1. Introduction
Pulmonary Sarcoidosis is a multisystem granulomatous interstitial lung disease (ILD) with variable presentation and prognosis. Although the disease may involve any organ, the lung is most commonly involved, at a rate of 90% in most series [
1,
2]. On average, a diagnosis of pulmonary sarcoidosis is made after 3 months of symptoms. In 20% of cases, pulmonary sarcoidosis patients experience symptoms up to 12-months before a diagnosis is made [
3]. Currently, the diagnosis of ILD relies on a multidisciplinary approach which includes three major components: clinical presentation, chest imaging, and lung histologic findings [
4,
5,
6,
7], wherein, both clinically and radiologically, the disease may mimic malignancies and infections [
8,
9,
10]. Although chest-trained radiologists are familiar with the radiographic manifestations of pulmonary sarcoidosis, geographically remote and underserved locations may not have access to such radiologists. Therefore, increasing the speed and diagnostic accuracy of pulmonary sarcoidosis using imaging features has great potential to improve clinically-important outcomes by directing these patients to expert care in a timelier fashion.
Both the chest radiograph and chest CT may be used to evaluate for pulmonary sarcoidosis. However, the chest CT scan is vastly superior to the chest radiograph in this regard. Several chest CT scan features are regarded as highly specific for pulmonary sarcoidosis [
11,
12,
13] and these are often undetectable on the chest radiograph.
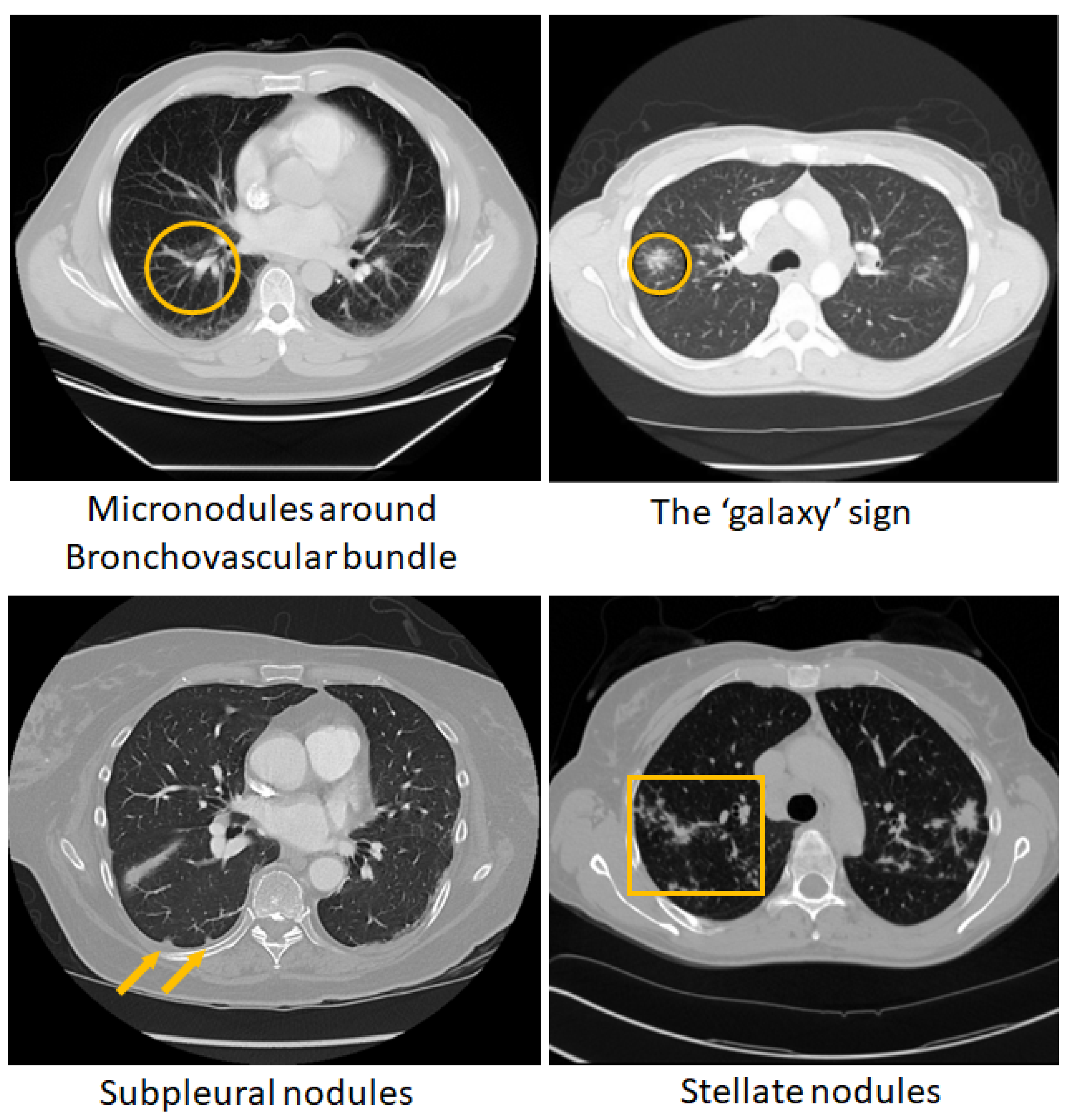
Figure 1 shows some of these chest CT patterns. Although such chest CT features of pulmonary sarcoidosis are regarded as highly specific for the disease and their diagnostic power was demonstrated in small cohorts [
14,
15], they have not been formally tested in diverse populations. Currently, there is no algorithmic diagnostic tool available that can leverage the characteristic CT findings of pulmonary sarcoidosis other than clinical diagnostic algorithms or guidelines [
7,
16].
Recent studies have shown that use of AI has significantly increased the efficiency of pulmonologists to distinguish respiratory diseases identified on chest CT or radiographs [
17,
18,
19] with a limited body of research on diagnosing pulmonary sarcoidosis from chest radiographs [
20,
21]. There is also increasing evidence AI has the potential to democratize radiology by enabling less-experienced radiologists in underserved areas to tap into subspecialty expertise [
22]. Therefore, development of an AI algorithm that can reliably diagnose pulmonary sarcoidosis at the level of a subspecialty thoracic radiologist from CT, would be a major advancement, an incredible asset to underserved regions, and could serve as a valued assistant for any radiologist.
Radiomics have been used extensively to build methods to automatically diagnose lung diseases and characterize lung nodules (benign vs. malignant) from CT [
23,
24,
25,
26,
27,
28,
29]. Radiomics are hand-crafted features/mathematical descriptors extracted from radiology images that are relatively straightforward to define, conceptualize, and interpret, and are both standardized and reproducible.These features are used to train a machine learning classifier, and predictions are made based on the trained model. Specifically, radiomics and machine learning approaches have been used to classify or diagnose ILDs [
30,
31,
32,
33,
34]. On the other hand, CNNs have an inherent capability to learn discriminative features within convolutional blocks for the diagnosis and classification of lung diseases [
35,
36,
37]. These features are abstract and often difficult to interpret multiscale features that are learned automatically. CNN features are unique to each input dataset, which allows considerable versatility but also introduces susceptibility to overfitting and lack of reproducibility [
38].
Extraction of radiomic features typically involve defining a precise region-of-interest, which is difficult for diffuse lung diseases such as ILDs and pulmonary sarcoidosis, while CNNs operate on entire images or sub-images. Attempts to combine both radiomics and CNNs also have been made in several ways. For example, radiomic features were extracted from CT image and used in a deep learning network, which is an example of early fusion and then further combined with clinical features at a late stage for prediction of EGFR gene mutation status for non-small cell lung carcinoma [
39]. Similarly, radiomics features were extracted separately and then combined with features derived from CNN and fused at an intermediate stage before classification of COPD staging [
40] and lung nodule classification [
28,
41]. All methods that extracted radiomic features however depended on defining a region of interest, except in Liang et al. [
42], where radiomics features were extracted from the entire lung, although the lung parenchyma was segmented. A comprehensive review of methods involving CNN and/or radiomics for ILDs is provided in Barnes et al. [
43].
With more recent advancements in deep learning, vision transformers (ViTs) [
44] have become popular in building robust classification models, sometimes outperforming CNNs [
45,
46]. The multi-headed, self-attention mechanism in ViT learns rich representations between the sequence of image patches, thus capturing global representation of an image. However, ViT requires large number of labeled images to train, limiting its application in studies with deficient data, particularly with medical imaging data. ViT also emphasizes on low-resolution features because of the consecutive down sampling, and this results in the lack of detailed localized information [
47]. On the other hand, due to strong inductive bias, CNNs can learn localized features such as edges, corners, and shapes, which may be common across different images, and can often achieve good performance with fewer training samples compared to ViT. To address the limitations of the CNN or ViT frameworks, a recent trend is to combine the ViT and CNN to sample both global and local information in an image for improved classification and segmentation tasks [
48,
49,
50,
51,
52]. This combination is important for differentiating between diffuse lung diseases such as pulmonary sarcoidosis and others, as there may be some similarity in the local features of the diseases - but the relative position where the features appear within the lung becomes an important differentiating factor in classification of the disease.
Based on the distinct advantages of using a radiomics, CNN or ViT, in this work, we present a novel approach using radiomics and CT-guided multichannel CNNViT ensemble classification framework to classify pulmonary sarcoidosis vs. lung cancer (LCa). The novel aspects in our framework are as follows:
Combination of 3D CNNs with 3D ViT that will allow capturing local information within convolutional blocks and the complex relationship between spatial positions of patches within a CT volume.
Extraction of radiomic texture features from the chest CT without defining any region-of-interest, and introducing multichannel CNN-ViT network architecture with a radiomic texture map and the CT volume as inputs, thus referring to the framework as RadCT-CNNViT.
Our framework also provides visual explainability for classification of pulmonary sarcoidosis vs lung malignancies (LCa), that suggests regions of interest that are considered important by the network for making the prediction.
Finally, an ablation study is performed to show that our method can leverage the strengths of both hand-crafted radiomics, CT imaging features, and learned CNN+ViT features to provide improved prediction performance compared to a CNN or ViT alone, and radiomics or CT alone.
2. Materials and Methods
In this section, we provide overviews of the data collection process and the preprocessing steps involved in the development of our method. We subsequently explore the multichannel ensemble AI framework comprising a CNN and a ViT architecture, extraction of radiomic texture features and combining the CNN and ViT architectures for classification. Additionally, we describe the details of the methods utilized to generate visual explanations based on the model predictions. Finally, we discuss the metrics used to evaluate the performance of presented methods.
2.1. Data and Pre-Processing
The chest CT images for clinically confirmed pulmonary sarcoidosis (PS) (n=126) were obtained from an IRB-approved study at Albany Medical College (AMC) (refer to the Compliance and Ethical Standards section for details). Chest CT exams for outpatients at AMC were performed using GE Revolution 256 CT scanner and GE VCT Lightspeed 16 slice scanner with a variety of protocols with an in-plane (
xy) (512x512 matrix) resolution between 0.625mm -1mm, and
z-resolution of 1.25mm - 5mm. Patients in the pulmonary sarcoidosis database were, on average, 48.9 years of age (22yrs-84yrs), female (n=77), male (n=48), unspecified (n=1), and white (n=101), black (n=17), Asian (n=2), unspecified race (n=6). Images of lung cancer (LCa) cases (n=93), comprising both primary (n=42) and metastatic (n=51) instances, were sourced from the TCIA (LIDC-IDRI) public archive, as described in previous studies [
53,
54]. The 3D CT volumes were center cropped in axial view to focus on the lung region, and then resized to 256x256x64.
2.2. The Multichannel Ensemble AI Framework for Classification
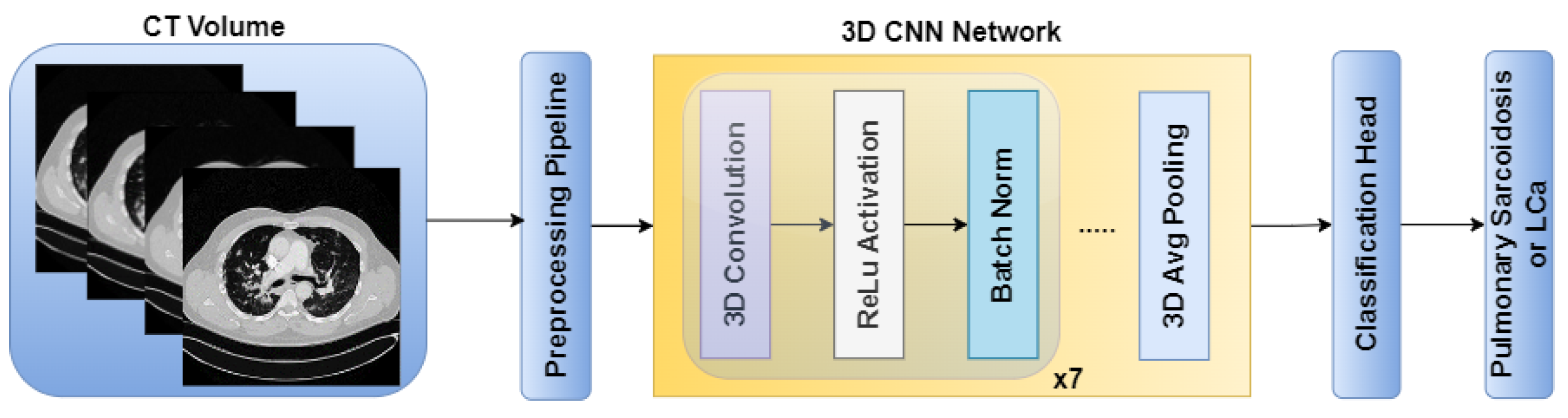
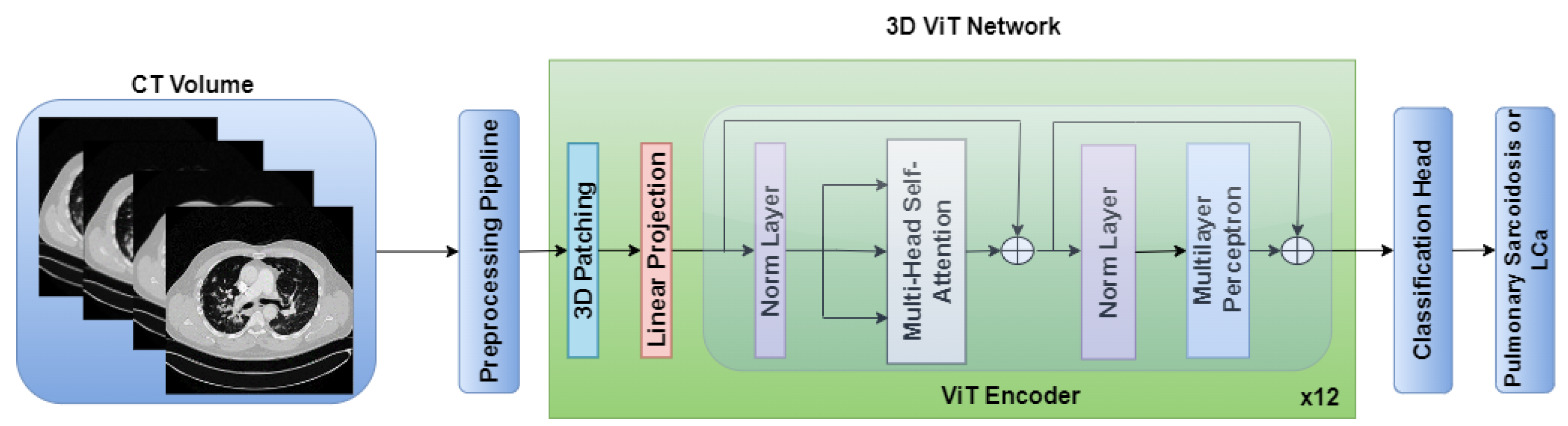
The standalone architectures of the CNN and ViT networks using only the CT volume as input are shown in
Figure 2 and
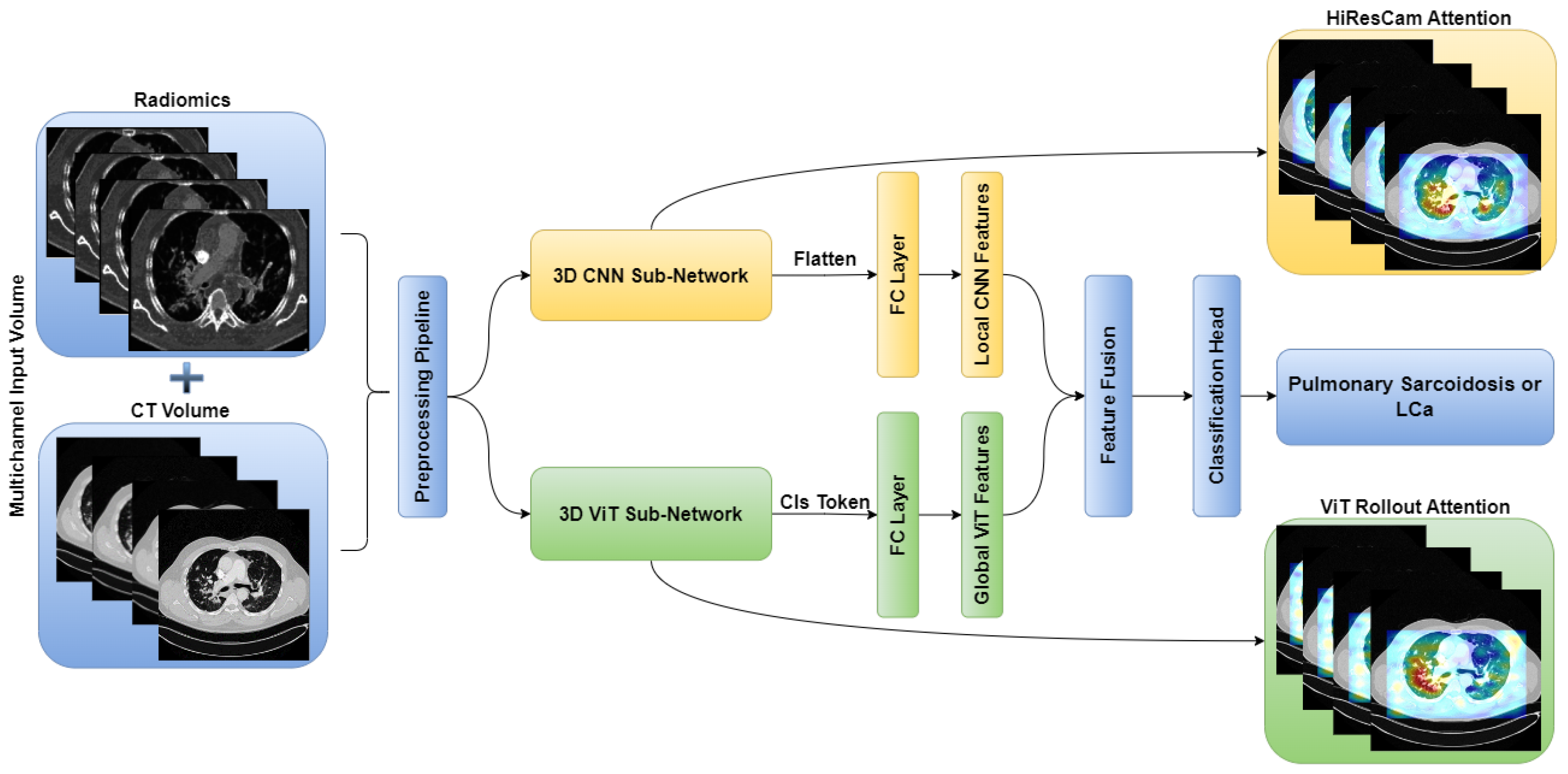
Figure 3, respectively. Subsequently, these networks are combined in an ensemble network, incorporating both the CT volume and radiomics feature, to construct the multichannel CT and radiomics-guided CNN-ViT (RadCT-CNNVIT) network. The architecture of the RadCT-CNNViT network is illustrated in
Figure 4.
2.2.1. Extracting Radiomics Texture
The input radiomics texture map for the framework was chosen based on our previous work [
55], where feature selection was performed using random forest (RF) on a subset of confirmed pulmonary sarcoidosis (n=61) and the MosMed public dataset [
56] of other ILDs that were not Covid-19 (n=154). Haralick texture features [
57] such as Cluster Prominence, Cluster Shade, Correlation, Energy, Entropy, Haralick Correlation, Inertia, and Inverse Difference Moment with an offset of 1 (3x3x3 window) were computed for each CT volume and then averaged to produce one feature map per texture feature. Each radiomic texture volume was then divided into 16x16x16 patches. Patch mean and standard deviation for each of the 8 texture features were computed, resulting in a feature vector of size 16, and each patch was treated as a sample with the image label. The feature vectors from the patches were used to fit a Random Forest (RF) classifier [
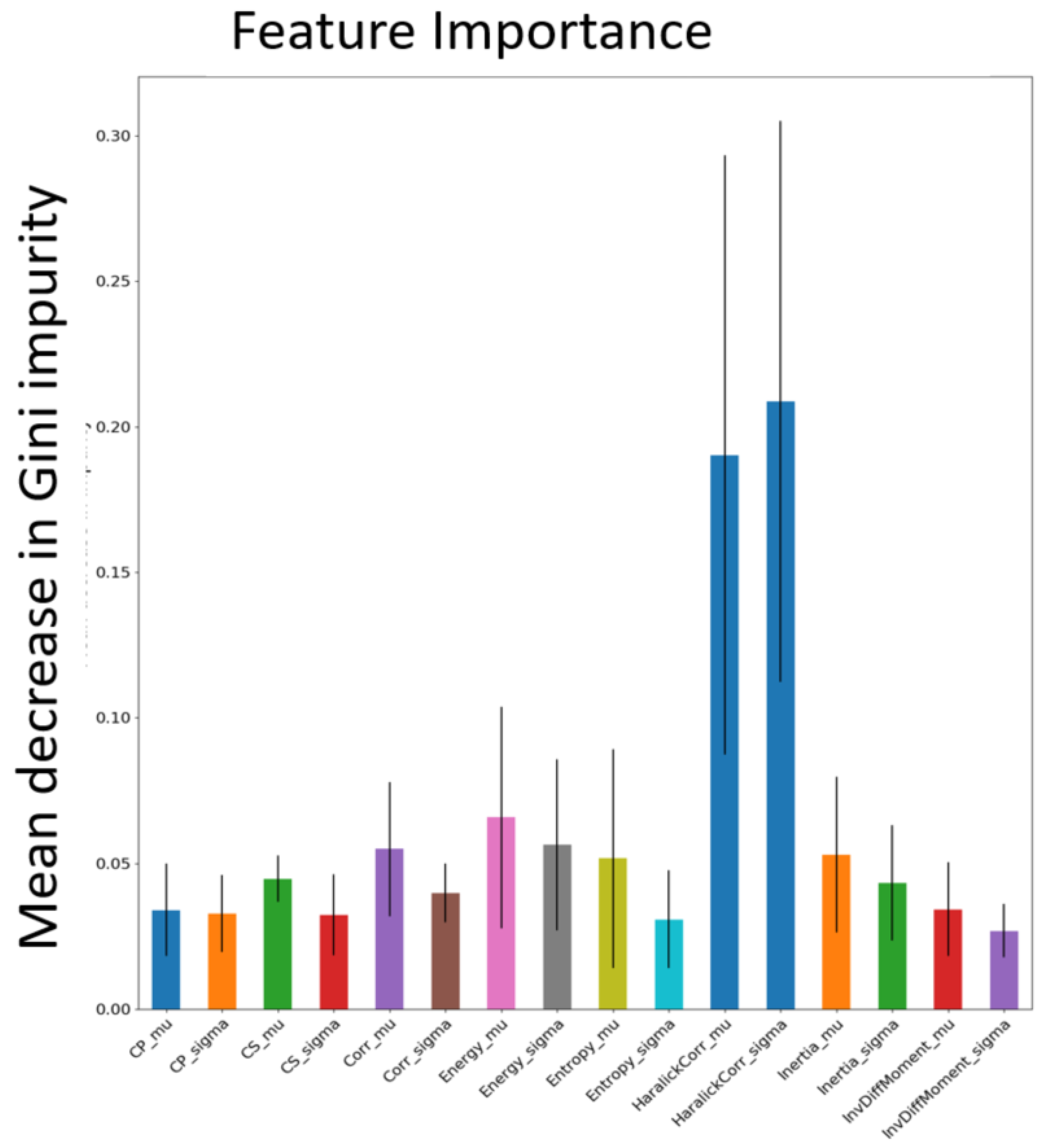
58] with 100 trees, where each patch was classified as pulmonary sarcoidosis or other ILD. The mean decrease in Gini impurity was computed as the average of feature importance scores over all trees in the RF in a 5-fold cross-validation strategy. The feature map corresponding to the highest score was chosen as input to the network architecture.
Figure 5 shows all the features and their mean Gini-impurity scores after avergaing across 5-folds.
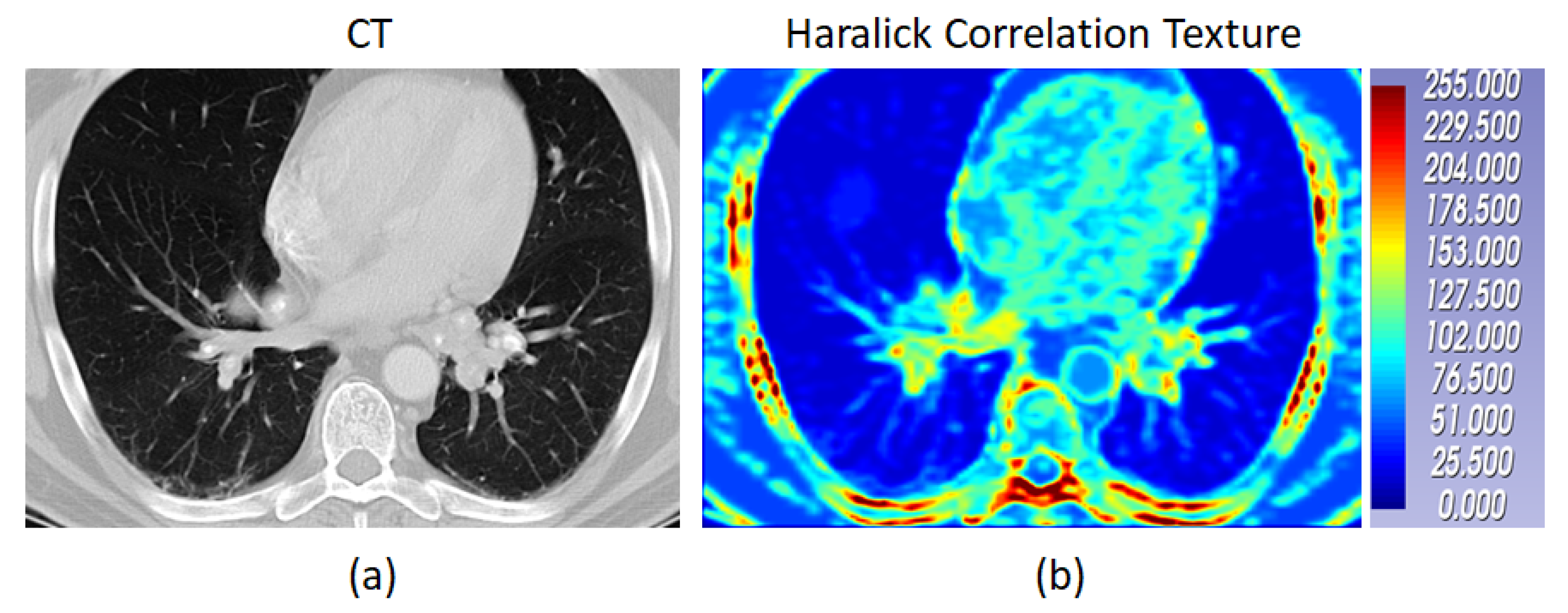
Figure 6 shows a case of pulmonary sarcoidosis and its corresponding Haralick correlation texture map.
2.2.2. The RadCT-CNNViT Architecture
Based on
Figure 5, the Haralick correlation maps were computed for pulmonary sarcoidosis and LCa cases and used as input to the RadCT-CNNViT framework with min-max intensity normalization for each 3D texture volume along with the CT volume clipped to a lung window of (-1000, 400) intensity range. The RadCT-CNNViT is a 3D multichannel ensemble network, which consists of two input channels feeding into two subnetworks: a 3D CNN feature extractor and a 3D ViT encoder. The 3D CNN feature extractor is responsible for learning local features from the volumetric radiomic & CT feature inputs. It consists of 7 convolution blocks, where each block comprises a 3D convolution layer, ReLU activation, and batch normalization. These convolution blocks employ 3D convolutional filters to capture spatial patterns and extract relevant features from the input data. The numbers of filters in each of the convolution blocks are 16, 32, 64, 128, 256 and 512 respectively. The first convolution block utilizes a kernel size of 3x3x3 and a stride number of 1. For downsampling, the subsequent convolution blocks use a kernel size of 4x4x4 and a stride number of 2. The last layer of the 3D CNN is followed by a 3D average pooling operation and fully connected layer, which help to reduce the spatial dimensions of the CNN features to 768.
On the contrary, the 3D ViT encoder focuses on capturing global features by treating the input as a sequence of 3D patches, each with a size of 16x16x16. The 3D ViT encoder consists of 12 transformer blocks with a hidden layer dimension of 768, and each block utilizes multi-head self-attention with 6 heads. The outputs from the 3D CNN feature extractor and the 3D ViT encoder are finally concatenated and fed into a fully connected (FC) layer with sigmoid activation for the classification of pulmonary sarcoidosis vs. LCa. Binary cross-entropy was used as a loss function with AdamW optimization; a learning rate of 1e-5 and 50 epochs were used to train the network. Additionally, we ultilized random flip, random noise, and random affine transformations from TorchIO [
59], a Python library designed for medical imaging augmentation, to augment the 3D data during training. The overview of the combined CNN-ViT network architecture is shown in
Figure 4.
2.3. Generating Visual Explanations for Predictions
To generate visual explanations and enhance the interpretability of our model’s predictions, we applied two techniques: HiResCAM [
60] and Attention Rollout [
61]. These methods offer crucial insights by generating visual attention maps for both CNN and ViT sub-networks, particularly beneficial for understanding complex deep learning models applied to medical imaging, such as chest CT scans. The overarching goal is to localize relevant disease features within the chest CT volume.
HiResCAM utilizes attention mechanisms to selectively weigh the contributions of different features within the CNN subnetwork. The computation of HiResCAM is described by Equation (
1). The process begins by computing the gradient of the raw score
corresponding to class
m with respect to a specific CNN feature map
A. This gradient, represented as
, highlights the significance of various features in influencing the prediction. Subsequently, an attention map is generated by element-wise multiplication between the computed gradient and the CNN feature map, followed by summation over the feature dimension
F. This attention map
provides visual cues, aiding in the localization of relevant disease features within the chest CT volume.
In contrast, Attention Rollout offers a distinct approach by tracing the path of attention from an initial region of interest to all other patches in the image. This recursive method dynamically visualizes how the ViT sub-network distributes its attention across different parts of the image. By quantifying the attention flow, Attention Rollout provides profound insights into how the ViT sub-network distributes its attention across various parts of the image, facilitating a deeper understanding of the underlying mechanisms driving predictions. The computation of Attention Rollout at layer
L is described by Equation (
2), where
represents the average of the multi-head self-attention matrix at layer
L, and
I denotes the identity matrix.
2.4. Performance Metrics
The performance metrics for evaluation of all methods in this ablation study included sensitivity, specificity, precision, accuracy, F1-score, and combined AUC, computed across 5-folds of cross-validation. These metrics were computed based on a confusion matrix which contains four parameters: TP (true positive), TN (true negative), FP (false positive), and FN (false negative). TP indicates correctly predicted pulmonary sarcoidosis, TN denotes correctly predicted LCa, FP represents incorrectly predicted pulmonary sarcoidosis, and FN indicates incorrectly predicted LCa. Sensitivity, specificity, precision, accuracy, and F1-score values were derived from these parameters using Equations (
3)–(
7):
3. Experiments and Results
We conducted a comprehensive ablation study to evaluate the performance of different network architectures (CNN, ViT, and CNNViT) using CT, radiomics, and multichannel CT-radiomics data. In this study, we performed a 5-fold cross-validation with a dataset of clinically confirmed cases of pulmonary sarcoidosis (n=126) and lung cancer (n=93).
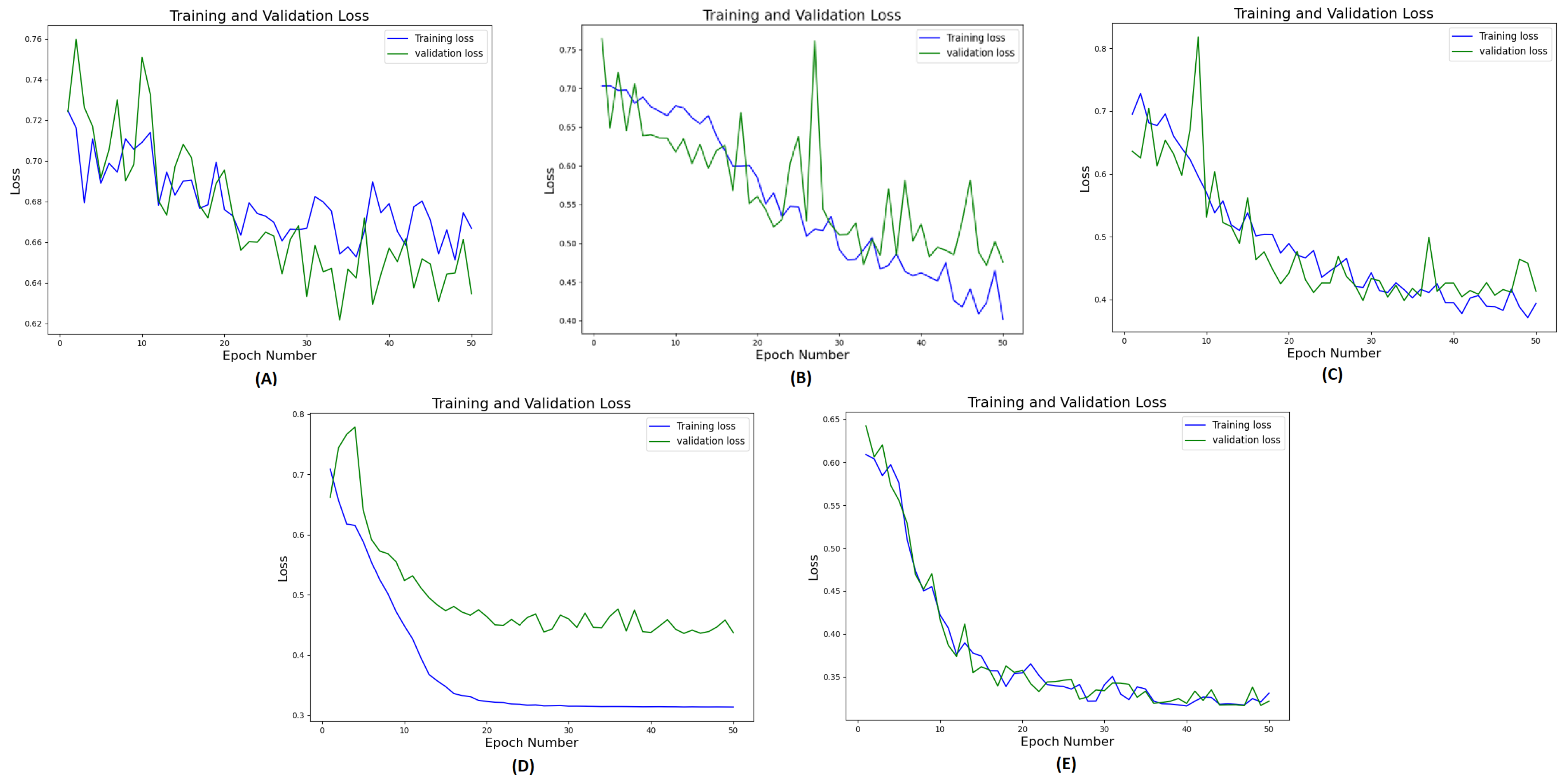
Figure 7 illustrates the training and validation loss curves of a single fold over 50 epochs for all different methods compared. It demonstrates that 3D ViT failed to converge due to the limited training dataset, while 3D CNN showed slower convergence with unstable loss. Conversely, 3D CNN-ViT ensemble network demonstrated improved convergence due to combination of global and local features. Moreover, RadCT-CNNViT achieved the lowest loss, and best converged training and validation losses in differentiating the diseases; further demonstrating the effectiveness of leveraging radiomics texture maps as input along with CT. The normalized confusion matrices for all experiments in this ablation study are shown in
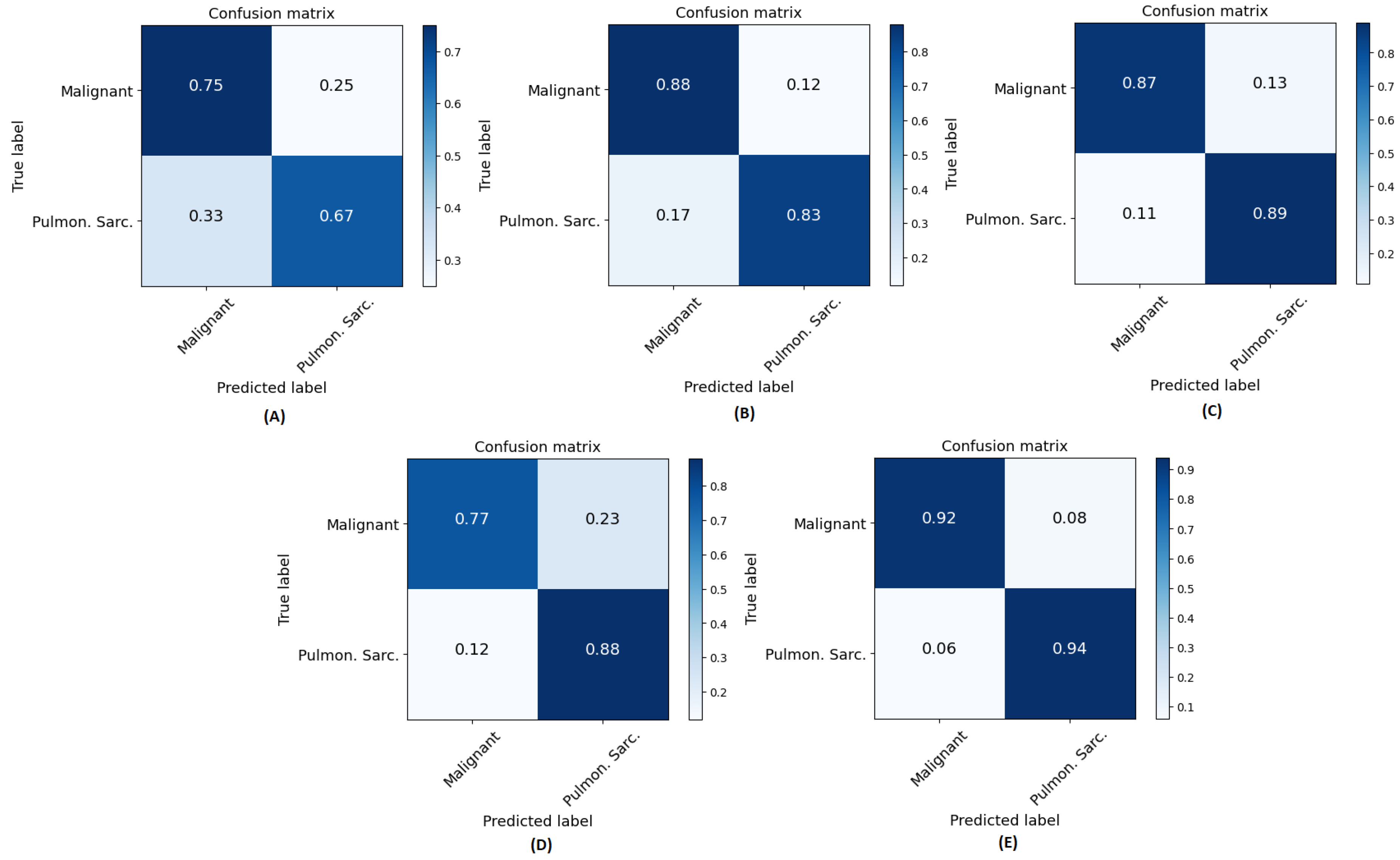
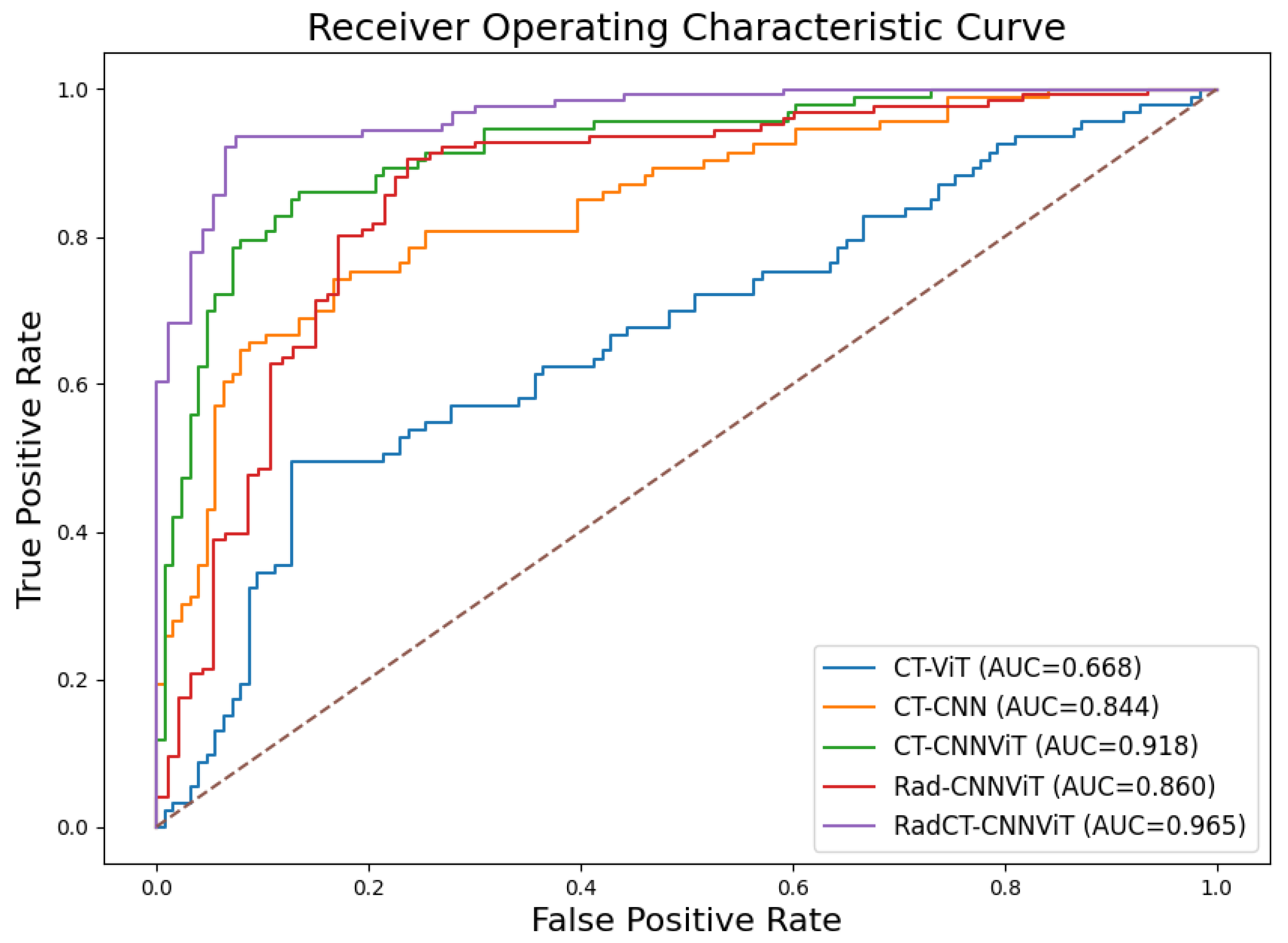
Figure 8. The confusion matrices show that true prediction rate for pulmonary sarcoidosis went higher than LCa when the CNN and ViT networks were combined, suggesting the value of combining global and local features for pulmonary sarcoidosis. Performance metrics for these experiments were derived from the confusion matrices and are summarized in
Table 1. Additionally, the corresponding ROC curves are depicted in
Figure 9. The RadCT-CNNViT model demonstrated the best performance with accuracy, sensitivity, specificity, precision, F1-score, and combined AUC of 0.93±0.04, 0.94±0.04, 0.93±0.08, 0.95±0.05, 0.94±0.04 and 0.97, respectively, compared to other variations in the ablation study, with statistical significance of
.
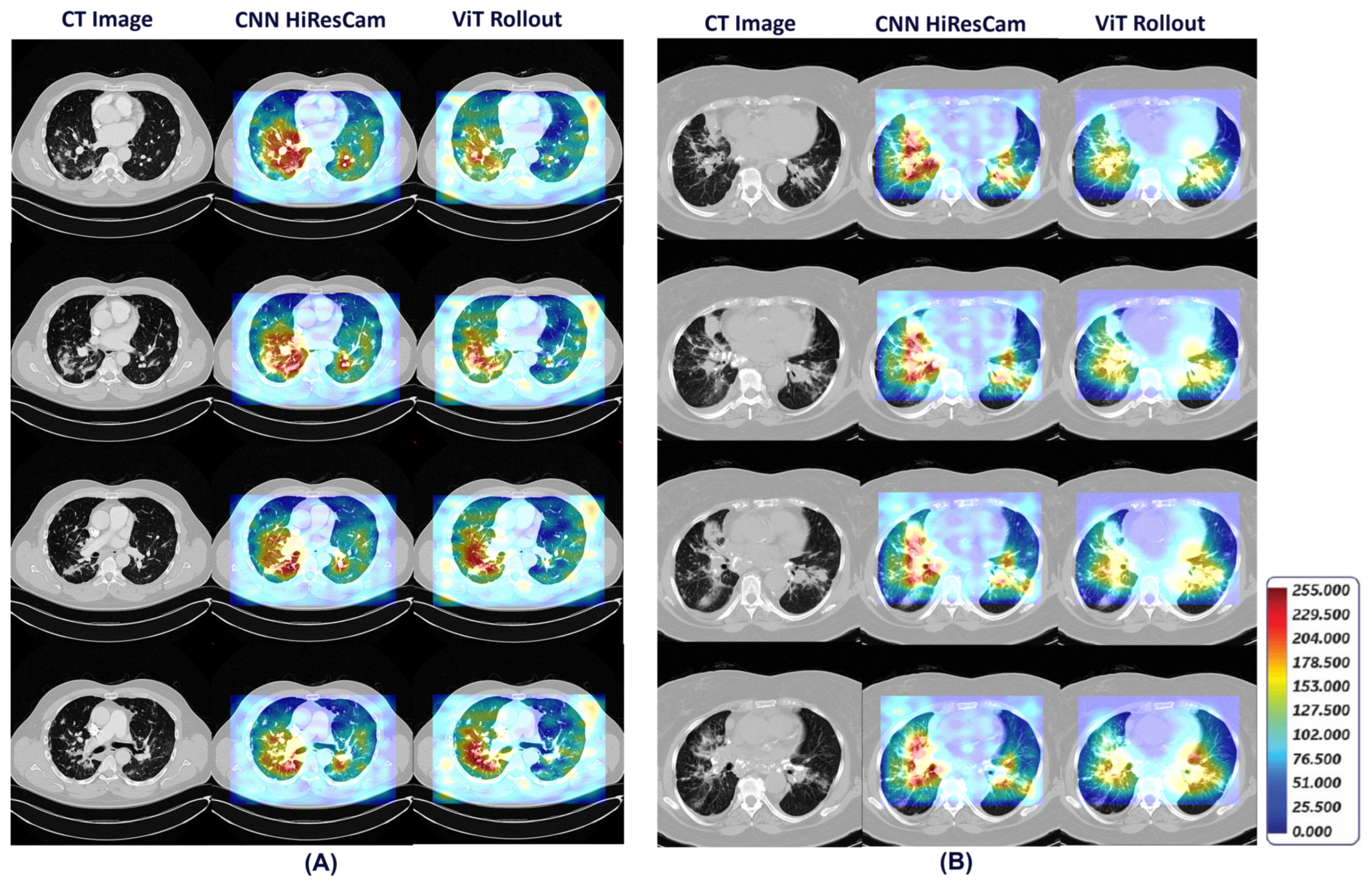
Figure 10 shows detailed visual explanations utilizing HiResCAM and ViT Attention Rollout techniques for both pulmonary sarcoidosis and LCa. The computed visual attention maps were overlaid onto the CT images to emphasize the regions-of-interest. We observed that features from the CNN subnetworks had denser visual representations (color maps) within local regions, while ViT showed overall global representations, as expected. These visual cues highlight features in specific regions-of-interest contributing to pulmonary sarcoidosis and lung cancer diagnoses.
4. Discussions
We presented a method to diagnose pulmonary sarcoidosis from LCa through a combination of CNN and ViT in two parallel branches of the network, retaining both local and global representations, along with a radiomics map as an additional input channel with CT volume. Although there have been previous attempts in combining CNN and ViT in various forms for disease diagnosis, we believe this is one of the first use cases of using radiomics texture maps and CT as 3D volumetric, multichannel inputs in a CNN-ViT framework. Previous studies have typically shown combination of radiomics and CNN features for lung disease classification, prognosis and staging using late fusion techniques i.e., radiomics features and CNN features were combined just before the classsification layer, which demonstrated improved performance compared to CNN features or radiomics features based classification only [
39,
40,
62]. In our previous work [
55], we showed how radiomics texture features used as input to a CNN-ViT framework had improved performance over using radiomics features with a traditional machine learning classifier to classify pulmonary sarcoidosis from other ILDs.
In this work, we showed that compared to a CNN or ViT alone, or using CT or radiomics only in a CNN-ViT ensemble network, a CT & radiomics-guided deep learning approach provides improved feature representation. Specifically, it highlights the effectiveness of feature fusion, in both early and intermediate stages i.e., proper utilization of radiomics texture maps, which are also 3D volumes extracted from CT as input features along with CT imaging features, and combining features extracted from CNN and ViT sub-networks before classification. The strong inductive bias of CNNs is necessary to reach the desired classification accuracy with less data. However, for diffuse lung diseases with no specific location within the lung, the global, long-range context offered by ViT is more adept at identifying/embedding interactions between image patches. Unfortunately, ViT does not provide as much local context compared to CNNs. Nevertheless, the problem of precisely embedding the local and global representations into one another remains. Hence, in this work, a dual structure of CNN-ViT is created to capture the respective feature representations for enhanced representation learning.
Radiomics texture features are computationally well-defined compared to abstract, hierarchical, and difficult-to-interpret CNN features. Haralick texture (correlation map), used in this work, captures features from the CT images that are not perceptible for the human eye [
63]. In essence, it describes how often one grey tone will appear in a specified spatial relationship to another gray tone on the image [
64]. As a result, subtle differentiation between different granulomatous disorders such as between the `galaxy sign’ of pulmonary sarcoidosis and that mimicing metastatic lung cancer [
65,
66], is possible using such radiomics texture features. Our experiments showed that although inclusion of radiomics feature as multichannel input with CT did improve all performance metrics in differentiating pulmonary sarcoidosis from LCa, neither CT nor radiomics alone could provide similar accuracies; leading to the confirmation of hypothesis that radiomics texture indeed was complementary to CT imaging features. However, we acknowledge that an extensive set of radiomics texture maps were not computed in our experiments, and only the Haralick textures were computed, which is a limitation of this work. The types of radiomics texture features are myriad, and the computation of all features, down-selecting the best features to remove noisy representations, is an intensive process. In future, we plan to include transform-based texture features in our experiments. The radiomics texture map extraction for pulmonary sarcoidosis or LCa did not involve annotation of regions-of-interest in the CT volume, which makes our AI framework further suitable for differentiating between other types of diffuse lung diseases.
The major limitations of this work include training and validation using a cross-validation approach due to limited sample size, and unavailability of a separate validation set from a multicenter study, which may affect the generalizability of the method. However, this being a pilot study to choose the best performing method between the radiomics, CNN and ViT combinations, improvement of one method over the other has been observed in the results without testing on a separate cohort. Additionally, our method only addresses a two-class problem i.e., diagnosing pulmonary sarcoidosis vs. LCa, however, in clinical settings, differentiating between pulmonary sarcoidosis and other forms of ILDs would be necessary, which is part of our future work.
We also acknowledge that the presented RadCT-CNNViT is complex in terms of training the network as it requires a GPU compute, we do not however think this limits the adoption of the method in a clinical setting as based on our experinece, 3D network inferences can be often performed on CPUs with advanced Intel optimization techniques. Additionally, we used a vanilla CNN and a standard ViT network in our implementation without trying different CNN or Vision Transformer versions such as ResNets or Swin Transformers because of limited training sample size as complex deep learning networks need a lot more data for model convergence during training. One of the hypotheses of this work was to show that while ViTs provide rich global feature representations, they do not outperform CNNs in low data setting, and by combining CNN and ViT, a reasonably acceptable classification performance can be achieved. Although, there is no prior literature on the sensitivity and specificty of diagnosing pulmonary sarcoidosis from chest CT, prior work [
15] suggests the performance of our method is similar to that of expert radiologists in diagnosing cardiac sarcoidosis with pulmonary and mediastinal involvement. The performance of our method is also higher than previous works that used chest x-ray to diagnose pulmonary sarcoidosis from healthy [
21] or patients with pneumonia [
20] involving deep learning or radiomics respectively with much smaller cohorts.
5. Conclusions
Pulmonary sarcoidosis is a diffuse lung disease, which is difficult to diagnose from CT imaging without a multidisciplinary clinical team, specifically in geographically underserved locations. The visual attention maps and intelligent network architecture from CNN and ViT used in our method are likely to reduce the burden of radiologists and provide a timely and reliable probability of pulmonary sarcoidosis diagnoses. Clinicians may also use this information directly to adjust their diagnostic probabilities in patients with diffuse lung disease. As our AI method to diagnose pulmonary sarcoidosis does not depend on input from radiologists, it may truly augment the radiologist’s impression, as the approaches of the radiologist and our method most probably will be different. This suggests that our method may not only increase the speed of the radiographic assessment of diffuse lung disease but may surpass current chest imaging diagnostic standards. Finally, although our method was applied to pulmonary sarcoidosis in this instance, it could be adapted to any interstitial lung disease. We therefore believe that our method ultimately has the capability to be used as a general diagnostic tool for all interstitial lung diseases as well as localized lung diseases.
Author Contributions
Conceptualization, Mitra, J. and Qiu, J. and Ghose, S.; methodology, Mitra, J. and Qiu, J. ; software, Qiu, J. and Mitra, J. and Ghose, S.; validation, Qiu, J. and Dumas, C. and Judson, M.; formal analysis, Mitra, J.; investigation, Judson, M.; data curation, Yang, J and Dumas, C. and Sarachan, B.; writing—original draft preparation, Qiu, J. and Mitra, J.; writing—review and editing, Judson, M and, Dumas, C. and Ghose, S. and Yang, J. and Sarachan, B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding
Institutional Review Board Statement
This research study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Institutional Review Board of Albany Medical Center, study number 6039, approved 7 December, 2020.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Retrospective data for pulmonary sarcoidosis were collected at Albany Medical College and cannot be shared publicly.
Acknowledgments
The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health, and their critical role in the creation of the free publicly available LIDC/IDRI Database used in this study.
Conflicts of Interest
Mitra, J.; Qiu, J.; Ghose, S.; and Sarachan, B are employees of GE HealthCare.
References
- Judson, M.A.; Boan, A.D.; Lackland, D.T. The clinical course of sarcoidosis: presentation, diagnosis, and treatment in a large white and black cohort in the United States. Sarcoidosis, vasculitis, and diffuse lung diseases: official journal of WASOG 2012, 29, 119–127. [Google Scholar] [PubMed]
- Baughman, R.P.; Teirstein, A.S.; Judson, M.A.; Rossman, M.D.; Yeager, H.J.; Bresnitz, E.A.; DePalo, L.; Hunninghake, G.; Iannuzzi, M.C.; Johns, C.J.; others. Clinical characteristics of patients in a case control study of sarcoidosis. American journal of respiratory and critical care medicine 2001, 164, 1885–1889. [Google Scholar] [CrossRef] [PubMed]
- Judson, M.A.; Thompson, B.W.; Rabin, D.L.; Steimel, J.; Knattereud, G.L.; Lackland, D.T.; Rose, C.; Rand, C.S.; Baughman, R.P.; Teirstein, A.S. The diagnostic pathway to sarcoidosis. Chest 2003, 123, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Crouser, E.D.; Maier, L.A.; Wilson, K.C.; Bonham, C.A.; Morgenthau, A.S.; Patterson, K.C.; Abston, E.; Bernstein, R.C.; Blankstein, R.; Chen, E.S.; others. Diagnosis and detection of sarcoidosis. An official American Thoracic Society clinical practice guideline. American journal of respiratory and critical care medicine 2020, 201, e26–e51. [Google Scholar] [CrossRef]
- Teoh, A.K.Y.; Holland, A.E.; Morisset, J.; Flaherty, K.R.; Wells, A.U.; Walsh, S.L.F.; Glaspole, I.; Wuyts, W.A.; Corte, T.J.; Collaborators, I.M.D. Essential Features of an Interstitial Lung Disease Multidisciplinary Meeting: An International Delphi Survey. Ann Am Thorac Soc. 2022, 19, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.T. Multidisciplinary Meetings in Interstitial Lung Disease: Polishing the Gold Standard. Ann Am Thorac Soc. 2022, 19, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Grutters, J.C. Establishing a Diagnosis of Pulmonary Sarcoidosis. J Clin Med. 2023, 12, 6898. [Google Scholar] [CrossRef] [PubMed]
- van’t Hoog, A.H.; Meme, H.K.; Van Deutekom, H.; Mithika, A.M.; Olunga, C.; Onyino, F.; Borgdorff, M.W. High sensitivity of chest radiograph reading by clinical officers in a tuberculosis prevalence survey. The International journal of tuberculosis and lung disease 2011, 15, 1308–1314. [Google Scholar] [CrossRef] [PubMed]
- Mortaz, E.; Adcock, I.M.; Barnes, P.J. Sarcoidosis: role of non-tuberculosis mycobacteria and Mycobacterium tuberculosis. Int J Mycobacteriol. 2014, 3, 225–9. [Google Scholar] [CrossRef]
- El Jammal, T.; Pavic, M.; Gerfaud-Valenti, M.; Jamilloux, Y.; Sève, P. Sarcoidosis and Cancer: A Complex Relationship. Front Med 2020, 24, 594118. [Google Scholar] [CrossRef]
- Abehsera, M.; Valeyre, D.; Grenier, P.; Jaillet, H.; Battesti, J.P.; Brauner, M.W. Sarcoidosis with pulmonary fibrosis: CT patterns and correlation with pulmonary function. AJR. American journal of roentgenology 2000, 174, 1751–1757. [Google Scholar] [CrossRef] [PubMed]
- Tana, C.; Donatiello, I.; Coppola, M.G.; Ricci, F.; Maccarone, M.T.; Ciarambino, T.; Cipollone, F.; Giamberardino, M.A. CT Findings in Pulmonary and Abdominal Sarcoidosis. Implications for Diagnosis and Classification. J Clin Med. 2020, 9, 3028. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, M.; Hatabu, H.; Morikawa, K.; Uematsu, H.; Ohno, Y.; Nishimura, K.; Nagai, S.; Izumi, T.; Konishi, J.; Itoh, H. Large coalescent parenchymal nodules in pulmonary sarcoidosis: “sarcoid galaxy” sign. AJR. American journal of roentgenology 2002, 178, 1389–1393. [Google Scholar] [CrossRef] [PubMed]
- Koide, T.; Saraya, T.; Tsukahara, Y.; Bonella, F.; Börner, E.; Ishida, M.; Ogawa, Y.; Hirukawa, I.; Oda, M.; Shimoda, M.; others. Clinical significance of the “galaxy sign” in patients with pulmonary sarcoidosis in a Japanese single-center cohort. Sarcoidosis Vasc Diffuse Lung Dis. 2016, 33, 247–252. [Google Scholar]
- Russo, J.J.; Nery, P.B.; Ha, A.C.; Healey, J.S.; Juneau, D.; Rivard, L.; Friedrich, M.G.; Gula, L.; Wisenberg, G.; deKemp, R.; others. Sensitivity and specificity of chest imaging for sarcoidosis screening in patients with cardiac presentations. Sarcoidosis Vasc Diffuse Lung Dis. 2019, 36, 18–24. [Google Scholar] [PubMed]
- Judson, M.A.; Costabel, U.; Drent, M.; Wells, A.; Maier, L.; Koth, L.; Shigemitsu, H.; Culver, D.A.; Gelfand, J.; Valeyre, D. ; others. The WASOG Sarcoidosis Organ Assessment Instrument: An update of a previous clinical tool. Sarcoidosis, vasculitis, and diffuse lung diseases : official journal of WASOG.
- İn, E.; Geçkil, A.A.; Kavuran, G.; Şahin, M.; Berber, N.K.; Kuluöztürk, M. Using artificial intelligence to improve the diagnostic efficiency of pulmonologists in differentiating COVID-19 pneumonia from community-acquired pneumonia. J Med Virol. 2022, 94, 3698–3705. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.; Cao, H.; FitzGerald, J.M.; Iannotti, N.; Yang, E.; Kocks, J.W.H.; Kostikas, K.; Price, D.; Reddel, H.K.; Tsiligianni, I.; others. Artificial Intelligence/Machine Learning in Respiratory Medicine and Potential Role in Asthma and COPD Diagnosis. The Journal of Allergy and Clinical Immunology: In Practice 2021, 9, 2255–2261. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.; Auffermann, W.F. Artificial Intelligence in the Imaging of Diffuse Lung Disease. Radiologic Clinics 2022, 60, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, N.; Maklad, A.S.; Malki, A.; Deif, M.A. Reliable Sarcoidosis Detection Using Chest X-rays with EfficientNets and Stain-Normalization Techniques. Sensors 2022, 22, 3846. [Google Scholar] [CrossRef]
- Prokop, P. Computer-aided Diagnosis of Sarcoidosis Based on X-Ray Images. Procedia Computer Science 2023, 225, 4611–4620. [Google Scholar] [CrossRef]
- Langlotz, C.P. Will Artificial Intelligence Replace Radiologists? Radiol Artif Intell. 2019, 1, e190058. [Google Scholar] [CrossRef] [PubMed]
- Frix, A.N.; Cousin, F.; Refaee, T.; Bottari, F.; Vaidyanathan, A.; Desir, C.; Vos, W.; Walsh, S.; Occhipinti, M.; Lovinfosse, P.; others. Radiomics in Lung Diseases Imaging: State-of-the-Art for Clinicians. J Pers Med. 2021, 11, 602–621. [Google Scholar] [CrossRef] [PubMed]
- Padmakumari, L.T.; Guido, G.; Caruso, D.; Nacci, I.; Gaudio, A.D.; Zerunian, M.; Polici, M.; Gopalakrishnan, R.; Mohamed, A.K.S.; De Santis, D.; others. The Role of Chest CT Radiomics in Diagnosis of Lung Cancer or Tuberculosis: A Pilot Study. Diagnostics 2022, 12, 739. [Google Scholar] [CrossRef] [PubMed]
- Hunter, B.; Chen, M.; Ratnakumar, P.; Alemu, E.; Logan, A.; Linton-Reid, K.; Tong, D.; Senthivel, N.; Bhamani, A.; Bloch, S.; others. A radiomics-based decision support tool improves lung cancer diagnosis in combination with the Herder score in large lung nodules. eBioMedicine 2022, 86, 104344. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; Bogowicz, M.; others. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Wu, Y.J.; Wu, F.Z.; Yang, S.C.; Tang, E.K.; Liang, C.H. Radiomics in Early Lung Cancer Diagnosis: From Diagnosis to Clinical Decision Support and Education. Diagnostics 2022, 12, 1064. [Google Scholar] [CrossRef]
- Astaraki, M.; Yang, G.; Zakko, Y.; Toma-Dasu, I.; Smedby, O.; Wang, C. A Comparative Study of Radiomics and Deep-Learning Based Methods for Pulmonary Nodule Malignancy Prediction in Low Dose CT Images. Frontiers in Oncology 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Wang, J.; Li, J.; Wang, X.; Li, B.; Xue, F.; Shao, G.; Xue, H. A wavelet features derived radiomics nomogram for prediction of malignant and benign early stage lung nodules. Sci. Rep. 2021, 11, 22330. [Google Scholar] [CrossRef]
- Rosas, I.O.; Yao, J.; Avila, N.A.; Chow, C.K.; Gahl, W.A.; Gochuico, B.R. Automated quantification of high-resolution CT scan findings in individuals at risk for pulmonary fibrosis. Chest 2011, 140, 1590–1597. [Google Scholar] [CrossRef]
- Chang, Y.; Lim, J.; Kim, N.; Seo, J.B.; Lynch, D.A. A support vector machine classifier reduces interscanner variation in the HRCT classification of regional disease pattern in diffuse lung disease: comparison to a Bayesian classifier. Med Phys. 2013, 40, 051912. [Google Scholar] [CrossRef]
- Depeursinge, A.; Chin, A.S.; Leung, A.N.; Terrone, D.; Bristow, M.; Rosen, G.; Rubin, D.L. Automated classification of usual interstitial pneumonia using regional volumetric texture analysis in high-resolution computed tomography. Invest Radiol. 2015, 50, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Chong, D.Y.; Kim, H.J.; Lo, P.; Young, S.; McNitt-Gray, M.F.; Abtin, F.; Goldin, J.G.; Brown, M.S. Robustness-Driven Feature Selection in Classification of Fibrotic Interstitial Lung Disease Patterns in Computed Tomography Using 3D Texture Features. IEEE Trans Med Imaging 2016, 35, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Budzikowski, J.D.; Foy, J.J.; Rashid, A.A.; Chung, J.H.; Noth, I.; Armato, S.G.r. Radiomics-based assessment of idiopathic pulmonary fibrosis is associated with genetic mutations and patient survival. J Med Imaging 2021, 8, 031903. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.B.; Jung, K.H.; Lee, Y.; Kim, H.J.; Kim, N.; Jun, S.; Seo, J.B.; Lynch, D.A. Comparison of Shallow and Deep Learning Methods on Classifying the Regional Pattern of Diffuse Lung Disease. J Digit Imaging 2018, 31, 415–424. [Google Scholar] [CrossRef]
- Walsh, S.L.F.; Calandriello, L.; Silva, M.; Sverzellati, N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med. 2018, 6, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Oyama, S.; Yokota, H.; Kondoh, Y.; Kataoka, K.; Johkoh, T.; Fukuoka, J.; Hashimoto, N.; Sakamoto, K.; Shiratori, Y.; Hasegawa, Y. A comprehensible machine learning tool to differentially diagnose idiopathic pulmonary fibrosis from other chronic interstitial lung diseases. Respirology 2022, 27, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Rawat, W.; Wang, Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Qi, S.; Pan, X.; Li, C.; Yao, Y.; Qian, W.; Guan, Y. Deep CNN Model Using CT Radiomics Feature Mapping Recognizes EGFR Gene Mutation Status of Lung Adenocarcinoma. Front Oncol. 2021, 0, 598721. [Google Scholar] [CrossRef]
- Yang, Y.; Zeng, N.; Chen, Z.; Li, W.; Guo, Y.; Wang, S.; Duan, W.; Liu, Y.; Chen, R.; Kang, Y. Multi-Layer Perceptron Classifier with the Proposed Combined Feature Vector of 3D CNN Features and Lung Radiomics Features for COPD Stage Classification. J Healthc Eng. 2023, 2023, 3715603. [Google Scholar] [CrossRef]
- Lin, C.Y.; Guo, S.M.; Lien, J.J.; Lin, W.T.; Liu, Y.S.; Lai, C.H.; Hsu, I.L.; Chang, C.C.; Tseng, Y.L. Combined model integrating deep learning, radiomics, and clinical data to classify lung nodules at chest CT. Radiol med 2024, 129, 56–69. [Google Scholar] [CrossRef]
- Liang, C.H.; Liu, Y.C.; Wan, Y.L.; Yun, C.H.; Wu, W.J.; López-González, R.; Huang, W.M. Quantification of Cancer-Developing Idiopathic Pulmonary Fibrosis Using Whole-Lung Texture Analysis of HRCT Images. Cancers 2021, 13, 5600. [Google Scholar] [CrossRef] [PubMed]
- Barnes, H.; Humphries, S.M.; George, P.M.; Assayag, D.; Glaspole, I.; Mackintosh, J.A.; Corte, T.J.; Glassberg, M.; Johannson, K.A.; Calandriello, L.; others. Machine learning in radiology: the new frontier in interstitial lung diseases. Lancet Digital Health 2023, 5, e41–50. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gellyand, S.o. An image is worth 16x16 words: Transformers for image recognition at scale. Proceedings of ICLR, 2021.
- Umejiaku, A.P.; Dhakal, P.; Sheng, V.S. Detecting COVID-19 Effectively with Transformers and CNN-Based Deep Learning Mechanisms. Applied Sciences 2023, 13, 4050–4059. [Google Scholar] [CrossRef]
- Okolo, G.I.; Katsigiannis, S.; Ramzan, N. IEViT: An enhanced vision transformer architecture for chest X-ray image classification. Computer Methods and Programs in Biomedicine 2022, 226, 107141. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; He, Y.; Frey, E.C. ; others. Vit-v-net: Vision transformer for unsupervised volumetric medical image registration. arXiv:2104.06468, arXiv:2104.06468 2021.
- Wang, T.; Lan, J.; Han, Z.; Hu, Z.; Huang, Y.; Deng, Y.; Zhang, H.; Wang, J.; Chen, M.; Jiangand, H.o. O-Net: a novel framework with deep fusion of CNN and transformer for simultaneous segmentation and classification. Frontiers in Neuroscience 2022, 16. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.D.; Rahman, M.M.; Ali, M.S.; Mahim, S.M.; Miah, M.S. Enhancing lung abnormalities diagnosis using hybrid DCNN-ViT-GRU model with explainable AI: A deep learning approach. Image and Vision Computing 2024, 142, 104918. [Google Scholar] [CrossRef]
- Cao, K.; Deng, T.; Zhang, C.; Lu, L.; Li, L. A CNN-transformer fusion network for COVID-19 CXR image classification. PLoS One 2022, 17, e0276758. [Google Scholar] [CrossRef] [PubMed]
- Mabrouk, A.; Díaz Redondo, R.P.; Dahou, A.; Abd Elaziz, M.; Kayed, M. Pneumonia Detection on Chest X-ray Images Using Ensemble of Deep Convolutional Neural Networks. Applied Sciences 2022, 12. [Google Scholar] [CrossRef]
- Ukwuoma, C.C.; Qin, Z.; Belal Bin Heyat, M.; Akhtar, F.; Bamisile, O.; Muaad, A.Y.; Addo, D.; Al-antari, M.A. A hybrid explainable ensemble transformer encoder for pneumonia identification from chest X-ray images. Journal of Advanced Research 2023, 48, 191–211. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; others. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Armato III, S.G.; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; others. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Medical Physics 2011, 38, 915–931. [Google Scholar] [CrossRef]
- Qiu, J.; Mitra, J.; Dumas, C.; Sarachan, B.; Ghose, S.; Judson, M. Radiomics-guided 3D CNN-Vision Transformer (Rad-CNNViT) ensemble to diagnose pulmonary sarcoidosis from CT. SPIE Medical Imaging: Image Processing, 2024. accepted.
- Morozov, S.; Chernina, V.; Blokhin, I.V.G. Chest computed tomography for outcome prediction in laboratory-confirmed covid-19: a retrospective analysis of 38,051 cases. Digital Diagnostics 2020, 1, 27–36. [Google Scholar] [CrossRef]
- Conners, R.W.; Trivedi, M.M.; Harlow, C.A. Segmentation of a High-Resolution Urban Scene using Texture Operators. Computer Vision, Graphics and Image Processing 1984, 25, 273–310. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Machine Learning 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Pérez-García, F.; Sparks, R.; Ourselin, S. TorchIO: a Python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. Computer Methods and Programs in Biomedicine 2021, 208, 106236. [Google Scholar] [CrossRef] [PubMed]
- Draelos, R.L.; Carin, L. Hirescam: Faithful location representation in visual attention for explainable 3d medical image classification. arXiv:2011.08891, arXiv:2011.08891 2020.
- Abnar, S.; Zuidema, W. Quantifying attention flow in transformers. arXiv:2005.00928, arXiv:2005.00928 2020.
- Cho, H.; Lee, H.Y.; Kim, E.; Lee, G.; Kim, J.; Kwon, J.; Park, H. Radiomics-guided deep neural networks stratify lung adenocarcinoma prognosis from CT scans. Commun Biol 2021, 4, 1286. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M. Statistical and Structural Approaches to Texture. Proceedings of the IEEE, 1979, Vol. 67, pp. 786–804.
- Haralick, R.M.; Shanmuga, K.; Dinstein, I. Textural Features for Image Classification. IEEE Transactions on Systems Man and Cybernetics Smc., 1973, Vol. 3, pp. 610–621.
- Kuhlman, J.E.; Fishman, E.K.; Hamper, U.M.; Knowles, M.; Siegelman, S.S. The computed tomographic spectrum of thoracic sarcoidosis. Radio Graphics 1989, 9, 449–466. [Google Scholar] [CrossRef]
- Kumazoe, H.; Matsunaga, K.; Nagata, N.; Komori, M.; Wakamatsu, K.; Kajiki, A.; Nakazono, T.; Kudo, S. “Reversed halo sign" of high-resolution computed tomography in pulmonary sarcoidosis. J Thorac Imaging 2009, 24, 66–68. [Google Scholar] [CrossRef]
Figure 1.
Visible patterns of pulmonary sarcoidosis on chest CT marked in `yellow’ circles, arrows and boxes.
Figure 1.
Visible patterns of pulmonary sarcoidosis on chest CT marked in `yellow’ circles, arrows and boxes.
Figure 2.
CNN architecture for pulmonary sarcoidosis vs. lung cancer (LCa) classification using chest CT images.
Figure 2.
CNN architecture for pulmonary sarcoidosis vs. lung cancer (LCa) classification using chest CT images.
Figure 3.
ViT architecture for pulmonary sarcoidosis vs. lung cancer (LCa) classification using chest CT images.
Figure 3.
ViT architecture for pulmonary sarcoidosis vs. lung cancer (LCa) classification using chest CT images.
Figure 4.
Multichannel RadCT-CNNViT architecture for pulmonary sarcoidosis vs. lung cancer (LCa) classification using chest CT images.
Figure 4.
Multichannel RadCT-CNNViT architecture for pulmonary sarcoidosis vs. lung cancer (LCa) classification using chest CT images.
Figure 5.
Feature importance was computed based on the mean decrease in Gini impurity for each of the Haralick texture features in discriminating pulmonary sarcoidosis from other ILDs. The mean and standard deviation of the Haralick correlation texture map were higher than those of other texture features.
Figure 5.
Feature importance was computed based on the mean decrease in Gini impurity for each of the Haralick texture features in discriminating pulmonary sarcoidosis from other ILDs. The mean and standard deviation of the Haralick correlation texture map were higher than those of other texture features.
Figure 6.
The CT of a case of pulmonary sarcoidosis and its corresponding Haralick correlation teture map are shown. The color bar shows the radiomic values normlaized between 0 to 255.
Figure 6.
The CT of a case of pulmonary sarcoidosis and its corresponding Haralick correlation teture map are shown. The color bar shows the radiomic values normlaized between 0 to 255.
Figure 7.
Training and validation loss curves (one-fold) for 50 epochs for the methods in ablation study: A) CT-ViT, B) CT-CNN, C) CT-CNNViT, D) Rad-CNNViT, E) RadCT-CNNViT.
Figure 7.
Training and validation loss curves (one-fold) for 50 epochs for the methods in ablation study: A) CT-ViT, B) CT-CNN, C) CT-CNNViT, D) Rad-CNNViT, E) RadCT-CNNViT.
Figure 8.
Normalized confusion matrices for all methods across all folds: A) CT-ViT, B) CT-CNN, C) CT-CNNViT, D) Rad-CNNViT, and E) RadCT-CNNViT. `Pulmon. Sarc.’ in axes labels is the abbreviation for pulmonary sarcoidosis and `malignant’ relates to LCa.
Figure 8.
Normalized confusion matrices for all methods across all folds: A) CT-ViT, B) CT-CNN, C) CT-CNNViT, D) Rad-CNNViT, and E) RadCT-CNNViT. `Pulmon. Sarc.’ in axes labels is the abbreviation for pulmonary sarcoidosis and `malignant’ relates to LCa.
Figure 9.
Combined receiver operating characteristic (ROC) curves for CT-ViT, CT-CNN, CT-CNNViT, Rad-CNNViT, and RadCT-CNNViT.
Figure 9.
Combined receiver operating characteristic (ROC) curves for CT-ViT, CT-CNN, CT-CNNViT, Rad-CNNViT, and RadCT-CNNViT.
Figure 10.
HiResCAM and ViT Attention Rollout visual explanations that highlight the regions-of-interest on CT scan associated with diagnosis of pulmonary sarcoidosis (A) and lung cancer (B).
Figure 10.
HiResCAM and ViT Attention Rollout visual explanations that highlight the regions-of-interest on CT scan associated with diagnosis of pulmonary sarcoidosis (A) and lung cancer (B).
Table 1.
Performance statistics (Sensitivity, Specificity, Accuracy and combined area under curve (AUC)) for CT-ViT, CT-CNN, CT-CNNViT, Rad-CNNViT, and RadCT-CNNViT.
Table 1.
Performance statistics (Sensitivity, Specificity, Accuracy and combined area under curve (AUC)) for CT-ViT, CT-CNN, CT-CNNViT, Rad-CNNViT, and RadCT-CNNViT.
| Network |
Sensitivity |
Specificity |
Precision |
Accuracy |
F1-Score |
AUC |
| CT-ViT |
0.68±0.09 |
0.66±0.02 |
0.72±0.08 |
0.67±0.05 |
0.70±0.08 |
0.67 |
| CT-CNN |
0.83±0.04 |
0.88±0.05 |
0.89±0.06 |
0.85±0.04 |
0.86±0.05 |
0.84 |
| CT-CNNViT |
0.87±0.05 |
0.89±0.06 |
0.92±0.05 |
0.88±0.04 |
0.89±0.05 |
0.92 |
| Rad-CNNViT |
0.88±0.06 |
0.77±0.09 |
0.84±0.06 |
0.84±0.05 |
0.86±0.06 |
0.86 |
| RadCT-CNNViT |
0.94±0.04 |
0.93±0.08 |
0.95±0.05 |
0.93±0.04 |
0.94±0.04 |
0.97 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).