Submitted:
01 May 2024
Posted:
03 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Human Trabecular Meshwork Culture and Characterisation
2.2. TGFβ Treatment and RNA Extraction
2.3. Small RNA Sequencing and Data Analysis
2.4. Functional and Pathway Enrichment Analysis
2.5. miRNA RT-qPCR Validation of miRNA-Seq Data
3. Results
3.1. Descriptive Features of Small RNA-Seq Data
3.2. Differential Expression of TGFβ1-Responsive miRNAs in TM Cells
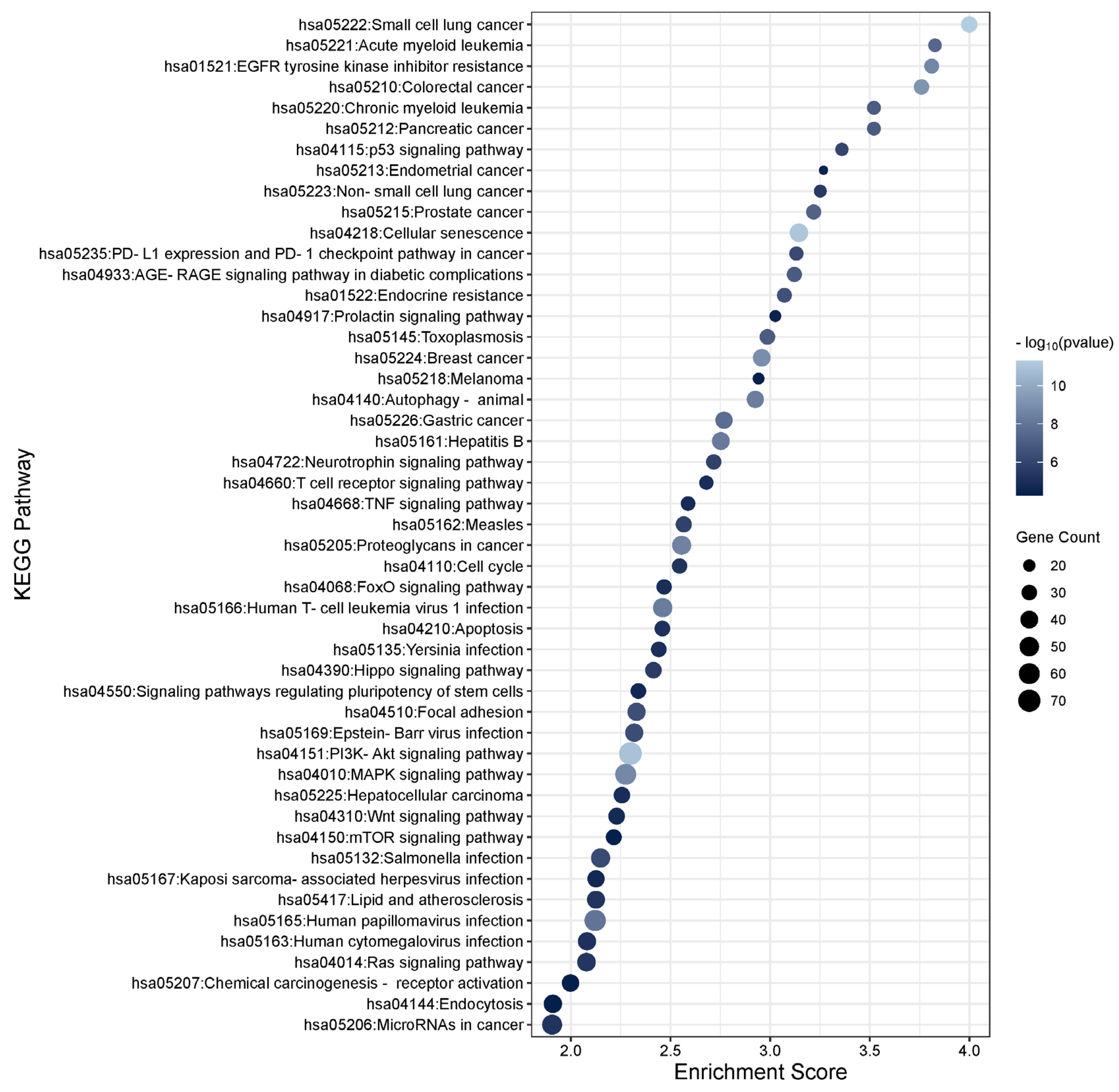
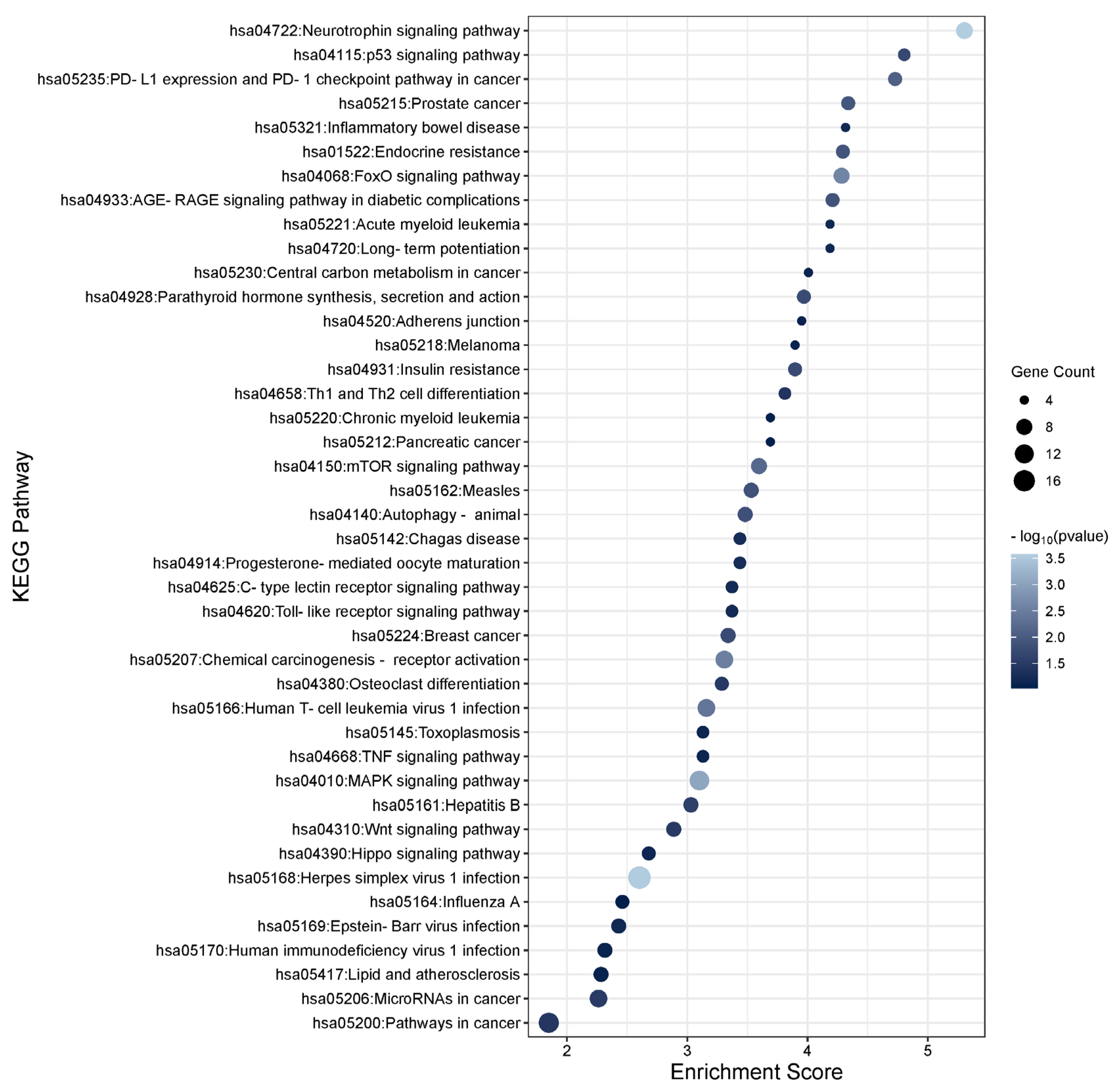
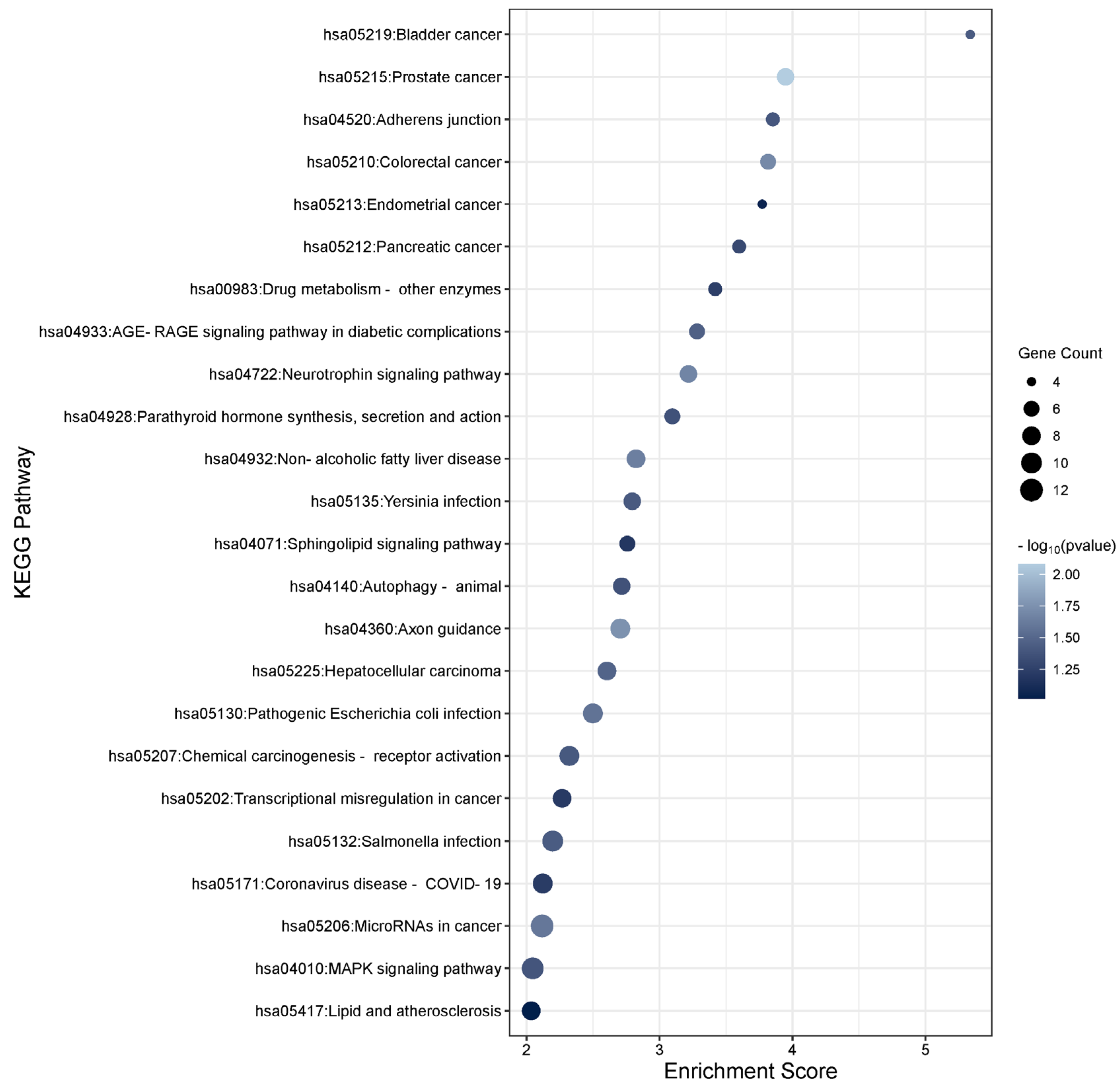
3.3. Functional Enrichment Analysis of the TGFβ1 Differentially Expressed Genes
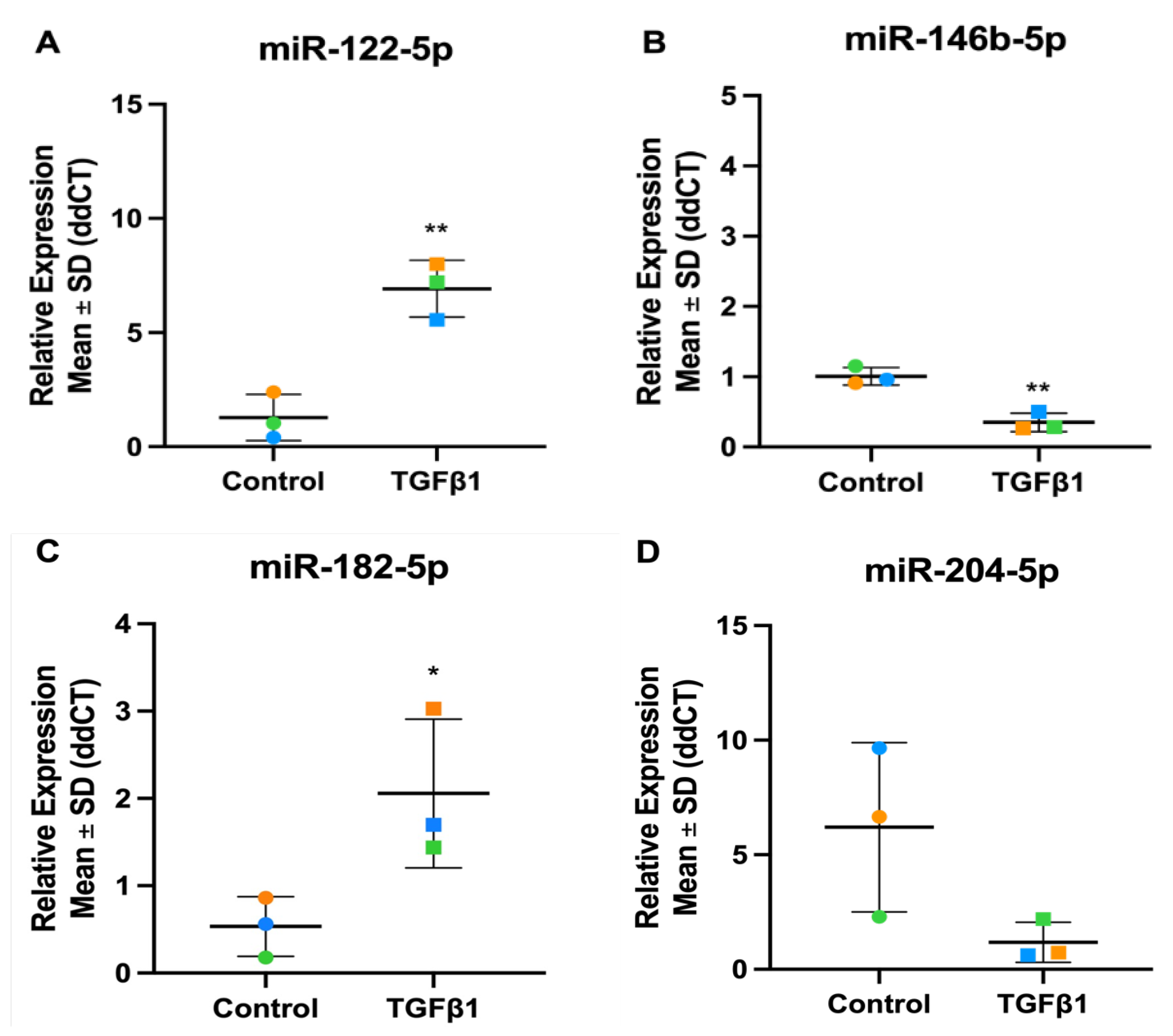
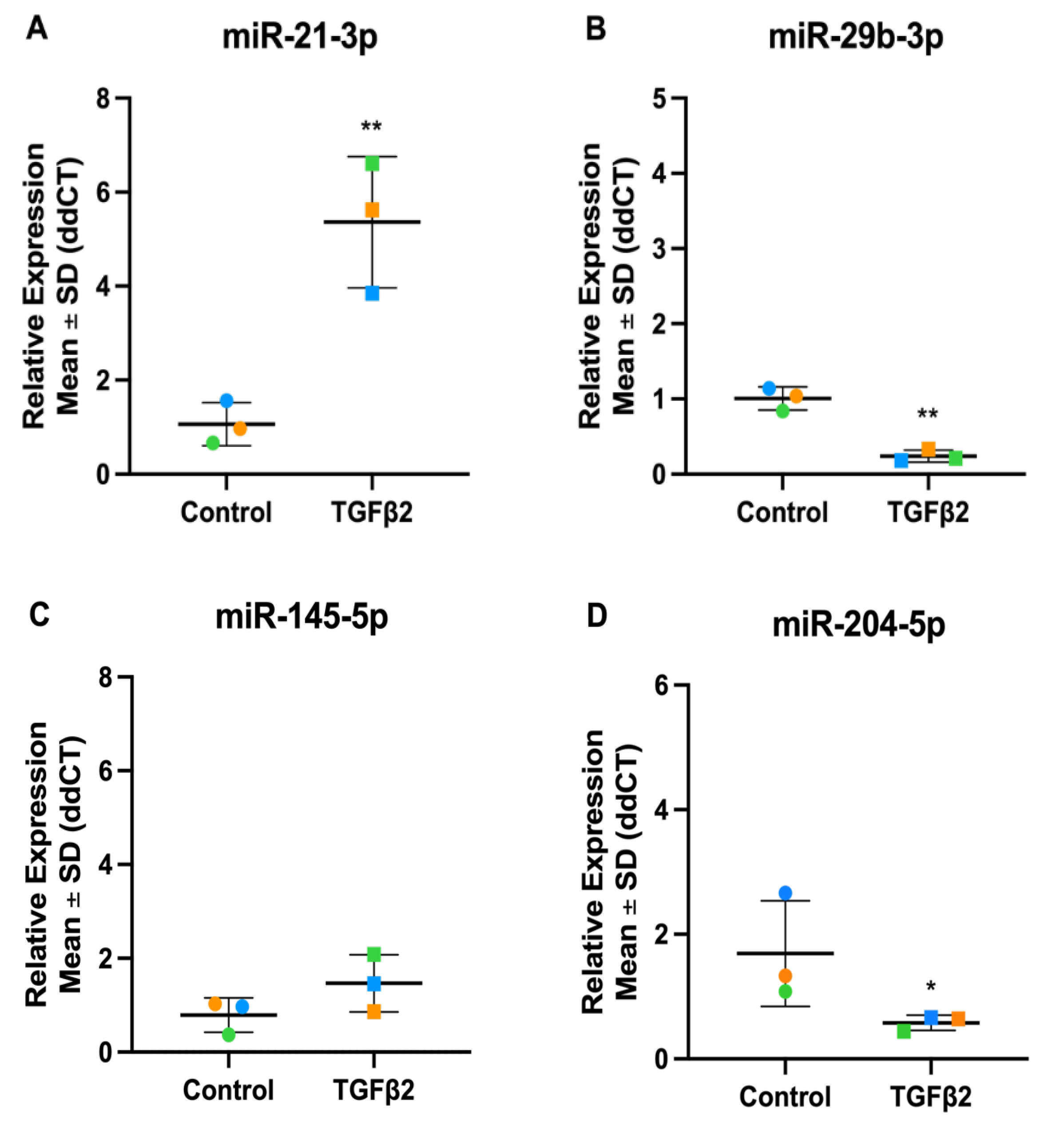
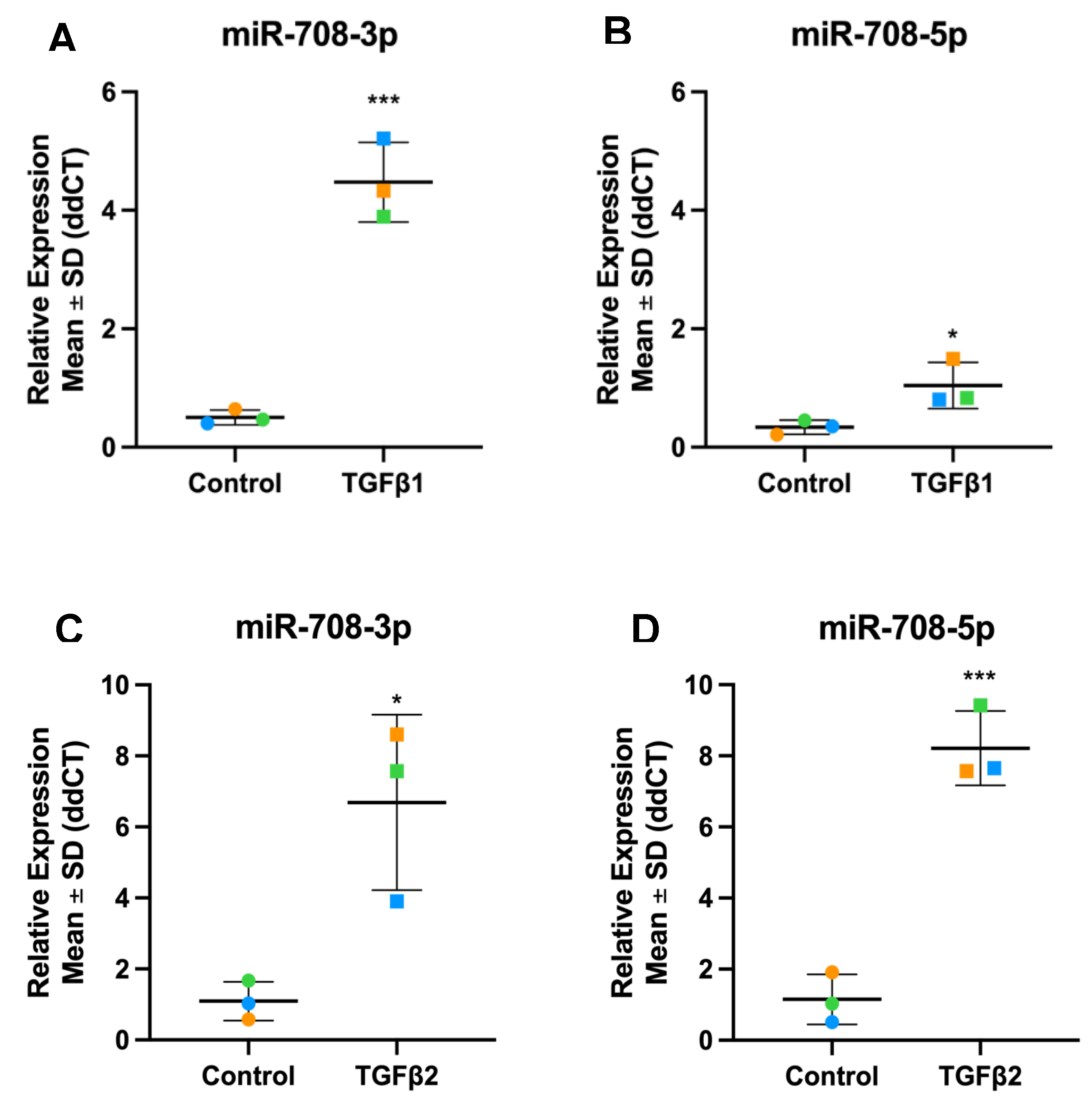
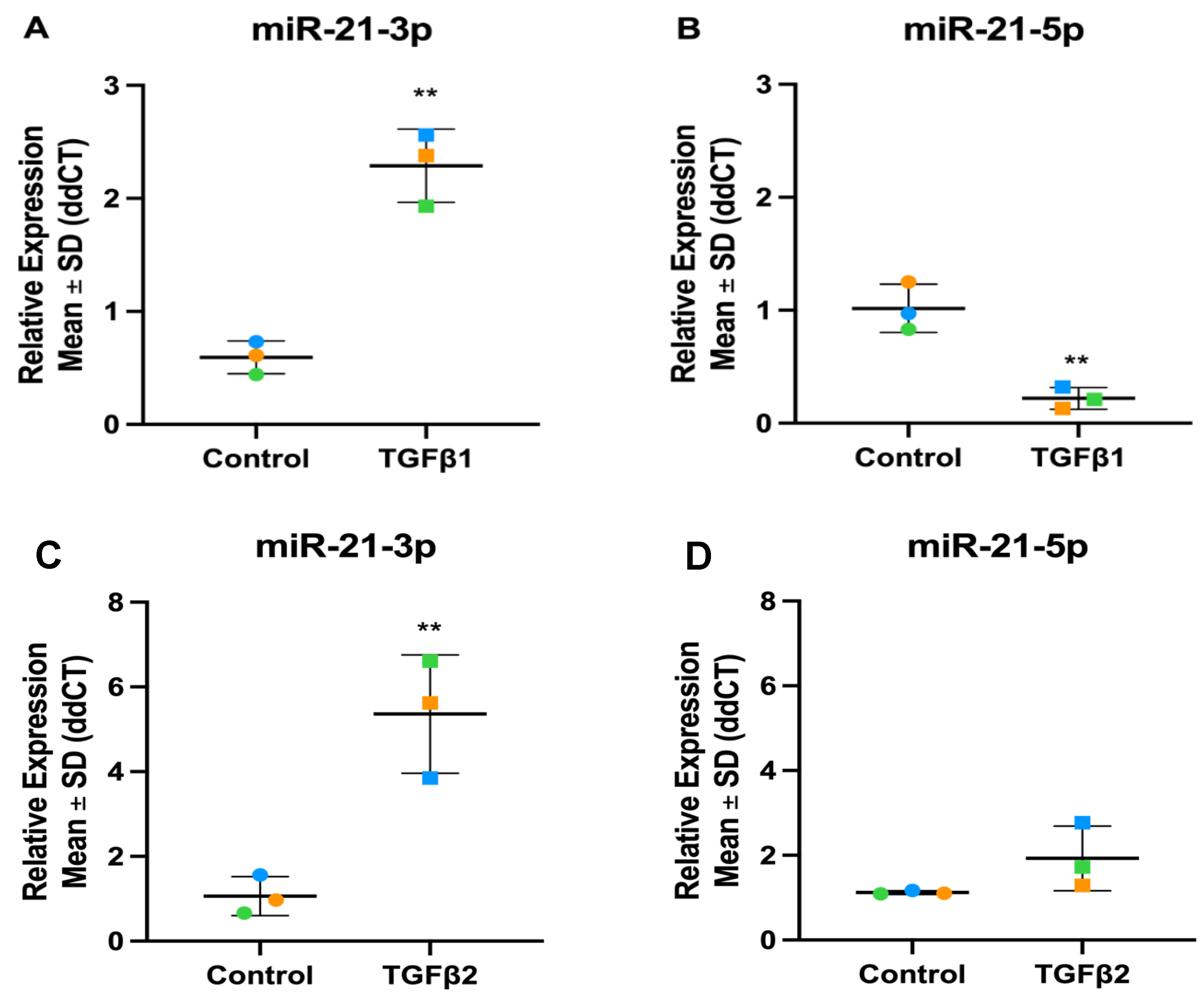
3.4. Validation of TGFβ1 Responsive DEmiRs by RT-qPCR
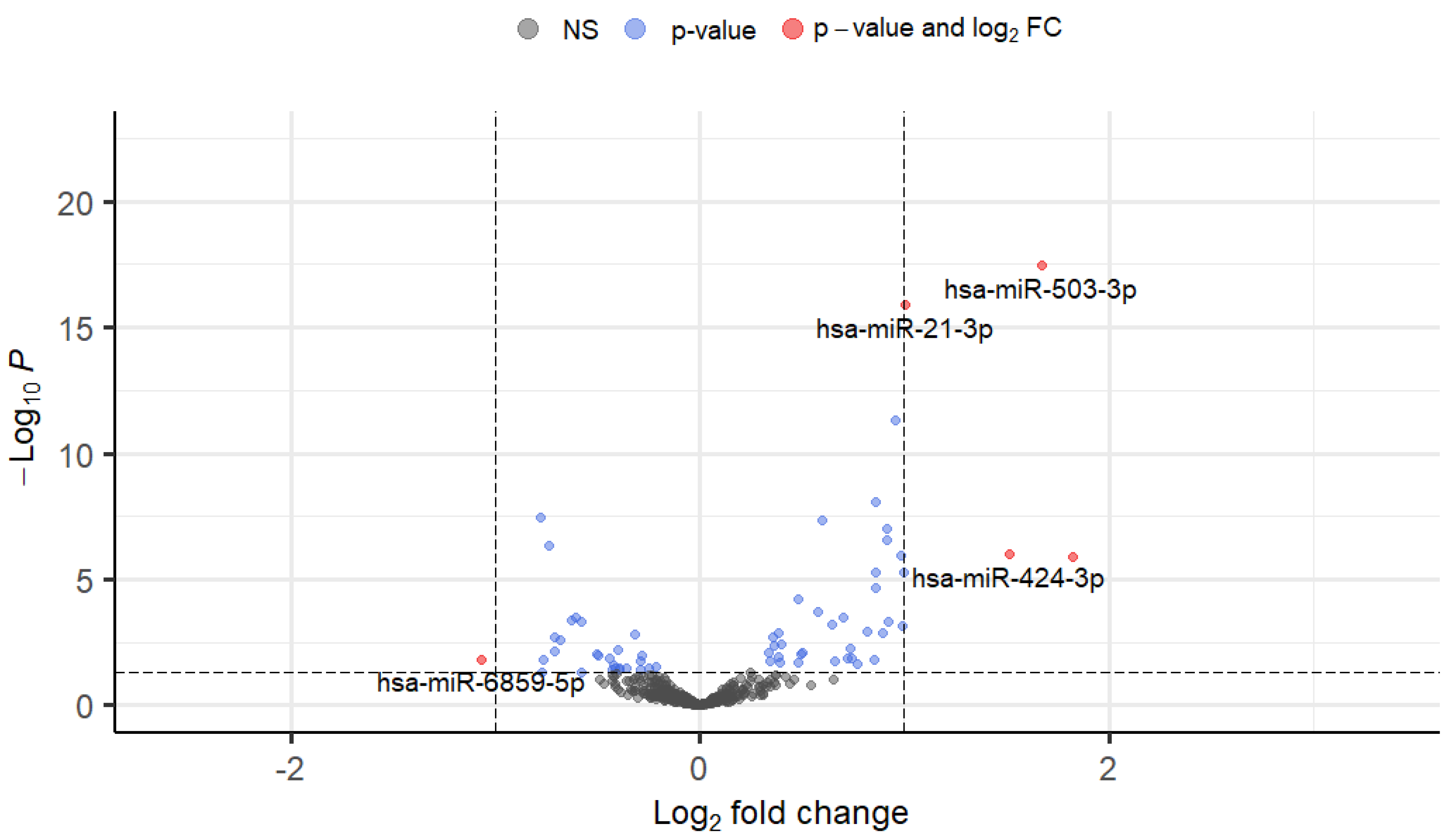
3.5. Differential Expression of TGFβ2-Responsive miRNAs in TM Cells
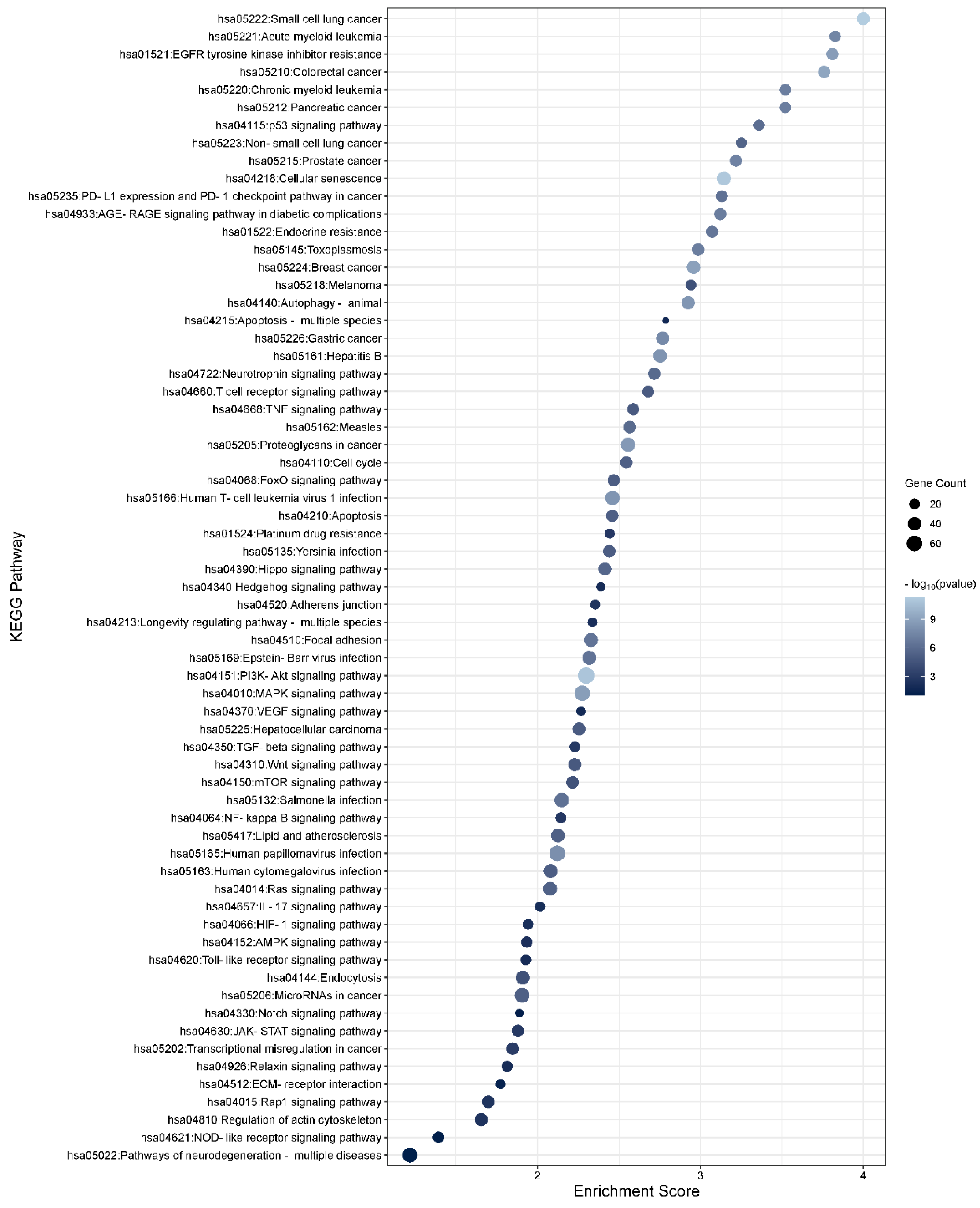
3.6. Functional Enrichment Analysis of the TGFβ2 Responsive Differentially Expressed Genes
3.7. Validation of TGFβ2 Responsive DEmiRs by RT-qPCR
3.8. Altered miRNA Expression Grouped by miRNA Families
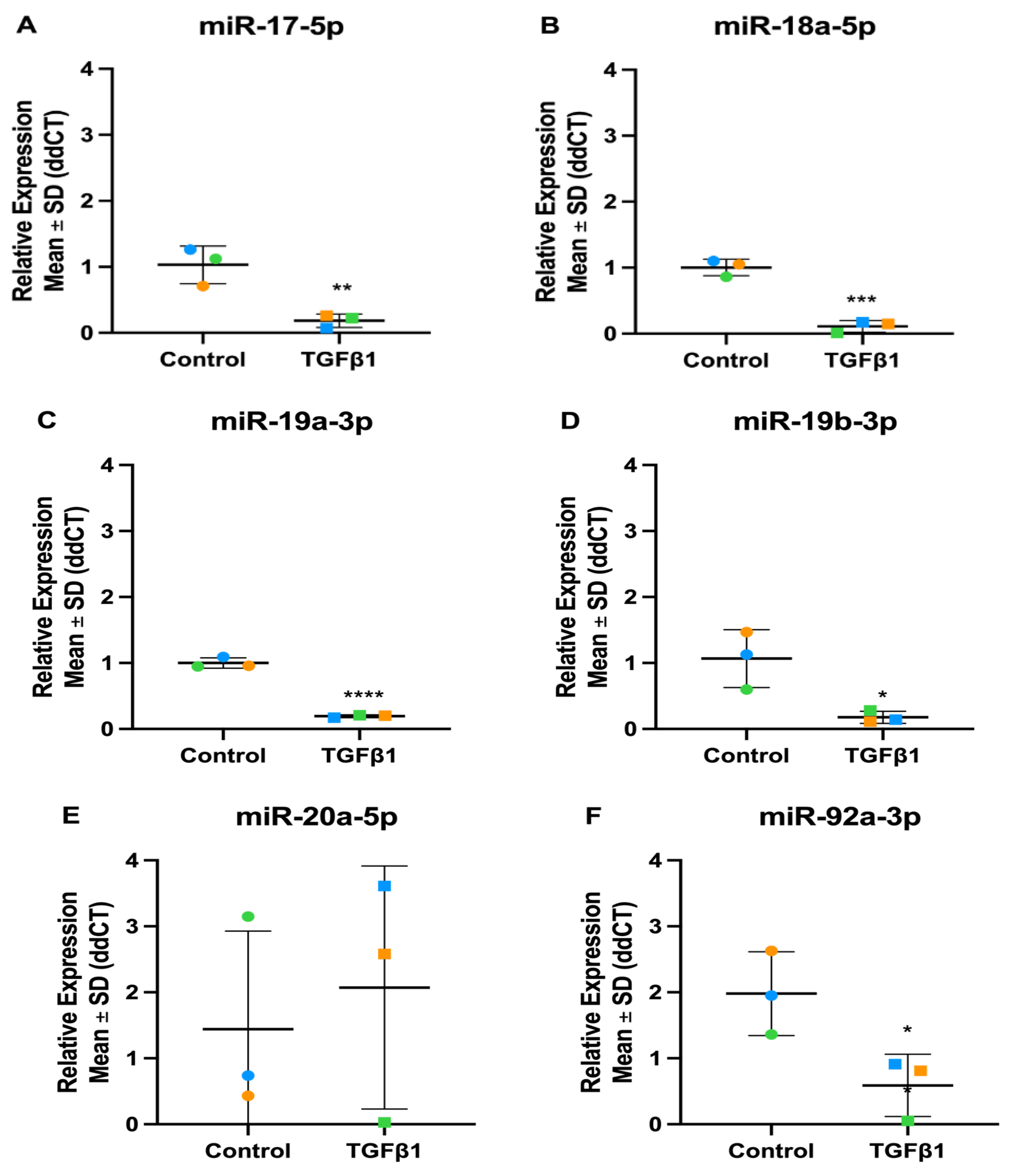
3.8.1. miR-17-92 Family
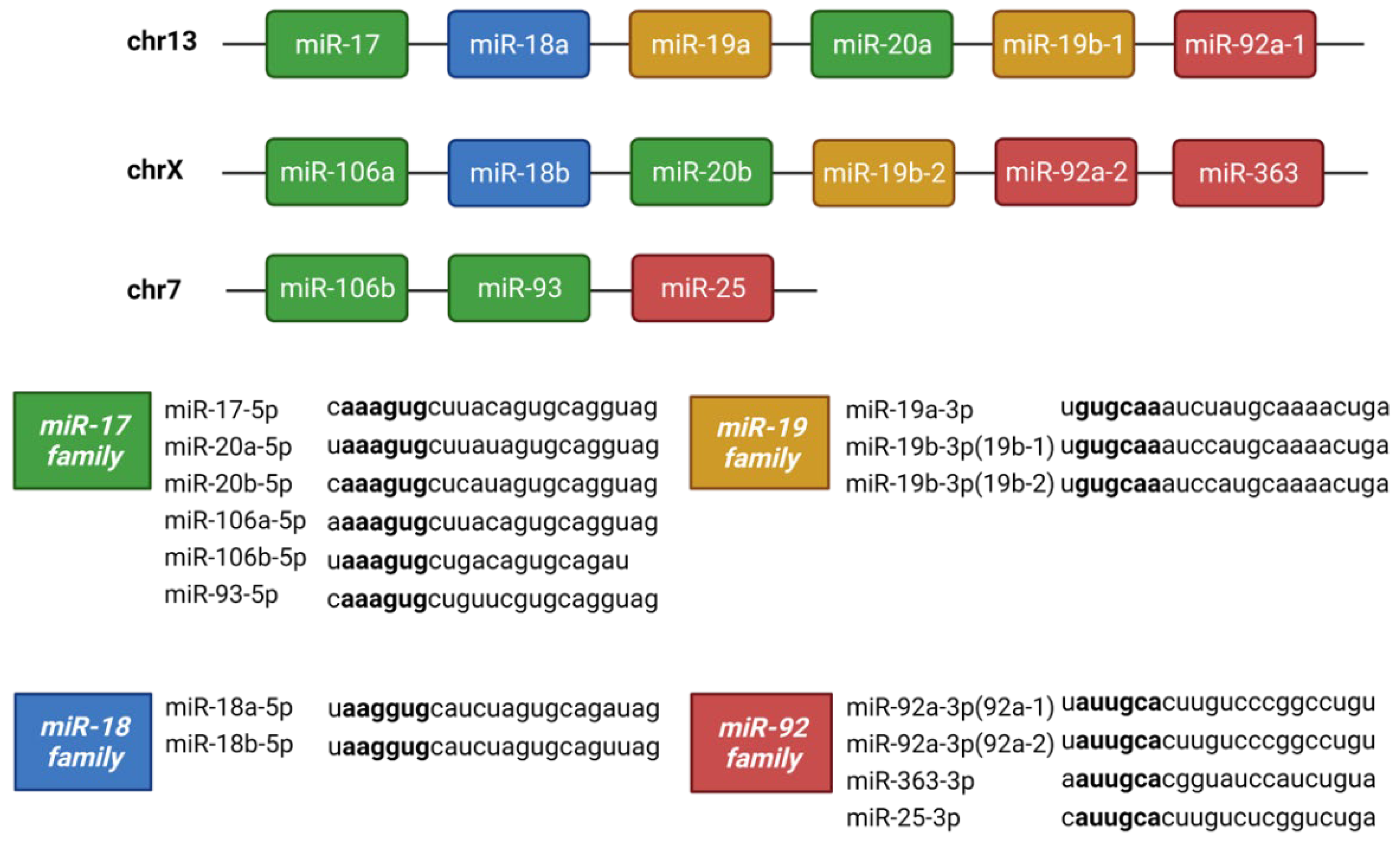
| miRNA | TGFβ1 dataset | TGFβ2 dataset | ||
| Fold Change | P-value | Fold Change | P-value | |
| hsa-miR-17-5p | 0.77 | 0.060 | 1.07 | 0.510 |
| hsa-miR-18a-5p | 0.67 | 0.007 | 1.13 | 0.222 |
| hsa-miR-19a-3p | 0.63 | 0.002 | 0.89 | 0.360 |
| hsa-miR-19b-3p | 0.64 | 0.001 | 0.96 | 0.765 |
| hsa-miR-20a-5p | 0.67 | 0.002 | 0.91 | 0.255 |
| hsa-miR-92a-3p | 0.83 | 0.160 | 0.92 | 0.361 |

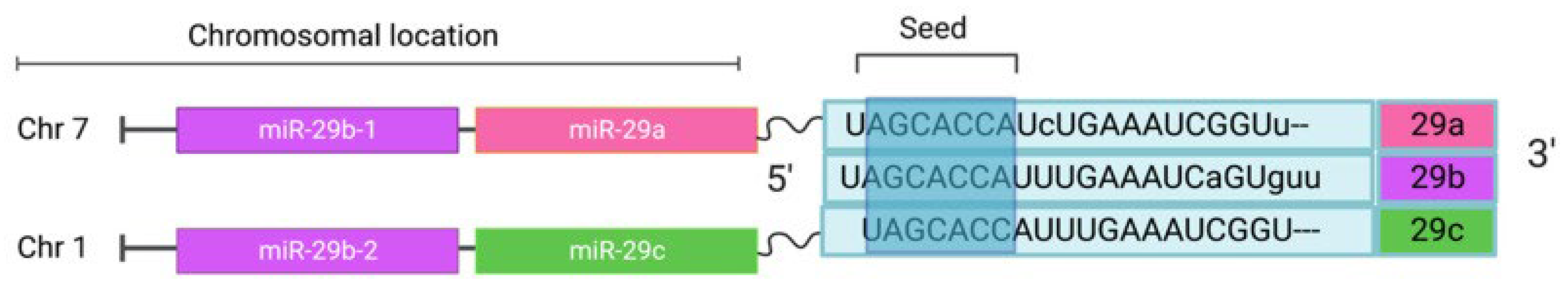
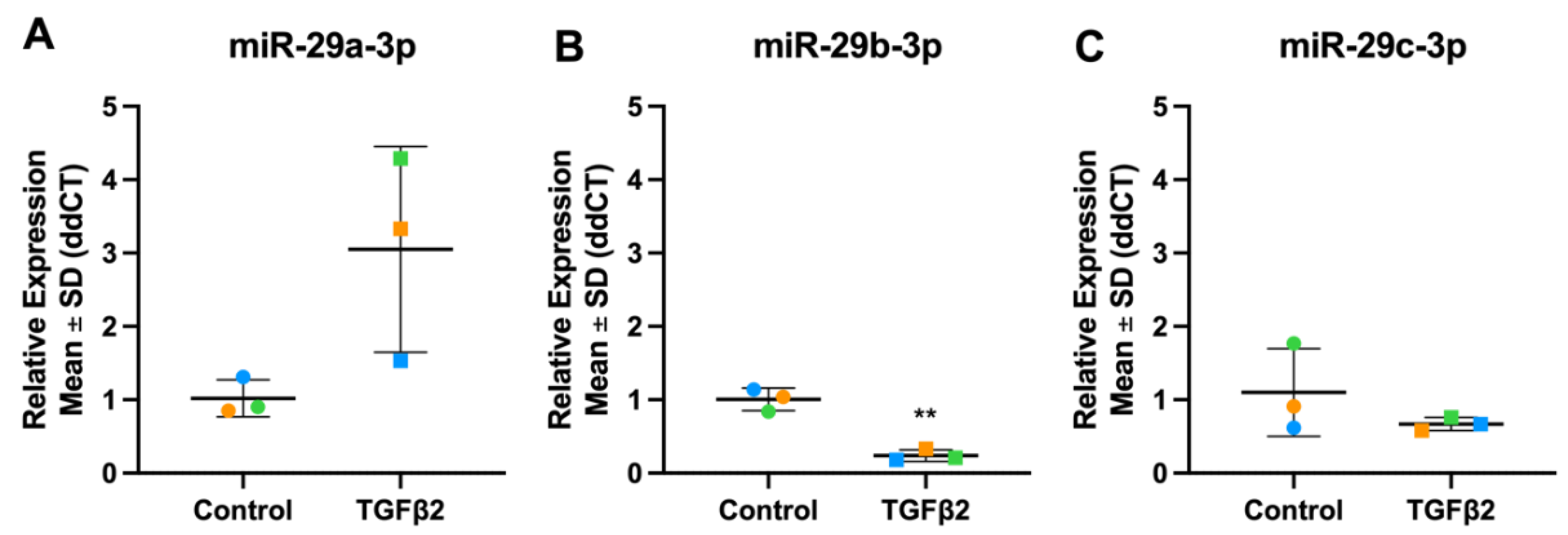
3.8.2. miR-29 Family
3.9. miRNA Strands
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weinreb, R.N.; Leung, C.K.S.; Crowston, J.G.; Medeiros, F.A.; Friedman, D.S.; Wiggs, J.L.; Martin, K.R. Primary Open-Angle Glaucoma. Nat Rev Dis Primers 2016, 2, 16067. [Google Scholar] [CrossRef] [PubMed]
- Tham, Y.-C.C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.-Y.Y. Global Prevalence of Glaucoma and Projections of Glaucoma Burden through 2040. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, W.S.; Shankar, V.; Razeghinejad, R.; Katz, L.J. Current and New Pharmacotherapeutic Approaches for Glaucoma. Expert Opin Pharmacother 2020, 21, 2027–2040. [Google Scholar] [CrossRef] [PubMed]
- Cvenkel, B.; Kolko, M. Current Medical Therapy and Future Trends in the Management of Glaucoma Treatment. J Ophthalmol 2020, 2020, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ang, A. Long Term Effect of Latanoprost on Intraocular Pressure in Normal Tension Glaucoma. British Journal of Ophthalmology 2004, 88, 630–634. [Google Scholar] [CrossRef]
- Scherer, W.J. A Retrospective Review of Non-Responders to Latanoprost. Journal of Ocular Pharmacology and Therapeutics 2002, 18, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Cao, M.; Liu, K.; Duan, X. Analysis of the Responsiveness of Latanoprost, Travoprost, Bimatoprost, and Tafluprost in the Treatment of OAG/OHT Patients. J Ophthalmol 2021, 2021, 1–12. [Google Scholar] [CrossRef]
- Dikopf, M.S.; Vajaranant, T.S.; Edward, D.P. Topical Treatment of Glaucoma: Established and Emerging Pharmacology. Expert Opin Pharmacother 2017, 18, 885–898. [Google Scholar] [CrossRef]
- Rao, P.V.; Pattabiraman, P.P.; Kopczynski, C. Role of the Rho GTPase/Rho Kinase Signaling Pathway in Pathogenesis and Treatment of Glaucoma: Bench to Bedside Research. Exp Eye Res 2017, 158, 23–32. [Google Scholar] [CrossRef]
- Tanna, A.P.; Johnson, M. Rho Kinase Inhibitors as a Novel Treatment for Glaucoma and Ocular Hypertension. Ophthalmology 2018, 125, 1741–1756. [Google Scholar] [CrossRef]
- O’Callaghan, J.; Delaney, C.; O’Connor, M.; van Batenburg-Sherwood, J.; Schicht, M.; Lütjen-Drecoll, E.; Hudson, N.; Ni Dhubhghaill, S.; Humphries, P.; Stanley, C.; et al. Matrix Metalloproteinase-3 (MMP-3)-Mediated Gene Therapy for Glaucoma. Sci Adv 2023, 9, eadf6537. [Google Scholar] [CrossRef] [PubMed]
- Fuchshofer, R.; Tamm, E.R. The Role of TGF-β in the Pathogenesis of Primary Open-Angle Glaucoma. Cell Tissue Res 2012, 347, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.F. The Cell and Molecular Biology of Glaucoma: Biomechanical Factors in Glaucoma. Investigative Opthalmology & Visual Science 2012, 53, 2473. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, R.H.; Nutaitis, M.; Vroman, D.; Crosson, C.E. Influence of Race and Age on Aqueous Humor Levels of Transforming Growth Factor-Beta 2 in Glaucomatous and Nonglaucomatous Eyes. Journal of Ocular Pharmacology and Therapeutics 2011, 27, 477–480. [Google Scholar] [CrossRef]
- Fleenor, D.L.; Shepard, A.R.; Hellberg, P.E.; Jacobson, N.; Pang, I.-H.; Clark, A.F. TGFβ2-Induced Changes in Human Trabecular Meshwork: Implications for Intraocular Pressure. Investigative Opthalmology & Visual Science 2006, 47, 226. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.C.; Li, J.; Chan, W.A.; Tripathi, B.J. Aqueous Humor in Glaucomatous Eyes Contains an Increased Level of TGF-Β2. Exp Eye Res 1994, 59, 723–728. [Google Scholar] [CrossRef]
- Prendes, M.A.; Harris, A.; Wirostko, B.M.; Gerber, A.L.; Siesky, B. The Role of Transforming Growth Factor b in Glaucoma and the Therapeutic Implications. Br. J. Ophthalmol. 2013, 97, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Wordinger, R.J.; Sharma, T.; Clark, A.F. The Role of TGF-Β2 and Bone Morphogenetic Proteins in the Trabecular Meshwork and Glaucoma. J Ocul Pharmacol Ther 2014, 30, 154–162. [Google Scholar] [CrossRef]
- Tovar-Vidales, T.; Clark, A.F.; Wordinger, R.J. Transforming Growth Factor-Beta2 Utilizes the Canonical Smad-Signaling Pathway to Regulate Tissue Transglutaminase Expression in Human Trabecular Meshwork Cells. Exp Eye Res 2011, 93, 442–451. [Google Scholar] [CrossRef]
- Gottanka, J. Effects of TGF- 2 in Perfused Human Eyes. Invest Ophthalmol Vis Sci 2004, 45, 153–158. [Google Scholar] [CrossRef]
- Schlötzer-Schrehardt, U.; Zenkel, M.; Küchle, M.; Sakai, L.Y.; Naumann, G.O.H. Role of Transforming Growth Factor-Β1 and Its Latent Form Binding Protein in Pseudoexfoliation Syndrome. Exp Eye Res 2001, 73, 765–780. [Google Scholar] [CrossRef] [PubMed]
- Kara, S.; Yildirim, N.; Ozer, A.; Colak, O.; Sahin, A. Matrix Metalloproteinase-2, Tissue Inhibitor of Matrix Metalloproteinase-2, and Transforming Growth Factor Beta 1 in the Aqueous Humor and Serum of Patients with Pseudoexfoliation Syndrome. Clinical Ophthalmology 2014, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Garweg, J.G.; Zandi, S.; Pfister, I.B.; Skowronska, M.; Gerhardt, C. Comparison of Cytokine Profiles in the Aqueous Humor of Eyes with Pseudoexfoliation Syndrome and Glaucoma. PLoS One 2017, 12, e0182571. [Google Scholar] [CrossRef] [PubMed]
- Clayton, S.W.; Ban, G.I.; Liu, C.; Serra, R. Canonical and Noncanonical TGF-β Signaling Regulate Fibrous Tissue Differentiation in the Axial Skeleton. Sci Rep 2020, 10, 21364. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Chen, Y.-G. TGF-β Signaling from Receptors to Smads. Cold Spring Harb Perspect Biol 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.J.; Blobe, G.C. Role of Transforming Growth Factor-β Superfamily Signaling Pathways in Human Disease. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2008, 1782, 197–228. [Google Scholar] [CrossRef]
- Hill, C.S. Nucleocytoplasmic Shuttling of Smad Proteins. Cell Res 2009, 19, 36–46. [Google Scholar] [CrossRef]
- Kubiczkova, L.; Sedlarikova, L.; Hajek, R.; Sevcikova, S. TGF-β – an Excellent Servant but a Bad Master. J Transl Med 2012, 10, 183. [Google Scholar] [CrossRef] [PubMed]
- Attisano, L.; Tuen Lee-Hoeflich, S. The Smads. Genome Biol 2001, 2, reviews3010.1. [Google Scholar] [CrossRef]
- Heldin, C.-H.; Moustakas, A. Signaling Receptors for TGF-β Family Members. Cold Spring Harb Perspect Biol 2016, 8, a022053. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of MicroRNA Biogenesis. Nat Rev Mol Cell Biol 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Kang, H. Role of MicroRNAs in TGF-β Signaling Pathway-Mediated Pulmonary Fibrosis. Int J Mol Sci 2017, 18, 2527. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, H.; Hagiwara, S.; Kantharidis, P.; Gohda, T.; Suzuki, Y. Potential Targeting of Renal Fibrosis in Diabetic Kidney Disease Using MicroRNAs. Front Pharmacol 2020, 11, 1797. [Google Scholar] [CrossRef] [PubMed]
- Gallant-Behm, C.L.; Piper, J.; Lynch, J.M.; Seto, A.G.; Hong, S.J.; Mustoe, T.A.; Maari, C.; Pestano, L.A.; Dalby, C.M.; Jackson, A.L.; et al. A MicroRNA-29 Mimic (Remlarsen) Represses Extracellular Matrix Expression and Fibroplasia in the Skin. J Invest Dermatol 2019, 139, 1073–1081. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.-S. Therapeutic Advances of MiRNAs: A Preclinical and Clinical Update. J Adv Res 2021, 28, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.L.; Yu, G.; Latimer, P.A.; Stack, C.; Robinson, K.; Dalby, C.M.; Kaminski, N.; van Rooij, E. Micro <scp>RNA</Scp> Mimicry Blocks Pulmonary Fibrosis. EMBO Mol Med 2014, 6, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Drewry, M.; Helwa, I.; Allingham, R.R.; Hauser, M.A.; Liu, Y. MiRNA Profile in Three Different Normal Human Ocular Tissues by MiRNA-Seq. Investigative Opthalmology & Visual Science 2016, 57, 3731. [Google Scholar] [CrossRef] [PubMed]
- Keller, K.E.; Bhattacharya, S.K.; Borrás, T.; Brunner, T.M.; Chansangpetch, S.; Clark, A.F.; Dismuke, W.M.; Du, Y.; Elliott, M.H.; Ethier, C.R.; et al. Consensus Recommendations for Trabecular Meshwork Cell Isolation, Characterization and Culture. Exp Eye Res 2018, 171, 164–173. [Google Scholar] [CrossRef]
- Callaghan, B.; Lester, K.; Lane, B.; Fan, X.; Goljanek-Whysall, K.; Simpson, D.A.; Sheridan, C.; Willoughby, C.E. Genome-Wide Transcriptome Profiling of Human Trabecular Meshwork Cells Treated with TGF-Β2. Sci Rep 2022, 12, 9564. [Google Scholar] [CrossRef]
- Babraham Bioinformatics Fastqc. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 September 2022).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J 2011, 17, 10. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R Package Rsubread Is Easier, Faster, Cheaper and Better for Alignment and Quantification of RNA Sequencing Reads. Nucleic Acids Res 2019, 47, e47–e47. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. GENCODE 2021. Nucleic Acids Res 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. MiRBase: MicroRNA Sequences, Targets and Gene Nomenclature. Nucleic Acids Res 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano Version 1.10.0: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. R-Package 2021. [Google Scholar]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat Protoc 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective MicroRNA Target Sites in Mammalian MRNAs. Elife 2015, 4. [Google Scholar] [CrossRef]
- Skoufos, G.; Kakoulidis, P.; Tastsoglou, S.; Zacharopoulou, E.; Kotsira, V.; Miliotis, M.; Mavromati, G.; Grigoriadis, D.; Zioga, M.; Velli, A.; et al. TarBase-v9.0 Extends Experimentally Supported MiRNA–Gene Interactions to Cell-Types and Virally Encoded MiRNAs. Nucleic Acids Res 2024, 52, D304–D310. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhao, Y.; Zhang, H.; Yang, S.; Chen, F. Integrated Evolutionary Analysis of Human MiRNA Gene Clusters and Families Implicates Evolutionary Relationships. Gene 2014, 534, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.L.; Wang, X.H.; Sun, B.F.; Zhang, X.D.; Zhu, X.H.; Yu, Z.J.; Luo, H. Expression, Regulation and Mechanism of Action of the MiR-17-92 Cluster in Tumor Cells. Int J Mol Med 2017, 40, 1624–1630. [Google Scholar] [CrossRef] [PubMed]
- Concepcion, C.P.; Bonetti, C.; Ventura, A. The MicroRNA-17-92 Family of MicroRNA Clusters in Development and Disease. The Cancer Journal 2012, 18, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Smyth, A.; Callaghan, B.; Willoughby, C.E.; O’Brien, C. The Role of MiR-29 Family in TGF-β Driven Fibrosis in Glaucomatous Optic Neuropathy. Int J Mol Sci 2022, 23, 10216. [Google Scholar] [CrossRef]
- Olive, V.; Li, Q.; He, L. Mir-17-92: A Polycistronic Oncomir with Pleiotropic Functions. Immunol Rev 2013, 253, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Eyholzer, M.; Schmid, S.; Wilkens, L.; Mueller, B.U.; Pabst, T. The Tumour-Suppressive MiR-29a/B1 Cluster Is Regulated by CEBPA and Blocked in Human AML. Br J Cancer 2010, 103, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Kriegel, A.J.; Liu, Y.; Fang, Y.; Ding, X.; Liang, M. The MiR-29 Family: Genomics, Cell Biology, and Relevance to Renal and Cardiovascular Injury. Physiol Genomics 2012, 44, 237–244. [Google Scholar] [CrossRef]
- Horita, M.; Farquharson, C.; Stephen, L.A. The Role of MiR-29 Family in Disease. J Cell Biochem 2021, 122, 696–715. [Google Scholar] [CrossRef] [PubMed]
- Smyth, A.; Callaghan, B.; Willoughby, C.E.; O’Brien, C. The Role of MiR-29 Family in TGF-β Driven Fibrosis in Glaucomatous Optic Neuropathy. Int J Mol Sci 2022, 23, 10216. [Google Scholar] [CrossRef]
- Medley, J.C.; Panzade, G.; Zinovyeva, A.Y. MicroRNA Strand Selection: Unwinding the Rules. WIREs RNA 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis Elegans. Science (1979) 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA Biogenesis: Coordinated Cropping and Dicing. Nat Rev Mol Cell Biol 2005, 6, 376–385. [Google Scholar] [CrossRef]
- Guo, L.; Lu, Z. The Fate of MiRNA* Strand through Evolutionary Analysis: Implication for Degradation As Merely Carrier Strand or Potential Regulatory Molecule? PLoS One 2010, 5, e11387. [Google Scholar] [CrossRef]
- Okamura, K.; Liu, N.; Lai, E.C. Distinct Mechanisms for MicroRNA Strand Selection by Drosophila Argonautes. Mol Cell 2009, 36, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.; Parker, M.; Challa, P.; Gonzalez, P. Long-Term Decrease of Intraocular Pressure in Rats by Viral Delivery of MiR-146a. Transl Vis Sci Technol 2021, 10, 14. [Google Scholar] [CrossRef]
- Li, X.; Zhao, F.; Xin, M.; Li, G.; Luna, C.; Li, G.; Zhou, Q.; He, Y.; Yu, B.; Olson, E.; et al. Regulation of Intraocular Pressure by MicroRNA Cluster MiR-143/145. Sci Rep 2017, 7, 915. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.; Li, G.; Huang, J.; Qiu, J.; Wu, J.; Yuan, F.; Epstein, D.L.; Gonzalez, P. Regulation of Trabecular Meshwork Cell Contraction and Intraocular Pressure by MiR-200c. PLoS One 2012, 7. [Google Scholar] [CrossRef]
- Fuchshofer, R.; Tamm, E.R. The Role of TGF-β in the Pathogenesis of Primary Open-Angle Glaucoma. Cell Tissue Res 2012, 347, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Keller, K.E.; Aga, M.; Bradley, J.M.; Kelley, M.J.; Acott, T.S. Extracellular Matrix Turnover and Outflow Resistance. Exp Eye Res 2009, 88, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Prendes, M.A.; Harris, A.; Wirostko, B.M.; Gerber, A.L.; Siesky, B. The Role of Transforming Growth Factor β in Glaucoma and the Therapeutic Implications. Br J Ophthalmol 2013, 97, 680–686. [Google Scholar] [CrossRef]
- Takahashi, E.; Inoue, T.; Fujimoto, T.; Kojima, S.; Tanihara, H. Epithelial Mesenchymal Transition-like Phenomenon in Trabecular Meshwork Cells. Exp Eye Res 2014, 118, 72–79. [Google Scholar] [CrossRef]
- Chakraborthy, M.; Rao, A. A Feedback Loop between TGF-Β1 and ATG5 Mediated by MiR-122-5p Regulates Fibrosis and EMT in Human Trabecular Meshwork Cells. Curr Issues Mol Biol 2023, 45, 2381–2392. [Google Scholar] [CrossRef] [PubMed]
- Drewry, M.D.; Challa, P.; Kuchtey, J.G.; Navarro, I.; Helwa, I.; Hu, Y.; Mu, H.; Stamer, W.D.; Kuchtey, R.W.; Liu, Y. Differentially Expressed MicroRNAs in the Aqueous Humor of Patients with Exfoliation Glaucoma or Primary Open-Angle Glaucoma. Hum Mol Genet 2018, 27, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Zhu, Q.; Lin, W.; Wang, L. MicroRNA-122 Inhibits Epithelial-mesenchymal Transition of Hepatic Stellate Cells Induced by the TGF-β1/Smad Signaling Pathway. Exp Ther Med 2018. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, J.-W.; Lin, J.-Y.; Miao, R.; Zhong, J.-C. Roles of MicroRNA-122 in Cardiovascular Fibrosis and Related Diseases. Cardiovasc Toxicol 2020, 20, 463–473. [Google Scholar] [CrossRef]
- Li, G.; Luna, C.; Qiu, J.; Epstein, D.L.; Gonzalez, P. Alterations in MicroRNA Expression in Stress-Induced Cellular Senescence. Mech Ageing Dev 2009, 130, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bailey, J.C.; Helwa, I.; Dismuke, W.M.; Cai, J.; Drewry, M.; Brilliant, M.H.; Budenz, D.L.; Christen, W.G.; Chasman, D.I.; et al. A Common Variant in MIR182 Is Associated With Primary Open-Angle Glaucoma in the NEIGHBORHOOD Consortium. Investigative Opthalmology & Visual Science 2016, 57, 4528. [Google Scholar] [CrossRef]
- Stafford, M.Y.C.; McKenna, D.J. MiR-182 Is Upregulated in Prostate Cancer and Contributes to Tumor Progression by Targeting MITF. Int J Mol Sci 2023, 24, 1824. [Google Scholar] [CrossRef]
- Ichiyama, K.; Dong, C. The Role of MiR-183 Cluster in Immunity. Cancer Lett 2019, 443, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Lei, R.; Hu, G. Roles of MiR-182 in Sensory Organ Development and Cancer. Thorac Cancer 2015, 6, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Q.; Zhou, Y.; Yang, Z.; Tan, M. Inhibition of MiR-182-5p Attenuates Pulmonary Fibrosis via TGF-β/Smad Pathway. Hum Exp Toxicol 2020, 39, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lei, R.; Zhuang, X.; Li, X.; Li, G.; Lev, S.; Segura, M.F.; Zhang, X.; Hu, G. MicroRNA-182 Targets SMAD7 to Potentiate TGFβ-Induced Epithelial-Mesenchymal Transition and Metastasis of Cancer Cells. Nat Commun 2016, 7, 13884. [Google Scholar] [CrossRef] [PubMed]
- Bhattachariya, A.; Dahan, D.; Ekman, M.; Boettger, T.; Braun, T.; Swärd, K.; Hellstrand, P.; Albinsson, S. Spontaneous Activity and Stretch-Induced Contractile Differentiation Are Reduced in Vascular Smooth Muscle of MiR-143/145 Knockout Mice. Acta Physiologica 2015, 215, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Jayaram, H.; Phillips, J.I.; Lozano, D.C.; Choe, T.E.; Cepurna, W.O.; Johnson, E.C.; Morrison, J.C.; Gattey, D.M.; Saugstad, J.A.; Keller, K.E. Comparison of MicroRNA Expression in Aqueous Humor of Normal and Primary Open-Angle Glaucoma Patients Using PCR Arrays: A Pilot Study. Investigative Opthalmology & Visual Science 2017, 58, 2884. [Google Scholar] [CrossRef] [PubMed]
- Paterson, M.R.; Kriegel, A.J. MiR-146a/b: A Family with Shared Seeds and Different Roots. Physiol Genomics 2017, 49, 243–252. [Google Scholar] [CrossRef]
- Li, G.; Luna, C.; Qiu, J.; Epstein, D.L.; Gonzalez, P. Modulation of Inflammatory Markers by MiR-146a during Replicative Senescence in Trabecular Meshwork Cells. Investigative Opthalmology & Visual Science 2010, 51, 2976. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-L.; Wang, X.; Mann, M.; Adamus, T.P.; Wang, D.; Moreira, D.F.; Zhang, Z.; Ouyang, C.; He, X.; Zhang, B.; et al. Myeloid Cell–Targeted MiR-146a Mimic Inhibits NF-ΚB–Driven Inflammation and Leukemia Progression in Vivo. Blood 2020, 135, 167–180. [Google Scholar] [CrossRef]
- Xiang, M.; Birkbak, N.J.; Vafaizadeh, V.; Walker, S.R.; Yeh, J.E.; Liu, S.; Kroll, Y.; Boldin, M.; Taganov, K.; Groner, B.; et al. STAT3 Induction of MiR-146b Forms a Feedback Loop to Inhibit the NF-ΚB to IL-6 Signaling Axis and STAT3-Driven Cancer Phenotypes. Sci Signal 2014, 7. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Gregorova, J.; Vychytilova-Faltejskova, P.; Sevcikova, S. Epigenetic Regulation of MicroRNA Clusters and Families during Tumor Development. Cancers 2021, 13, 1333. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.L.; Wang, X.H.; Sun, B.F.; Zhang, X.D.; Zhu, X.H.; Yu, Z.J.; Luo, H. Expression, Regulation and Mechanism of Action of the MiR-17-92 Cluster in Tumor Cells (Review). Int J Mol Med 2017, 40, 1624–1630. [Google Scholar] [CrossRef] [PubMed]
- Petrocca, F.; Vecchione, A.; Croce, C.M. Emerging Role of MiR-106b-25/MiR-17-92 Clusters in the Control of Transforming Growth Factor β Signaling. Cancer Res 2008, 68, 8191–8194. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Boström, A.-K.; Impens, F.; Fredlund, E.; Van Peer, G.; De Antonellis, P.; von Stedingk, K.; Ghesquière, B.; Schulte, S.; Dews, M.; et al. The MiR-17-92 MicroRNA Cluster Regulates Multiple Components of the TGF-β Pathway in Neuroblastoma. Mol Cell 2010, 40, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Z.; Bai, J.; Song, W.; Zhang, F. MiR-17-5p Regulates the Proliferation and Apoptosis of Human Trabecular Meshwork Cells by Targeting Phosphatase and Tensin Homolog. Mol Med Rep 2019, 19, 3132–3138. [Google Scholar] [CrossRef]
- Tellios, N.; Belrose, J.C.; Tokarewicz, A.C.; Hutnik, C.; Liu, H.; Leask, A.; Motolko, M.; Iijima, M.; Parapuram, S.K. TGF-β Induces Phosphorylation of Phosphatase and Tensin Homolog: Implications for Fibrosis of the Trabecular Meshwork Tissue in Glaucoma. Sci Rep 2017, 7, 812. [Google Scholar] [CrossRef]
- Knox, J.; Bou-Gharios, G.; Hamill, K.J.; Willoughby, C.E. MiR-18a-5p Targets Connective Tissue Growth Factor Expression and Inhibits Transforming Growth Factor Β2-Induced Trabecular Meshwork Cell Contractility. Genes 2022, 13, 1500. [Google Scholar] [CrossRef] [PubMed]
- Browne, J.G.; Ho, S.L.; Kane, R.; Oliver, N.; Clark, A.F.; O’Brien, C.J.; Crean, J.K. Connective Tissue Growth Factor Is Increased in Pseudoexfoliation Glaucoma. Investigative Opthalmology & Visual Science 2011, 52, 3660. [Google Scholar] [CrossRef]
- Junglas, B.; Kuespert, S.; Seleem, A.A.; Struller, T.; Ullmann, S.; Bösl, M.; Bosserhoff, A.; Köstler, J.; Wagner, R.; Tamm, E.R.; et al. Connective Tissue Growth Factor Causes Glaucoma by Modifying the Actin Cytoskeleton of the Trabecular Meshwork. Am J Pathol 2012, 180, 2386–2403. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.P.; Ai, W.B.; Wan, L.Y.; Zhang, Y.Q.; Wu, J.F. The Roles of MicroRNA Families in Hepatic Fibrosis. Cell Biosci 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Cushing, L.; Kuang, P.; Lü, J. The Role of MiR-29 in Pulmonary Fibrosis. Biochemistry and Cell Biology 2015, 93, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.; Li, G.; Qiu, J.; Epstein, D.L.; Gonzalez, P. Cross-Talk between MiR-29 and Transforming Growth Factor-Betas in Trabecular Meshwork Cells. Investigative Opthalmology & Visual Science 2011, 52, 3567. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, G.; Oh, D.-J.; Kang, M.H.; Rhee, D.J. Coordinated Regulation of Extracellular Matrix Synthesis by the MicroRNA-29 Family in the Trabecular Meshwork. Investigative Opthalmology & Visual Science 2011, 52, 3391. [Google Scholar] [CrossRef] [PubMed]
- Luna, C.; Li, G.; Qiu, J.; Epstein, D.L.; Gonzalez, P. Role of MiR-29b on the Regulation of the Extracellular Matrix in Human Trabecular Meshwork Cells under Chronic Oxidative Stress. Mol Vis 2009, 15, 2488–2497. [Google Scholar] [PubMed]
- MacDonald, W.W.; Swaminathan, S.S.; Heo, J.Y.; Castillejos, A.; Hsueh, J.; Liu, B.J.; Jo, D.; Du, A.; Lee, H.; Kang, M.H.; et al. Effect of SPARC Suppression in Mice, Perfused Human Anterior Segments, and Trabecular Meshwork Cells. Investigative Opthalmology & Visual Science 2022, 63, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Cui, J.; Duan, X.; Chen, H.; Fan, F. Suppression of Type I Collagen Expression by MiR-29b via PI3K, Akt, and Sp1 Pathway in Human Tenon’s Fibroblasts. Investigative Opthalmology & Visual Science 2012, 53, 1670. [Google Scholar] [CrossRef]
- Deng, Z.; He, Y.; Yang, X.; Shi, H.; Shi, A.; Lu, L.; He, L. MicroRNA-29: A Crucial Player in Fibrotic Disease. Mol Diagn Ther 2017, 21, 285–294. [Google Scholar] [CrossRef]
- Chioccioli, M.; Roy, S.; Newell, R.; Pestano, L.; Dickinson, B.; Rigby, K.; Herazo-Maya, J.; Jenkins, G.; Ian, S.; Saini, G.; et al. A Lung Targeted MiR-29 Mimic as a Therapy for Pulmonary Fibrosis. EBioMedicine 2022, 85, 104304. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018, 9. [Google Scholar] [CrossRef]
- Yoke-Kqueen, C. Differential MicroRNA Expression and Identification of Putative MiRNA Targets and Pathways in Head and Neck Cancers. Int J Mol Med 2011. [Google Scholar] [CrossRef] [PubMed]
- Amirfallah, A.; Knutsdottir, H.; Arason, A.; Hilmarsdottir, B.; Johannsson, O.T.; Agnarsson, B.A.; Barkardottir, R.B.; Reynisdottir, I. Hsa-MiR-21-3p Associates with Breast Cancer Patient Survival and Targets Genes in Tumor Suppressive Pathways. PLoS One 2021, 16, e0260327. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, Z.; Kusumanchi, P.; Han, S.; Liangpunsakul, S. Critical Role of MicroRNA-21 in the Pathogenesis of Liver Diseases. Front Med (Lausanne) 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Sen, C.K.; Roy, S. MicroRNA 21 in Tissue Injury and Inflammation. Cardiovasc Res 2012, 96, 230–233. [Google Scholar] [CrossRef]
- Hong, Y.; Ye, M.; Wang, F.; Fang, J.; Wang, C.; Luo, J.; Liu, J.; Liu, J.; Liu, L.; Zhao, Q.; et al. MiR-21-3p Promotes Hepatocellular Carcinoma Progression via SMAD7/YAP1 Regulation. Front Oncol 2021, 11. [Google Scholar] [CrossRef]
- Krichevsky, A.M.; Gabriely, G. MiR-21: A Small Multi-faceted RNA. J Cell Mol Med 2009, 13, 39–53. [Google Scholar] [CrossRef]
- Pink, R.C.; Samuel, P.; Massa, D.; Caley, D.P.; Brooks, S.A.; Carter, D.R.F. The Passenger Strand, MiR-21-3p, Plays a Role in Mediating Cisplatin Resistance in Ovarian Cancer Cells. Gynecol Oncol 2015, 137, 143–151. [Google Scholar] [CrossRef]
- Shi, P.; Zhao, X.-D.; Shi, K.-H.; Ding, X.-S.; Tao, H. MiR-21–3p Triggers Cardiac Fibroblasts Pyroptosis in Diabetic Cardiac Fibrosis via Inhibiting Androgen Receptor. Exp Cell Res 2021, 399, 112464. [Google Scholar] [CrossRef]
- Zhu, H.; Luo, H.; Li, Y.; Zhou, Y.; Jiang, Y.; Chai, J.; Xiao, X.; You, Y.; Zuo, X. MicroRNA-21 in Scleroderma Fibrosis and Its Function in TGF-β- Regulated Fibrosis-Related Genes Expression. J Clin Immunol 2013, 33, 1100–1109. [Google Scholar] [CrossRef]
- Yuan, J.; Chen, H.; Ge, D.; Xu, Y.; Xu, H.; Yang, Y.; Gu, M.; Zhou, Y.; Zhu, J.; Ge, T.; et al. Mir-21 Promotes Cardiac Fibrosis after Myocardial Infarction Via Targeting Smad7. Cellular Physiology and Biochemistry 2017, 42, 2207–2219. [Google Scholar] [CrossRef] [PubMed]
- LIU, R.-H.; NING, B.; MA, X.-E.; GONG, W.-M.; JIA, T.-H. Regulatory Roles of MicroRNA-21 in Fibrosis through Interaction with Diverse Pathways (Review). Mol Med Rep 2016, 13, 2359–2366. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Song, M.; Stamer, W.D.; Qiao, Y.; Chen, X.; Sun, X.; Lei, Y.; Chen, J. MiR-21-5p: A Viable Therapeutic Strategy for Regulating Intraocular Pressure. Exp Eye Res 2020, 200. [Google Scholar] [CrossRef]
- Jiao, W.; Leng, X.; Zhou, Q.; Wu, Y.; Sun, L.; Tan, Y.; Ni, H.; Dong, X.; Shen, T.; Liu, Y.; et al. Different MiR-21-3p Isoforms and Their Different Features in Colorectal Cancer. Int J Cancer 2017, 141, 2103–2111. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zeng, Z.; Starkuviene, V.; Erfle, H.; Kan, K.; Zhang, J.; Gunkel, M.; Sticht, C.; Rahbari, N.; Keese, M. MicroRNAs Influence the Migratory Ability of Human Umbilical Vein Endothelial Cells. Genes (Basel) 2022, 13, 640. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Ye, C.; Ge, R.; Wang, Y.; Tian, X.-L.; Chen, Q.; Li, Y.-H.; Zhu, G.-Q.; Zhou, B. MiR-21-3p in Extracellular Vesicles from Vascular Fibroblasts of Spontaneously Hypertensive Rat Promotes Proliferation and Migration of Vascular Smooth Muscle Cells. Life Sci 2023, 330, 122023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; He, C.; Li, R.; Ke, Y.; Sun, K.; Wang, J. MiR-708 and MiR-335-3p Inhibit the Apoptosis of Retinal Ganglion Cells Through Suppressing Autophagy. Journal of Molecular Neuroscience 2021, 71, 284–292. [Google Scholar] [CrossRef]
- Carvalho de Oliveira, J.; Mathias, C.; Oliveira, V.C.; Pezuk, J.A.; Brassesco, M.S. The Double Face of MiR-708: A Pan-Cancer Player with Dissociative Identity Disorder. Genes 2022, 13, 2375. [Google Scholar] [CrossRef]
- Monteleone, N.J.; Lutz, C.S. MiR-708-5p: A MicroRNA with Emerging Roles in Cancer. Oncotarget 2017, 8, 71292–71316. [Google Scholar] [CrossRef]
- Kefaloyianni, E.; Muthu, M.L.; Kaeppler, J.; Sun, X.; Sabbisetti, V.; Chalaris, A.; Rose-John, S.; Wong, E.; Sagi, I.; Waikar, S.S.; et al. ADAM17 Substrate Release in Proximal Tubule Drives Kidney Fibrosis. JCI Insight 2016, 1. [Google Scholar] [CrossRef]
- Arribas, J.; Esselens, C. ADAM17 as a Therapeutic Target in Multiple Diseases. Curr Pharm Des 2009, 15, 2319–2335. [Google Scholar] [CrossRef] [PubMed]
- Scheller, J.; Chalaris, A.; Garbers, C.; Rose-John, S. ADAM17: A Molecular Switch to Control Inflammation and Tissue Regeneration. Trends Immunol 2011, 32, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.A.; Karnam, S.; Skiba, N.; Maddala, R.; Challa, P.; Vann, R.; Rao, V. Vasorin, an Antagonist of TGF-β Is Decreased in Glaucoma Patients. Invest Ophthalmol Vis Sci 2021, 62, 1638–1638. [Google Scholar]
- Ashok, A.; Chaudhary, S.; Rana, N.A.; Kritikos, A.; Ayyagari, R.V.; Singh, N. The Prion Protein as a Mediator of Amyloid-β Toxicity in Retinal and Glaucomatous Degeneration Associated with Alzheimer’s Disease. Invest Ophthalmol Vis Sci 2020, 61, 244–244. [Google Scholar]
- Liu, B.; Li, R.; Zhang, J.; Meng, C.; Zhang, J.; Song, X.; Lv, C. MicroRNA-708-3p as a Potential Therapeutic Target via the ADAM17-GATA/STAT3 Axis in Idiopathic Pulmonary Fibrosis. Exp Mol Med 2018, 50, e465–e465. [Google Scholar] [CrossRef] [PubMed]
- Leppäranta, O.; Pulkkinen, V.; Koli, K.; Vähätalo, R.; Salmenkivi, K.; Kinnula, V.L.; Heikinheimo, M.; Myllärniemi, M. Transcription Factor GATA-6 Is Expressed in Quiescent Myofibroblasts in Idiopathic Pulmonary Fibrosis. Am J Respir Cell Mol Biol 2010, 42, 626–632. [Google Scholar] [CrossRef]
- Kimura, T.; Ishii, Y.; Yoh, K.; Morishima, Y.; Iizuka, T.; Kiwamoto, T.; Matsuno, Y.; Homma, S.; Nomura, A.; Sakamoto, T.; et al. Overexpression of the Transcription Factor GATA-3 Enhances the Development of Pulmonary Fibrosis. Am J Pathol 2006, 169, 96–104. [Google Scholar] [CrossRef]
- Pedroza, M.; Le, T.T.; Lewis, K.; Karmouty-Quintana, H.; To, S.; George, A.T.; Blackburn, M.R.; Tweardy, D.J.; Agarwal, S.K. STAT-3 Contributes to Pulmonary Fibrosis through Epithelial Injury and Fibroblast-myofibroblast Differentiation. The FASEB Journal 2016, 30, 129–140. [Google Scholar] [CrossRef]
- Lee, J.; Guan, W.; Han, S.; Hong, D.; Kim, L.; Kim, H. Micro <scp>RNA</scp>-708-3p Mediates Metastasis and Chemoresistance through Inhibition of Epithelial-to-mesenchymal Transition in Breast Cancer. Cancer Sci 2018, 109, 1404–1413. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stöckel, D.; Meese, E.; Lenhof, H.-P.; Keller, A. MiRPathDB 2.0: A Novel Release of the MiRNA Pathway Dictionary Database. Nucleic Acids Res 2020, 48, D142–D147. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H. MicroRNA Control of TGF-β Signaling. Int J Mol Sci 2018, 19, 1901. [Google Scholar] [CrossRef]
- Zhang, Q.; Ye, H.; Xiang, F.; Song, L.-J.; Zhou, L.-L.; Cai, P.-C.; Zhang, J.-C.; Yu, F.; Shi, H.-Z.; Su, Y.; et al. MiR-18a-5p Inhibits Sub-Pleural Pulmonary Fibrosis by Targeting TGF-β Receptor II. Molecular Therapy 2017, 25, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.; Tao, T.; Su, P.; Liao, Z.; Wang, X.; Lei, Y.; Zhao, P.; Liu, L. Targeting MiR-18a Sensitizes Chondrocytes to Anticytokine Therapy to Prevent Osteoarthritis Progression. Cell Death Dis 2020, 11, 947. [Google Scholar] [CrossRef] [PubMed]
- Krutilina, R.; Sun, W.; Sethuraman, A.; Brown, M.; Seagroves, T.N.; Pfeffer, L.M.; Ignatova, T.; Fan, M. MicroRNA-18a Inhibits Hypoxia-Inducible Factor 1α Activity and Lung Metastasis in Basal Breast Cancers. Breast Cancer Research 2014, 16, R78. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Li, Z.; Wang, Y.; Yu, X.; Shao, X.; Li, Y.; Peng, C.; Zhao, Y.; Wang, Y.-L. MiR-18a Contributes to Preeclampsia by Downregulating Smad2 (Full Length) and Reducing TGF-β Signaling. Mol Ther Nucleic Acids 2020, 22, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wu, Y.; Liu, D.; Zhuang, L. MicroRNA-18a-5p Represses Scar Fibroblast Proliferation and Extracellular Matrix Deposition through Regulating Smad2 Expression. Exp Ther Med 2021, 22, 1318. [Google Scholar] [CrossRef]
- Lee, M.; Goraya, N.; Kim, S.; Cho, S. Hippo-yap Signaling in Ocular Development and Disease. Developmental Dynamics 2018, 247, 794–806. [Google Scholar] [CrossRef]
- Zhu, J.; Lin, S.; Ye, J. YAP and TAZ, the Conductors That Orchestrate Eye Development, Homeostasis, and Disease. J Cell Physiol 2019, 234, 246–258. [Google Scholar] [CrossRef]
- Rezaei, T.; Amini, M.; Hashemi, Z.S.; Mansoori, B.; Rezaei, S.; Karami, H.; Mosafer, J.; Mokhtarzadeh, A.; Baradaran, B. MicroRNA-181 Serves as a Dual-Role Regulator in the Development of Human Cancers. Free Radic Biol Med 2020, 152, 432–454. [Google Scholar] [CrossRef]
- Chen, M.; Wang, M.; Xu, S.; Guo, X.; Jiang, J. Upregulation of MiR-181c Contributes to Chemoresistance in Pancreatic Cancer by Inactivating the Hippo Signaling Pathway. Oncotarget 2015, 6, 44466–44479. [Google Scholar] [CrossRef] [PubMed]
- Kubelac, P.; Braicu, C.; Raduly, L.; Chiroi, P.; Nutu, A.; Cojocneanu, R.; Budisan, L.; Berindan-Neagoe, I.; Achimas-Cadariu, P. Comprehensive Analysis of the Expression of Key Genes Related to Hippo Signaling and Their Prognosis Impact in Ovarian Cancer. Diagnostics 2021, 11, 344. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. Hippo Encodes a Ste-20 Family Protein Kinase That Restricts Cell Proliferation and Promotes Apoptosis in Conjunction with Salvador and Warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Wei, X.; Li, W.; Udan, R.S.; Yang, Q.; Kim, J.; Xie, J.; Ikenoue, T.; Yu, J.; Li, L.; et al. Inactivation of YAP Oncoprotein by the Hippo Pathway Is Involved in Cell Contact Inhibition and Tissue Growth Control. Genes Dev 2007, 21, 2747–2761. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Maglic, D.; Dill, M.T.; Mojumdar, K.; Ng, P.K.-S.; Jeong, K.J.; Tsang, Y.H.; Moreno, D.; Bhavana, V.H.; et al. Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer. Cell Rep 2018, 25, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Xia, W.; Fisher, G.J.; Voorhees, J.J.; Quan, T. YAP/TAZ Regulates TGF-β/Smad3 Signaling by Induction of Smad7 via AP-1 in Human Skin Dermal Fibroblasts. Cell Communication and Signaling 2018, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Hiemer, S.E.; Szymaniak, A.D.; Varelas, X. The Transcriptional Regulators TAZ and YAP Direct Transforming Growth Factor β-Induced Tumorigenic Phenotypes in Breast Cancer Cells. Journal of Biological Chemistry 2014, 289, 13461–13474. [Google Scholar] [CrossRef]
- Varelas, X.; Sakuma, R.; Samavarchi-Tehrani, P.; Peerani, R.; Rao, B.M.; Dembowy, J.; Yaffe, M.B.; Zandstra, P.W.; Wrana, J.L. TAZ Controls Smad Nucleocytoplasmic Shuttling and Regulates Human Embryonic Stem-Cell Self-Renewal. Nat Cell Biol 2008, 10, 837–848. [Google Scholar] [CrossRef]
- Li, H.; Raghunathan, V.; Stamer, W.D.; Ganapathy, P.S.; Herberg, S. Extracellular Matrix Stiffness and TGFβ2 Regulate YAP/TAZ Activity in Human Trabecular Meshwork Cells. Front Cell Dev Biol 2022, 10. [Google Scholar] [CrossRef]
- Browne, J.G.; Ho, S.L.; Kane, R.; Oliver, N.; Clark, A.F.; O’Brien, C.J.; Crean, J.K. Connective Tissue Growth Factor Is Increased in Pseudoexfoliation Glaucoma. Investigative Opthalmology & Visual Science 2011, 52, 3660. [Google Scholar] [CrossRef]
- Junglas, B.; Kuespert, S.; Seleem, A.A.; Struller, T.; Ullmann, S.; Bösl, M.; Bosserhoff, A.; Köstler, J.; Wagner, R.; Tamm, E.R.; et al. Connective Tissue Growth Factor Causes Glaucoma by Modifying the Actin Cytoskeleton of the Trabecular Meshwork. American Journal of Pathology 2012, 180, 2386–2403. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhao, X.; Liu, X.; Wang, Y.; Huang, J.; Jiang, B.; Chen, Q.; Yu, J. MiR-146a Functions as a Tumor Suppressor in Prostate Cancer by Targeting Rac1. Prostate 2014, 74, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yuan, B.; Chen, G.; Zhang, L.; Zhuang, Y.; Niu, H.; Zeng, Z. Circular RNA RSF1 Promotes Inflammatory and Fibrotic Phenotypes of Irradiated Hepatic Stellate Cell by Modulating MiR-146a-5p. J Cell Physiol 2020, 235, 8270–8282. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Li, K.; Zheng, P.; Wang, Y.; Lv, Y.; Shen, L.; Chen, Y.; Xue, Z.; Li, B.; Jin, L.; et al. Weighted Gene Coexpression Network Analysis Identified MicroRNA Coexpression Modules and Related Pathways in Type 2 Diabetes Mellitus. Oxid Med Cell Longev 2019, 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.E. Non-Smad Pathways in TGF-β Signaling. Cell Res 2009, 19, 128–139. [Google Scholar] [CrossRef]
- Han, H.; Wecker, T.; Grehn, F.; Schlunck, G. Elasticity-Dependent Modulation of TGF-β Responses in Human Trabecular Meshwork Cells. Investigative Opthalmology & Visual Science 2011, 52, 2889. [Google Scholar] [CrossRef]
- Pervan, C.L. Smad-Independent TGF-Β2 Signaling Pathways in Human Trabecular Meshwork Cells. Exp Eye Res 2017, 158, 137–145. [Google Scholar] [CrossRef]
- Bradshaw, A.D.; Sage, E.H. SPARC, a Matricellular Protein That Functions in Cellular Differentiation and Tissue Response to Injury. Journal of Clinical Investigation 2001, 107, 1049–1054. [Google Scholar] [CrossRef]
- Kang, M.H.; Oh, D.-J.; Kang, J.; Rhee, D.J. Regulation of SPARC by Transforming Growth Factor Β2 in Human Trabecular Meshwork. Investigative Opthalmology & Visual Science 2013, 54, 2523. [Google Scholar] [CrossRef]
- Inoue-Mochita, M.; Inoue, T.; Kojima, S.; Futakuchi, A.; Fujimoto, T.; Sato-Ohira, S.; Tsutsumi, U.; Tanihara, H. Interleukin-6–Mediated Trans-Signaling Inhibits Transforming Growth Factor-β Signaling in Trabecular Meshwork Cells. Journal of Biological Chemistry 2018, 293, 10975–10984. [Google Scholar] [CrossRef]
- Liton, P.B.; Li, G.; Luna, C.; Gonzalez, P.; Epstein, D.L. Cross-Talk between TGF-Beta1 and IL-6 in Human Trabecular Meshwork Cells. Mol Vis 2009, 15, 326–334. [Google Scholar] [PubMed]
- Naidu, S.; Shi, L.; Magee, P.; Middleton, J.D.; Laganá, A.; Sahoo, S.; Leong, H.S.; Galvin, M.; Frese, K.; Dive, C.; et al. PDGFR-Modulated MiR-23b Cluster and MiR-125a-5p Suppress Lung Tumorigenesis by Targeting Multiple Components of KRAS and NF-KB Pathways. Sci Rep 2017, 7, 15441. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, A.C.; Liu, J. Epigenetics and Signaling Pathways in Glaucoma. Biomed Res Int 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wecker, T.; Grehn, F.; Schlunck, G. Elasticity-Dependent Modulation of TGF-β Responses in Human Trabecular Meshwork Cells. Investigative Opthalmology & Visual Science 2011, 52, 2889. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Ouyang, G.; Bao, S. The Activation of Akt/PKB Signaling Pathway and Cell Survival. J Cell Mol Med 2005, 9, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, J.; Murphy, C.; Juster, R. Trabecular Meshwork Cellularity in Primary Open-Angle Glaucoma and Nonglaucomatous Normals. Ophthalmology 1984, 91, 564–579. [Google Scholar] [CrossRef] [PubMed]
- Baleriola, J.; García-Feijoo, J.; Martínez-de-la-Casa, J.M.; Fernández-Cruz, A.; de la Rosa, E.J.; Fernández-Durango, R. Apoptosis in the Trabecular Meshwork of Glaucomatous Patients. Mol Vis 2008, 14, 1513–1516. [Google Scholar]
- Kim, E.H.; Jo, Y.; Sai, S.; Park, M.-J.; Kim, J.-Y.; Kim, J.S.; Lee, Y.-J.; Cho, J.-M.; Kwak, S.-Y.; Baek, J.-H.; et al. Tumor-Treating Fields Induce Autophagy by Blocking the Akt2/MiR29b Axis in Glioblastoma Cells. Oncogene 2019, 38, 6630–6646. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, W.; Chen, J.; Li, Y.; Xu, M.; Cai, Y. MiR-122 and MiR-199 Synergistically Promote Autophagy In oral Lichen Planus by Targeting the Akt/MTOR Pathway. Int J Mol Med 2019. [Google Scholar] [CrossRef]
- Ganesan, H.; Nandy, S.K.; Banerjee, A.; Pathak, S.; Zhang, H.; Sun, X.-F. RNA-Interference-Mediated MiR-122-Based Gene Regulation in Colon Cancer, a Structural In Silico Analysis. Int J Mol Sci 2022, 23, 15257. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, L.; Chen, Z.; Diao, S.; He, Y.; Zhang, Z.; Zhang, H.; Yuan, X.; Li, J. MIR143 Inhibits Steroidogenesis and Induces Apoptosis Repressed by H3K27me3 in Granulosa Cells. Front Cell Dev Biol 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Weeraratne, S.D.; Amani, V.; Teider, N.; Pierre-Francois, J.; Winter, D.; Kye, M.J.; Sengupta, S.; Archer, T.; Remke, M.; Bai, A.H.C.; et al. Pleiotropic Effects of MiR-183~96~182 Converge to Regulate Cell Survival, Proliferation and Migration in Medulloblastoma. Acta Neuropathol 2012, 123, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Bernstock, J.D.; Klimanis, D.; Hallenbeck, J.M. Akt Protein Kinase, MiR-200/MiR-182 Expression and Epithelial-Mesenchymal Transition Proteins in Hibernating Ground Squirrels. Front Mol Neurosci 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Honardoost, M.; Soleimani, M.; Arefian, E.; Sarookhani, M. reza Expression Change of MiR-214 and MiR-135 during Muscle Differentiation. Cell J 2015, 17, 461–470. [Google Scholar]
- Guo, P.; Lan, J.; Ge, J.; Nie, Q.; Mao, Q.; Qiu, Y. MiR-708 Acts as a Tumor Suppressor in Human Glioblastoma Cells. Oncol Rep 2013, 30, 870–876. [Google Scholar] [CrossRef]
- Saini, S.; Majid, S.; Shahryari, V.; Arora, S.; Yamamura, S.; Chang, I.; Zaman, M.S.; Deng, G.; Tanaka, Y.; Dahiya, R. MiRNA-708 Control of CD44(+) Prostate Cancer-Initiating Cells. Cancer Res 2012, 72, 3618–3630. [Google Scholar] [CrossRef] [PubMed]
- Ronen, S.; Abbott, D.W.; Kravtsov, O.; Abdelkader, A.; Xu, Y.; Banerjee, A.; Iczkowski, K.A. PTEN Loss and P27 Loss Differ among Morphologic Patterns of Prostate Cancer, Including Cribriform. Hum Pathol 2017, 65, 85–91. [Google Scholar] [CrossRef]
- Feng, Y.; Zou, W.; Hu, C.; Li, G.; Zhou, S.; He, Y.; Ma, F.; Deng, C.; Sun, L. Modulation of CASC2/MiR-21/PTEN Pathway Sensitizes Cervical Cancer to Cisplatin. Arch Biochem Biophys 2017, 623–624, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qin, T.; Mao, J.; Zhang, J.; Fan, S.; Lu, Y.; Sun, Z.; Zhang, Q.; Song, B.; Li, L. PTENP1/MiR-20a/PTEN Axis Contributes to Breast Cancer Progression by Regulating PTEN via PI3K/AKT Pathway. J Exp Clin Cancer Res 2019, 38, 256. [Google Scholar] [CrossRef]
- Yuan, J.; Su, Z.; Gu, W.; Shen, X.; Zhao, Q.; Shi, L.; Jin, C.; Wang, X.; Cong, H.; Ju, S. MiR-19b and MiR-20a Suppress Apoptosis, Promote Proliferation and Induce Tumorigenicity of Multiple Myeloma Cells by Targeting PTEN. Cancer Biomark 2019, 24, 279–289. [Google Scholar] [CrossRef]
- Shi, L.; Zhu, W.; Huang, Y.; Zhuo, L.; Wang, S.; Chen, S.; Zhang, B.; Ke, B. Cancer-Associated Fibroblast-Derived Exosomal MicroRNA-20a Suppresses the PTEN/PI3K-AKT Pathway to Promote the Progression and Chemoresistance of Non-Small Cell Lung Cancer. Clin Transl Med 2022, 12, e989. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Xiang, Y.; Tang, Y.; Ge, Z.; Li, Q.; Zhang, Y. Circ-ADAM9 Targeting PTEN and ATG7 Promotes Autophagy and Apoptosis of Diabetic Endothelial Progenitor Cells by Sponging Mir-20a-5p. Cell Death Dis 2020, 11, 526. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, L.; Li, E.; Zhang, G.; Hou, Y.; Yuan, W.; Qu, W.; Ding, L. Long-Chain Non-Coding RNA HOTAIR Promotes the Progression of Osteoarthritis via Sponging MiR-20b/PTEN Axis. Life Sci 2020, 253, 117685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jiang, Y.; Guo, X.; Zhang, B.; Wu, J.; Sun, J.; Liang, H.; Shan, H.; Zhang, Y.; Liu, J.; et al. Long Non-Coding RNA Cardiac Hypertrophy-Associated Regulator Governs Cardiac Hypertrophy via Regulating MiR-20b and the Downstream PTEN/AKT Pathway. J Cell Mol Med 2019, 23, 7685–7698. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Dong, S.; Tian, J. MiR-29b Promotes the Osteogenic Differentiation of Mesenchymal Stem Cells Derived from Human Adipose Tissue via the PTEN/AKT/Β-catenin Signaling Pathway. Int J Mol Med 2020, 46, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Li, W.; Xu, Z.; Sun, Z.; Ye, H.; Wu, Y.; Zhang, Y.; Xie, L.; Jiang, D.; Jia, R.; et al. Extracellular Vesicles from Human Umbilical Cord Mesenchymal Stem Cells Reduce Lipopolysaccharide-Induced Spinal Cord Injury Neuronal Apoptosis by Mediating MiR-29b-3p/PTEN. Connect Tissue Res 2022, 63, 634–649. [Google Scholar] [CrossRef]
- Suo, H.-B.; Zhang, K.-C.; Zhao, J. MiR-200a Promotes Cell Invasion and Migration of Ovarian Carcinoma by Targeting PTEN. Eur Rev Med Pharmacol Sci 2018, 22, 4080–4089. [Google Scholar] [CrossRef]
- Xu, M.; Wang, G.; Zhou, H.; Cai, J.; Li, P.; Zhou, M.; Lu, Y.; Jiang, X.; Huang, H.; Zhang, Y.; et al. TGF-Β1-MiR-200a-PTEN Induces Epithelial-Mesenchymal Transition and Fibrosis of Pancreatic Stellate Cells. Mol Cell Biochem 2017, 431, 161–168. [Google Scholar] [CrossRef]
- Li, Y.; Sun, J.; Cai, Y.; Jiang, Y.; Wang, X.; Huang, X.; Yin, Y.; Li, H. MiR-200a Acts as an Oncogene in Colorectal Carcinoma by Targeting PTEN. Exp Mol Pathol 2016, 101, 308–313. [Google Scholar] [CrossRef]
- Morgan, J.T.; Raghunathan, V.K.; Chang, Y.-R.; Murphy, C.J.; Russell, P. Wnt Inhibition Induces Persistent Increases in Intrinsic Stiffness of Human Trabecular Meshwork Cells. Exp Eye Res 2015, 132, 174–178. [Google Scholar] [CrossRef]
- Raghunathan, V.K.; Morgan, J.T.; Dreier, B.; Reilly, C.M.; Thomasy, S.M.; Wood, J.A.; Ly, I.; Tuyen, B.C.; Hughbanks, M.; Murphy, C.J.; et al. Role of Substratum Stiffness in Modulating Genes Associated with Extracellular Matrix and Mechanotransducers YAP and TAZ. Investigative Opthalmology & Visual Science 2013, 54, 378. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Millar, J.C.; Wang, W.-H.; Silverman, S.M.; Liu, Y.; Wordinger, R.J.; Rubin, J.S.; Pang, I.-H.; Clark, A.F. Existence of the Canonical Wnt Signaling Pathway in the Human Trabecular Meshwork. Investigative Opthalmology & Visual Science 2012, 53, 7043. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, J.; Zhu, J.; Zhang, S. Hypoxic Non-Small-Cell Lung Cancer Cell-Secreted Exosomal MicroRNA-582-3p Drives Cancer Cell Malignant Phenotypes by Targeting Secreted Frizzled-Related Protein 1. Cancer Manag Res 2020, 12, 10151–10161. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ju, L.; Li, C.; Cheng, H.; Li, N.; Zhang, Q.; Sun, S.; Ding, L.; Sui, X.; Zhang, C.; et al. MiR-582-3p Participates in the Regulation of Biological Behaviors of A549 Cells by Ambient PM2.5 Exposure. Environmental Science and Pollution Research 2022, 29, 13624–13634. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Cai, J.; Chen, B.; Wu, S.; Li, R.; Xu, X.; Yang, Y.; Guan, H.; Zhu, X.; Zhang, L.; et al. Aberrantly Expressed MiR-582-3p Maintains Lung Cancer Stem Cell-like Traits by Activating Wnt/β-Catenin Signalling. Nat Commun 2015, 6, 8640. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; McNatt, L.G.; Pang, I.H.; Millar, J.C.; Hellberg, P.E.; Hellberg, M.H.; Steely, H.T.; Rubin, J.S.; Fingert, J.H.; Sheffield, V.C.; et al. Increased Expression of the WNT Antagonist SFRP-1 in Glaucoma Elevates Intraocular Pressure. J Clin Invest 2008, 118, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Dhamodaran, K.; Baidouri, H.; Sandoval, L.; Raghunathan, V. Wnt Activation After Inhibition Restores Trabecular Meshwork Cells Toward a Normal Phenotype. Investigative Opthalmology & Visual Science 2020, 61, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sanchez, L.U.; Zheng, J.J. Small-Molecules Wnt Inhibitors Modulate the Effect of Dexamethasone on MYOC Expression in Primary Human Trabecular Meshwork Cells. Invest Ophthalmol Vis Sci 2020, 61, 3466. [Google Scholar]
- Dobrzycka, M.; Sulewska, A.; Biecek, P.; Charkiewicz, R.; Karabowicz, P.; Charkiewicz, A.; Golaszewska, K.; Milewska, P.; Michalska-Falkowska, A.; Nowak, K.; et al. MiRNA Studies in Glaucoma: A Comprehensive Review of Current Knowledge and Future Perspectives. Int J Mol Sci 2023, 24, 14699. [Google Scholar] [CrossRef]
- Tan, C.; Jia, F.; Zhang, P.; Sun, X.; Qiao, Y.; Chen, X.; Wang, Y.; Chen, J.; Lei, Y. A MiRNA Stabilizing Polydopamine Nano-Platform for Intraocular Delivery of MiR-21-5p in Glaucoma Therapy. J Mater Chem B 2021, 9. [Google Scholar] [CrossRef]
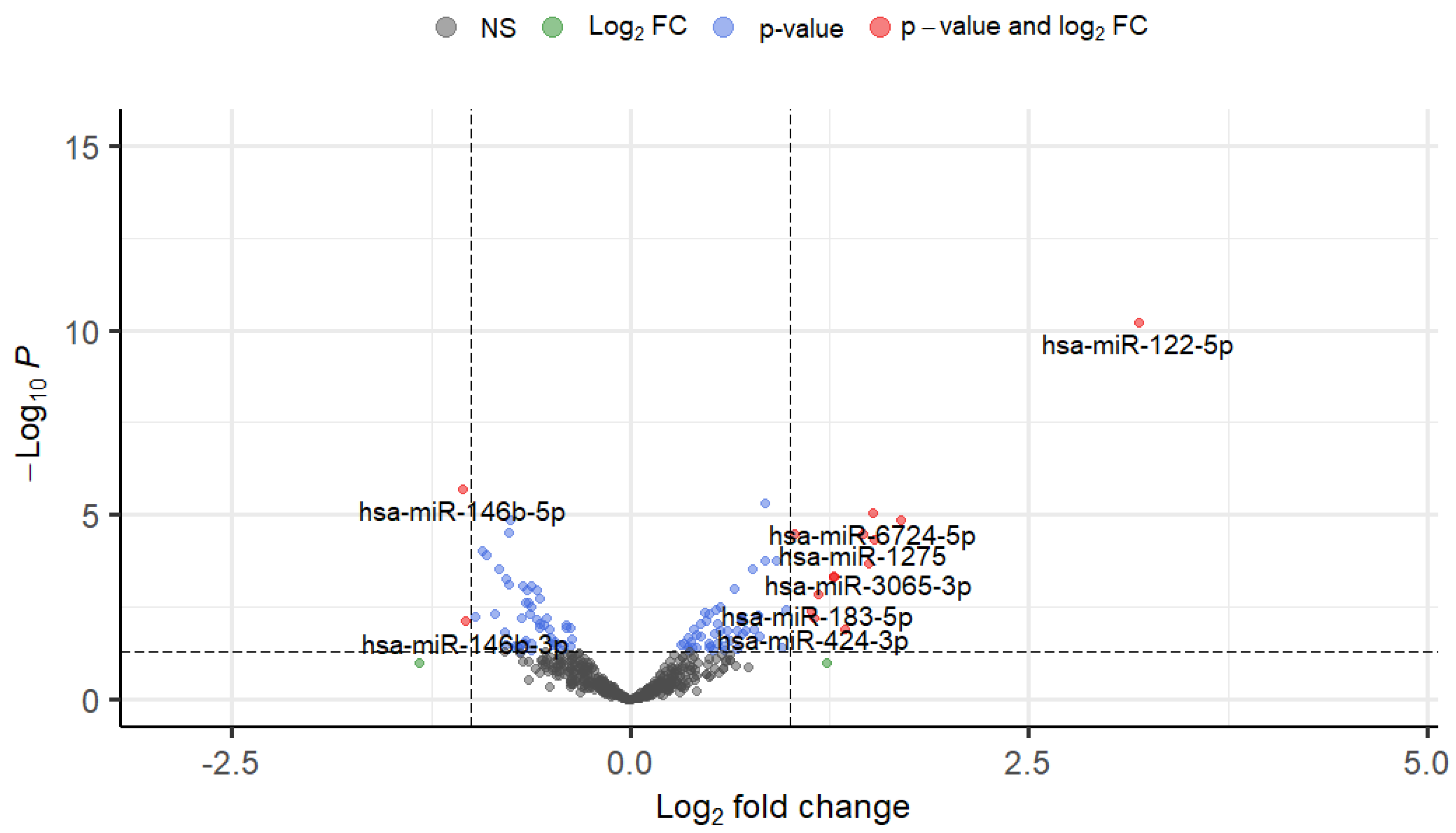
| miRNAs | Fold Change | Log2FC | P-value |
|---|---|---|---|
| hsa-miR-122-5p | 9.13380754 | 3.19121639 | 6.15E-11 |
| hsa-miR-139-5p | 3.25102103 | 1.70089289 | 1.38E-05 |
| hsa-miR-3065-5p | 2.89575179 | 1.53393795 | 4.57E-05 |
| hsa-miR-6724-5p | 2.86974573 | 1.52092291 | 8.51E-06 |
| hsa-miR-3065-3p | 2.8198025 | 1.49559412 | 0.00020201 |
| hsa-miR-1275 | 2.74691918 | 1.45781447 | 3.19E-05 |
| hsa-miR-122b-5p | 2.53489937 | 1.34192848 | 0.01283611 |
| hsa-miR-10395-5p | 2.4233795 | 1.27702035 | 0.00046603 |
| hsa-miR-10395-3p | 2.42334707 | 1.27700104 | 0.00048254 |
| hsa-miR-183-5p | 2.25763024 | 1.17480922 | 0.00134539 |
| hsa-miR-424-3p | 2.22458455 | 1.15353593 | 0.00616844 |
| hsa-miR-10401-5p | 2.19097642 | 1.13157396 | 0.00394718 |
| hsa-miR-23a-5p | 2.04351302 | 1.03105144 | 3.24E-05 |
| hsa-miR-6716-3p | 1.96658773 | 0.97569454 | 0.00379127 |
| hsa-miR-503-3p | 1.92984739 | 0.94848677 | 0.03910098 |
| hsa-miR-146a-5p | 1.89191456 | 0.91984694 | 0.00017454 |
| hsa-miR-181b-5p | 1.79978193 | 0.84782212 | 4.79E-06 |
| hsa-miR-708-5p | 1.79528708 | 0.84421456 | 0.0001733 |
| hsa-miR-182-5p | 1.75251953 | 0.80943052 | 0.01995341 |
| hsa-miR-543 | 1.73760333 | 0.79709877 | 0.00498442 |
| hsa-miR-3679-5p | 1.70656765 | 0.77109761 | 0.01196514 |
| hsa-miR-210-3p | 1.6996398 | 0.76522903 | 0.00029772 |
| hsa-miR-216a-5p | 1.64469194 | 0.71781738 | 0.01385089 |
| hsa-miR-365a-5p | 1.63395301 | 0.7083665 | 0.01578449 |
| hsa-miR-129-5p | 1.61816529 | 0.69435898 | 0.0061433 |
| hsa-miR-10401-3p | 1.61801867 | 0.69422826 | 0.00669733 |
| hsa-miR-10527-5p | 1.59469424 | 0.67327984 | 0.0141122 |
| hsa-miR-181b-3p | 1.59404885 | 0.67269584 | 0.04147052 |
| hsa-miR-24-2-5p | 1.57231126 | 0.65288685 | 0.00099304 |
| hsa-miR-495-3p | 1.54712784 | 0.62959242 | 0.02393389 |
| miRNAs | Fold Change | Log2FC | P-value |
|---|---|---|---|
| hsa-miR-146b-5p | 0.4822713 | -1.0520831 | 1.96E-06 |
| hsa-miR-146b-3p | 0.48914069 | -1.0316786 | 0.00766537 |
| hsa-miR-651-5p | 0.50861592 | -0.9753515 | 0.00545332 |
| hsa-miR-204-5p | 0.52483108 | -0.9300749 | 9.46E-05 |
| hsa-miR-99a-5p | 0.53341448 | -0.9066711 | 0.00011864 |
| hsa-miR-218-1-3p | 0.55523124 | -0.8488393 | 0.00496279 |
| hsa-miR-660-5p | 0.56382312 | -0.8266855 | 0.00028811 |
| hsa-miR-549a-3p | 0.57752493 | -0.7920449 | 0.01521947 |
| hsa-miR-26a-1-3p | 0.58078639 | -0.7839205 | 0.00053314 |
| hsa-miR-500b-3p | 0.58162831 | -0.7818306 | 0.03273988 |
| hsa-miR-15a-5p | 0.58925697 | -0.7630312 | 2.90E-05 |
| hsa-miR-452-3p | 0.59020601 | -0.7607095 | 0.00077017 |
| hsa-miR-218-5p | 0.59343236 | -0.7528445 | 1.37E-05 |
| hsa-miR-26a-2-3p | 0.60264724 | -0.7306143 | 0.03565943 |
| hsa-miR-1255a | 0.61107737 | -0.710573 | 0.0361198 |
| hsa-miR-502-5p | 0.61558839 | -0.6999621 | 0.03998012 |
| hsa-miR-99a-3p | 0.61886292 | -0.6923082 | 0.03455279 |
| hsa-miR-20b-5p | 0.61932422 | -0.6912332 | 0.04897627 |
| hsa-miR-9985 | 0.62109457 | -0.6871151 | 0.00625545 |
| hsa-miR-195-5p | 0.62558473 | -0.6767228 | 0.00080717 |
| hsa-let-7c-3p | 0.63013432 | -0.6662687 | 0.03065585 |
| hsa-miR-19a-3p | 0.63378238 | -0.6579405 | 0.00243884 |
| hsa-miR-335-3p | 0.63422802 | -0.6569265 | 0.02491361 |
| hsa-miR-19b-3p | 0.6385444 | -0.6471412 | 0.00104799 |
| hsa-miR-335-5p | 0.64169946 | -0.6400303 | 0.00237981 |
| hsa-miR-29b-3p | 0.64655536 | -0.6291542 | 0.00472803 |
| hsa-miR-497-5p | 0.6492189 | -0.6232231 | 0.00294707 |
| hsa-miR-20a-3p | 0.65002719 | -0.621428 | 0.04420571 |
| hsa-miR-454-5p | 0.65124762 | -0.6187219 | 0.03029567 |
| hsa-miR-101-3p | 0.65134387 | -0.6185087 | 0.00085122 |
| miRNA Name | Fold Change | Log2FC | P-value |
|---|---|---|---|
| hsa-miR-181b-2-3p | 3.53991903 | 1.82371636 | 1.27E-06 |
| hsa-miR-503-3p | 3.18394765 | 1.67081662 | 3.60E-18 |
| hsa-miR-424-3p | 2.85950834 | 1.51576711 | 1.05E-06 |
| hsa-miR-21-3p | 2.00735084 | 1.00529279 | 1.26E-16 |
| hsa-miR-708-3p | 1.99270313 | 0.99472679 | 5.17E-06 |
| hsa-miR-6716-3p | 1.99202337 | 0.99423458 | 0.00072026 |
| hsa-miR-503-5p | 1.98194499 | 0.98691692 | 1.18E-06 |
| hsa-miR-27a-5p | 1.93586762 | 0.9529803 | 4.55E-12 |
| hsa-miR-6724-5p | 1.89734963 | 0.92398555 | 0.00050886 |
| hsa-miR-145-3p | 1.88867966 | 0.91737802 | 2.71E-07 |
| hsa-miR-181a-2-3p | 1.88789033 | 0.91677496 | 9.80E-08 |
| hsa-miR-3187-3p | 1.85686868 | 0.89287179 | 0.00135669 |
| hsa-miR-24-2-5p | 1.8166234 | 0.86125937 | 5.52E-06 |
| hsa-miR-143-5p | 1.81551189 | 0.86037638 | 8.45E-09 |
| hsa-miR-708-5p | 1.81499426 | 0.85996499 | 2.20E-05 |
| hsa-miR-3179 | 1.80771879 | 0.85417027 | 0.01538545 |
| hsa-miR-3065-3p | 1.76608959 | 0.82055853 | 0.00122102 |
| hsa-miR-10401-5p | 1.70851951 | 0.77274672 | 0.02374628 |
| hsa-miR-5701 | 1.67320751 | 0.74261638 | 0.01320843 |
| hsa-miR-216a-5p | 1.66610529 | 0.73647958 | 0.00576722 |
| hsa-miR-135a-5p | 1.65188519 | 0.72411342 | 0.01440031 |
| hsa-miR-214-5p | 1.62635075 | 0.70163843 | 0.00031568 |
| hsa-miR-23a-5p | 1.58158263 | 0.66136893 | 0.0171346 |
| hsa-miR-3065-5p | 1.56823909 | 0.64914552 | 0.00064297 |
| hsa-miR-145-5p | 1.51098725 | 0.59549148 | 4.34E-08 |
| hsa-miR-181b-5p | 1.49487096 | 0.58002095 | 0.00019705 |
| hsa-miR-199b-5p | 1.41320621 | 0.49897199 | 0.0083763 |
| hsa-miR-424-5p | 1.41198062 | 0.49772029 | 0.00883417 |
| hsa-miR-210-3p | 1.39737981 | 0.4827242 | 6.48E-05 |
| hsa-miR-6511a-3p | 1.39588043 | 0.48117537 | 0.01910117 |
| miRNA Name | Fold Change | Log2FC | P-value |
|---|---|---|---|
| hsa-miR-6859-5p | 0.47775733 | -1.0656501 | 0.01471161 |
| hsa-miR-218-1-3p | 0.5838476 | -0.7763363 | 3.56E-08 |
| hsa-miR-20b-5p | 0.5851248 | -0.7731837 | 0.04888215 |
| hsa-miR-15a-3p | 0.58819277 | -0.765639 | 0.01533516 |
| hsa-miR-26a-1-3p | 0.60051027 | -0.7357392 | 4.51E-07 |
| hsa-miR-744-3p | 0.61105129 | -0.7106346 | 0.00203953 |
| hsa-miR-760 | 0.61267545 | -0.7068051 | 0.00696877 |
| hsa-miR-3613-5p | 0.62166996 | -0.6857792 | 0.00247998 |
| hsa-miR-485-5p | 0.64873167 | -0.6243062 | 0.00044685 |
| hsa-miR-3618 | 0.6551584 | -0.6100843 | 0.00031305 |
| hsa-miR-96-5p | 0.66784058 | -0.5824243 | 0.04879471 |
| hsa-let-7c-3p | 0.66958237 | -0.5786666 | 0.00046092 |
| hsa-miR-26a-2-3p | 0.70516762 | -0.5039619 | 0.00985704 |
| hsa-miR-454-5p | 0.70792943 | -0.4983225 | 0.01136527 |
| hsa-miR-452-3p | 0.73641685 | -0.4414055 | 0.01341871 |
| hsa-miR-302b-3p | 0.74394926 | -0.4267239 | 0.03858273 |
| hsa-miR-379-3p | 0.74729697 | -0.4202464 | 0.02575367 |
| hsa-miR-200a-3p | 0.75177354 | -0.41163 | 0.03409936 |
| hsa-miR-330-3p | 0.75655443 | -0.4024842 | 0.00629521 |
| hsa-miR-652-3p | 0.76101822 | -0.3939971 | 0.04107676 |
| hsa-miR-330-5p | 0.76266111 | -0.390886 | 0.0358708 |
| hsa-miR-582-3p | 0.7793746 | -0.3596112 | 0.03470739 |
| hsa-miR-218-5p | 0.80206106 | -0.318216 | 0.00150533 |
| hsa-miR-324-3p | 0.81716595 | -0.291299 | 0.0171506 |
| hsa-miR-29b-3p | 0.8178589 | -0.2900761 | 0.03715585 |
| hsa-miR-204-5p | 0.82376963 | -0.2796872 | 0.01111315 |
| hsa-miR-887-3p | 0.84285826 | -0.2466381 | 0.03219643 |
| hsa-miR-138-5p | 0.86349689 | -0.2117371 | 0.03055703 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





