1. Introduction
The DEAD-box (DDX) family proteins, characterized by their conserved motifs and ATP-dependent RNA helicase activity, are pivotal in various cellular processes, including RNA metabolism, ribosome biogenesis, and the response to stress[
1,
2].
Recent studies show that some DDX family members may play important roles in the dynamic process of liquid-liquid phase separation (LLPS), which is integral to the formation of membrane-less organelles such as stress granules and processing bodies[
3,
4]. The association of these DDX proteins with LLPS highlights a novel aspect of cellular organization and regulation, where biochemical reactions are compartmentalized without membrane boundaries[
5,
6]. This compartmentalization is crucial for understanding how cells organize molecules in space and time, particularly in response to environmental cues and stress[
7].
Mutations and dysregulations in DDX3X and related proteins have been linked to severe human pathologies, including neurodevelopmental disorders, cancers, and viral infections[
8,
9,
10]. This association underscores the clinical importance of understanding DDX protein functions and their mechanistic roles in LLPS. In the context of LLPS, the roles of specific sequence motifs such as the RG/RGG motifs present in some DDX family proteins are of particular interest[
1,
11]. RG/RGG motifs are characterized by its repeating arginine (R) and glycine (G) amino acids[
11,
12]. These motifs are known for their ability to mediate protein-protein and protein-RNA interactions, which are essential for the formation of dynamic assemblies like RNA granules[
13,
14]. However, whether and how the number and distribution of RG/RGG motifs impact the proteins’ phase separation behavior and, consequently, their biological functionality, are still poorly addressed.
As an important member of the DDX family, the human DDX3X is a multifunctional helicase exhibiting broader roles in RNA metabolism, cellular stress responses, and innate immunity[
15,
16]. DDX3X's involvement in LLPS is particularly significant, as it influences the assembly and dynamics of stress granules, which are critical under conditions of cellular stress[
17,
18]. Its yeast homolog, DED1, is essential for mRNA splicing and translation initiation[
19,
20,
21]. DBP1 is another yeast paralogue of DDX3X that is involved in mRNA decay and ribosome biogenesis[
22].
Using a cell-free system, the present study analyzed how variations in the number and locations of RG/RGG motifs affect LLPS. Additionally, through bioinformatic and statistical analyses, we have investigated the mutations within these motifs and their associations with various cancers, offering insights into how these alterations may disrupt normal cellular processes and contribute to disease pathogenesis.
2. Results and Discussion
2.1. DEAD-Box Family Proteins Contain Multiple RG/RGG Motifs in the Disordered Regions That Are Important Phase Separation Capacity
Utilizing phase separation databases such as PhaSepDB, PhaSePro, LLPSDB v2.0, and DrLLPS, we have identified multiple RG/RGG repeat sequences within the protein sequences of the DEAD-box family (Supplemental
Table S1). To further investigate the impact of RG/RGG repeats on the proteins’ phase separation capacities, we have focused on the human DDX3X, its yeast ortholog DED1 and paralog DBP1 genes.
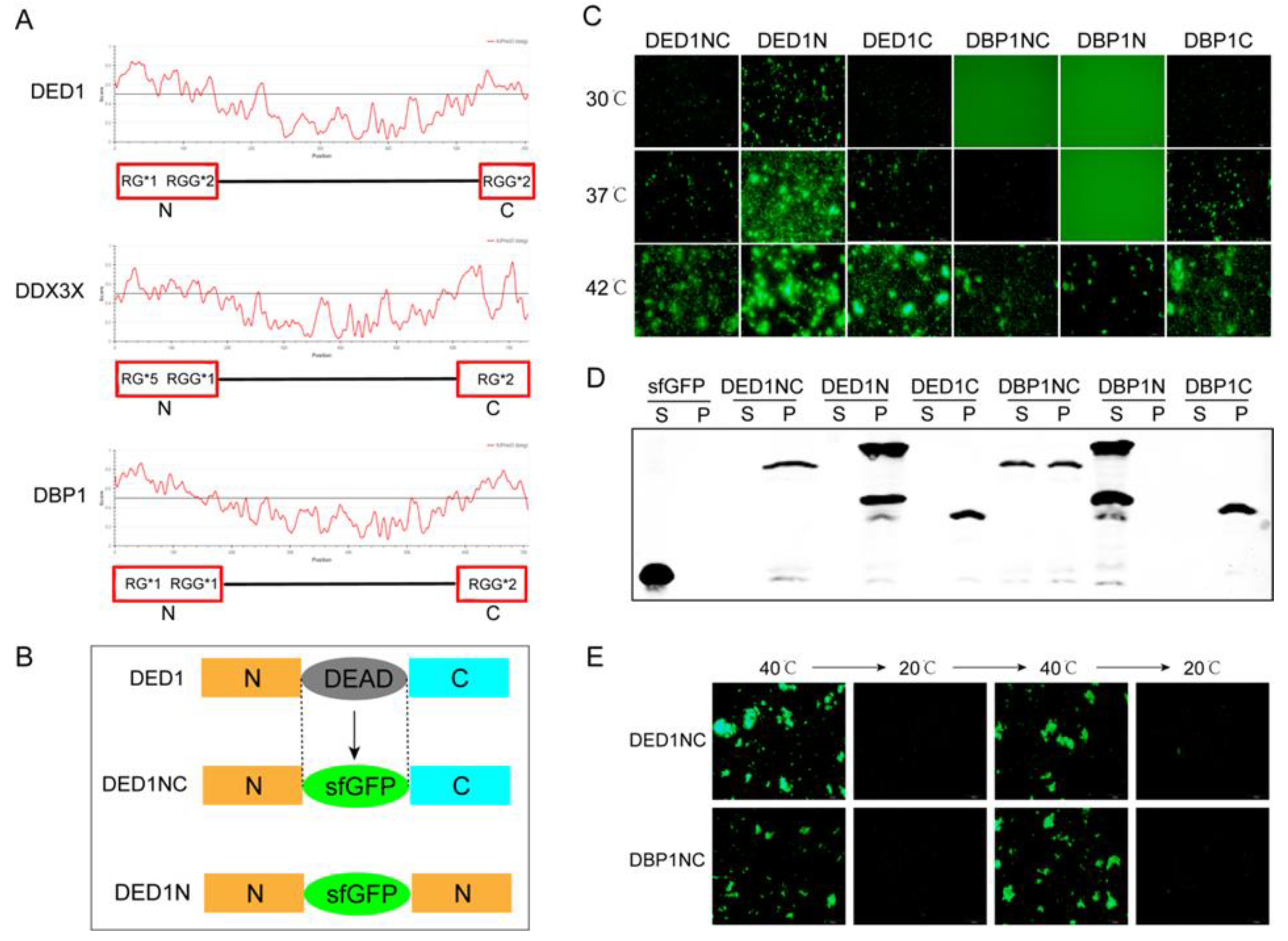
Comparative analysis using the Intrinsically Unstructured Protein Prediction (IUPRED) algorithm indicated that all three proteins exhibit disordered sequences that form unstable three-dimensional structures under physiological conditions, particularly at both the N and C termini, with scores exceeding 0.5 (
Figure 1A). All these regions contain RG/RGG structures with varying numbers of RG/RGG motifs, suggesting their potential role in influencing phase separation dynamics.
2.2. Determination of Phase Separation Capacity of Specific Protein Sequences Using Cell-Free System
To understand whether variations of RG/RGG motifs affect the LLPS behavior of DDX family proteins, we developed a cell-free protein synthesis and aggregation assay. Unlike difficult protein purification, the cell-free system enables rapid and tunable protein expression with a linear DNA template, serving as an efficient platform in examining how RGG motifs impact protein phase separation and aggregation. Protein aggregation was monitored using fluorescence by replacing the conserved DEAD box motif with sfGFP (Superfold Green Fluorescent Protein) [
3]. As shown in
Figure 1B, three different configurations of fusion protein sequences containing sfGFP were constructed with different combinations of the N and C terminals to analyze the impact of RGG motif location on protein self-aggregation and phase separation.
Using a linear DNA template and cell extracts, protein expression was induced for 4 hours to reach the concentration similar to cellular levels (~ 5 μM). Protein aggregation was observed by varying the temperatures. As shown in
Figure 1C, self-aggregation was more profoundly at higher temperatures, with all constructs of DED1 and DBP1 displaying self-aggregation at 42°C. At a lower temperature of 30°C, DED1N still showed evident self-aggregation, while the self-aggregation capability of DED1NC, DED1C and DBP1C decreased. DBP1NC self-aggregation was observed after temperature was increased to 37°C, but DBP1N aggregation was not observed even at this temperature (
Figure 1C). For DDX3X constructs, self-aggregation was more difficult with only DDX3XN occurred at 42°C, while both DDX3XNC and DDX3XC were unable to undergo self-aggregation (Supplemental
Figure S1A). Considering that DDX3X has only one RGG at its N terminal, these results indicate that the more the RGG numbers the easier the self-aggregation. The results also showed that the phase separation could be enhanced when having the same sequences on both N and C terminal as observed in DED1 (
Figure 1B), which may be due to additional electrostatic interactions, π-π or hydrogen bonding force between the same RGGs structures [
23,
24,
25]. Compared to RGG motifs, the impact of RG motifs on self-aggregation is not profound, as DDX3X has more RGs than DED1 and DPB1 but displayed a smaller aggregation capacity.
2.3. The Protein Solubility Was Decreased during Phase Separation but Reversible when Increasing Temperature
To investigate whether self-aggregation alters protein solubility during phase separation, we analyzed the supernatant phase (S phase) and precipitation phase (P phase) after inducing protein synthesis at 37°C in a cell-free system followed by ultracentrifugation. Western blotting results showed that the protein solubility was reduced in samples with visible self-aggregation, indicating that most proteins were present in the P phase (
Figure 1D and
Figure S1B). In contrast, proteins that did not undergo self-aggregation retained higher solubility and remained in the S phase (
Figure 1D and
Supplemental Figure S1B). Additionally, protein aggregation and solubility showed a temperature-dependent reversibility. As shown in
Figure 1E, DED1NC and DBP1NC both formed liquid condensates and self-aggregation above a certain temperature threshold (40°C) and unraveled into phase separation below this temperature. These results suggest that phase separation/self-aggregation is a dynamic process that fluctuates with temperature changes. Further studies with either cell extracts or purified proteins in the cell-free system revealed a similar temperature threshold for the onset of self-aggregation (
Figure S2).
2.4. More RGG Repeats Augmented Phase Separation and Self-Aggregation
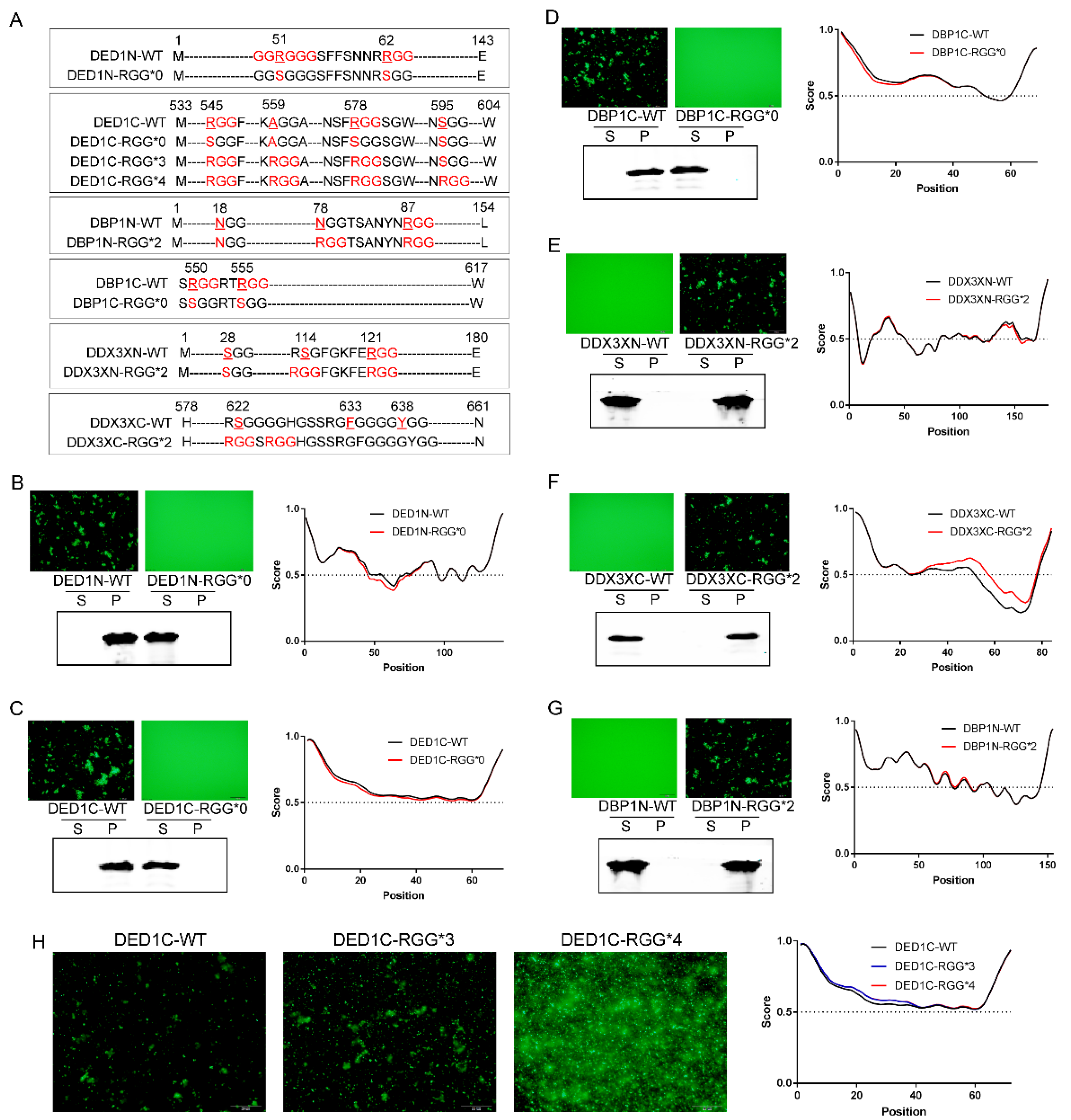
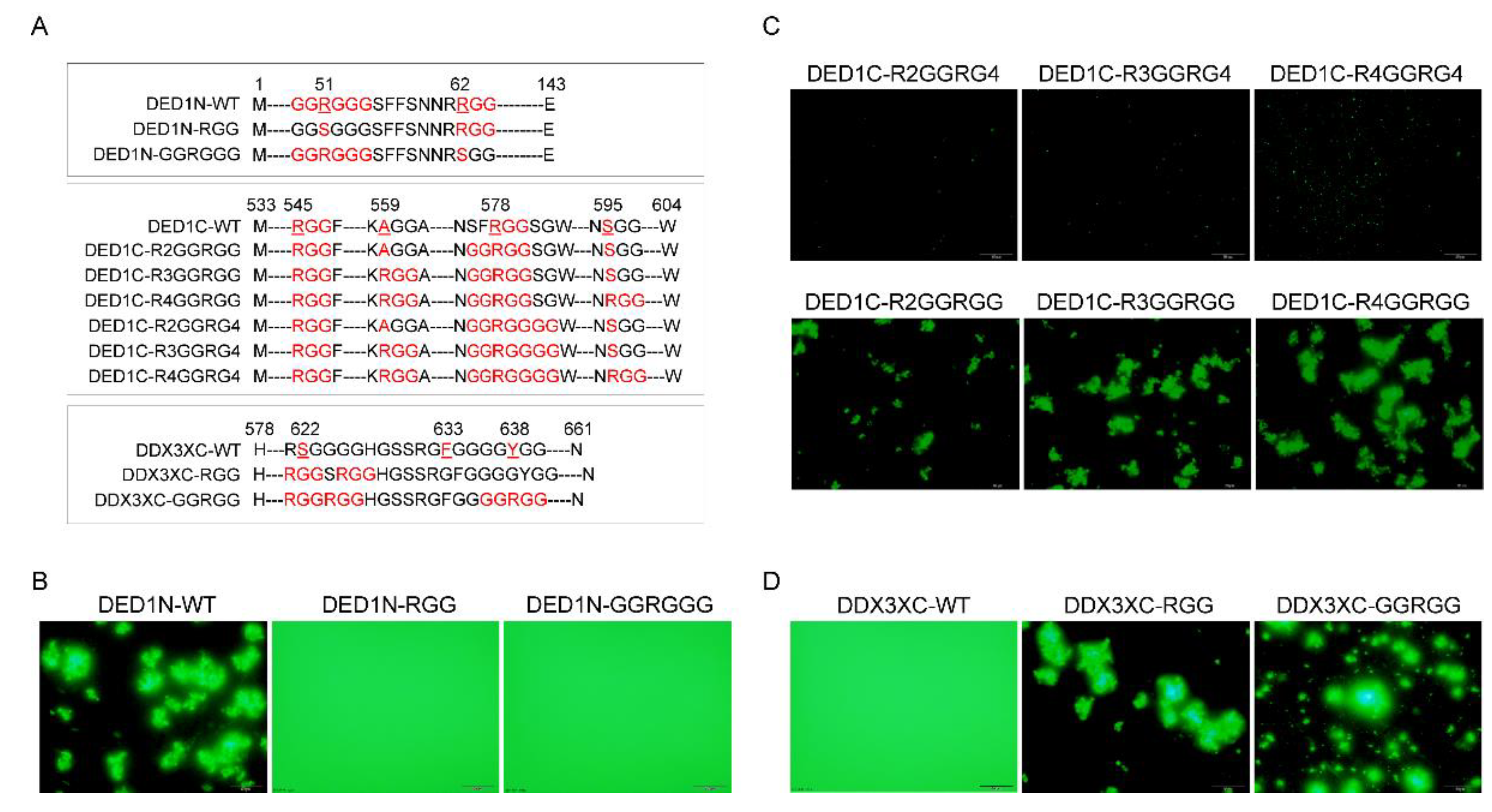
To demonstrate how RGG motif patterns influence self-aggregative phase separation, we systematically mutated the protein-sfGFP constructs to alter the numbers of RGG repeats - (RGG)n, the spacing between RGG motifs - RGG-Xn-RGG, and the number of glycine residues in R(G)n stretches (
Figure 2A). As shown in
Figure 2, when RGG was mutated to SGG in protein constructs with evident self-aggregation capacity (DED1N, DED1C and DBP1C), their phase separation was abolished, as determined by fluorescence microscopy and the solubility analysis using Western blotting. In addition, IUPRED analysis showed a corresponding decrease in the degree of amino acid sequence disorder (
Figure 2B–D). In contrast, adding 2 RGG repeats to the initially non-aggregating proteins (DDX3XN, DDX3XC and DBP1N) changed their phase separation capacity, leading to self-aggregation and precipitation into condensates, and enhanced the degree of amino acid disorder (
Figure 2E–G). Similarly, adding more RGG repeats augmented the aggregation of DED1C at 30℃ (
Figure 2H).
2.5. The Distance between RGG Motifs Affects Phase Separation Capacity
Having established that the number of RGG repeats positively enhances phase separation, we next investigated the impact of the distance between two RGGs. For the three proteins that showed evident self-aggregation, the number of amino acids inserted between two RGGs in DED1N is 7, 30 in DED1C, and 2 in DBP1C, respectively. Therefore, the RGG repeats with a distance of up to 30 amino can still interact with each other to affect phase separation. To further determine the distance limit, we generated several proteins mutations with different numbers of amino acids between RGG motifs by point mutation to convert xGG motifs into RGG (
Figure 3A). For example, DBP1N-N18R is a mutated form of DBP1N in which the N-terminal 18th amino acid, N, is converted into R, leading to the generation of two RGGs with a distance of 63 amino acids (designated as RGG-(X)
63-RGG). Similarly, DDX3XN-S28R is a mutated form of DDX3X in which the N-terminal 28th amino acid, S, is converted into R, leading to the generation of two RGGs with a distance of 90 amino acids (RGG-(X)
90-RGG). As shown in
Figure 3, the increased number of RGG in DBP1N-N18R and DDX3XN-S28R did not change the non-aggregating status of wild-type protein, indicating that a distance greater than 63 amino acids between two RGGs cannot increase phase separation. However, two other mutations DBP1N-N78R (RGG-(X)
6-RGG) and DDX3XN-S114G (RGG-(X)
5-RGG), with RGG spacings of ≤6 amino acids, enabled self-aggregation (
Figure 3B,C). The fewer the number of amino acids between RGGs, the stronger the self-aggregation ability of phase separation.
2.6. The Impact of Glycine Numbers in R(G)n Motifs on Phase Separation
As RG repeats showed a weak effect on protein self-aggregation compared to RGG repeats, how about increasing the number of glycine? Therefore, we mutated the DDX3XC and DED1C protein sequences at certain locations and generated R(G)
n motifs with varied numbers of glycine residues. As shown in
Figure 3, the results demonstrated that, similar to R(G)
1 which does not enhance self-aggregate (
Figure 1C and
Figure S2A), glycine repeats with numbers greater than 5 also do not enhance self-aggregation. However, phase separation was observed when the number of glycine is between 2 and 4, with no significant differences in aggregation capacity among them (
Figure 3D,E). These results suggest that a balanced combination of arginine and glycine residues is required to enhance the π-π stacking forces needed for phase separation.
2.7. GGRGG Motif Enhances Phase Separation
The impact of glycine on phase separation is not only affected by its number but also its locations relative to the amino acid arginine (R). Noticing that DED1N exhibited greater self-aggregative phase separation capacity than DED1C and DBP1C at an equal temperature threshold (
Figure 1C), we hypothesized that this may be due to the presence of a unique GGRGGG motif besides an RGG motif with a distance of 8 amino acids between them (
Figure 4A). After mutating either the GGRGGG motif to keep only the RGG (DED1N-RGG) or the RGG motif to keep only GGRGGG (DED1N-GGRGG), results showed that neither one was able to form self-aggregates (
Figure 4B). The effect of GGRGG motif on self-aggregation was further investigated by mutation analysis of DED1C and DDX3XC (
Figure 4C,D). Results showed that the phase separation was enhanced with 2 glycine residues located on C termini of R, whereas phase separation was abolished when the glycine number increased to 4. These results indicate that the arginine asymmetry may enable distinct protein-protein interactions not achieved by regular RGG repeats.
2.8. Bioinformatic Analysis Reveals DDX3X Mutations in RG/RGG Motifs across Cancers
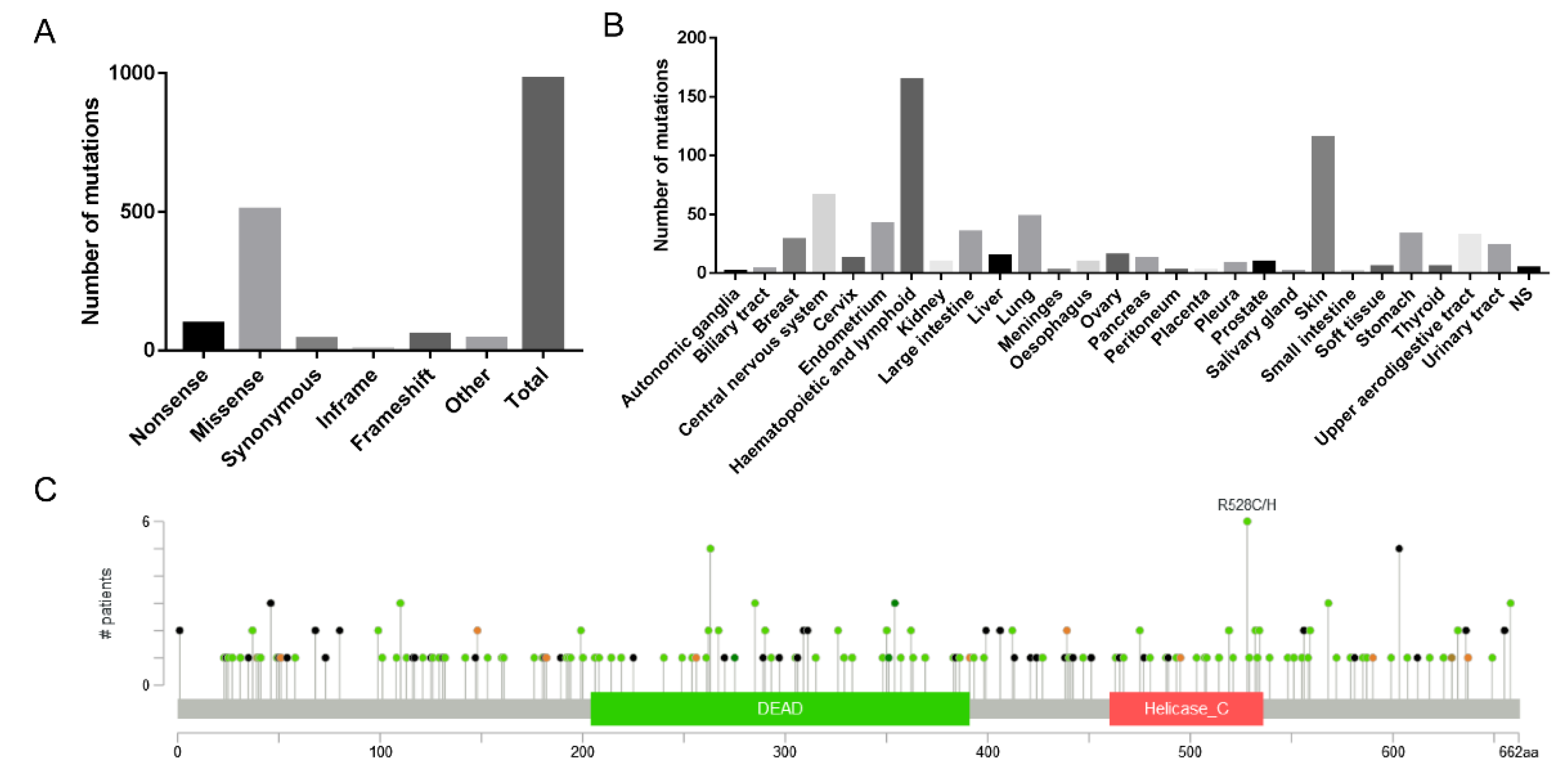
To further investigate whether the influence of RG/RGG motifs on protein phase separation affects the protein's function and its potential association with cancer development, we analyzed the mutation distribution of DDX3X gene using the COSMIC and cBioPortal databases.
First, we used the COSMIC database to analyze 980 reported cancer-related mutation types in the DDX3X gene, among which 509 were missense mutations, accounting for 51.94% of the total mutation types, making it the primary type of gene mutation (
Figure 5A). By analyzing 628 positive samples out of 5,411 mutation samples, we found that DDX3X mutations mainly occurred in lymphoid (164) and skin (115) tissues (
Figure 5B). Notably, when analyzing the mutations in RG/RGG motifs across the DDX3X gene, we discovered that mutations in both arginine and glycine residues within each motif could influence tissue carcinogenesis (
Table 1).
Next, we used the cBioPortal database to analyze 32 studies (10,967 samples). The results showed that the mapping of DDX3X mutation sites and amino acid sequences in cancer indicated that mutations were primarily concentrated at the N-terminus (
Figure 5C), suggesting that the main active sites of DDX3X associated with carcinogenesis might be located at the N-terminus. Interestingly, RG motifs in DDX3X are also mainly distributed at the N-terminus, implying a potential connection between these motifs and cancer development. Furthermore, we found that even the absence of a single RG/RGG repeat sequence was associated with tumor occurrence in tissues such as the stomach, skin, cervix, and colon (
Table 2), demonstrating the functional importance of these gene sequences.
These bioinformatic analyses highlight the importance of RG/RGG motifs in DDX3X and other DDX family members in the context of cancer development. The concentration of mutations at the N-terminus of DDX3X, where RG motifs are primarily located, suggests a potential link between these motifs and carcinogenesis. Moreover, the observation that even single RG/RGG repeat sequence alterations are associated with tumor occurrence in various tissues underscores the functional significance of these motifs. Further research is needed to elucidate the precise mechanisms by which RG/RGG motif mutations contribute to cancer development and to explore potential therapeutic targets based on these findings.
2.9. The Expression Profile of DDX3X in Different Cancers and Its Co-Expression with G3BP1
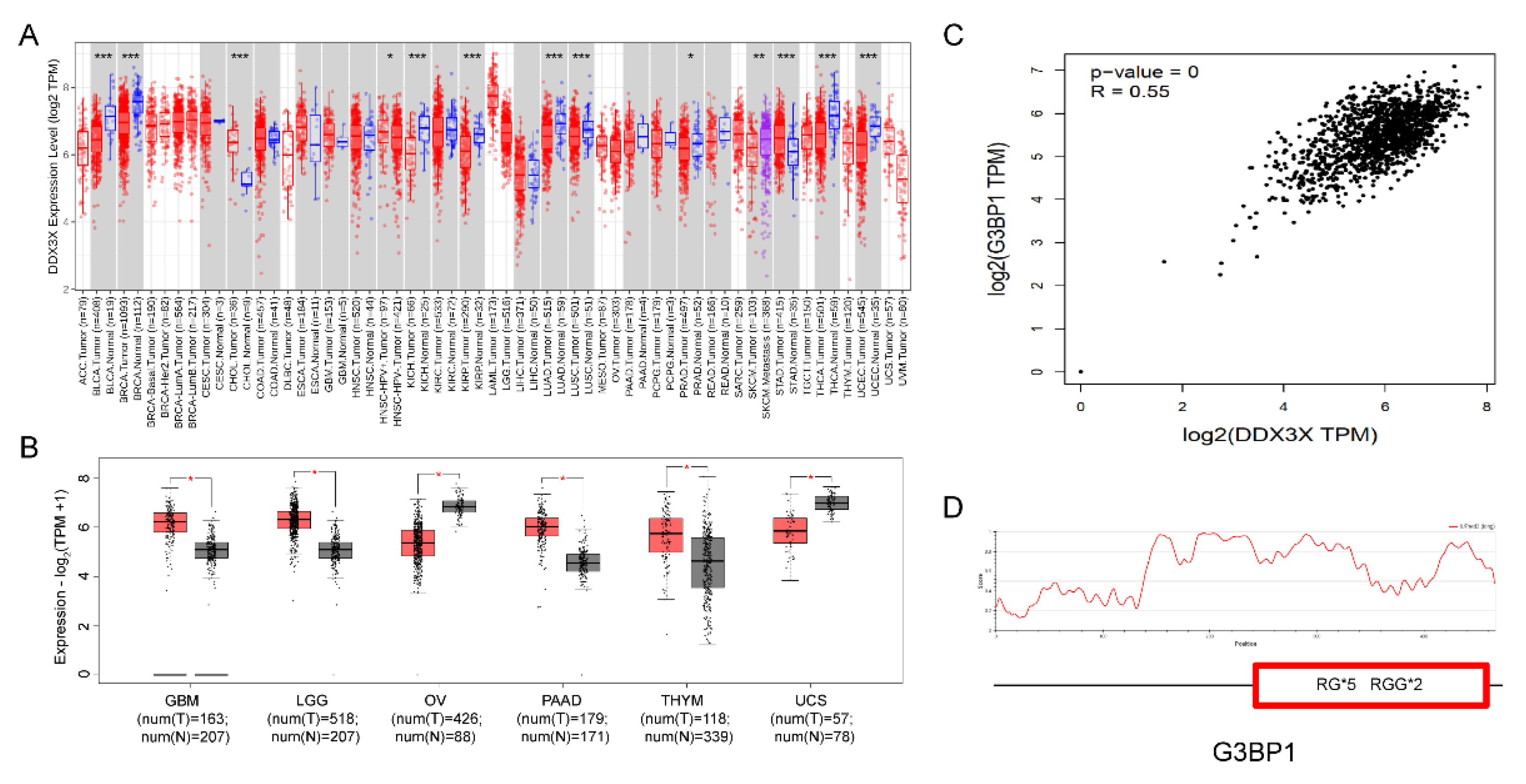
To further investigate the expression patterns of the DDX3X gene in different cancer tissues, we performed a pan-cancer analysis of DDX3X mRNA expression data using the TIMER database from TCGA. The results showed that compared to normal tissues, DDX3X expression was significantly upregulated in 4 out of 23 paired cancer tissues and adjacent normal tissues, while it was significantly downregulated in 2 tissues (
Figure 6A). Using the GEPIA database, we further analyzed the mRNA expression levels of DDX3X in the 6 tissues with significant changes. The results demonstrated that DDX3X was significantly upregulated in GBM (Glioblastoma multiforme), LGG (Brain Lower Grade Glioma), PAAD (Pancreatic adenocarcinoma), and THYM (Thymoma), while it was significantly downregulated in OV (Ovarian serous cystadenocarcinoma) and UCS (Uterine Carcinosarcoma) (
Figure 6B).
Interestingly, in these 6 cancer tissues, DDX3X was found to be significantly co-expressed with G3BP1, an RNA-binding protein (
Figure 6C). G3BP1 can form stress granules through phase separation and is a potential therapeutic target for cancer [
26,
27]. Our analysis revealed that G3BP1 contains multiple RG/RGG sequences (
Figure 6D), suggesting that DDX3X may interact with G3BP1 through their RG/RGG sequences.
2.10. Co-Phase Separation of DDX3X and G3BP1 in a Cell-Free System
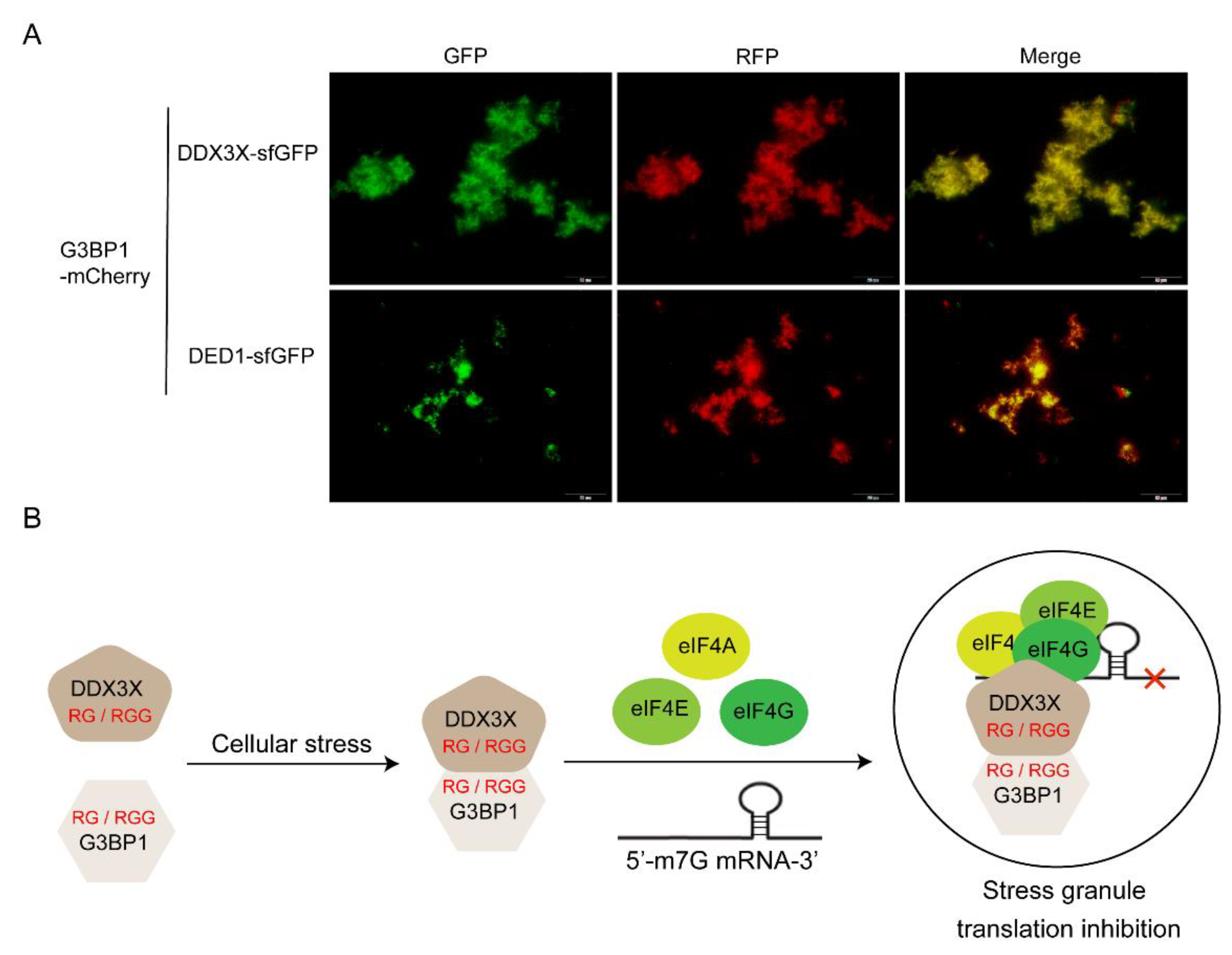
To verify whether DDX3X and G3BP1 can interact with each other through their RGG motifs and undergo co-phase separation, the human G3BP1 gene sequence was fused with the mCherry (a red fluorescent protein) sequence to construct an encoding gene for the G3BP1-mCherry fusion protein. In a cell-free system, G3BP1-mCherry DNA template was added together with either DDX3X-sfGFP or DED1-sfGFP DNA template, and the mixture was incubated at 30°C for 16 hours to synthesize both proteins. Subsequently, the mixture was incubated at 40°C for 30 minutes to induce protein phase separation and aggregation. Fluorescence microscopy was used to detect the aggregation and spatial localization of sfGFP and mCherry fluorescence in different fluorescence channels. The results showed that both DDX3X and G3BP1 could undergo phase separation and aggregation under high-temperature induction, and they co-localized spatially. DED1 also exhibited similar behavior (
Figure 7A). These findings suggest that the cellular functions of G3BP1 and DDX3X may be closely related to their co-phase separation and aggregation.
3. Materials and Methods
3.1. Construction of DNA Sequences Expressing sfGFP with Different RG/RGG Motifs
The gene sequences for DDX3X, DED1, and DBP1 were obtained from human cells or the yeast S. cerevisiae. The sequence of sfGFP (Superfold Green Fluorescent Protein) was obtained from the pET23a-sfGFP plasmid. Using the DDX3X, DED1, and DBP1 sequences as templates, the N-terminal or C-terminal sequences (DDX3XN, DDX3XC, DED1N, DED1C, DBP1N, DBP1C) were PCR-amplified. Then, the N-terminal sequence, sfGFP sequence (replacing the DEAD domain), and the corresponding C-terminal sequence were connected in order by overlap PCR, and the related plasmids were constructed using pET23a as the vector (
Table S2).
Different RG/RGG motifs were constructed using a combination of PCR amplification and site-directed mutagenesis. Primers used for mutagenesis are listed in Supplemental
Table S3. The PCR site-directed mutagenesis reactions were performed using high-fidelity FastPfu DNA Polymerase (Cat.AP221, Transgen) to mutate the amino acid coding sequences at specific positions, converting RxG to RGG sequences (Supplemental
Figure S3E). The PCR products were then treated with DpnI restriction enzyme (Cat.FD1703, Thermo Scientific) to digest the parental DNA template and selectively retain the mutated DNA sequences. The mutated DNA fragments were purified using a PCR purification kit (Cat.DC301, Vazyme). The assembled DNA constructs were cloned into the expression vector pET23a.
3.2. Cell-Free Expression of Protein with Different RG/RGG Motifs
Different DNA sequences encoding proteins with different RG/RGG motifs were obtained using PCR and site-directed mutagenesis. For in vitro protein expression, 15 μL of the cell-free synthesis complex was prepared on ice, and 6 nM of linear DNA templates encoding proteins with different RG/RGG motifs were added. The samples were then incubated in a 30°C incubator for 4 hours to synthesize proteins through transcription and translation (reaching a concentration of approximately 5 μM). Components in the cell-free complex were as follows: 1.2 mM ATP, 0.85 mM GTP/UTP/CTP, 31.5 μg/ml folinic acid, 170.6 μg/ml tRNA, 0.4 mM NAD, 0.27 mM coenzyme A, 4 mM oxalic acid, 1 mM putrescine, 1.5 mM spermidine, 57.33 mM HEPES buffer, 12 mM magnesium glutamate, 10 mM ammonium glutamate, 130 mM potassium glutamate, 2 mM of 20 amino acids, 30 mM PEP, and 5 μl of E. coli cell extract.
3.3. Phase Separation Analysis
Following cell-free protein expression, phase separation and aggregation were induced by incubating the samples at different temperatures (30, 37 or 42°C) for 30 minutes. The expressed protein mixtures were loaded into custom-made flow chambers consisting of glass coverslips separated by a double-sided tape spacer. Fluorescence imaging of the samples was performed at room temperature using an upright fluorescence microscope (Olympus BX53) or confocal microscope (Zeiss LSM980 Airyscan 2). Image analysis was conducted using Fiji (ImageJ) software. Fluorescence microscopy images with a Scale bar of 20 μm.
3.4. Solubility and Sequence Association Analysis
Following phase separation and aggregation, the protein samples were separated into supernatant and pellet fractions by centrifugation at 12,000 g for 10 minutes at 4°C. The supernatant and pellet fractions were solubilized with SDS-PAGE loading buffer and 200 mM DTT at 80°C for 10 min, then subjected to PAGE analysis. RGG motifs was detected by Western blotting using a primary anti-6×His monoclonal antibody (Cat.66005, Proteintech) and a secondary anti-mouse antibody conjugated to IRDye 800CW (Cat.925-32210, LI-COR). Disorder analysis of the RGG motifs was performed using the IUPred (
https://iupred.elte.hu/) database, which identifies intrinsically disordered regions (IDRs) based on a biophysical model that calculates the probability (between 0 and 1) of a given residue being disordered. Residues with scores greater than 0.5 are considered to be part of a disordered region.
3.5. Mutation Analysis of RG/RGG Motifs from Public Databases
Proteins containing RG/RGG motifs were first identified using the UniProt database (
https://www.uniprot.org/). The arginine residues within RG/RGG motifs and their potential association with cancer were then analyzed using Catalogue Of Somatic Mutations In Cancer (COSMIC) database (
https://cancer.sanger.ac.uk/) and the cBioPortal database (
https://www.cbioportal.org/). The analysis included mutation types, the number of mutations in each tissue (COSMIC), the distribution of mutations along the protein sequence (cBioPortal), and the tumor types associated with these mutations. The statistical significance of the association between arginine mutations and specific cancer types was assessed using Fisher's exact test, with a p-value < 0.05 considered significant.
3.6. Gene Expression Analysis in Pan-Cancer Using TIMER and GEPIA 2 Databases
The expression levels of DDX family genes were analyzed across various tissues for pan-cancer analysis using the TIMER database (
http://cistrome.dfci.harvard.edu/TIMER/) and TCGA data. In order to overcome the lack of normal tissue data for certain genes in TCGA, we also utilized the GEPIA 2 database (
http://gepia2.cancer-pku.cn/), which integrates both TCGA and Genotype-Tissue Expression (GTEx) data.
4. Conclusions
In this study, we investigated the role of RG/RGG motifs in the phase separation and self-aggregation properties of the DEAD-box RNA helicase DDX3X/ DED1/DBP1 using a cell-free system. Our results demonstrate that the number, location, and composition of RG/RGG motifs significantly influence the ability of these proteins to undergo phase separation and form self-aggregates. Further bioinformatic analysis revealed a significant association between DDX3X mutations in RG/RGG motifs and various types of cancer, suggesting a potential role for these motifs in cancer pathogenesis. Furthermore, we found that DDX3X is co-expressed with the stress granule protein G3BP1 in several cancer types and undergo co-phase separation with G3BP1 in a cell-free system, suggesting a potential functional interaction between these proteins in phase-separated structures.
The RG/RGG motifs have been previously reported playing important roles in the aggregation of several proteins, including the RNA-binding proteins FUS and hnRNPA1[
28,
29]. Our findings extend these observations to DDX3X, demonstrating that the presence of multiple RG/RGG motifs in the N-terminal and C-terminal regions of the protein facilitates its phase separation and self-aggregation. Moreover, we demonstrate that the spacing between RG/RGG motifs and the number of glycine residues within each motif appear to be critical factors in determining the extent of phase separation, as evidenced by mutational analysis. These results highlighted the importance of the spatial arrangement and composition of low-complexity domains in driving phase separation, which are consistent with previous studies [
30,
31].
The phase separation and self-aggregation of DDX3X mediated by RG/RGG motifs may have important functional implications. DDX3X has been shown to localize to stress granules, which are phase-separated structures that form in response to cellular stress [
32]. The RG/RGG motifs in DDX3X may facilitate its recruitment to stress granules and promote their assembly or stability. Additionally, the phase separation of DDX3X may modulate its interactions with RNA and other protein partners, thereby regulating its role in various aspects of RNA metabolism, such as translation initiation and mRNA export [
33,
34].
Our bioinformatic analysis revealed a significant enrichment of DDX3X mutations in RG/RGG motifs across multiple cancer types, particularly in the N-terminal region of the protein. These mutations may disrupt the phase separation properties of DDX3X, leading to altered stress granule dynamics or dysregulated RNA metabolism, which could contribute to cancer development. Indeed, previous studies have implicated DDX3X in various aspects of cancer biology, including cell cycle regulation, apoptosis, and metastasis [
35,
36,
37]. Our findings suggest that mutations in RG/RGG motifs may represent a novel mechanism by which DDX3X contributes to cancer pathogenesis.
Intriguingly, we also found that DDX3X is co-expressed with the stress granule protein G3BP1 in several cancer types. G3BP1 is a key component of stress granules and has been shown to promote their assembly through phase separation [
38]. The co-expression of DDX3X and G3BP1 in cancer may reflect a functional interaction between these proteins in stress granule formation or other phase-separated structures. Both proteins contain multiple RG/RGG motifs, which may facilitate their co-aggregation and contribute to aberrant stress granule dynamics in cancer cells. Our proposed interaction mechanism suggests that DDX3X and G3BP1 may interact through their RG/RGG domains and subsequently bind to translation factors eIF4A/E/G to form stress granules (
Figure 7B)[
33,
39,
40]. These stress granules associate with mRNA and regulate the translation of downstream genes by influencing the function of the 43S preinitiation complex (PIC), ultimately affecting cellular physiological functions [
41,
42,
43]. Targeting the interaction between DDX3X and G3BP1, or their individual phase separation properties, may represent a potential therapeutic strategy for cancers in which these proteins are dysregulated.
In conclusion, our study demonstrates that RG/RGG motifs play a critical role in the phase separation and self-aggregation properties of DDX3X. Mutations in these motifs are significantly associated with various types of cancer, suggesting a potential mechanism by which DDX3X may contribute to cancer pathogenesis. The co-expression of DDX3X with the stress granule protein G3BP1 in cancer further highlights the potential importance of phase separation in DDX3X-related cancer biology. These findings provide a foundation for future studies aimed at elucidating the mechanistic details of DDX3X phase separation and its role in cancer development, as well as exploring potential therapeutic strategies targeting this process.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, H.C. and W.M; methodology, H.C., S.Z, and B.L; software, H.C.; validation, H.C. and X.Z; formal analysis, H.C.; data curation, H.C., B.L., X.Z. C.Y., S.Z. and W.M.; writing—original draft preparation, review and editing, H.C. and W.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0901702), and the National Science Foundation of Shandong (ZR2022MC057).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-Box ATPases Are Global Regulators of Phase-Separated Organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef]
- Linder, P.; Jankowsky, E. From Unwinding to Clamping - the DEAD Box RNA Helicase Family. Nat Rev Mol Cell Biol 2011, 12, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase Transition of a Disordered Nuage Protein Generates Environmentally Responsive Membraneless Organelles. Mol Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Cui, B.C.; Sikirzhytski, V.; Aksenova, M.; Lucius, M.D.; Levon, G.H.; Mack, Z.T.; Pollack, C.; Odhiambo, D.; Broude, E.; Lizarraga, S.B.; et al. Pharmacological Inhibition of DEAD-Box RNA Helicase 3 Attenuates Stress Granule Assembly. Biochem Pharmacol 2020, 182, 114280. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Zhang, J. Liquid-Liquid Phase Separation Drives Cellular Function and Dysfunction in Cancer. Nat Rev Cancer 2022, 22, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-Y.; Dyakov, B.J.A.; Zhang, J.; Knight, J.D.R.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.-C. Properties of Stress Granule and P-Body Proteomes. Mol Cell 2019, 76, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Valentin-Vega, Y.A.; Wang, Y.-D.; Parker, M.; Patmore, D.M.; Kanagaraj, A.; Moore, J.; Rusch, M.; Finkelstein, D.; Ellison, D.W.; Gilbertson, R.J.; et al. Cancer-Associated DDX3X Mutations Drive Stress Granule Assembly and Impair Global Translation. Sci Rep 2016, 6, 25996. [Google Scholar] [CrossRef] [PubMed]
- Cargill, M.J.; Morales, A.; Ravishankar, S.; Warren, E.H. RNA Helicase, DDX3X, Is Actively Recruited to Sites of DNA Damage in Live Cells. DNA Repair (Amst) 2021, 103, 103137. [Google Scholar] [CrossRef]
- Heerma van Voss, M.R.; Brilliant, J.D.; Vesuna, F.; Bol, G.M.; van der Wall, E.; van Diest, P.J.; Raman, V. Combination Treatment Using DDX3 and PARP Inhibitors Induces Synthetic Lethality in BRCA1-Proficient Breast Cancer. Med Oncol 2017, 34, 33. [Google Scholar] [CrossRef]
- Chowdhury, M.N.; Jin, H. The RGG Motif Proteins: Interactions, Functions, and Regulations. Wiley Interdiscip Rev RNA 2023, 14, e1748. [Google Scholar] [CrossRef]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG Motif. Mol Cell 2013, 50, 613–623. [Google Scholar] [CrossRef]
- Rajyaguru, P.; Parker, R. RGG Motif Proteins: Modulators of mRNA Functional States. Cell Cycle 2012, 11, 2594–2599. [Google Scholar] [CrossRef]
- Poornima, G.; Mythili, R.; Nag, P.; Parbin, S.; Verma, P.K.; Hussain, T.; Rajyaguru, P.I. RGG-Motif Self-Association Regulates eIF4G-Binding Translation Repressor Protein Scd6. RNA Biol 2019, 16, 1215–1227. [Google Scholar] [CrossRef]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, Physiologic Functions and Cancer. Mol Cancer 2021, 20, 38. [Google Scholar] [CrossRef]
- Shih, J.-W.; Wang, W.-T.; Tsai, T.-Y.; Kuo, C.-Y.; Li, H.-K.; Wu Lee, Y.-H. Critical Roles of RNA Helicase DDX3 and Its Interactions with eIF4E/PABP1 in Stress Granule Assembly and Stress Response. Biochem J 2012, 441, 119–129. [Google Scholar] [CrossRef]
- Shen, H.; Yanas, A.; Owens, M.C.; Zhang, C.; Fritsch, C.; Fare, C.M.; Copley, K.E.; Shorter, J.; Goldman, Y.E.; Liu, K.F. Sexually Dimorphic RNA Helicases DDX3X and DDX3Y Differentially Regulate RNA Metabolism through Phase Separation. Mol Cell 2022, 82, 2588–2603. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Hess, D.; Eglinger, J.; Fritsch, A.W.; Kreysing, M.; Weinert, B.T.; Choudhary, C.; Matthias, P. Acetylation of Intrinsically Disordered Regions Regulates Phase Separation. Nat Chem Biol 2019, 15, 51–61. [Google Scholar] [CrossRef]
- Aryanpur, P.P.; Mittelmeier, T.M.; Bolger, T.A. The RNA Helicase Ded1 Regulates Translation and Granule Formation during Multiple Phases of Cellular Stress Responses. Mol Cell Biol 2022, 42, e0024421. [Google Scholar] [CrossRef]
- Hilliker, A.; Gao, Z.; Jankowsky, E.; Parker, R. The DEAD-Box Protein Ded1 Modulates Translation by the Formation and Resolution of an eIF4F-mRNA Complex. Mol Cell 2011, 43, 962–972. [Google Scholar] [CrossRef]
- Iserman, C.; Desroches Altamirano, C.; Jegers, C.; Friedrich, U.; Zarin, T.; Fritsch, A.W.; Mittasch, M.; Domingues, A.; Hersemann, L.; Jahnel, M.; et al. Condensation of Ded1p Promotes a Translational Switch from Housekeeping to Stress Protein Production. Cell 2020, 181, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Sen, N.D.; Gupta, N.; K Archer, S.; Preiss, T.; Lorsch, J.R.; Hinnebusch, A.G. Functional Interplay between DEAD-Box RNA Helicases Ded1 and Dbp1 in Preinitiation Complex Attachment and Scanning on Structured mRNAs in Vivo. Nucleic Acids Res 2019, 47, 8785–8806. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.A.; Vernon, R.M.; Forman-Kay, J.D. RGG/RG Motif Regions in RNA Binding and Phase Separation. J Mol Biol 2018, 430, 4650–4665. [Google Scholar] [CrossRef] [PubMed]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi Contacts Are an Overlooked Protein Feature Relevant to Phase Separation. Elife 2018, 7, e31486. [Google Scholar] [CrossRef] [PubMed]
- Arribas-Layton, M.; Dennis, J.; Bennett, E.J.; Damgaard, C.K.; Lykke-Andersen, J. The C-Terminal RGG Domain of Human Lsm4 Promotes Processing Body Formation Stimulated by Arginine Dimethylation. Mol Cell Biol 2016, 36, 2226–2235. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Mathieu, C.; Kolaitis, R.-M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch That Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Zhu, X.; Liu, Z.; Song, Y.; Wu, S.; Lu, X.; Yang, J.; Jin, C. Aluminum Exposure Induces Central Nervous System Impairment via Activating NLRP3-Medicated Pyroptosis Pathway. Ecotoxicol Environ Saf 2023, 264, 115401. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Singh, M. Structure Specific Recognition of Telomeric Repeats Containing RNA by the RGG-Box of hnRNPA1. Nucleic Acids Res 2020, 48, 4492–4506. [Google Scholar] [CrossRef]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.S.; Varela, J.A.; Lin, J.Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Ströhl, F.; et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell 2018, 173, 720–734. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat Rev Mol Cell Biol 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Kato, M.; Han, T.W.; Xie, S.; Shi, K.; Du, X.; Wu, L.C.; Mirzaei, H.; Goldsmith, E.J.; Longgood, J.; Pei, J.; et al. Cell-Free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell 2012, 149, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mañán-Mejías, P.M.; Miles, H.N.; Putnam, A.A.; MacGillivray, L.R.; Ricke, W.A. DDX3X and Stress Granules: Emerging Players in Cancer and Drug Resistance. Cancers (Basel) 2024, 16, 1131. [Google Scholar] [CrossRef] [PubMed]
- Ryan, C.S.; Schröder, M. The Human DEAD-Box Helicase DDX3X as a Regulator of mRNA Translation. Front Cell Dev Biol 2022, 10, 1033684. [Google Scholar] [CrossRef] [PubMed]
- De Colibus, L.; Stunnenberg, M.; Geijtenbeek, T.B.H. DDX3X Structural Analysis: Implications in the Pharmacology and Innate Immunity. Curr Res Immunol 2022, 3, 100–109. [Google Scholar] [CrossRef]
- Cargill, M.; Venkataraman, R.; Lee, S. DEAD-Box RNA Helicases and Genome Stability. Genes (Basel) 2021, 12, 1471. [Google Scholar] [CrossRef] [PubMed]
- Bol, G.M.; Vesuna, F.; Xie, M.; Zeng, J.; Aziz, K.; Gandhi, N.; Levine, A.; Irving, A.; Korz, D.; Tantravedi, S.; et al. Targeting DDX3 with a Small Molecule Inhibitor for Lung Cancer Therapy. EMBO Mol Med 2015, 7, 648–669. [Google Scholar] [CrossRef]
- Randolph, M.E.; Afifi, M.; Gorthi, A.; Weil, R.; Wilky, B.A.; Weinreb, J.; Ciero, P.; Hoeve, N.T.; van Diest, P.J.; Raman, V.; et al. RNA Helicase DDX3 Regulates RAD51 Localization and DNA Damage Repair in Ewing Sarcoma. iScience 2024, 27, 108925. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wang, Z.; Kang, Y.; Yi, Q.; Wang, T.; Bai, Y.; Liu, Y. Stress Granule Homeostasis Is Modulated by TRIM21-Mediated Ubiquitination of G3BP1 and Autophagy-Dependent Elimination of Stress Granules. Autophagy 2023, 19, 1934–1951. [Google Scholar] [CrossRef]
- Linsalata, A.E.; He, F.; Malik, A.M.; Glineburg, M.R.; Green, K.M.; Natla, S.; Flores, B.N.; Krans, A.; Archbold, H.C.; Fedak, S.J.; et al. DDX3X and Specific Initiation Factors Modulate FMR1 Repeat-Associated Non-AUG-Initiated Translation. EMBO Rep 2019, 20, e47498. [Google Scholar] [CrossRef]
- Lin, T.-C. DDX3X Multifunctionally Modulates Tumor Progression and Serves as a Prognostic Indicator to Predict Cancer Outcomes. Int J Mol Sci 2019, 21, 281. [Google Scholar] [CrossRef]
- Hu, R.; Qian, B.; Li, A.; Fang, Y. Role of Proteostasis Regulation in the Turnover of Stress Granules. Int J Mol Sci 2022, 23, 14565. [Google Scholar] [CrossRef] [PubMed]
- Aramburu-Núñez, M.; Custodia, A.; Pérez-Mato, M.; Iglesias-Rey, R.; Campos, F.; Castillo, J.; Ouro, A.; Romaus-Sanjurjo, D.; Sobrino, T. Stress Granules and Acute Ischemic Stroke: Beyond mRNA Translation. Int J Mol Sci 2022, 23, 3747. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Qing, Y.; Dong, L.; Han, L.; Wu, D.; Li, Y.; Li, W.; Xue, J.; Zhou, K.; Sun, M.; et al. QKI Shuttles Internal m7G-Modified Transcripts into Stress Granules and Modulates mRNA Metabolism. Cell 2023, 186, 3208–3226. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
The protein configurations and determination of phase separation using a cell-free system. A) Amino acid disorder of DED1, DDX3X, and DBP1 proteins predicted by the IUPRED algorithm, and the number of RG/RGG motifs in the disordered regions. B) Schematic representation of the protein constructs containing sfGFP (DED1 is shown as a representative example). C) Expression of DED1NC, DED1N, DED1C, DBP1NC, DBP1N, and DBP1C in a cell-free system (4 h at 30℃) followed by 30 min incubation at different temperatures (30, 37, and 42℃); D) Western blot analysis of the supernatant (S) and pellet (P) fractions at 37℃. E) Reversibility of protein aggregation of DED1NC/DBP1NC between high temperature (40℃) and low temperature (20℃).
Figure 1.
The protein configurations and determination of phase separation using a cell-free system. A) Amino acid disorder of DED1, DDX3X, and DBP1 proteins predicted by the IUPRED algorithm, and the number of RG/RGG motifs in the disordered regions. B) Schematic representation of the protein constructs containing sfGFP (DED1 is shown as a representative example). C) Expression of DED1NC, DED1N, DED1C, DBP1NC, DBP1N, and DBP1C in a cell-free system (4 h at 30℃) followed by 30 min incubation at different temperatures (30, 37, and 42℃); D) Western blot analysis of the supernatant (S) and pellet (P) fractions at 37℃. E) Reversibility of protein aggregation of DED1NC/DBP1NC between high temperature (40℃) and low temperature (20℃).
Figure 2.
The impact of RGG motif numbers on phase separation. A) Schematic representation of the point mutation sequence. B-D) Mutation of RGG to SGG in protein constructs with evident self-aggregation capacity (DED1N, DED1C, and DBP1C) abolished phase separation (induced at 37 ℃ for 30 min). E-G) Addition of 2 RGG repeats to initially non-aggregating proteins (DDX3XN, DDX3XC, and DBP1N) changed their phase separation capacity, leading to self-aggregation and precipitation into condensates (induced at 37 ℃ for 30 min). H) Increasing the number of RGG repeats augments the aggregation of DED1C at 30℃. Phase separation and aggregation were assessed by fluorescence microscopy, solubility analysis using Western blotting, and amino acid disorder predictions using IUPRED. S: supernatant; P: pellet.
Figure 2.
The impact of RGG motif numbers on phase separation. A) Schematic representation of the point mutation sequence. B-D) Mutation of RGG to SGG in protein constructs with evident self-aggregation capacity (DED1N, DED1C, and DBP1C) abolished phase separation (induced at 37 ℃ for 30 min). E-G) Addition of 2 RGG repeats to initially non-aggregating proteins (DDX3XN, DDX3XC, and DBP1N) changed their phase separation capacity, leading to self-aggregation and precipitation into condensates (induced at 37 ℃ for 30 min). H) Increasing the number of RGG repeats augments the aggregation of DED1C at 30℃. Phase separation and aggregation were assessed by fluorescence microscopy, solubility analysis using Western blotting, and amino acid disorder predictions using IUPRED. S: supernatant; P: pellet.
Figure 3.
The impact of RGG spacing and glycine number in R(G)n on phase separation capacity. A) Schematic representation of the point mutation sequence. B) Introducing two RGG motifs with a spacing of 6 amino acids (RGG-(X)6-RGG) in the DBP1N-N78R mutant enables phase separation (induced at 37 ℃ for 30 min), while a spacing of 63 amino acids (RGG-(X)63-RGG) in the DBP1N-N18R mutant does not change the non-aggregating status of the wild-type protein (DBP1N-WT). C) Similarly, introducing two RGG motifs with a spacing of 5 amino acids (RGG-(X)5-RGG) in DDX3XN-S114G enables phase separation, while a spacing of 90 amino acids (RGG-(X)90-RGG) in DDX3XN-S28R does not. D-E) Increasing the number of glycine residues to >5 does not enhance self-aggregation, while glycine repeats of 2-4 promote phase separation with no significant differences in aggregation capacity. Phase separation and aggregation were assessed by fluorescence microscopy, solubility analysis using Western blotting, and amino acid disorder predictions using IUPRED . S: supernatant; P: pellet.
Figure 3.
The impact of RGG spacing and glycine number in R(G)n on phase separation capacity. A) Schematic representation of the point mutation sequence. B) Introducing two RGG motifs with a spacing of 6 amino acids (RGG-(X)6-RGG) in the DBP1N-N78R mutant enables phase separation (induced at 37 ℃ for 30 min), while a spacing of 63 amino acids (RGG-(X)63-RGG) in the DBP1N-N18R mutant does not change the non-aggregating status of the wild-type protein (DBP1N-WT). C) Similarly, introducing two RGG motifs with a spacing of 5 amino acids (RGG-(X)5-RGG) in DDX3XN-S114G enables phase separation, while a spacing of 90 amino acids (RGG-(X)90-RGG) in DDX3XN-S28R does not. D-E) Increasing the number of glycine residues to >5 does not enhance self-aggregation, while glycine repeats of 2-4 promote phase separation with no significant differences in aggregation capacity. Phase separation and aggregation were assessed by fluorescence microscopy, solubility analysis using Western blotting, and amino acid disorder predictions using IUPRED . S: supernatant; P: pellet.
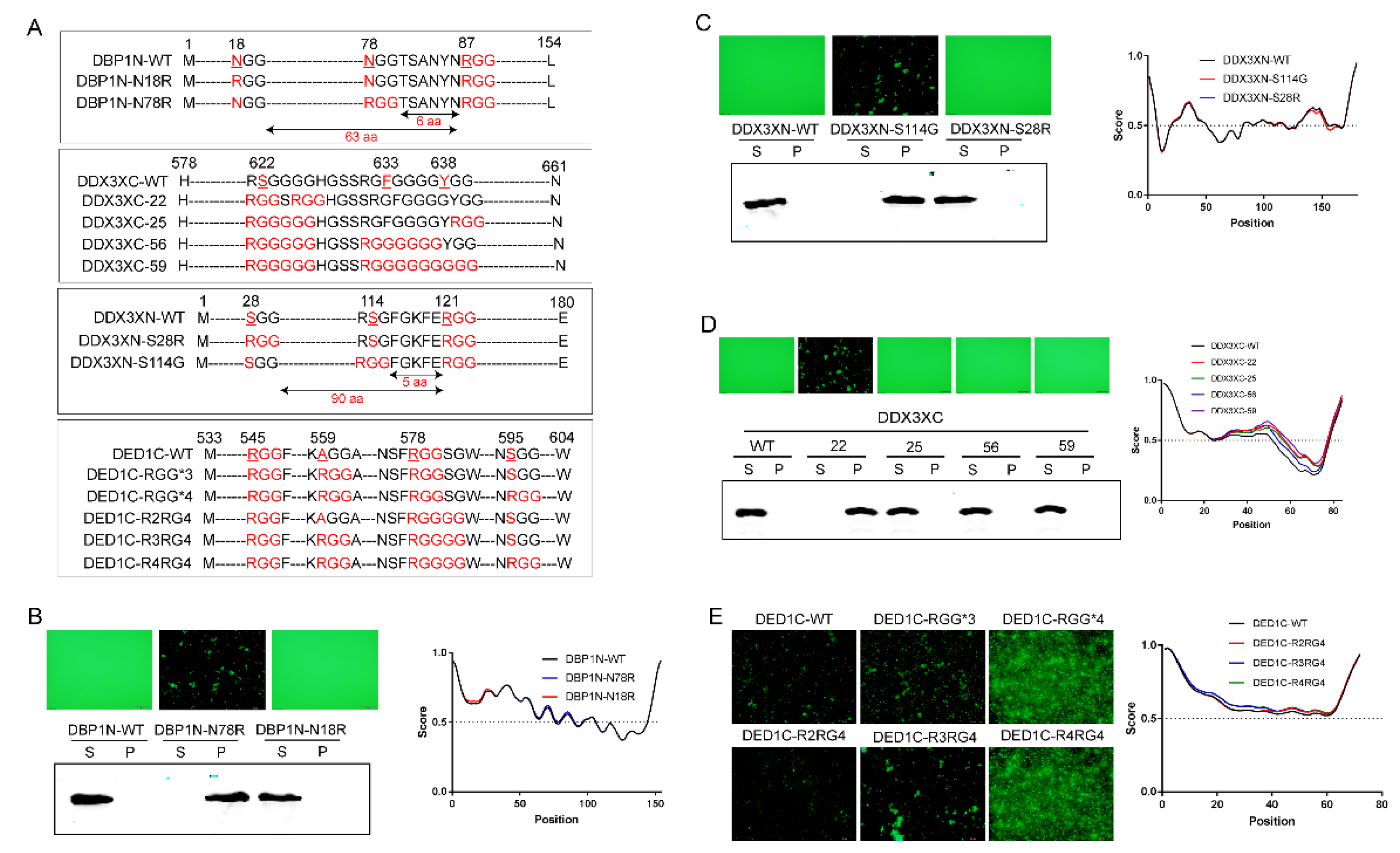
Figure 4.
The impact of GGRGG motif on phase separation. A) Schematic representation of the DED1N protein construct, highlighting the presence of a unique GGRGGG motif and an RGG motif with a certain. B) Mutating either the GGRGGG motif to keep only the RGG (DED1N-RGG) or the RGG motif to keep only GGRGGG (DED1N-GGRGG) abolishes self-aggregation (induced at 37℃) in DED1N. C-D) The effect of the GGRGG motif on self-aggregation in DED1C and DDX3XC. Introducing 2 glycine residues at the C-terminus of R enhances phase separation, while increasing the glycine number to 4 abolishes phase separation (induced at 30℃ for 30 min).
Figure 4.
The impact of GGRGG motif on phase separation. A) Schematic representation of the DED1N protein construct, highlighting the presence of a unique GGRGGG motif and an RGG motif with a certain. B) Mutating either the GGRGGG motif to keep only the RGG (DED1N-RGG) or the RGG motif to keep only GGRGGG (DED1N-GGRGG) abolishes self-aggregation (induced at 37℃) in DED1N. C-D) The effect of the GGRGG motif on self-aggregation in DED1C and DDX3XC. Introducing 2 glycine residues at the C-terminus of R enhances phase separation, while increasing the glycine number to 4 abolishes phase separation (induced at 30℃ for 30 min).
Figure 5.
Bioinformatic analysis of DDX3X mutations in RG/RGG motifs across cancers. A) Distribution of DDX3X mutation types in cancer-related samples from the COSMIC database. Missense mutations account for 51.94% of the total mutation types, making it the primary type of gene mutation. B) Distribution of DDX3X mutations across different tissues based on the analysis of 628 positive samples out of 5,411 mutation samples from the COSMIC database. DDX3X mutations mainly occurred in lymphoid (164) and skin (115) tissues. C) Mapping of DDX3X mutation sites and amino acid sequences in cancer samples from the cBioPortal database (32 studies, 10,967 samples).
Figure 5.
Bioinformatic analysis of DDX3X mutations in RG/RGG motifs across cancers. A) Distribution of DDX3X mutation types in cancer-related samples from the COSMIC database. Missense mutations account for 51.94% of the total mutation types, making it the primary type of gene mutation. B) Distribution of DDX3X mutations across different tissues based on the analysis of 628 positive samples out of 5,411 mutation samples from the COSMIC database. DDX3X mutations mainly occurred in lymphoid (164) and skin (115) tissues. C) Mapping of DDX3X mutation sites and amino acid sequences in cancer samples from the cBioPortal database (32 studies, 10,967 samples).
Figure 6.
DDX3X expression profiles in different cancers and its co-expression with G3BP1. A) Pan-cancer analysis of DDX3X mRNA expression using the TIMER database from TCGA. B) mRNA expression levels of DDX3X in different tissues analyzed using the GEPIA database. C) DDX3X is significantly co-expressed with G3BP1, an RNA-binding protein that forms stress granules. D) The presence of multiple RG/RGG sequences in G3BP1 protein, and amino acid disorder prediction using IUPRED.
Figure 6.
DDX3X expression profiles in different cancers and its co-expression with G3BP1. A) Pan-cancer analysis of DDX3X mRNA expression using the TIMER database from TCGA. B) mRNA expression levels of DDX3X in different tissues analyzed using the GEPIA database. C) DDX3X is significantly co-expressed with G3BP1, an RNA-binding protein that forms stress granules. D) The presence of multiple RG/RGG sequences in G3BP1 protein, and amino acid disorder prediction using IUPRED.
Figure 7.
Interaction between DDX3X and G3BP1 proteins. A) Fluorescence co-localization analysis reveals that green fluorescent protein-labeled DDX3X/DED1 co-localizes with red fluorescent protein-labeled G3BP1. B) Schematic representation of the proposed interaction mechanism between DDX3X and G3BP1. The two proteins may interact through their RG/RGG domains under situations of cellular stress, and subsequently bind to translation factors eIF4A/E/G to form stress granules. These stress granules associate with mRNA and regulate the translation of downstream, ultimately affecting cellular physiological functions.
Figure 7.
Interaction between DDX3X and G3BP1 proteins. A) Fluorescence co-localization analysis reveals that green fluorescent protein-labeled DDX3X/DED1 co-localizes with red fluorescent protein-labeled G3BP1. B) Schematic representation of the proposed interaction mechanism between DDX3X and G3BP1. The two proteins may interact through their RG/RGG domains under situations of cellular stress, and subsequently bind to translation factors eIF4A/E/G to form stress granules. These stress granules associate with mRNA and regulate the translation of downstream, ultimately affecting cellular physiological functions.
Table 1.
Mutations in arginine and glycine residues within RG/RGG motifs of DDX3X and their association with tissue carcinogenesis.
Table 1.
Mutations in arginine and glycine residues within RG/RGG motifs of DDX3X and their association with tissue carcinogenesis.
| Protein Mutation |
CDS Mutation |
Primary Tissue |
Histology |
| p.R79K |
c.236G>A |
Upper aerodigestive tract |
Carcinoma |
| p.G80= |
c.240G>A |
Skin |
Carcinoma |
| p.G80R |
c.238G>A |
Upper aerodigestive tract |
Carcinoma |
| p.R88C |
c.262C>T |
Liver |
Other |
| p.R88C |
c.262C>T |
Skin |
Malignant melanoma |
| p.G89R |
c.265G>A |
Upper aerodigestive tract |
Carcinoma |
| p.R93K |
c.278G>A |
Upper aerodigestive tract |
Carcinoma |
| p.R99C |
c.295C>T |
Stomach |
Carcinoma |
| p.R99H |
c.296G>A |
Stomach |
Carcinoma |
| p.R110C |
c.328C>T |
Endometrium |
Carcinoma |
| p.R110C |
c.328C>T |
Stomach |
Carcinoma |
| p.R110S |
c.328C>A |
Skin |
Malignant melanoma |
| p.R121C |
c.361C>T |
Pancreas |
Carcinoma |
| p.R121H |
c.362G>A |
Large intestine |
Carcinoma |
| p.R315C |
c.943C>T |
Stomach |
Carcinoma |
| p.R315H |
c.944G>A |
Endometrium |
Carcinoma |
| p.R315S |
c.943C>A |
Cervix |
Carcinoma |
| p.G316E |
c.947G>A |
Lung |
Carcinoma |
| p.G316V |
c.947G>T |
Breast |
Carcinoma |
| p.R333G |
c.997A>G |
Haematopoietic and lymphoid |
Lymphoid neoplasm |
| p.R333T |
c.998G>C |
Upper aerodigestive tract |
Carcinoma |
| p.R503K |
c.1508G>A |
Haematopoietic and lymphoid |
Lymphoid neoplasm |
| p.R503T |
c.1508G>C |
Cervix |
Carcinoma |
| p.R585P |
c.1754G>C |
Large intestine |
Carcinoma |
| p.R585P |
c.1754G>C |
Upper aerodigestive tract |
Carcinoma |
| p.R632I |
c.1895G>T |
Endometrium |
Carcinoma |
| p.R632K |
c.1895G>A |
Urinary tract |
Carcinoma |
| p.R632T |
c.1895G>C |
Lung |
Carcinoma |
| p.R632T |
c.1895G>C |
Skin |
Carcinoma |
Table 2.
Association between the absence of single RG/RGG repeat sequences in DDX3X and tumor occurrence in various tissues.
Table 2.
Association between the absence of single RG/RGG repeat sequences in DDX3X and tumor occurrence in various tissues.
| Protein Mutation |
Cancer Type |
| R99C |
Diffuse Type Stomach Adenocarcinoma |
| R99H |
Papillary Stomach Adenocarcinoma |
| R110C |
Uterine Endometrioid Carcinoma |
| R110C |
Stomach Adenocarcinoma |
| R110S |
Cutaneous Melanoma |
| R121H |
Colon Adenocarcinoma |
| R315H |
Uterine Endometrioid Carcinoma |
| R333T |
Head and Neck Squamous Cell Carcinoma |
| R503T |
Cervical Squamous Cell Carcinoma |
| R585C |
Uterine Endometrioid Carcinoma |
| R632I |
Uterine Endometrioid Carcinoma |
| R632K |
Bladder Urothelial Carcinoma |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).