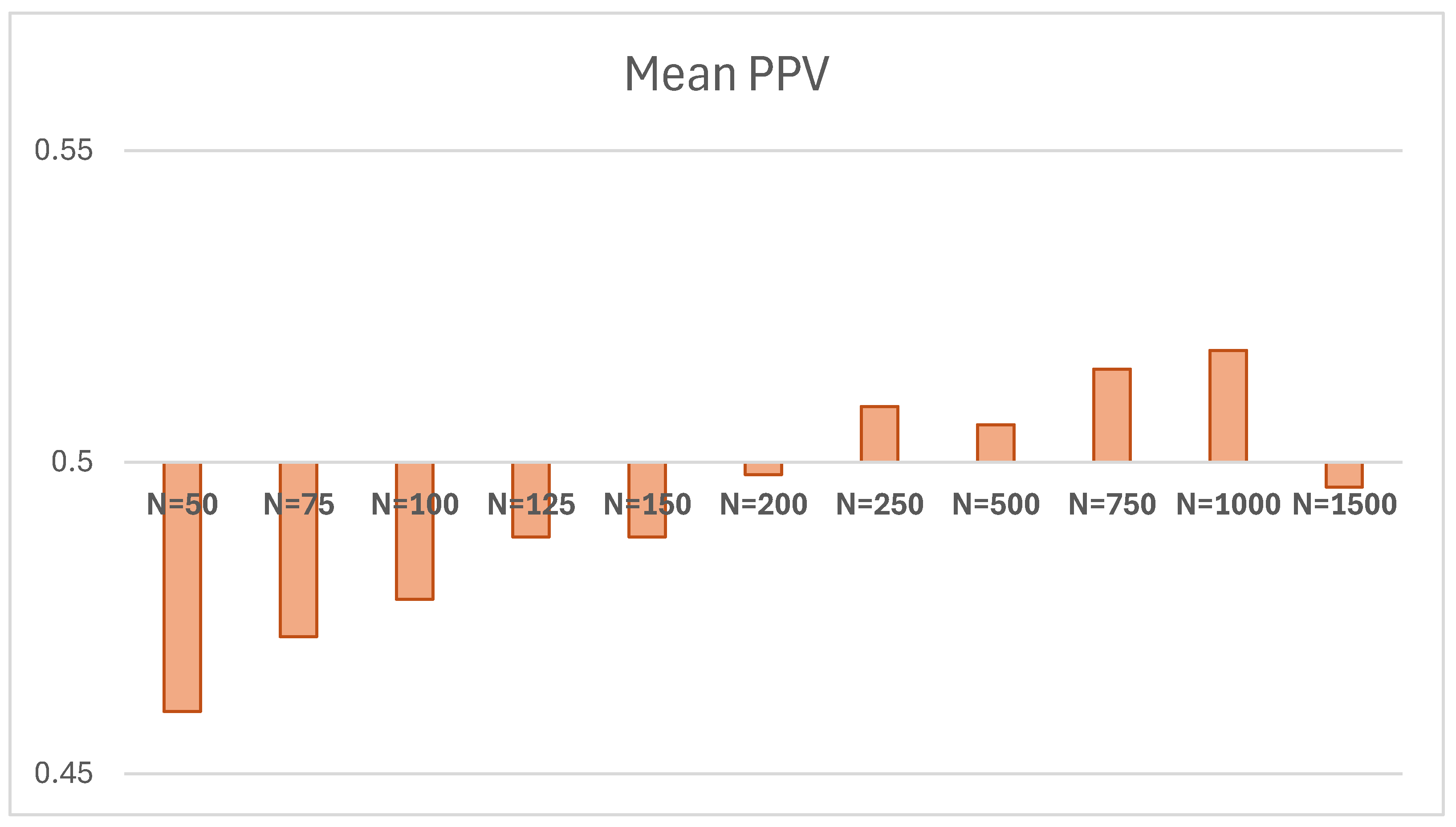
| 50 | 0.460 | -0.040 | 0.220 | 500 | 57.161 | 1.14322 | 11.989 | 498 |
| 75 | 0.472 | -0.028 | 0.212 | 500 | 54.311 | 1.08622 | 10.724 | 500 |
| 100 | 0.478 | -0.022 | 0.207 | 500 | 52.803 | 1.05606 | 10.484 | 500 |
| 125 | 0.488 | -0.012 | 0.220 | 500 | 52.401 | 1.04802 | 10.248 | 500 |
| 150 | 0.488 | -0.012 | 0.223 | 500 | 51.393 | 1.02786 | 10.628 | 500 |
| 200 | 0.498 | -0.002 | 0.215 | 500 | 51.428 | 1.02856 | 10.274 | 500 |
| 250 | 0.509 | 0.009 | 0.217 | 500 | 50.694 | 1.01388 | 10.068 | 500 |
| 500 | 0.506 | 0.006 | 0.224 | 500 | 50.010 | 1.0002 | 9.866 | 500 |
| 750 | 0.515 | 0.015 | 0.217 | 500 | 50.299 | 1.00458 | 9.938 | 500 |
| 1000 | 0.518 | 0.018 | 0.222 | 500 | 50.487 | 1.00974 | 10.210 | 500 |
| 1500 | 0.496 | -0.004 | 0.222 | 500 | 49.926 | 0.99852 | 9.729 | 500 |
| Bayes | ML | Bayes Mean BIC – ML Mean BIC | |||
|---|---|---|---|---|---|
| N | Mean BIC | SD | Mean BIC | SD | |
| 50 | 2043.033 | 36.454 | 2039.316 | 35.157 | 3.717 |
| 75 | 3018.171 | 45.095 | 3017.153 | 43.054 | 1.018 |
| 100 | 3991.500 | 48.712 | 3990.543 | 48.387 | 0.957 |
| 125 | 4961.522 | 54.133 | 4961.807 | 54.398 | -0.285 |
| 150 | 5929.844 | 59.222 | 5929.626 | 56.952 | 0.218 |
| 200 | 7864.169 | 67.388 | 7864.701 | 66.513 | -0.532 |
| 250 | 9795.346 | 75.982 | 9794.467 | 74.758 | 0.879 |
| 500 | 19443.339 | 110.671 | 19439.265 | 105.014 | 4.074 |
| 750 | 29075.952 | 126.450 | 29072.427 | 124.156 | 3.525 |
| 1000 | 38703.213 | 153.895 | 38697.330 | 155.697 | 5.883 |
| 1500 | 57942.162 | 179.213 | 57943.171 | 183.588 | -1.009 |
| Bayes | ML | |||||
|---|---|---|---|---|---|---|
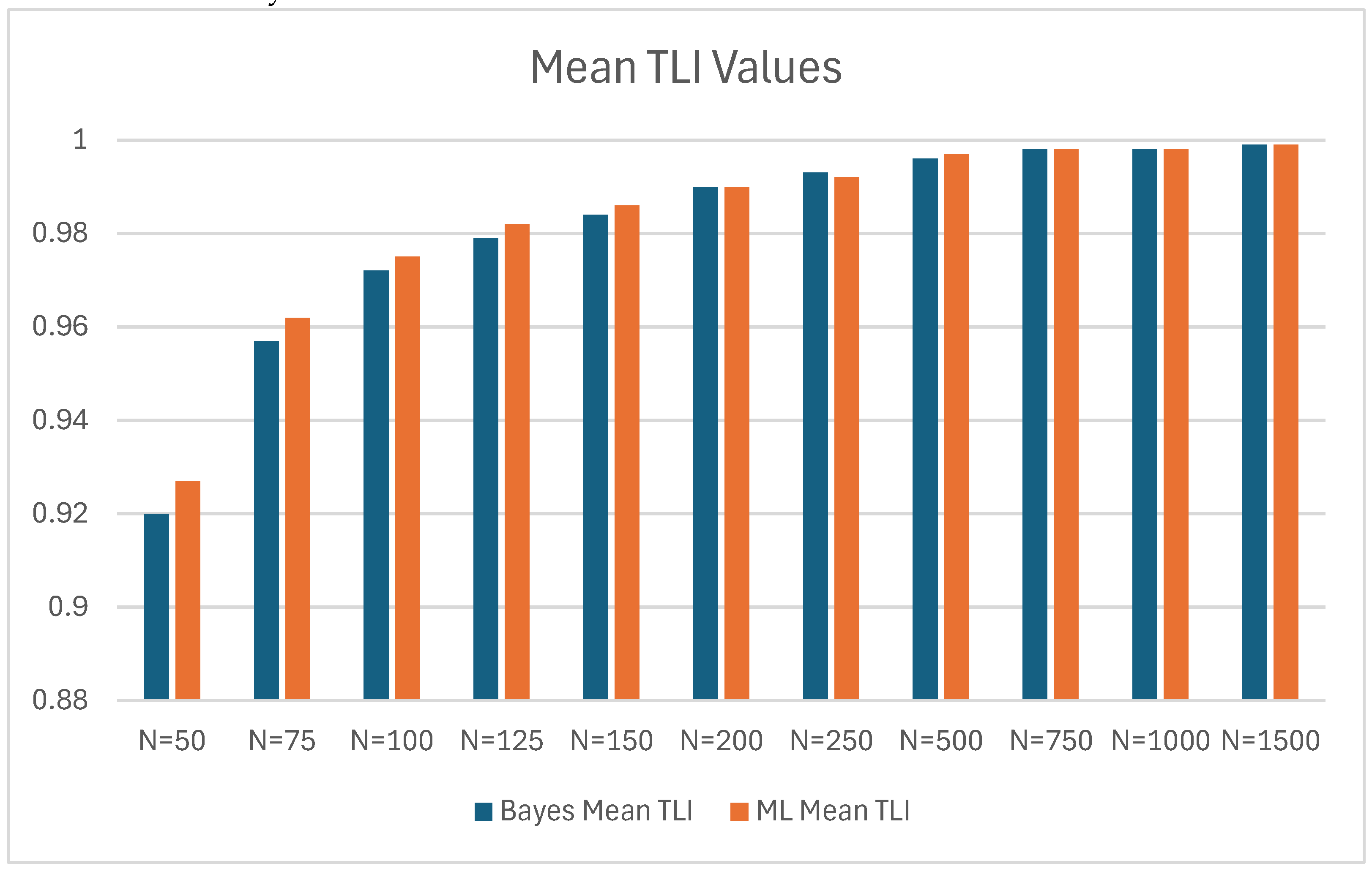
| N | Mean TLI | SD | Mean TLI | SD | Bayes Mean TLI - ML Mean TLI | |
| 50 | 0.920 | 0.073 | 0.927 | 0.080 | -0.007 | |
| 75 | 0.957 | 0.045 | 0.962 | 0.044 | -0.005 | |
| 100 | 0.972 | 0.032 | 0.975 | 0.033 | -0.003 | |
| 125 | 0.979 | 0.025 | 0.982 | 0.025 | -0.003 | |
| 150 | 0.984 | 0.021 | 0.986 | 0.021 | -0.002 | |
| 200 | 0.990 | 0.015 | 0.990 | 0.015 | 0 | |
| 250 | 0.993 | 0.011 | 0.992 | 0.011 | 0.001 | |
| 500 | 0.996 | 0.006 | 0.997 | 0.005 | -0.001 | |
| 750 | 0.998 | 0.004 | 0.998 | 0.004 | 0 | |
| 1000 | 0.998 | 0.003 | 0.998 | 0.003 | 0 | |
| 1500 | 0.999 | 0.002 | 0.999 | 0.002 | 0 | |
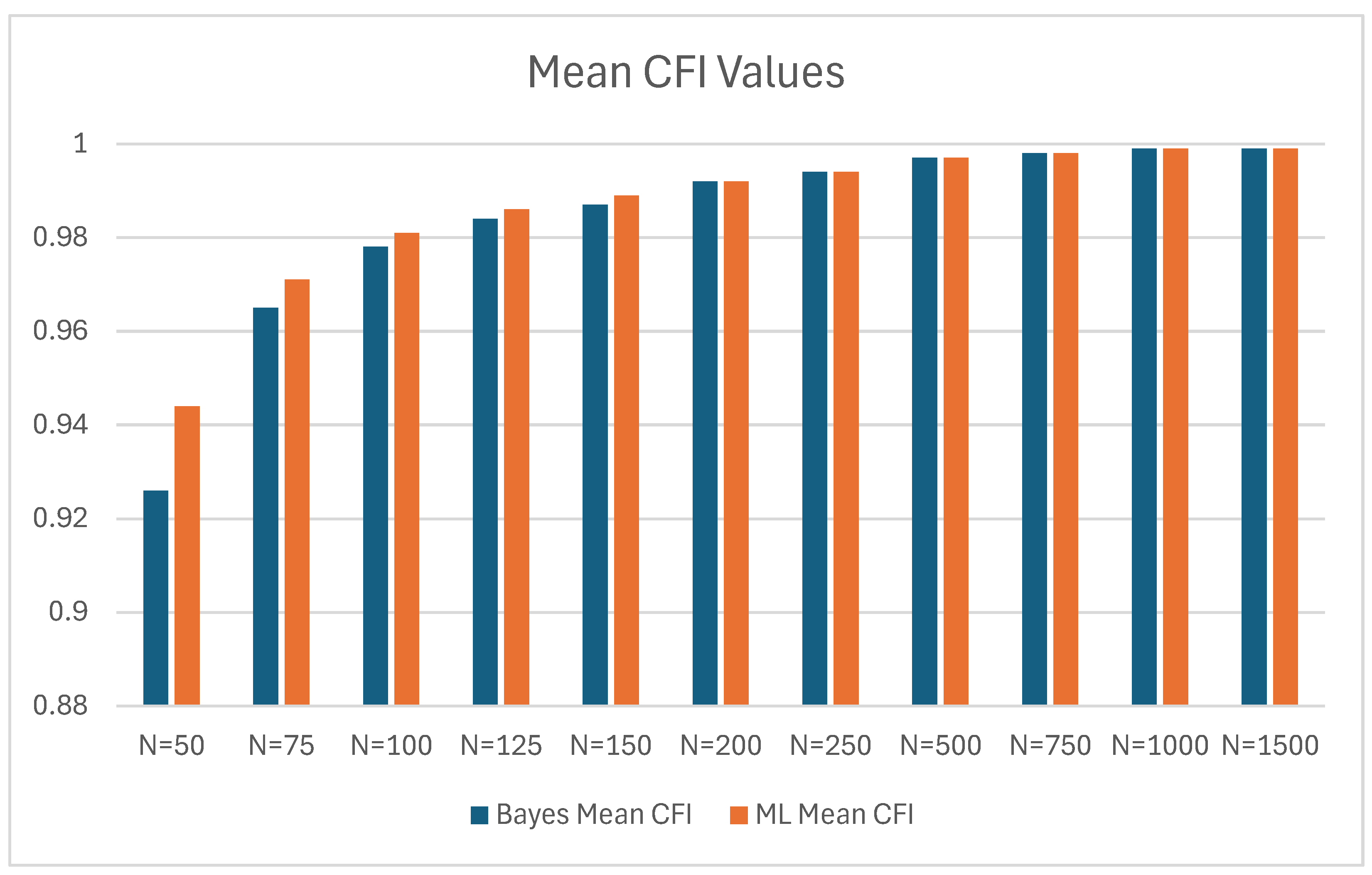
| Bayes | ML | Bayes Mean CFI - ML Mean CFI | |||
|---|---|---|---|---|---|
| N | Mean CFI | SD | Mean CFI | SD | |
| 50 | 0.926 | 0.066 | 0.944 | 0.060 | -0.018 |
| 75 | 0.965 | 0.037 | 0.971 | 0.034 | -0.006 |
| 100 | 0.978 | 0.025 | 0.981 | 0.025 | -0.003 |
| 125 | 0.984 | 0.020 | 0.986 | 0.019 | -0.002 |
| 150 | 0.987 | 0.017 | 0.989 | 0.016 | -0.002 |
| 200 | 0.992 | 0.012 | 0.992 | 0.012 | 0 |
| 250 | 0.994 | 0.008 | 0.994 | 0.009 | 0 |
| 500 | 0.997 | 0.004 | 0.997 | 0.004 | 0 |
| 750 | 0.998 | 0.003 | 0.998 | 0.003 | 0 |
| 1000 | 0.999 | 0.002 | 0.999 | 0.002 | 0 |
| 1500 | 0.999 | 0.001 | 0.999 | 0.001 | 0 |
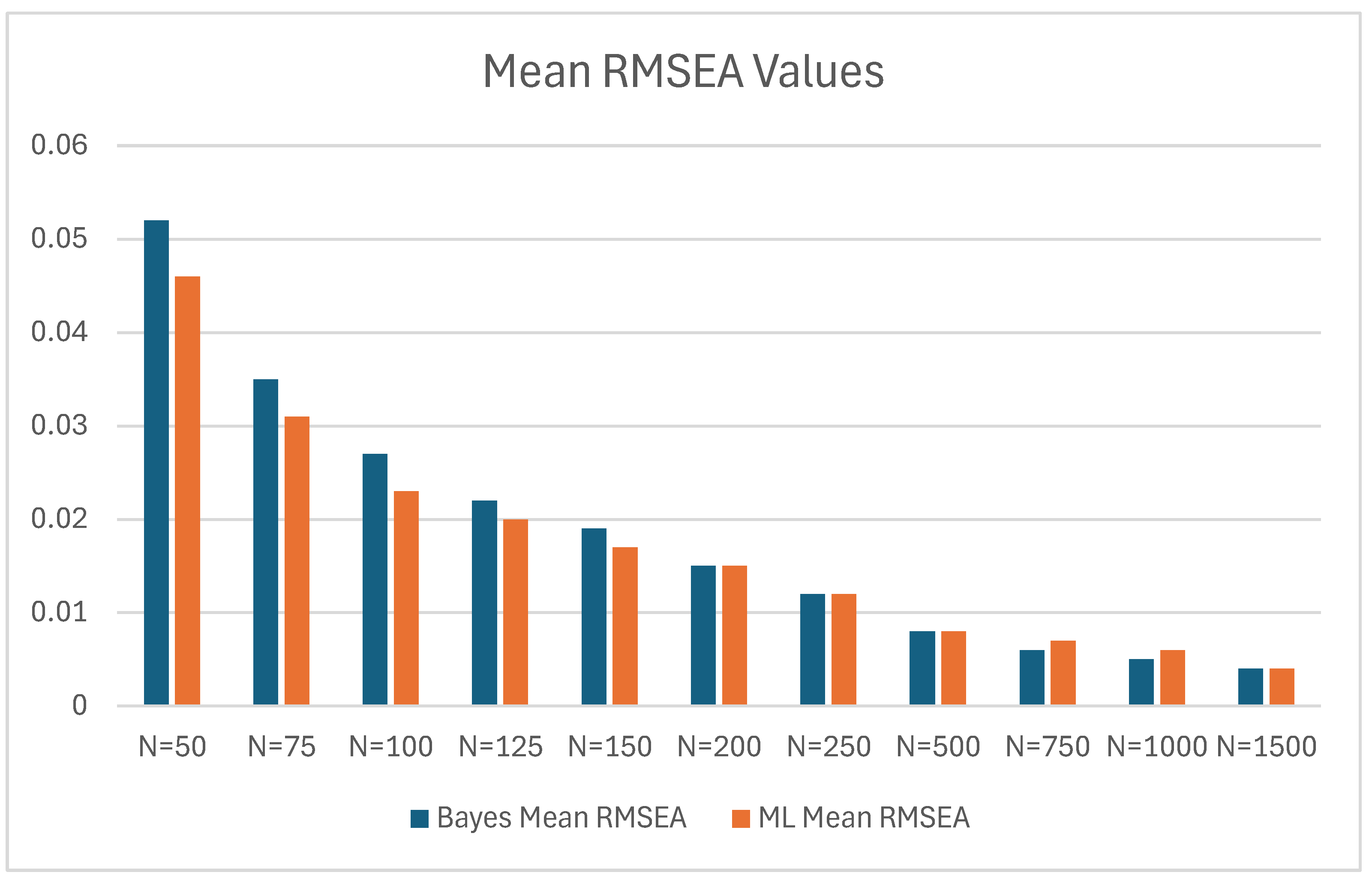
| Bayes | ML | Bayes Mean RMSEA – ML Mean RMSEA | |||
|---|---|---|---|---|---|
| N | Mean RMSEA | SD | Mean RMSEA | SD | |
| 50 | 0.052 | 0.034 | 0.046 | 0.038 | 0.006 |
| 75 | 0.035 | 0.028 | 0.031 | 0.028 | 0.004 |
| 100 | 0.027 | 0.023 | 0.023 | 0.024 | 0.004 |
| 125 | 0.022 | 0.021 | 0.020 | 0.021 | 0.002 |
| 150 | 0.019 | 0.019 | 0.017 | 0.019 | 0.002 |
| 200 | 0.015 | 0.016 | 0.015 | 0.016 | 0.000 |
| 250 | 0.012 | 0.014 | 0.012 | 0.014 | 0.000 |
| 500 | 0.008 | 0.010 | 0.008 | 0.010 | 0.000 |
| 750 | 0.006 | 0.008 | 0.007 | 0.008 | -0.001 |
| 1000 | 0.005 | 0.007 | 0.006 | 0.007 | -0.001 |
| 1500 | 0.004 | 0.006 | 0.004 | 0.006 | 0.000 |
| Sample Size | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 75 | 100 | 125 | 150 | 200 | 250 | 500 | 750 | 1000 | 1500 | |||
| Parameter | True Value | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | |
| F1 by | Y1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Y2 | 1.00 | 1.24 | 1.09 | 1.06 | 1.05 | 1.03 | 1.02 | 1.02 | 1.01 | 1.01 | 1.01 | 1.00 | |
| Y3 | 1.00 | 1.19 | 1.07 | 1.04 | 1.03 | 1.03 | 1.02 | 1.02 | 1.01 | 1.01 | 1.01 | 1.00 | |
| F2 by | Y4 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Y5 | 0.80 | 1.22 | 0.86 | 0.84 | 0.82 | 0.82 | 0.81 | 0.81 | 0.81 | 0.80 | 0.80 | 0.80 | |
| Y6 | 0.80 | 1.13 | 0.86 | 0.83 | 0.81 | 0.81 | 0.81 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | |
| F3 by | Y7 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Y8 | 0.90 | 0.93 | 0.92 | 0.92 | 0.91 | 0.91 | 0.90 | 0.90 | 0.90 | 0.90 | 0.90 | 0.90 | |
| Y9 | 0.90 | 0.92 | 0.92 | 0.91 | 0.91 | 0.91 | 0.90 | 0.90 | 0.90 | 0.90 | 0.90 | 0.90 | |
| F4 by | Y10 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Y11 | 0.70 | 0.71 | 0.70 | 0.70 | 0.70 | 0.71 | 0.71 | 0.70 | 0.70 | 0.70 | 0.70 | 0.70 | |
| Y12 | 0.70 | 0.73 | 0.73 | 0.72 | 0.71 | 0.71 | 0.71 | 0.71 | 0.70 | 0.70 | 0.70 | 0.70 | |
| F4 on | F3 | 0.60 | 0.57 | 0.58 | 0.59 | 0.59 | 0.59 | 0.59 | 0.59 | 0.60 | 0.60 | 0.60 | 0.60 |
| F3 on | F1 | 0.50 | 0.56 | 0.52 | 0.51 | 0.51 | 0.51 | 0.50 | 0.50 | 0.50 | 0.51 | 0.50 | 0.50 |
| F2 | 0.70 | 1.01 | 0.73 | 0.72 | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.70 | 0.70 | |
| Sample Size | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 75 | 100 | 125 | 150 | 200 | 250 | 500 | 750 | 1000 | 1500 | |||
| Parameter | TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | Mean PE -TV | |
| F1 by | Y1 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Y2 | 1.00 | 0.24 | 0.09 | 0.06 | 0.05 | 0.03 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | |
| Y3 | 1.00 | 0.19 | 0.07 | 0.04 | 0.03 | 0.03 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | |
| F2 by | Y4 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Y5 | 0.80 | 0.42 | 0.06 | 0.04 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | |
| Y6 | 0.80 | 0.33 | 0.06 | 0.03 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| F3 by | Y7 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Y8 | 0.90 | 0.03 | 0.02 | 0.02 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Y9 | 0.90 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| F4 by | Y10 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Y11 | 0.70 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Y12 | 0.70 | 0.03 | 0.03 | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | |
| F4 on | F3 | 0.60 | -0.03 | -0.02 | -0.01 | -0.01 | -0.01 | -0.01 | -0.01 | 0.00 | 0.00 | 0.00 | 0.00 |
| F3 on | F1 | 0.50 | 0.06 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.00 |
| F2 | 0.70 | 0.31 | 0.03 | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | |
| Sample Size | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 75 | 100 | 125 | 150 | 200 | 250 | 500 | 750 | 1000 | 1500 | ||
| Parameter | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | |
| F1 by | Y1 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Y2 | 98.8% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Y3 | 98.2% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| F2 by | Y4 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Y5 | 93.8% | 99.4% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Y6 | 93.4% | 99.6% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| F3 by | Y7 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Y8 | 99.4% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Y9 | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| F4 by | Y10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Y11 | 88.4% | 97.6% | 99.8% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Y12 | 90.8% | 99.0% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| F4 on | F3 | 92.8% | 98.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| F3 on | F1 | 69.6% | 87.0% | 95.4% | 99.0% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| F2 | 89.2% | 98.0% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Sample Size | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 75 | 100 | 125 | 150 | 200 | 250 | 500 | 750 | 1000 | 1500 | |||
| Parameter | True Value | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | Mean PE | |
| F1 by | Y1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Y2 | 1.00 | 1.10 | 1.05 | 1.03 | 1.02 | 1.02 | 1.01 | 1.01 | 1.01 | 1.00 | 1.00 | 1.00 | |
| Y3 | 1.00 | 1.08 | 1.05 | 1.03 | 1.01 | 1.01 | 1.01 | 1.01 | 1.00 | 1.00 | 1.00 | 1.00 | |
| F2 by | Y4 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Y5 | 0.80 | 0.85 | 0.83 | 0.82 | 0.81 | 0.81 | 0.81 | 0.81 | 0.80 | 0.80 | 0.80 | 0.80 | |
| Y6 | 0.80 | 0.83 | 0.82 | 0.81 | 0.81 | 0.81 | 0.81 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | |
| F3 by | Y7 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Y8 | 0.90 | 0.93 | 0.92 | 0.92 | 0.92 | 0.92 | 0.91 | 0.91 | 0.91 | 0.91 | 0.91 | 0.90 | |
| Y9 | 0.90 | 0.93 | 0.93 | 0.92 | 0.92 | 0.92 | 0.91 | 0.91 | 0.91 | 0.90 | 0.90 | 0.90 | |
| F4 by | Y10 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Y11 | 0.70 | 0.79 | 0.73 | 0.72 | 0.72 | 0.72 | 0.71 | 0.71 | 0.70 | 0.70 | 0.70 | 0.70 | |
| Y12 | 0.70 | 0.75 | 0.73 | 0.71 | 0.71 | 0.71 | 0.70 | 0.70 | 0.70 | 0.70 | 0.70 | 0.70 | |
| F4 on | F3 | 0.60 | 0.60 | 0.61 | 0.61 | 0.61 | 0.61 | 0.61 | 0.61 | 0.61 | 0.61 | 0.60 | 0.60 |
| F3 on | F1 | 0.50 | 0.53 | 0.52 | 0.51 | 0.51 | 0.51 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 |
| F2 | 0.70 | 0.73 | 0.72 | 0.71 | 0.71 | 0.71 | 0.71 | 0.71 | 0.70 | 0.70 | 0.70 | 0.70 | |
| Sample Size | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 75 | 100 | 125 | 150 | 200 | 250 | 500 | 750 | 1000 | 1500 | |||
| Parameter | TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | Mean PE - TV | |
| F1 by | Y1 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Y2 | 1.00 | 0.10 | 0.05 | 0.03 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | |
| Y3 | 1.00 | 0.08 | 0.05 | 0.03 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | |
| F2 by | Y4 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Y5 | 0.80 | 0.05 | 0.03 | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Y6 | 0.80 | 0.03 | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| F3 by | Y7 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Y8 | 0.90 | 0.03 | 0.02 | 0.02 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | |
| Y9 | 0.90 | 0.03 | 0.03 | 0.02 | 0.02 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | |
| F4 by | Y10 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Y11 | 0.70 | 0.09 | 0.03 | 0.02 | 0.02 | 0.02 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Y12 | 0.70 | 0.05 | 0.03 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| F4 on | F3 | 0.60 | 0.00 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 |
| F3 on | F1 | 0.50 | 0.03 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| F2 | 0.70 | 0.03 | 0.02 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Sample Size | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 | 75 | 100 | 125 | 150 | 200 | 250 | 500 | 750 | 1000 | 1500 | ||
| Parameter | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | %Sig. PEs | |
| F1 by | Y1 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Y2 | 95.2% | 99.6% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Y3 | 96.0% | 99.4% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| F2 by | Y4 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Y5 | 92.8% | 99.6% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Y6 | 92.6% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| F3 by | Y7 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Y8 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Y9 | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| F4 by | Y10 | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Y11 | 83.5% | 95.8% | 99.6% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| Y12 | 83.7% | 95.0% | 98.8% | 99.8% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
| F4 on | F3 | 87.3% | 97.4% | 99.2% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| F3 on | F1 | 71.5% | 89.0% | 97.6% | 99.2% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% |
| F2 | 85.7% | 99.2% | 99.6% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





