Submitted:
02 May 2024
Posted:
02 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Virus Inoculations
2.2. Isolation of RNA and Reverse Transcription Quantitative PCR (RT-qPCR)
2.3. Determination of Relative Viral RNA Titer
3. Results
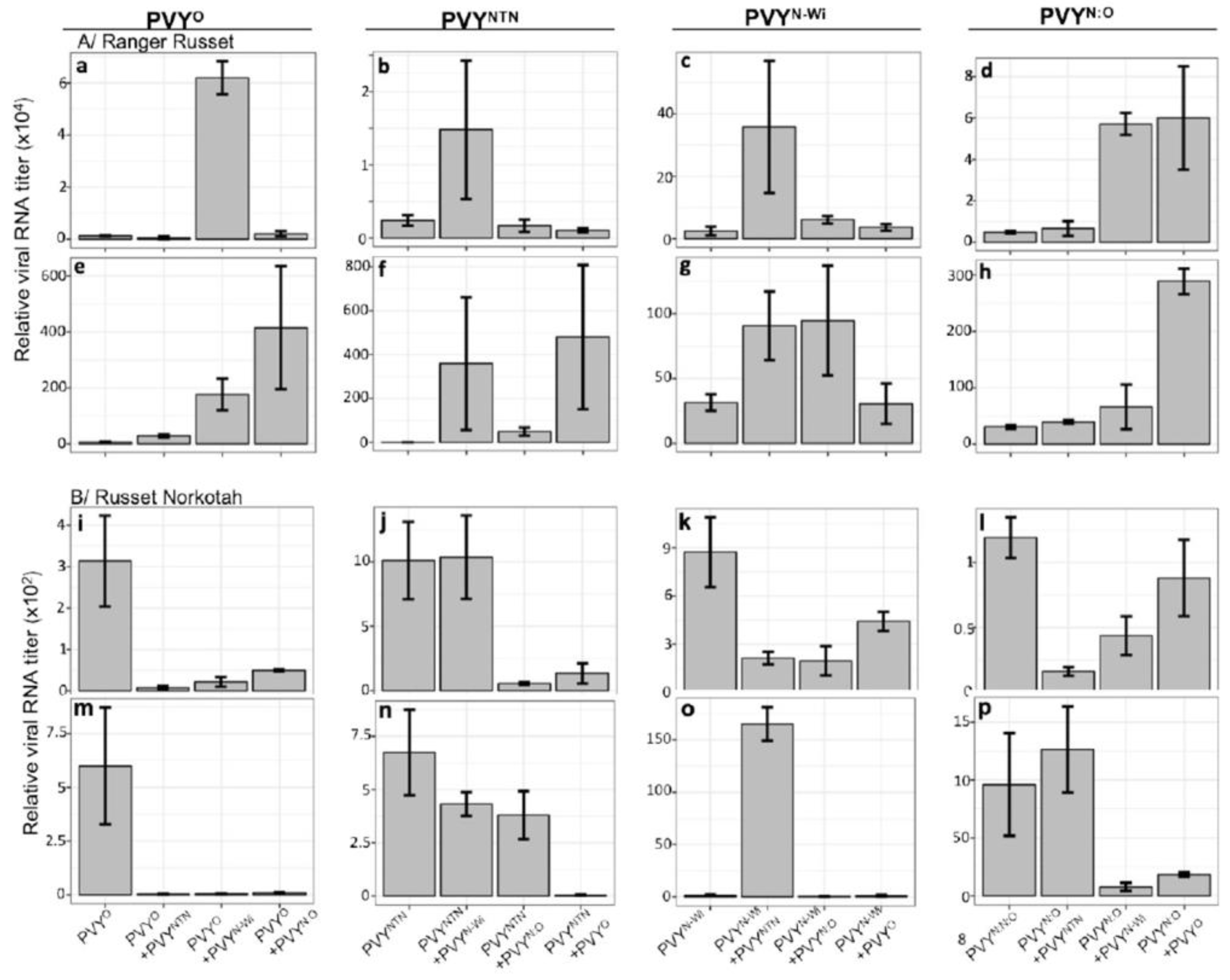
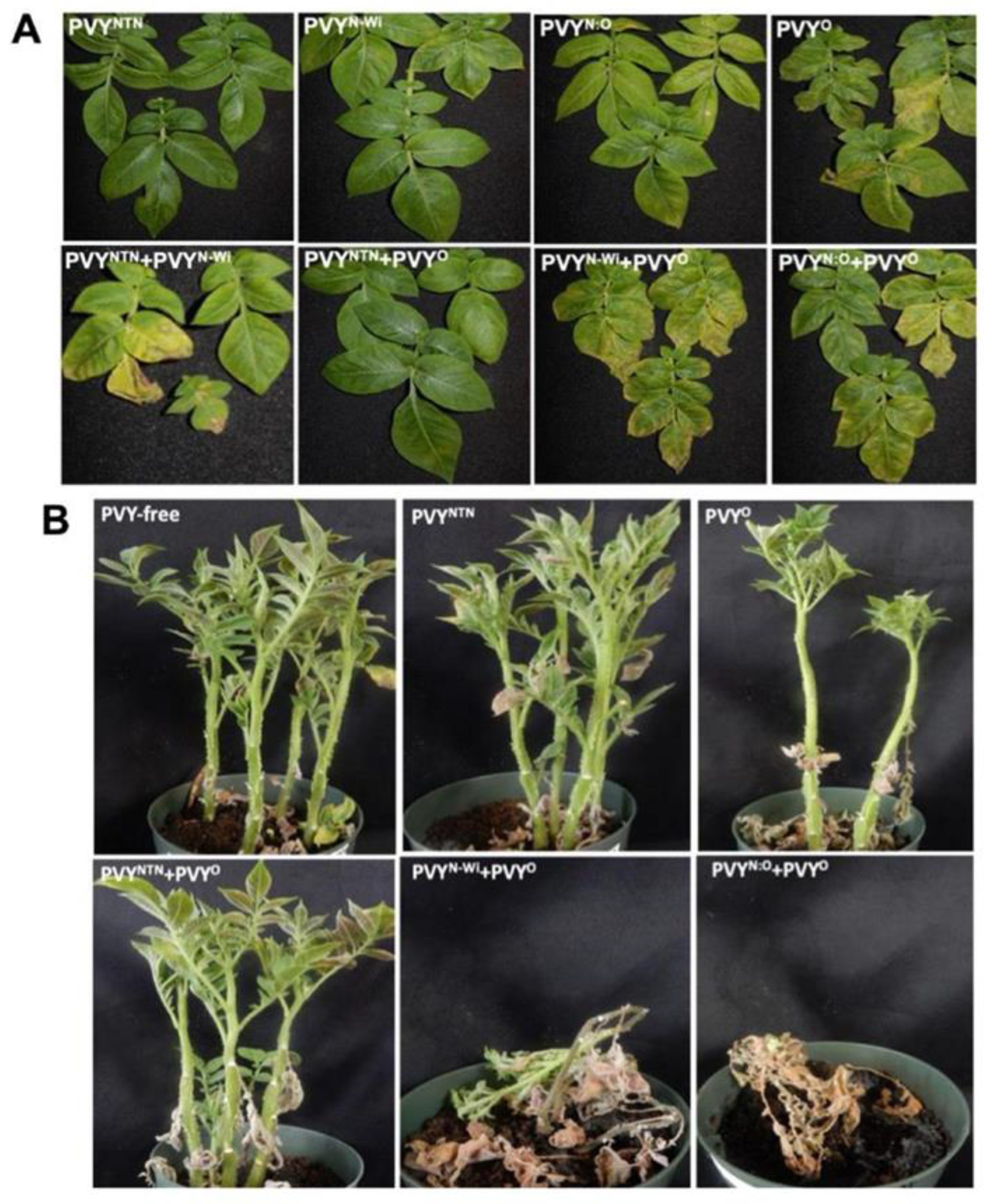
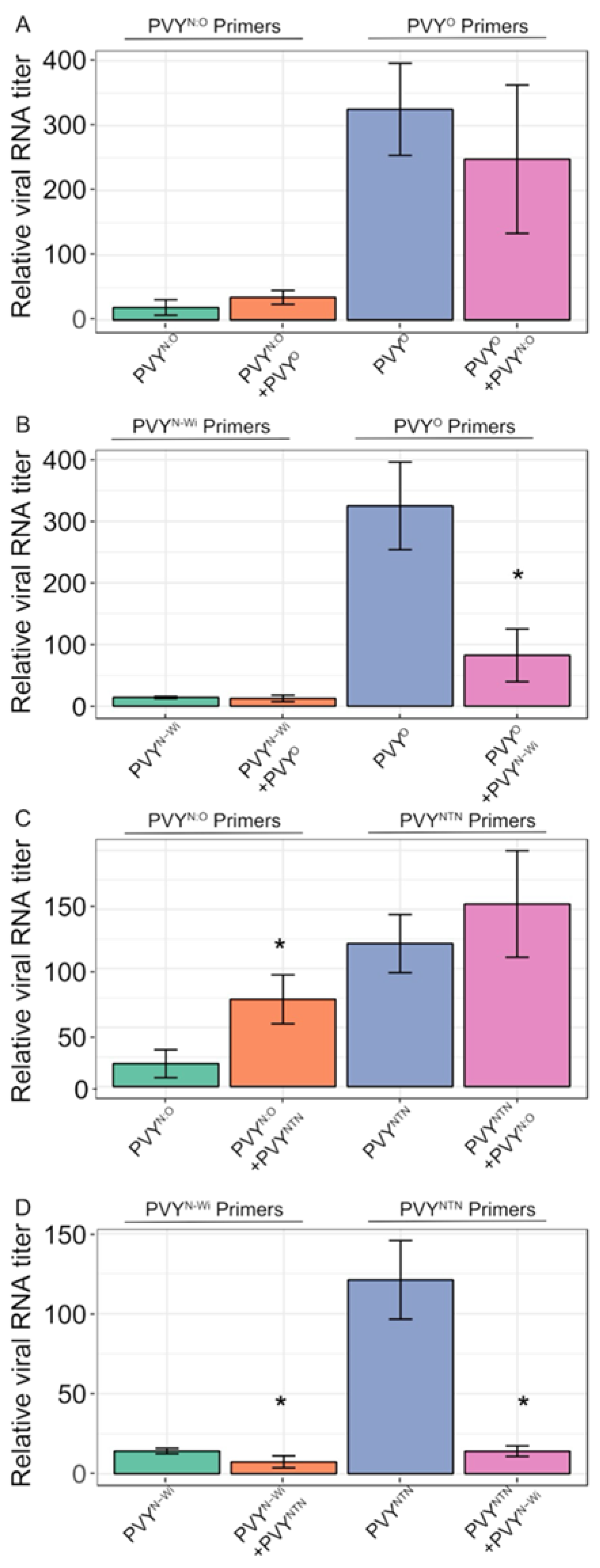
3.1. Antagonistic and Synergistic Interactions between PVY Strains in Potato
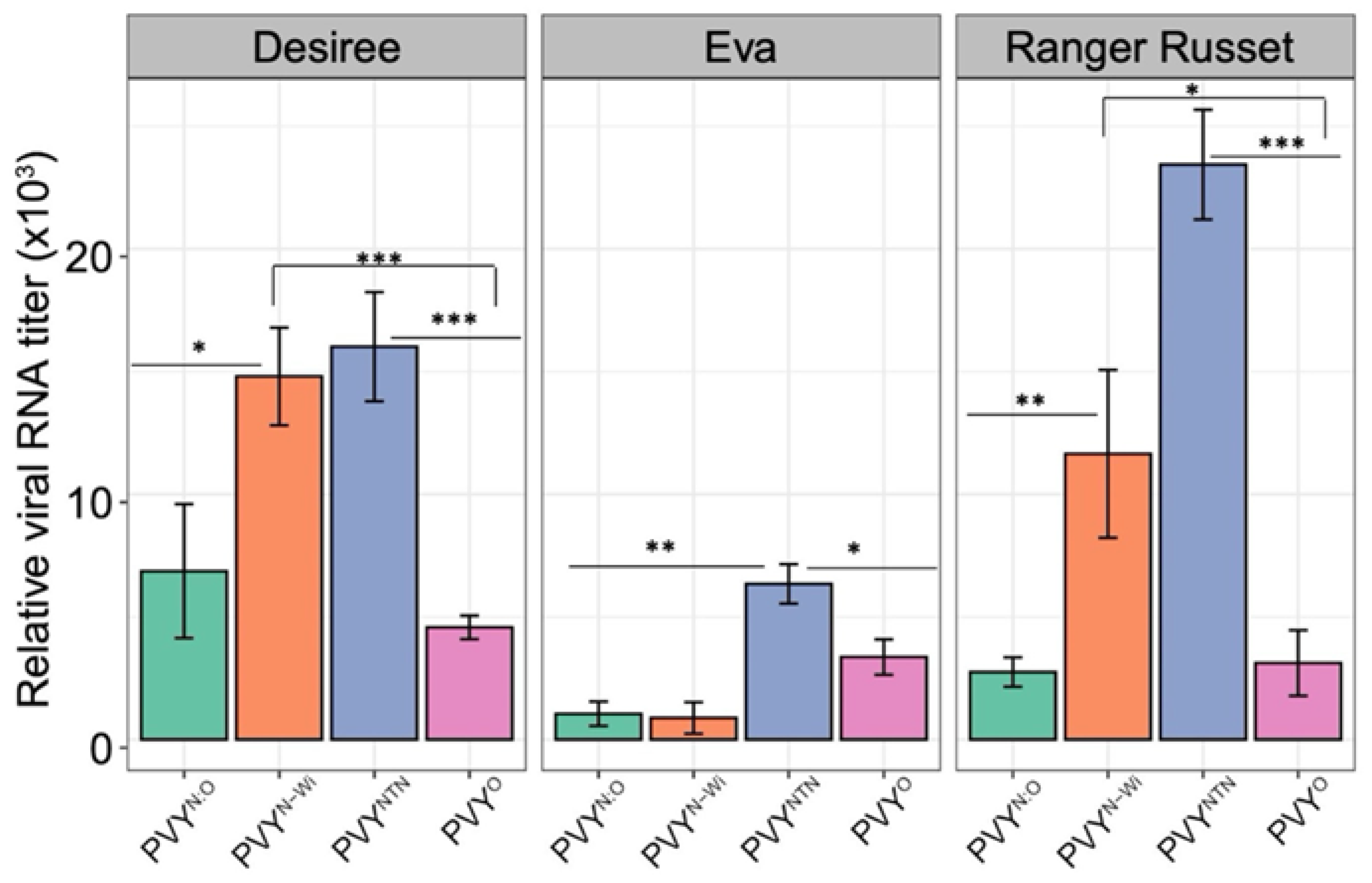
3.2. RT-qPCR Quantification of Viral RNA
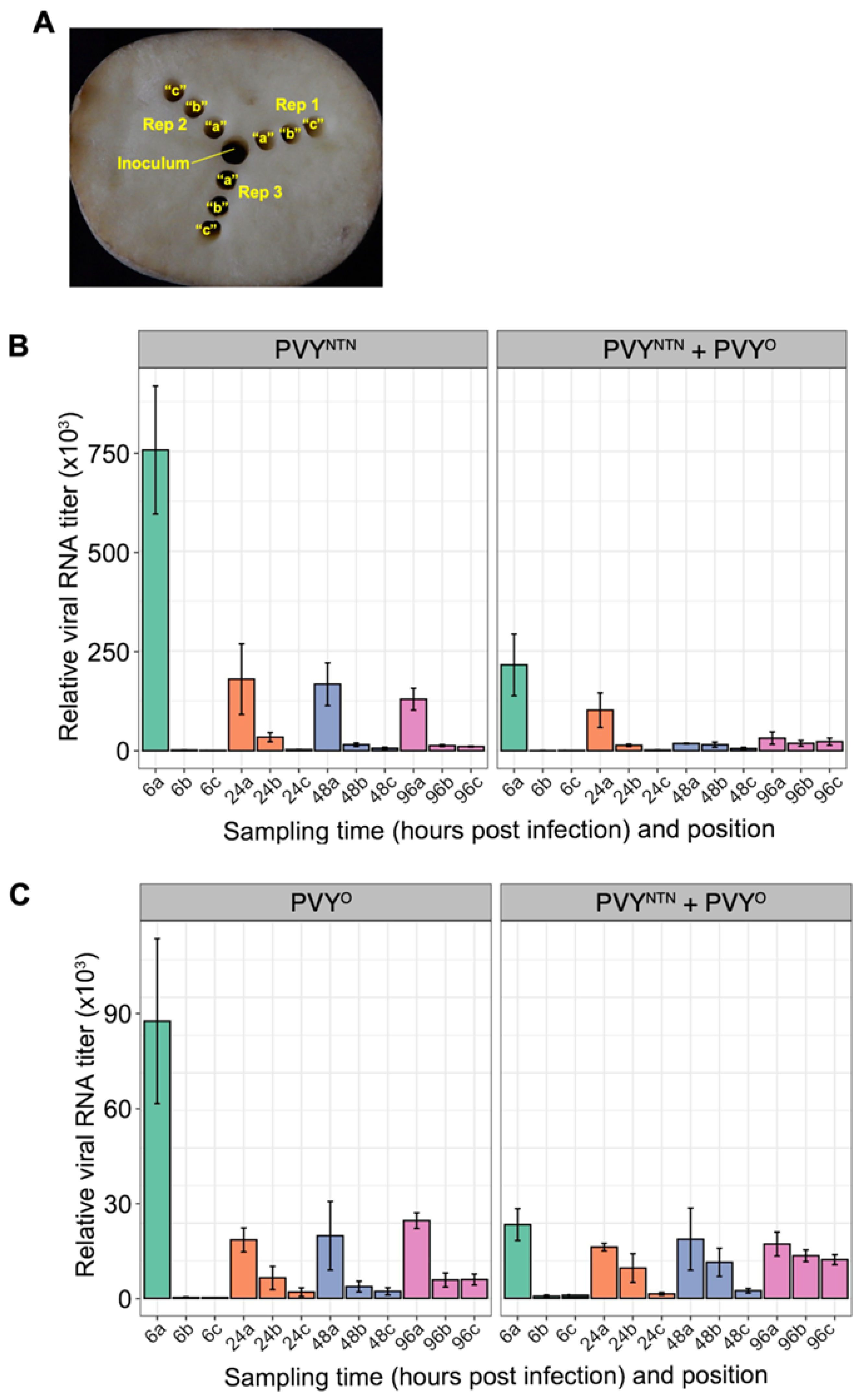
3.3. Replication and Cell-To-Cell Movement of PVY Strains
3.4. Interactions between PVY Strains in Potato Tubers
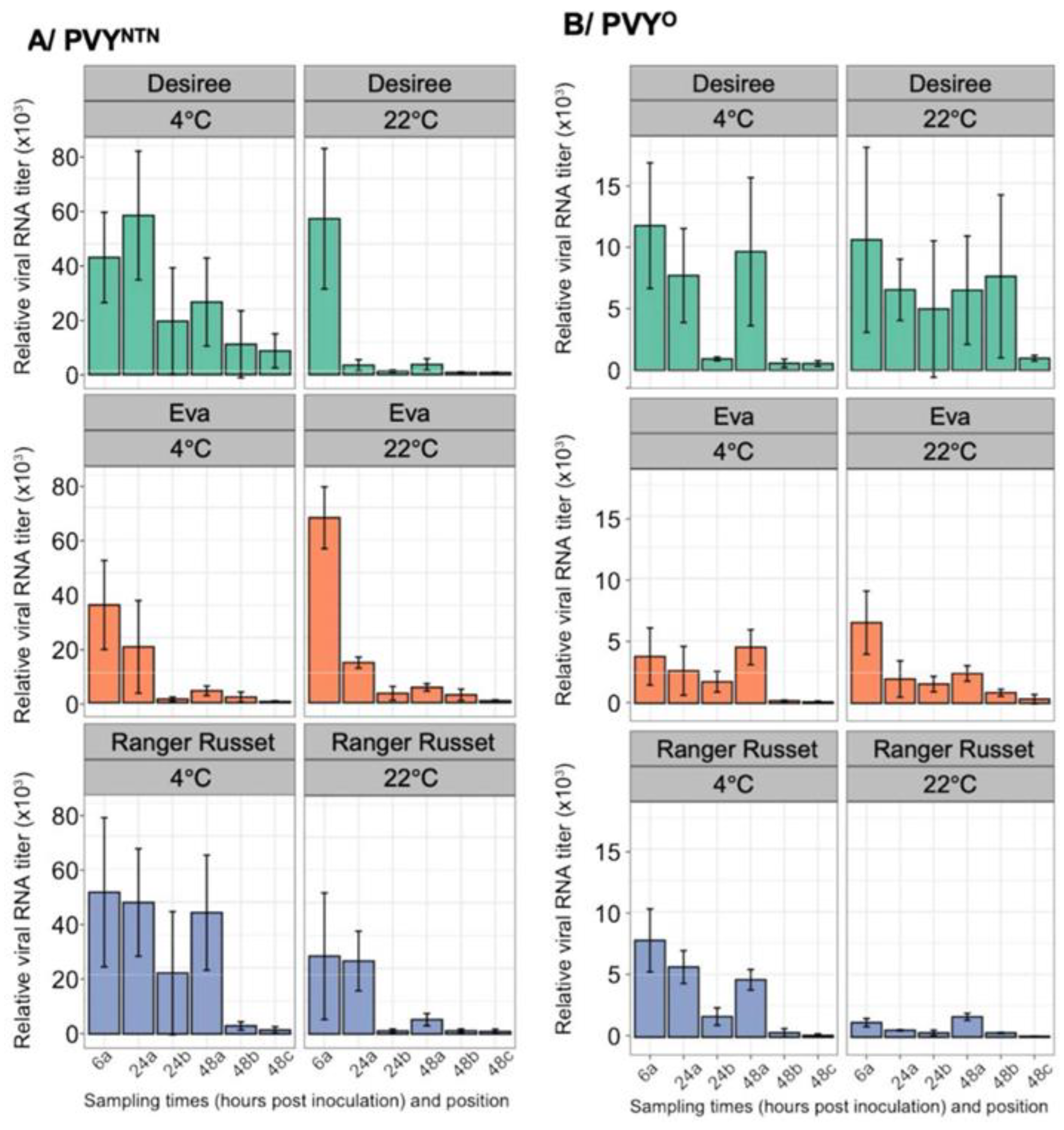
3.5. Effect of Temperature on PVY Cell-To-Cell Movement in Tubers
3.6. Evidence of Virus Replication in Potato Tubers
4. Discussion
Supplementary Materials
Funding
Acknowledgements
References
- Scholthof, K.-B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 Plant Viruses in Molecular Plant Pathology. Molecular Plant Pathology 2011, 12, 938–954. [CrossRef]
- Karasev, A. V.; Gray, S.M. Continuous and Emerging Challenges of Potato Virus Y in Potato. Annual Review of Phytopathology 2013, 51, 571–586.
- Gray, S.; De Boer, S.; Lorenzen, J.; Karasev, A.; Whitworth, J.; Nolte, P.; Singh, R.; Boucher, A.; Xu, H. Potato Virus Y: An Evolving Concern for Potato Crops in the United States and Canada. Plant Dis 2010, 94, 1384–1397. [CrossRef]
- Karasev, A.V.; Hu, X.; Brown, C.J.; Kerlan, C.; Nikolaeva, O.V.; Crosslin, J.M.; Gray, S.M. Genetic Diversity of the Ordinary Strain of Potato Virus Y (PVY) and Origin of Recombinant PVY Strains. Phytopathology 2011, 101, 778–785. [CrossRef]
- Green, K.J.; Brown, C.J.; Gray, S.M.; Karasev, A.V. Phylogenetic Study of Recombinant Strains of Potato Virus Y. Virology 2017, 507, 40–52. [CrossRef]
- MacKenzie, T.D.B.; Nie, X.; Bisht, V.; Singh, M. Proliferation of Recombinant PVY Strains in Two Potato-Producing Regions of Canada, and Symptom Expression in 30 Important Potato Varieties with Different PVY Strains. Plant Dis 2019, 103, 2221–2230. [CrossRef]
- Hu, X.; Karasev, A.; … C.B.-J. of G.; 2009, undefined Sequence Characteristics of Potato Virus Y Recombinants. microbiologyresearch.org 2009, 90, 3033–3041. [CrossRef]
- Rowley, J.S.; Gray, S.M.; Karasev, A. V. Screening Potato Cultivars for New Sources of Resistance to Potato Virus Y. American Journal of Potato Research 2014 92:1 2015, 92, 38–48. [CrossRef]
- Carroll, J.E.; Smith, D.M.; Gray, S.M. Preferential Acquisition and Inoculation of PVYNTN over PVYO in Potato by the Green Peach Aphid Myzus Persicae (Sulzer). J Gen Virol 2016, 97, 797–802. [CrossRef]
- Boonham, N.; Walsh, K.; Preston, S.; North, J.; Smith, P.; Barker, I. The Detection of Tuber Necrotic Isolates of Potato Virus Y, and the Accurate Discrimination of PVY(O), PVY(N) and PVY(C) Strains Using RT-PCR. J Virol Methods 2002, 102, 103–112. [CrossRef]
- Nie, B.; Singh, M.; Murphy, A.; Sullivan, A.; Xie, C.; Nie, X. Response of Potato Cultivars to Five Isolates Belonging to Four Strains of Potato Virus Y. Plant Dis 2012, 96, 1422–1429. [CrossRef]
- Gebhardt, C.; Valkonen, J.P. Organization of Genes Controlling Disease Resistance in the Potato Genome. Annu Rev Phytopathol 2001, 39, 79–102. [CrossRef]
- Mestre, P.; Brigneti, G.; Baulcombe, D.C. An Ry-Mediated Resistance Response in Potato Requires the Intact Active Site of the NIa Proteinase from Potato Virus Y. Plant J. 2000, 23, 653–661.
- Dupuis, B.; Bragard, C.; Schumpp, O. Resistance of Potato Cultivars as a Determinant Factor of Potato Virus Y (PVY) Epidemiology. Potato Res. 2019, 62, 123–138. [CrossRef]
- Funke, C.N.; Nikolaeva, O.V.; Green, K.J.; Tran, L.T.; Chikh-Ali, M.; Quintero-Ferrer, A.; Cating, R.A.; Frost, K.E.; Hamm, P.B.; Olsen, N.; et al. Strain-Specific Resistance to Potato Virus Y (PVY) in Potato and Its Effect on the Relative Abundance of PVY Strains in Commercial Potato Fields. Plant Dis 2017, 101, 20–28. [CrossRef]
- Glais, L.; Tribodet, M.; Kerlan, C. Genomic Variability in Potato Potyvirus Y (PVY): Evidence That PVY(N)W and PVY(NTN) Variants Are Single to Multiple Recombinants between PVY(O) and PVY(N) Isolates. Arch Virol 2002, 147, 363–378. [CrossRef]
- Hu, X.; Karasev, A.V.; Brown, C.J.; Lorenzen, J.H. Sequence Characteristics of Potato Virus Y Recombinants. J Gen Virol 2009, 90, 3033–3041. [CrossRef]
- Lorenzen, J.; Nolte, P.; Martin, D.; Pasche, J.S.; Gudmestad, N.C. NE-11 Represents a New Strain Variant Class of Potato Virus Y. Arch Virol 2008, 153, 517–525. [CrossRef]
- Ogawa, T.; Nakagawa, A.; Hataya, T.; Ohshima, K. The Genetic Structure of Populations of Potato Virus Y in Japan; Based on the Analysis of 20 Full Genomic Sequences. Journal of Phytopathology 2012, 160, 661–673. [CrossRef]
- Jones, R. a. C. Strain Group Specific and Virus Specific Hypersensitive Reactions to Infection with Potyviruses in Potato Cultivars. Annals of Applied Biology 1990, 117, 93–105. [CrossRef]
- Mondal, S.; Lin, Y.-H.; Carroll, J.E.; Wenninger, E.J.; Bosque-Pérez, N.A.; Whitworth, J.L.; Hutchinson, P.; Eigenbrode, S.; Gray, S.M. Potato Virus Y Transmission Efficiency from Potato Infected with Single or Multiple Virus Strains. Phytopathology 2017, 107, 491–498. [CrossRef]
- Hühnlein, A.; Drechsler, N.; Steinbach, P.; Thieme, T.; Schubert, J. Comparison of Three Methods for the Detection of Potato Virus Y in Seed Potato Certification. Journal of Plant Diseases and Protection 2013, 120, 57–69.
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A Practical Approach to RT-qPCR-Publishing Data That Conform to the MIQE Guidelines. Methods 2010, 50, S1-5. [CrossRef]
- Wilhelm, J.; Pingoud, A.; Hahn, M. Validation of an Algorithm for Automatic Quantification of Nucleic Acid Copy Numbers by Real-Time Polymerase Chain Reaction. Analytical Biochemistry 2003, 317, 218–225. [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [CrossRef]
- Karasev, A.V.; Gray, S.M. Continuous and Emerging Challenges of Potato Virus Y in Potato. Annu Rev Phytopathol 2013, 51, 571–586. [CrossRef]
- Jones, R.A.C.; Vincent, S.J. Strain-Specific Hypersensitive and Extreme Resistance Phenotypes Elicited by Potato Virus Y Among 39 Potato Cultivars Released in Three World Regions Over a 117-Year Period. Plant Disease 2018, 102, 185–196. [CrossRef]
- Caplan, J.L.; Mamillapalli, P.; Burch-Smith, T.M.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplastic Protein NRIP1 Mediates Innate Immune Receptor Recognition of a Viral Effector. Cell 2008, 132, 449–462. [CrossRef]
- Tian, Y.-P.; Valkonen, J.P.T. Recombination of Strain O Segments to HCpro-Encoding Sequence of Strain N of Potato Virus Y Modulates Necrosis Induced in Tobacco and in Potatoes Carrying Resistance Genes Ny or Nc. Molecular Plant Pathology 2015, 16, 735–747. [CrossRef]
- Jin, H.; Liu, Y.; Yang, K.-Y.; Kim, C.Y.; Baker, B.; Zhang, S. Function of a Mitogen-Activated Protein Kinase Pathway in N Gene-Mediated Resistance in Tobacco. Plant J 2003, 33, 719–731. [CrossRef]
- Komatsu, K.; Hashimoto, M.; Ozeki, J.; Yamaji, Y.; Maejima, K.; Senshu, H.; Himeno, M.; Okano, Y.; Kagiwada, S.; Namba, S. Viral-Induced Systemic Necrosis in Plants Involves Both Programmed Cell Death and the Inhibition of Viral Multiplication, Which Are Regulated by Independent Pathways. Mol Plant Microbe Interact 2010, 23, 283–293. [CrossRef]
- Valkonen, J.P.; Rokka, V.M.; Watanabe, K.N. Examination of the Leaf-Drop Symptom of Virus-Infected Potato Using Anther Culture-Derived Haploids. Phytopathology 1998, 88, 1073–1077. [CrossRef]
- Moury, B.; Caromel, B.; Johansen, E.; Simon, V.; Chauvin, L.; Jacquot, E.; Kerlan, C.; Lefebvre, V. The Helper Component Proteinase Cistron of Potato Virus Y Induces Hypersensitivity and Resistance in Potato Genotypes Carrying Dominant Resistance Genes on Chromosome IV. Mol. Plant Microbe Interact. 2011, 24, 787–797. [CrossRef]
- Rowley, J.S.; Gray, S.M.; Karasev, A.V. Screening Potato Cultivars for New Sources of Resistance to Potato Virus Y. Am. J. Potato Res. 2015, 92, 38–48. [CrossRef]
- Plaisted, R.L.; Halseth, D.E.; Brodie, B.B.; Slack, S.A.; Sieczka, J.B.; Christ, B.J.; Paddock, K.M.; Peck, M.W. Eva:A Midseason Golden Nematode- and Virus-Resistant Variety for Use as Tablestock or Chipstock. Am. J. Pot Res 2001, 78, 65–68. [CrossRef]
- Nazarian-Firouzabadi, F.; Visser, R.G.F. Potato Starch Synthases: Functions and Relationships. Biochemistry and Biophysics Reports 2017, 10, 7–16. [CrossRef]
- Kumar, G.N.M.; Iyer, S.; Knowles, N.R. Extraction of RNA from Fresh, Frozen, and Lyophilized Tuber and Root Tissues. J Agric Food Chem 2007, 55, 1674–1678. [CrossRef]
- Vasanthan, T.; Bergthaller, W.; Driedger, D.; Yeung, J.; Sporns, P. Starch from Alberta Potatoes: Wet-Isolation and Some Physicochemical Properties. Food Research International 1999, 32, 355–365. [CrossRef]
- Wischmann, null; Hamborg Nielsen T, null; Lindberg Moller B, null In Vitro Biosynthesis of Phosphorylated Starch in Intact Potato Amyloplasts. Plant Physiol 1999, 119, 455–462. [CrossRef]
- Mondal, S.; Ghanim, M.; Roberts, A.; Gray, S.M. Different Potato Virus Y Strains Frequently Co-Localize in Single Epidermal Leaf Cells and in the Aphid Stylet. J Gen Virol 2021, 102. [CrossRef]
- Singh, M.; Singh, R.P. Host Dependent Cross-Protection between PVYN, PVY°, and PVA in Potato Cultivars and Solarium Brachycarpum. Canadian Journal of Plant Pathology 1995, 17, 82–86. [CrossRef]
- Szajko, K.; Strzelczyk-Żyta, D.; Marczewski, W. Ny-1 and Ny-2 Genes Conferring Hypersensitive Response to Potato Virus Y (PVY) in Cultivated Potatoes: Mapping and Marker-Assisted Selection Validation for PVY Resistance in Potato Breeding. Mol Breed 2014, 34, 267–271. [CrossRef]
- Syller, J. Facilitative and Antagonistic Interactions between Plant Viruses in Mixed Infections. Mol Plant Pathol 2012, 13, 204–216. [CrossRef]
- Syller, J.; Grupa, A. Antagonistic Within-Host Interactions between Plant Viruses: Molecular Basis and Impact on Viral and Host Fitness. Mol Plant Pathol 2016, 17, 769–782. [CrossRef]
- Tatineni, S.; Riethoven, J.-J.M.; Graybosch, R.A.; French, R.; Mitra, A. Dynamics of Small RNA Profiles of Virus and Host Origin in Wheat Cultivars Synergistically Infected by Wheat Streak Mosaic Virus and Triticum Mosaic Virus: Virus Infection Caused a Drastic Shift in the Endogenous Small RNA Profile. PLoS ONE 2014, 9, e111577. [CrossRef]
- Glais, L.; Faurez, F.; Tribodet, M.; Boulard, F.; Jacquot, E. The Amino Acid 419 in HC-Pro Is Involved in the Ability of PVY Isolate N605 to Induce Necrotic Symptoms on Potato Tubers. Virus Res. 2015, 208, 110–119. [CrossRef]
- Abrams, M. Environmental Grain, Organism Fitness, and Type Fitness. Entangled Life 2014, 127–151. [CrossRef]
- Metcalf, C.J.E.; Birger, R.B.; Funk, S.; Kouyos, R.D.; Lloyd-Smith, J.O.; Jansen, V. a. A. Five Challenges in Evolution and Infectious Diseases. Epidemics 2015, 10, 40–44. [CrossRef]
- Barbour, J.D.; Grant, R.M. The Role of Viral Fitness in HIV Pathogenesis. Curr HIV/AIDS Rep 2005, 2, 29–34. [CrossRef]
- Domingo, E. Mechanisms of Viral Emergence. Vet Res 2010, 41, 38. [CrossRef]
- Wargo, A.R.; Kurath, G. Viral Fitness: Definitions, Measurement, and Current Insights. Curr Opin Virol 2012, 2, 538–545. [CrossRef]
- Elena, S.F.; Lalić, J. Plant RNA Virus Fitness Predictability: Contribution of Genetic and Environmental Factors. Plant Pathology 2013, 62, 10–18. [CrossRef]
- Milling, A.; Meng, F.; Denny, T.P.; Allen, C. Interactions with Hosts at Cool Temperatures, Not Cold Tolerance, Explain the Unique Epidemiology of Ralstonia Solanacearum Race 3 Biovar 2. Phytopathology 2009, 99, 1127–1134. [CrossRef]
- Gachango, E.; Hanson, L.E.; Rojas, A.; Hao, J.J.; Kirk, W.W. Fusarium Spp. Causing Dry Rot of Seed Potato Tubers in Michigan and Their Sensitivity to Fungicides. Plant Dis 2012, 96, 1767–1774. [CrossRef]
- Fiers, M.; Edel-Hermann, V.; Chatot, C.; Le Hingrat, Y.; Alabouvette, C.; Steinberg, C. Potato Soil-Borne Diseases. A Review. Agron. Sustain. Dev. 2012, 32, 93–132. [CrossRef]
- Tiwari, R.K.; Kumar, R.; Sharma, S.; Sagar, V.; Aggarwal, R.; Naga, K.C.; Lal, M.K.; Chourasia, K.N.; Kumar, D.; Kumar, M. Potato Dry Rot Disease: Current Status, Pathogenomics and Management. 3 Biotech 2020, 10, 503. [CrossRef]
- Magdalena, G.; Dariusz, M. Losses during Storage of Potato Varieties in Relation to Weather Conditions during the Vegetation Period and Temperatures during Long-Term Storage. Am. J. Potato Res. 2018, 95, 130–138. [CrossRef]
- Fox, A.; Evans, F.; Browning, I. Direct Tuber Testing for Potato Y Potyvirus by Real-Time RT-PCR and ELISA: Reliable Options for Post-Harvest Testing?*. EPPO Bulletin 2005, 35, 93–97. [CrossRef]
| A/ RT-PCR | |||
|---|---|---|---|
| Target | Designation | Sequence (5’-3’) | Length (bp) |
| *PVYNTN |
NTN7350-F
NTN8266-R |
ACATCACCGATGAGCAGG
GTACATACCCTCGATTAGCA |
918 |
| *PVYO |
O1962-F
O2296-R |
TCAACATTCTATCCACCAAC
ACGTTTGAGTGTCATGGT |
335 |
| PVYN:O |
N:O1008-F
N:O1703-R |
GCACGTTCCAAGGTTACC
TCGCTTAGCATGATATTCCCT |
695 |
| **PVYN-Wi |
N-Wi_YN5-1780-F
N-Wi_YO3-2558-R |
TCCGAATGGGACAAGAAAACTTG
AGGCTCATCTAACAGCAACTGTC |
778 |
| B/ RT-qPCR*** | |||
| Target | Designation | Sequence (5’-3’) | Length (bp) |
| PVYNTN |
NTNq-6515-F NTNq-6631-R |
TCCGAGCTCCAGTGCAGAAT AAGTGCTGCCCGGTACATTG |
116 |
| PVYO |
Oq-4-F Oq-138-R |
CGCAAAAACACTCATAAAAGCTCA TGGTTGGAAGTGATGAAATTGCT |
134 |
| PVYN:O |
NOq-6444-F NOq-6574-R |
GGATATCATCCTCATCAAATGCCG TCGACGATGCATACTTCTCCTG |
130 |
| PVYN-Wi |
N-Wiq-35-F N-Wiq-156-R |
TTCCTTGCAATTCTCTTAAACGGT ACGAACCGAAACAGATTGTTGAC |
121 |
| All strains |
Uni-q-9426-F
Uni-q-9549-R |
GTGGCAGGGTGATTTCGTCA
AGAATCGCAACATCACCTAATCG |
123 |
| 1-alpha EF1α |
EF1-F EF1-R |
TGGAGGCACTCCCTGGTGACA
TGTGGCAGTCGAGCACTGGT |
194 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




